Significance
Many bacteria contain large, circular DNA molecules, called plasmids, that encode physiologically, medically, and commercially important genes, including genes conferring virulence and drug resistance. The largest plasmids use active segregation systems to maintain themselves in the host bacterium. Such segregation systems provide remarkable insight into bacterial cell biology. The pLS32 plasmid, found in a commercially important strain of Bacillus subtilis, relies on a segregation system encoded by the alf operon. We show that DNA movement is driven by self-assembly of a polymeric protein, called AlfA, and that self-assembly of AlfA into a functional, DNA-segregating machine requires the activity of the accessory factor AlfB, which (unexpectedly) promotes both assembly and disassembly of AlfA polymers.
Keywords: DNA segregation, bacterial cytoskeleton, Bacillus subtilis, reconstitution
Abstract
In bacteria, some plasmids are partitioned to daughter cells by assembly of actin-like proteins (ALPs). The best understood ALP, ParM, has a core set of biochemical properties that contributes to its function, including dynamic instability, spontaneous nucleation, and bidirectional elongation. AlfA, an ALP that pushes plasmids apart in Bacillus, relies on a different set of underlying properties to segregate DNA. AlfA elongates unidirectionally and is not dynamically unstable; its assembly and disassembly are regulated by a cofactor, AlfB. Free AlfB breaks up AlfA bundles and promotes filament turnover. However, when AlfB is bound to the centromeric DNA sequence, parN, it forms a segrosome complex that nucleates and stabilizes AlfA filaments. When reconstituted in vitro, this system creates polarized, motile comet tails that associate by antiparallel filament bundling to form bipolar, DNA-segregating spindles.
The first filament-forming actin-like protein (ALP) was identified in bacteria in 2001 (1), and subsequent work identified more than 30 additional classes of ALPs in eubacteria and archaea (2). These proteins are involved in a variety of cellular processes, including assembly of the cell wall (1, 3–5), positioning of organelles (6), anchoring of cytokinesis machinery (7), and segregation of DNA (8). Most DNA-segregating ALPs participate in type II plasmid partitioning systems, which consist of three components: (i) a centromeric DNA sequence; (ii) a DNA-binding protein that interacts with the centromeric sequence to form a segrosome complex; and (iii) a polymer-forming ALP, whose self-assembly moves the segrosome through the cytoplasm (SI Appendix, Fig. S1). In the best understood type II segregation system, bidirectional polymerization of the ALP, ParM, pushes pairs of plasmids to opposite poles of rod-shaped cells. Extensive studies, both in vivo and in vitro, have produced detailed models of ParM-mediated DNA segregation (9, 10), but it is unclear to what extent these models can be generalized to describe other ALP-dependent DNA segregation systems.
To better understand the diversity of molecular mechanisms underlying plasmid segregation, we studied a type II plasmid partitioning system encoded by the alf operon from the Bacillus subtilis plasmid pLS32 (11). The alf operon was first identified based on its ability to maintain plasmids through the process of sporulation, presumably by actively pushing a plasmid into the forespore at one end of the cell. In addition, the alf operon confers approximately eightfold greater stability to plasmids in rapidly dividing cells. The alf operon itself contains a centromeric sequence, parN, with three short, repeated sequences, and it encodes both an ALP, AlfA, and a DNA-binding protein, AlfB. The AlfA protein shares 15% sequence identity with ParM (2), whereas AlfB shares only 8% identity with ParR and has no significant BLAST hits (expect value < 1). In addition to sequence differences, AlfA and ParM also differ biochemically. ParM filaments are dynamically unstable and do not form bundles in the absence of molecular crowding agents. AlfA, in contrast, displays no evidence of dynamic instability and forms stable filaments that self-assemble