Abstract
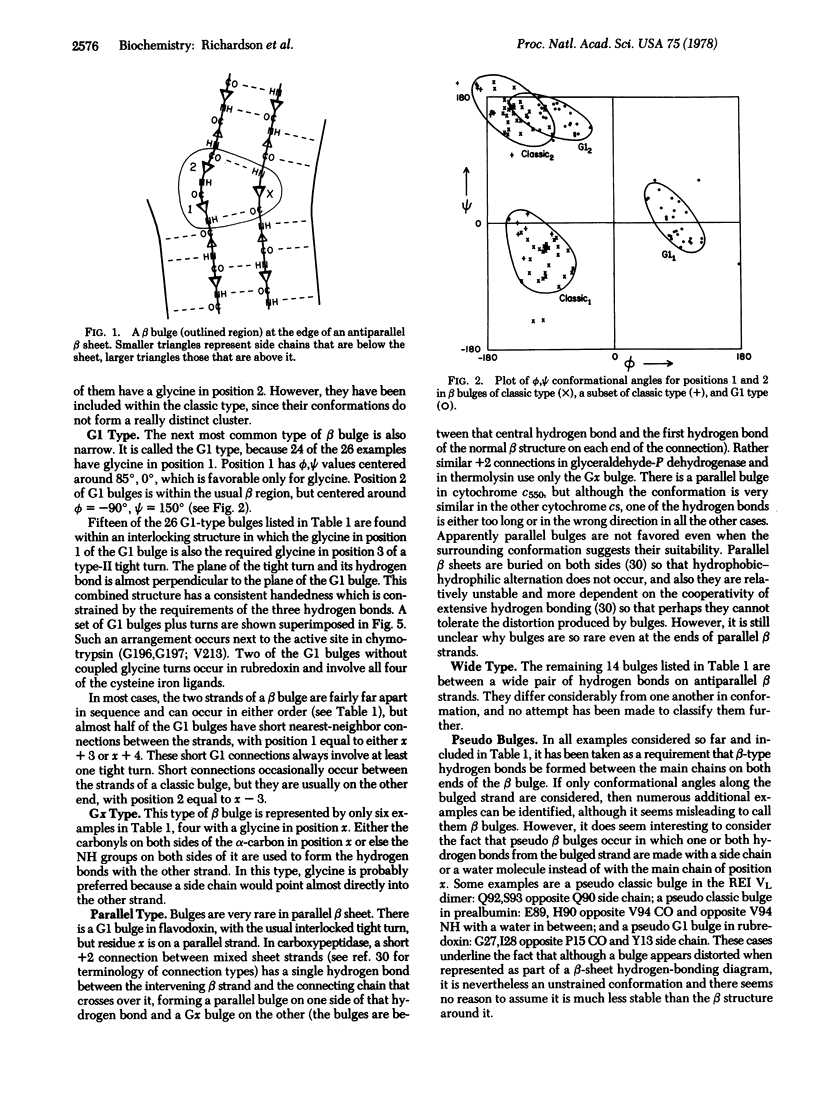
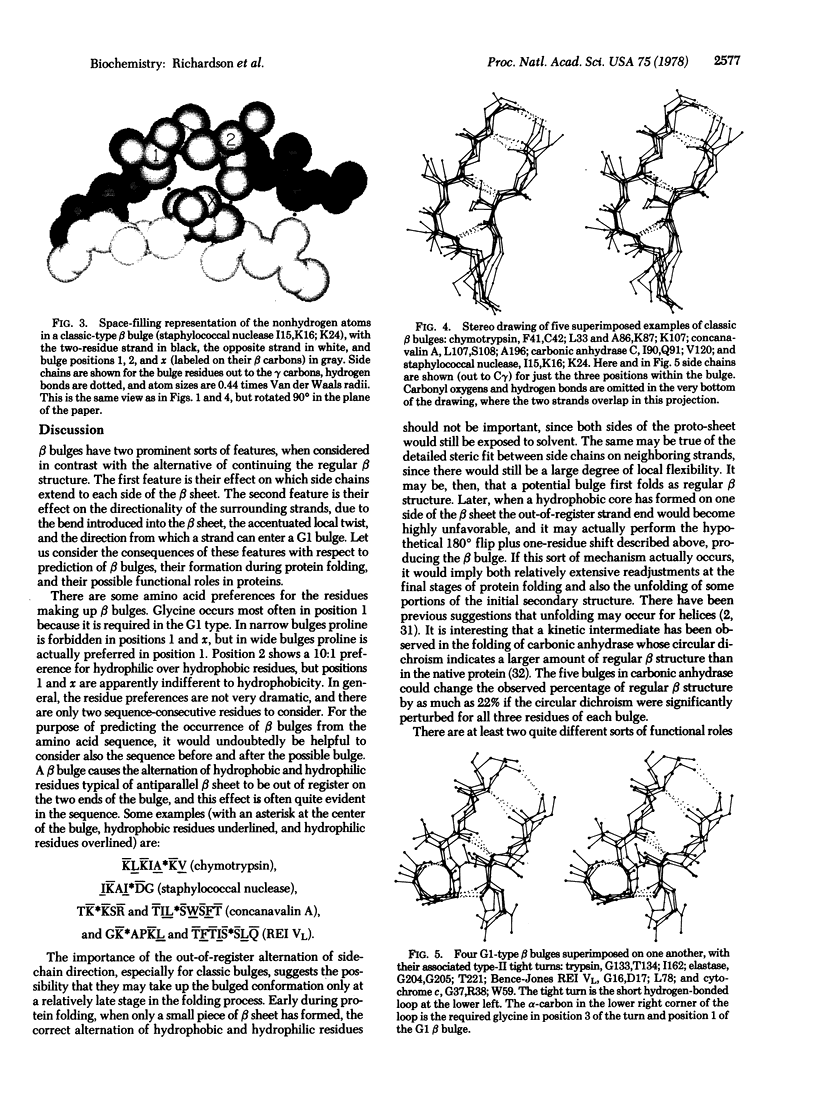
A beta bulge is a region between two consecutive beta-type hydrogen bonds which includes two residues (positions 1 and 2) on one strand opposite a single residue (position x) on the other strand. Compared to regular beta structure, a beta bulge puts the usual alternation of side-chain direction out of register on one of the strands, introduces a slight bend in the beta sheet, and locally accentuates the usual right-handed strand twist. Almost all beta bulges are between antiparallel strands, usually between a narrow rather than a wide pair of hydrogen bonds. Ninety-one examples are listed. The two commonest types are the "classic" beta bulge, with position 1 in approximately alpha-helical conformation, and the "G1" beta bulge, with a required glycine at position 1 in approximately left-handed alpha-helical conformation, G1 bulges almost always occur in combination with a type II tight turn. The functional roles of beta bulges probably include compensating for the effects of a single-residue insertion or deletion within beta structure and providing the strong local twist required for form closed beta barrel structures.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almassy R. J., Dickerson R. E. Pseudomonas cytochrome c551 at 2.0 A resolution: enlargement of the cytochrome c family. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2674–2678. doi: 10.1073/pnas.75.6.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birktoft J. J., Blow D. M. Structure of crystalline -chymotrypsin. V. The atomic structure of tosyl- -chymotrypsin at 2 A resolution. J Mol Biol. 1972 Jul 21;68(2):187–240. doi: 10.1016/0022-2836(72)90210-0. [DOI] [PubMed] [Google Scholar]
- Blake C. C., Oatley S. J. Protein-DNA and protein-hormone interactions in prealbumin: a model of the thyroid hormone nuclear receptor? Nature. 1977 Jul 14;268(5616):115–120. doi: 10.1038/268115a0. [DOI] [PubMed] [Google Scholar]
- Burnett R. M., Darling G. D., Kendall D. S., LeQuesne M. E., Mayhew S. G., Smith W. W., Ludwig M. L. The structure of the oxidized form of clostridial flavodoxin at 1.9-A resolution. J Biol Chem. 1974 Jul 25;249(14):4383–4392. [PubMed] [Google Scholar]
- Carter C. W., Jr, Kraut J., Freer S. T., Nguyen-Huu-Xuong, Alden R. A., Bartsch R. G. Two-Angstrom crystal structure of oxidized Chromatium high potential iron protein. J Biol Chem. 1974 Jul 10;249(13):4212–4225. [PubMed] [Google Scholar]