into mixed-polarity bundles, even in high salt concentrations (12, 13). The dynamic instability of ParM filaments is thought to play an important role in plasmid segregation, in part by providing the energy required to move the plasmids in a directed fashion. Energy must be expended to disassemble unneeded filaments and raise the concentration of monomers above the critical concentration required to elongate cargo-attached filaments. Otherwise, monomer and polymer reach a stable, steady-state ratio, and directed motion ceases. In eukaryotic cells, several proteins, including the actin-binding proteins cofilin and profilin, collaborate to maintain a high concentration of monomeric actin, orders of magnitude higher than the critical concentration for polymerization. In bacteria, the dynamic instability of unattached ParM filaments maintains the concentration of monomeric ParM approximately fourfold above the critical concentration of filaments attached to segrosomes (9, 14). If assembly of AlfA filaments drives plasmid movement, then how, in the absence of dynamic instability, is the concentration of monomeric AlfA maintained at a level sufficient to drive the growth of segrosome-attached filaments? To understand how AlfA filaments assemble and move DNA, we followed AlfA-dependent plasmid segregation in vivo and reconstituted the process in vitro using purified components. Like the par system, the alf system relies on assembly of actin-like polymers to push plasmids, but the two systems use different sets of underlying molecular mechanisms to favor the assembly of cargo-attached filaments over free filaments. AlfB binds to parN DNA, creating a segrosome complex that nucleates AlfA filaments and remains attached to their growing ends. Remarkably, however, the AlfB protein by itself also debundles and destabilizes AlfA filaments and maintains a high concentration of monomeric AlfA sufficient to drive plasmid movement. Once formed, AlfA filaments elongate in a polarized manner, from only one end, and binding of the segrosome complex increases filament stability, even in the presence of high concentrations of free AlfB. Together, these properties produce treadmilling AlfA “comet tails” that move DNA similar to the way polarized actin comet tails move cargo in eukaryotic cytoplasm. In sporulating cells, these monopolar comet tails would be sufficient to push plasmids into the forespore so that they would survive sporulation. In actively growing cells, antiparallel association of these comet tails can also segregate plasmids to opposite poles and decrease the rate of plasmid loss.
Results
AlfA Forms Dynamic Filaments in Vivo That Push Plasmids.
To study AlfA-driven cargo movement in B. subtilis cells, we constructed miniplasmids containing both the replication origin and alf operon from plasmid pLS32 (15). We inserted a second copy of the AlfA gene at the end of the native alf operon. The additional gene encodes an AlfA-GFP fusion protein and enables us to express both WT and fluorescently labeled AlfA in the same cells. In our construct, AlfA-GFP is present at 25% of the level of WT AlfA, whereas, in a similar, previously published strain (11), AlfA-GFP is present at 7% (SI Appendix, Fig. S2). Although expression of AlfA-GFP decreases the efficiency of the alf operon by approximately twofold, the modified alf operon remains functional and confers a fourfold increase in stability over plasmids lacking an alf operon altogether (SI Appendix, Table S4). We therefore used this construct to follow AlfA dynamics in vivo (Fig. 1). Consistent with previous reports (11), we found that AlfA forms filamentous structures that move, grow, and shorten on a timescale of approximately 1 min. The majority of cells do not contain a single, cell-spanning filament, but rather contain multiple, discrete bundles that frequently fragment, diffuse, and anneal with one another (Fig. 1A). These observations are surprising, given our previous demonstration that purified AlfA forms stable filament bundles (12), and they suggest that additional factors dynamize the filaments.
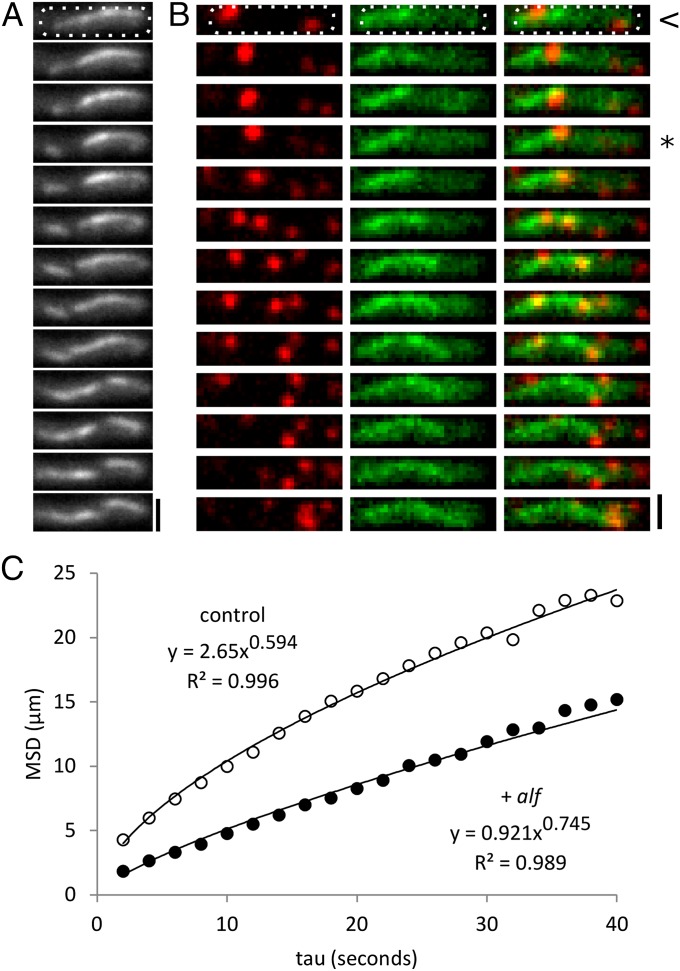
Fig. 1.
AlfA forms dynamic filaments in vivo that push plasmids. (A) AlfA bundles from pNCH106 in PY79 form a dynamic polymer network in the cell. This sequence contains examples of growth, shortening, fragmentation, and apparent annealing. Time between images: 10 s. (Scale bar, 1 µm.) Temperature, 30 °C. Cell outline shown in white dashed line. (B) pJKP06 plasmids (red) track along the sides and ends of growing AlfA bundles (green) in PY79 cells, and these motions can separate pairs of plasmids. Interval, 5 s. (Scale bar, 1 µm.) Temperature, 25 °C. Cell outline shown in white dashed line. Frames indicating transit along preexisting filaments begin with <, and those indicating tip surfing begin with *. (C) Mean squared displacement (MSD) of trajectories of plasmids with (pJKP06, n = 570) and without (pJKP02, n = 612) the alf system shows that alf plasmids have decreased mobility (a reduced apparent diffusion coefficient) but a more directed character of motion (an increased exponential scaling factor, α). Temperature, 25 °C.
We visualized plasmids containing a lacO array bound to chromosome-encoded mCherry-LacI and observed that some of them “surf” on the tips of elongating AlfA structures (Fig. 1B, frames beginning with *), similar to the way plasmids move on the ends of ParM spindles (16). Other plasmids, however, move linearly along preexisting AlfA polymers in a manner we never observed with ParM-driven plasmids (Fig. 1B, frames beginning with <). To quantify the population-level effects of the alf system on plasmid mobility, we used MicroTracker (17) to follow the movement of >500 plasmids over time. Surprisingly, plasmids carrying the alf operon are less mobile than controls, but the character of their motion is less diffusive and more directed. In plots of mean squared displacement vs. time (Fig. 1C), the exponential scaling factor that describes plasmid motion (α) is significantly increased by the presence of the alf operon, suggesting that plasmid motions are influenced by an active process (18). We next worked to figure out how accessory factors regulate the stability of AlfA filament bundles (12) and promote directed motion of plasmids.
Regulation of AlfA Polymer Dynamics by Elements of the alf Operon: AlfB and parN.
The alf operon contains three potential ORFs: AlfA, AlfB, and AlfC. Two have been shown to encode proteins important for plasmid segregation: AlfA and AlfB. Loss of the third, AlfC, has little effect on plasmid stability (19). The AlfB protein is thought to bind three DNA repeats in the parN locus and form a kinetochore-like segrosome structure (19) that interacts with AlfA filaments. If free AlfB helps regulate AlfA polymer dynamics, independent of its role in the segrosome, we would expect its cellular concentration to be much higher than that of the AlfB binding sites in parN, which we estimate to be ∼25 nM (15). We would, instead, expect the AlfB concentration to be comparable to that of AlfA. The concentration of AlfA, in turn, should be higher than the critical concentration for its polymerization (2.4 µM) (12). To find out whether this is case, we used quantitative immunoblotting to estimate the concentrations of AlfA and AlfB in Bacillus cells (SI Appendix, Fig. S3). Consistent with a role outside the segrosome, we found that the ratio of AlfB to AlfA is quite high (∼1:2). We estimate the AlfA concentration to be 20 µM in the B. subtilis cell, whereas AlfB is present at ∼8 µM. In contrast, the estimated concentrations of ParM and ParR are, respectively, ∼15 and <1 µM (8).