- Chothia C. Conformation of twisted beta-pleated sheets in proteins. J Mol Biol. 1973 Apr 5;75(2):295–302. doi: 10.1016/0022-2836(73)90022-3. [DOI] [PubMed] [Google Scholar]
- Chothia C., Levitt M., Richardson D. Structure of proteins: packing of alpha-helices and pleated sheets. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4130–4134. doi: 10.1073/pnas.74.10.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman P. M., Jansonius J. N., Matthews B. W. The structure of thermolysin: an electron density map at 2-3 A resolution. J Mol Biol. 1972 Oct 14;70(3):701–724. doi: 10.1016/0022-2836(72)90569-4. [DOI] [PubMed] [Google Scholar]
- Drenth J., Hol W. G., Jansonius J. N., Koekoek R. A comparison of the three-dimensional structures of subtilisin BPN' and subtilisin novo. Cold Spring Harb Symp Quant Biol. 1972;36:107–116. doi: 10.1101/sqb.1972.036.01.016. [DOI] [PubMed] [Google Scholar]
- Drenth J., Jansonius J. N., Koekoek R., Wolthers B. G. The structure of papain. Adv Protein Chem. 1971;25:79–115. doi: 10.1016/s0065-3233(08)60279-x. [DOI] [PubMed] [Google Scholar]
- Eklund H., Nordström B., Zeppezauer E., Söderlund G., Ohlsson I., Boiwe T., Söderberg B. O., Tapia O., Brändén C. I., Akeson A. Three-dimensional structure of horse liver alcohol dehydrogenase at 2-4 A resolution. J Mol Biol. 1976 Mar 25;102(1):27–59. doi: 10.1016/0022-2836(76)90072-3. [DOI] [PubMed] [Google Scholar]
- Epp O., Colman P., Fehlhammer H., Bode W., Schiffer M., Huber R., Palm W. Crystal and molecular structure of a dimer composed of the variable portions of the Bence-Jones protein REI. Eur J Biochem. 1974 Jun 15;45(2):513–524. doi: 10.1111/j.1432-1033.1974.tb03576.x. [DOI] [PubMed] [Google Scholar]
- Finkelstein A. V., Ptitsyn O. B. A theory of protein molecule self-organization. IV. Helical and irregular local structures of unfolded protein chains. J Mol Biol. 1976 May 5;103(1):15–24. doi: 10.1016/0022-2836(76)90049-8. [DOI] [PubMed] [Google Scholar]
- Huber R., Kukla D., Rühlmann A., Steigemann W. Pancreatic trypsin inhibitor (Kunitz). I. Structure and function. Cold Spring Harb Symp Quant Biol. 1972;36:141–148. doi: 10.1101/sqb.1972.036.01.019. [DOI] [PubMed] [Google Scholar]
- Ko B. P., Yazgan A., Yeagle P. L., Lottich S. C., Henkens R. W. Kinetics and mechanism of refolding of bovine carbonic anhydrase. A probe study of the formation of the active site. Biochemistry. 1977 Apr 19;16(8):1720–1725. doi: 10.1021/bi00627a031. [DOI] [PubMed] [Google Scholar]