We next used right-angle light scattering to follow kinetics of AlfA assembly in the presence of various concentrations of AlfB (Fig. 2A). Substoichiometric concentrations of AlfB (<12% of AlfA concentration) have little effect on the kinetics of polymer assembly, suggesting that AlfB by itself does not promote nucleation or severing of AlfA filaments. Low concentrations of AlfB do, however, dramatically reduce the intensity of the light scattering signal at steady state. Light scattering by AlfA is dominated by large filament bundles (12) so AlfB might decrease steady-state scattering because it antagonizes AlfA bundling, even at these low stoichiometries. We confirmed this by electron microscopy (Fig. 2D), which reveals both a dramatic decrease in the number of AlfA bundles (Fig. 2C) and an overall shortening of filament and bundle lengths in the presence of AlfB. How does AlfB reduce the bundling of AlfA filaments? Two simple possibilities are that AlfB either (i) binds to the sides of AlfA filaments and prevents their lateral association or (ii) decreases the lifetime of AlfA filaments so that they do not survive long enough to be incorporated into bundles. Consistent with both roles for AlfB, we find that it has no effect on AlfA bundles formed in the presence of the slowly hydrolyzable nucleotide analog ATPγS (SI Appendix, Fig. S5B). This insensitivity suggests that AlfB promotes disassembly of filaments that have hydrolyzed their bound ATP, or that AlfB cannot promote the disassembly of bundled filaments. Higher concentrations of AlfB (≥40% of the AlfA concentration) begin to reduce the rate of polymerization, suggesting that AlfB might—with low affinity—also cap AlfA filaments or sequester AlfA monomers. Consistent with low affinity sequestration, we found that AlfB increases the critical concentration of AlfA in a concentration-dependent manner (SI Appendix, Table S5) and that AlfB binds to monomeric AlfA (SI Appendix, Fig. S6).
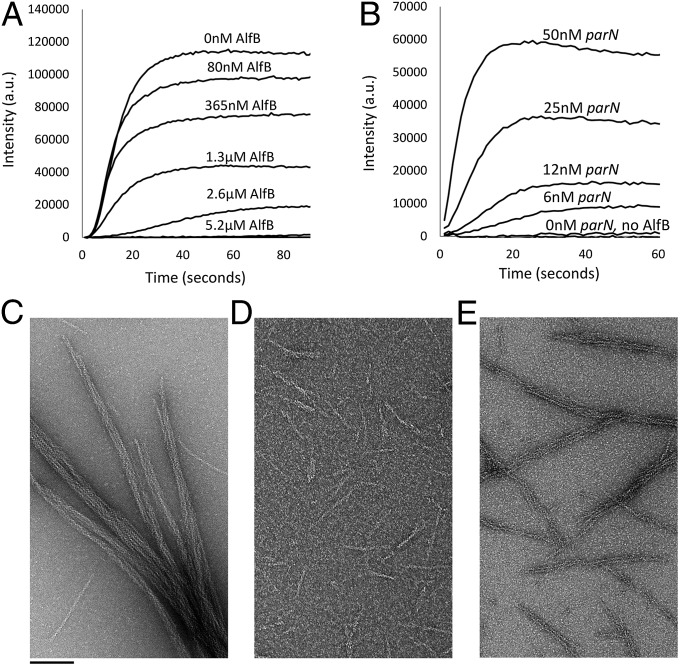
Fig. 2.
AlfB and the parN/AlfB segrosome complex regulate stability and self-association of AlfA filaments. (A) Increasing concentrations of AlfB destabilize 3.3 µM AlfA polymerized in the presence of 2.5 mM ATP. Buffer, 25 mM Tris, pH 7.5, 100 mM KCl, 1 mM MgCl2, and 1 mM DTT. (B) Increasing concentrations of parN promote polymerization of 2 µM AlfA in the presence of 1.3 µM AlfB. This concentration of AlfA does not support polymerization without parN even in the absence of AlfB (bottom trace; 5 mM ATP). Buffer as above. (C) AlfA alone forms regular bundles. Electron micrographs of negatively stained 4 µM AlfA polymerized with 2 mM ATP alone. Buffer as above. (D) AlfB debundles AlfA polymer and shortens the polymer length distribution (as above with 2 µM AlfB). Buffer as above. (E) The addition of both 2 µM AlfB and 60 nM parN restores bundles (as above with 60 nM parN). Buffer as above.
By contrast, addition of AlfB together with double-stranded DNA containing the centromeric parN sequence promotes AlfA polymerization at low concentrations, below those required for assembly of AlfA alone or in the presence of AlfB (Fig. 2B). That is, the parN/AlfB segrosome complex lowers the AlfA concentration required for filament assembly (SI Appendix, Table S5). In addition, parN promotes AlfA bundling in the presence of AlfB (Fig. 2E). Given that the number of AlfB binding sites introduced by parN (assuming binding of a dimer of AlfB to each of three repeats per molecule of parN) is a small fraction of the total concentration of AlfB (360 nM of 2 µM total), the increased bundling cannot be accounted for solely by a reduction in free AlfB. Because AlfB does not disrupt stable, preformed bundles (SI Appendix, Fig. S5B), parN-stabilized filaments likely form bundles because they survive long enough to interact with each other. These effects are observed only with parN-containing DNA, and not with nonspecific DNA (SI Appendix, Fig. S4).
Normalizing for the reduced critical concentration, we found that the segrosome complex abolishes the early lag phase of polymerization associated with spontaneous nucleation (Fig. 3A). On log-log plots (Fig. 3A, Inset), the segrosome decreases the slope of the initial phase of polymerization from ∼2 to ∼1, indicating that segrosome-mediated nucleation occurs in a single step (20) rather than the two steps required for spontaneous AlfA assembly (12). To test whether the number of AlfB binding sequences in parN is optimized for nucleation, we compared the rates of polymerization induced by WT parN with mutants containing various numbers of AlfB binding sites (SI Appendix, Table S6). Remarkably, parN constructs containing three tandem repeats induce the fastest rate of polymer assembly (Fig. 3B). Adding or removing binding sites significantly slows polymer assembly, even when the total concentration of all components (AlfA, AlfB, and AlfB binding sites) remains constant. Oddly, increasing the spacing between parN repeats further accelerates polymerization of AlfA and, judging from the steady-state light scattering signal, may increase polymer stability (SI Appendix, Fig. S5A). Decreasing the length of the linker or removing it entirely has only a small effect on assembly kinetics. These data suggest that, although the AlfB/parN segrosome complex nucleates rapid filament assembly, the parN locus has not evolved to promote the fastest assembly possible (21). Finally, we verified the sequence-specific binding of AlfB to parN (22) by electron microscopy of negatively stained samples (Fig. 3 C and D).
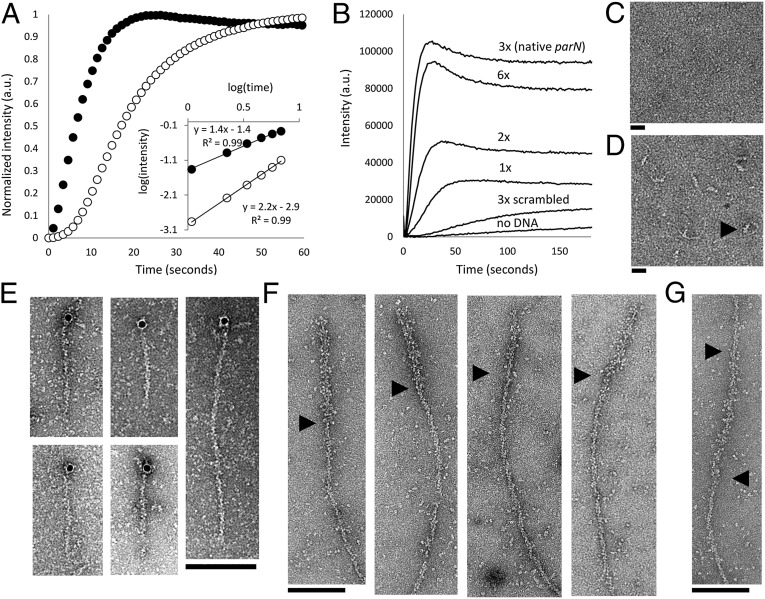
Fig. 3.