- Liljas A., Kannan K. K., Bergstén P. C., Waara I., Fridborg K., Strandberg B., Carlbom U., Järup L., Lövgren S., Petef M. Crystal structure of human carbonic anhydrase C. Nat New Biol. 1972 Feb 2;235(57):131–137. doi: 10.1038/newbio235131a0. [DOI] [PubMed] [Google Scholar]
- Mathews F. S., Levine M., Argos P. Three-dimensional Fourier synthesis of calf liver cytochrome b 5 at 2-8 A resolution. J Mol Biol. 1972 Mar 14;64(2):449–464. doi: 10.1016/0022-2836(72)90510-4. [DOI] [PubMed] [Google Scholar]
- Némethy G., Phillips D. C., Leach S. J., Scheraga H. A. A second right-handed helical structure with the parameters of the Pauling-Corey alpha-helix. Nature. 1967 Apr 22;214(5086):363–365. doi: 10.1038/214363a0. [DOI] [PubMed] [Google Scholar]
- Poljak R. J., Amzel L. M., Chen B. L., Phizackerley R. P., Saul F. The three-dimensional structure of the fab' fragment of a human myeloma immunoglobulin at 2.0-angstrom resolution. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3440–3444. doi: 10.1073/pnas.71.9.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiocho F. A., Lipscomb W. N. Carboxypeptidase A: a protein and an enzyme. Adv Protein Chem. 1971;25:1–78. doi: 10.1016/s0065-3233(08)60278-8. [DOI] [PubMed] [Google Scholar]
- Reeke G. N., Jr, Becker J. W., Edelman G. M. The covalent and three-dimensional structure of concanavalin A. IV. Atomic coordinates, hydrogen bonding, and quaternary structure. J Biol Chem. 1975 Feb 25;250(4):1525–1547. [PubMed] [Google Scholar]
- Richardson J. S. beta-Sheet topology and the relatedness of proteins. Nature. 1977 Aug 11;268(5620):495–500. doi: 10.1038/268495a0. [DOI] [PubMed] [Google Scholar]
- Srinivasan R., Balasubramanian R., Rajan S. S. Extended helical conformation newly observed in protein folding. Science. 1976 Nov 12;194(4266):720–722. doi: 10.1126/science.982035. [DOI] [PubMed] [Google Scholar]
- Takano T., Trus B. L., Mandel N., Mandel G., Kallai O. B., Swanson R., Dickerson R. E. Tuna cytochrome c at 2.0 A resolution. II. Ferrocytochrome structure analysis. J Biol Chem. 1977 Jan 25;252(2):776–785. [PubMed] [Google Scholar]
- Venkatachalam C. M. Stereochemical criteria for polypeptides and proteins. V. Conformation of a system of three linked peptide units. Biopolymers. 1968 Oct;6(10):1425–1436. doi: 10.1002/bip.1968.360061006. [DOI] [PubMed] [Google Scholar]
- Wyckoff H. W., Tsernoglou D., Hanson A. W., Knox J. R., Lee B., Richards F. M. The three-dimensional structure of ribonuclease-S. Interpretation of an electron density map at a nominal resolution of 2 A. J Biol Chem. 1970 Jan 25;245(2):305–328. [PubMed] [Google Scholar]