AlfB forms a complex on parN DNA that nucleates AlfA polymerization and binds to the ends of polar filaments. (A) Intensity-normalized light scattering of 300 nM of AlfA polymer formed in the absence (○) and presence (●) of 1.3 µM AlfB and 50 nM parN. The nucleation-dominated lag phase of polymerization is dramatically reduced with AlfB and parN, and the change in the slope of early time points on a log-log plot indicates that the number of steps of polymer assembly is reduced (5 mM ATP). Buffer, 25 mM Tris, pH 7.5, 100 mM KCl, 1 mM MgCl2, and 1 mM DTT. (B) Stabilization of AlfA by the parN/AlfB complex depends on the valency of AlfB-binding DNA repeats. Each reaction contains 2.1 µM AlfA, 1.3 µM AlfB, and 75 nM individual repeats. For example, the sequence containing a single repeat is present at 75 nM, whereas the native parN sequence, with three repeats, is present at 25 nM (2 mM ATP). Buffer as above. (C) 500 nM AlfB does not form a complex in the presence of 50 nM parC (the centromere of the ParM system). Buffer, 25 mM Tris, pH 7.5, 30 mM KCl, 10 mM (NH4)2SO4, 1 mM DTT, and 1 mM EDTA. (Scale bar, 20 nM.) (D) A complex is formed in the presence of parN. (Scale bar, 20 nM.) Black arrowhead points to a representative complex. Buffer as above. (E) AlfB-parN binds to the ends of filaments; 10-nm colloidal gold particles coated in streptavidin localize predominantly to the ends of 3.5 µM AlfA KK21AA KK101AA filaments in the presence of 1.3 µM AlfB and 120 nM parN-biotin (2 mM ATP). Buffer, 25 mM Tris, pH 7.5, 100 mM KCl, 1 mM MgCl2, and 1 mM DTT. (F) Filaments are polar. KK21AA KK101AA filaments (3 μM total monomer concentration) was polymerized off of AMP-PNP stabilized seeds composed of 50% KK21AA KK101AA AlfA and 50% AlfA-biotin decorated with an excess of streptavidin and diluted to 100 nM. Black arrowheads show boundaries between streptavidin-decorated seeds and normal filaments. Buffer as above. (G) A subset (21%) of these labeled filaments displays the seed in the middle. This configuration may be due to bundling of the WT polymer or streptavidin cross-linking within the seed. Buffer as above. Black arrowheads denote ends of the streptavidin seed.
The nucleating and stabilizing activities of the segrosome complex suggest that it might bind to the ends of AlfA filaments. To localize the parN/AlfB complex on AlfA filaments, we coupled 10-nm colloidal gold-streptavidin conjugates to parN-biotin (SI Appendix, Table S6) and, to improve the clarity of our images, we generated an AlfA mutant with reduced bundling (SI Appendix, Fig. S7). We observed some particles associated with the sides of AlfA filaments (15/66, or 23% of filaments) but, in the majority of cases where we observed filaments associated with gold particles (48/66, or 73% of filaments), the particles are attached to only one end of the filament (Fig. 3E). We rarely observed segrosome complexes decorating both ends of an AlfA filament (3/66 or 5% of filaments), and this may be due to residual bundling activity of the mutant (∼5% of observed structures on EM grids are bundles) rather than symmetrical association of the segrosome with the filaments. To test this idea, we constructed stable AlfA seeds that we could distinguish from normal AlfA filaments in electron micrographs by decorating short, stabilized AlfA filaments with streptavidin. Seeds were assembled from 50% bundling-deficient AlfA mutant and 50% biotinylated WT AlfA (12). When seeds are elongated by 100% bundling-deficient AlfA in the presence of adenosine 5′-(β,γ-imido)triphosphate lithium salt hydrate (AMP-PNP), the majority (11/14 or 79%) grow preferentially from one end (Fig. 3F). A small fraction of seeds grow from both ends (3/14 or 21%), possibly as a result of streptavidin cross-linking, residual bundling activity, or annealing (Fig. 3G).
Reconstitution of AlfA-Mediated DNA Movement and Segregation: Treadmilling of AlfA Bundles Produces Motile Comet Tails That Push DNA.
We used total internal reflection fluorescence (TIRF) microscopy to watch assembly of AlfA filaments in the presence of AlfB-parN segrosome complexes. Using multivalent, streptavidin parN-Cy3 conjugates, we found that a 1:2 ratio of AlfB to AlfA supported formation of dynamic, polarized, comet tail structures that drive directed motion of DNA (Fig. 4 A and B). The segrosome complexes associate with the thick ends of tapered filament bundles, whose width and density decrease from the parN-associated end to the distal tip. At lower DNA concentrations, comet tails are longer and adhere more tightly to the glass surface. The increased adhesion revealed that DNA surfs processively on the elongating tips of comet tails (Fig. 4C and Movie S1). The morphology and dynamics of these structures indicate that the DNA-associated end is continuously assembling, whereas the free end is continuously disassembling. Thus, the filament destabilizing effect of free AlfB and the stabilizing effect of AlfB/parN segrosomes work together to promote preferential elongation of DNA-attached AlfA filaments and bundles and to generate sustained treadmilling of AlfA bundles. In addition to surfing growing ends, some parN particles also move processively along their lengths, probably propelled by association with the end of a filament growing along the bundle. Individual bundles can also support bidirectional movement (Fig. 4C and Movie S2). At higher DNA concentrations, individual DNA foci occasionally split, with the two components moving away in opposite directions along the bundle (Fig. 4D and Movie S3). At higher parN concentrations, comet tails are shorter, probably because the higher number of segrosome-associated AlfA comets compete for AlfA monomers. We frequently observed motile AlfA/segrosome structures undergoing complex rearrangements, including capture of adjacent structures by bundling, fragmentation of DNA particles on bifurcating bundles, and bidirectional DNA segregation (Fig. 4E and Movies S4 and S5). We observed a distinct bias toward bidirectional segregation, in that pairs of DNA particles attached to the same structure move apart at three times the frequency that they move together (SI Appendix, Table S7). This bias, together with previously reported reference-free averages of AlfA filament pairs (12), suggests that AlfA filaments have an inherent preference for antiparallel bundling.
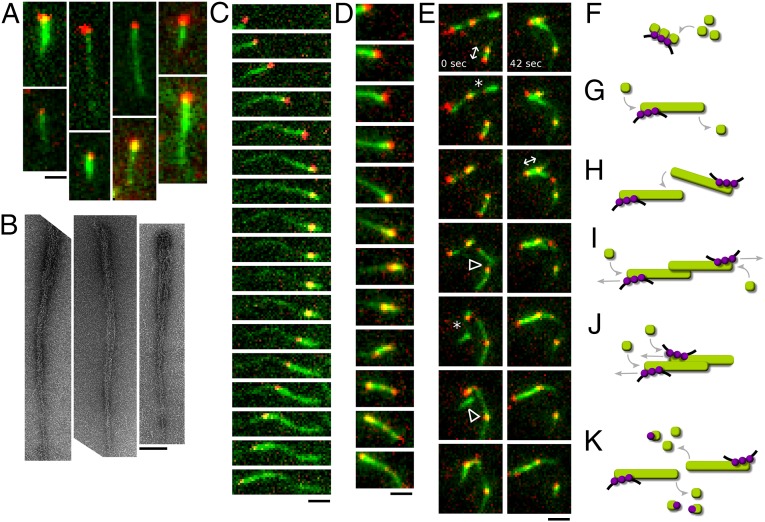
Fig. 4.
Reconstitution of AlfA-mediated DNA segregation. Bundling of AlfA comet tails organizes DNA movements along a single axis. (A and B) In the presence of AlfB, AlfA polymers form comet tails behind multivalent parN-streptavidin conjugates. (A) These asymmetric structures can be visualized by TIRF microscopy. (Scale bar, 1 µm.) Buffer, 25 mM Tris, pH 7.5, 100 mM KCl, 1 mM MgCl2, 1 mM DTT, 0.4% 400 CP methylcellulose, and 15 mg/mL BSA. (B) Asymmetric structures can also be seen with electron microscopy. (Scale bar, 100 nm.) Note the tapered appearance of the AlfA bundles. (3.2 µM AlfA, 1.6 µM AlfB, 60 nM parN). Buffer as above, without methylcellulose and BSA. (C) The growing end of an AlfA polymer pushes a parN conjugate. When the filament elongation ceases, the parN particle reverses direction and tracks backward along the original bundle ahead of new polymer growth, demonstrating that these bundles can support bidirectional movement (20 nM parN). (Scale bar, 1 µm.) Interval, 20 s. Buffer as in A. (D) A parN particle on the end of an AlfA polymer splits in two. One focus continues to track the growing filament tip, whereas the other reverses direction and moves backward along the bundle (60 nM parN). (Scale bar, 1 µm.) Interval, 12 s. Buffer as in A. (E) A group of AlfA bundles with multiple parN foci undergo complex rearrangements including bundle thinning and fragmenting (*), capture by annealing (∆), and segregation by polymerization (↔) (60 nM parN). (Scale bar, 1 µm.) Interval, 6 s. Buffer as in A. (F–K) Model for AlfA-mediated segregation. (F) AlfA (green) polymer assembly occurs preferentially at parN (black)-AlfB (purple) complexes, which nucleate filaments below the critical concentration by coordinating AlfA monomers. (G) The parN-AlfB complex also stabilizes filaments beneath the AlfA critical concentration. (H) When filaments encounter one another, they may capture one another by bundling together. (I) Insertional polymerization of antiparallel bundles will push plasmids apart from one another. (J) Parallel bundles will not bring plasmids together, unless only the leading filament stalls. If the lagging filament stalls, plasmids will still be segregated. Therefore, assuming parallel and antiparallel bundling are equally likely, plasmids will tend to be segregated. (K) The range of paired plasmid movement is limited by filament turnover at ends free of AlfB-parN, with the debundling and sequestration activities of free AlfB promoting the turnover of AlfA filaments.
Discussion
In our initial characterization of its assembly dynamics, we found that AlfA forms stable bundles, with no indication of dynamic instability (12). Given this, we wondered how segrosome-attached filaments could move cargo at steady state. That is, how is the assembly of segrosome-attached filaments privileged over that of unattached filaments. Here we demonstrate that this problem is solved by the Alf B protein, which not only couples AlfA filaments to DNA but also destabilizes unattached AlfA filaments and bundles. The net effect is to raise the steady state concentration of AlfA monomers and promote preferential elongation of DNA-associated filament ends.
In addition to stabilizing attached AlfA filaments, the AlfB-parN segrosome complex also nucleates new filament formation. The efficiency of nucleation depends strongly on the number of AlfB-binding repeats in the DNA sequence, with the WT number of three repeats being optimal. This number of repeats might be related to the fact that the critical AlfA nucleus is composed of three monomers (12), but because the AlfB protein is likely a dimer, further biophysical studies will be required to understand the mechanism of segrosome-mediated nucleation. Interestingly, we find that increasing the spacing between AlfB-binding repeats in parN increases the efficiency of nucleation. Either (i) there is no selective advantage to faster nucleation or (ii) other functions of AlfB, such as transcriptional repression (19), dictate its spacing along the DNA.
Although AlfB promotes dissociation of AlfA bundles, segrosome-bound AlfA filaments form bundles even in the presence of excess AlfB. Because substoichiometric concentrations of parN can produce this effect, it is unlikely to be caused by depletion of free AlfB. Furthermore, because we only rarely observe gold-labeled parN conjugates decorating the sides of AlfA filaments, it is unlikely that the segrosome complex bundles filaments by cross-linking them along their lengths. Instead, we propose that bundling may be related to filament stability. We previously observed formation of bundles in vitro by slow, lateral association of AlfA filaments (12), and therefore we suggest that long-lived filaments have more time to find each other and form bundles. AlfB appears to destabilize AlfA filaments after they have hydrolyzed their bound ATP. This destabilization would undermine the stability of existing bundles and decrease the frequency with which filaments interact to form bundles in the first place. The AlfB/parN segrosome increases the stability of bound filaments and probably preserves them long enough to permit bundle formation. Based on our data, it is unclear whether bundling itself also increases the stability of AlfA polymers, but this is likely. Broadly, the role of bundling and molecular crowding in plasmid segregation systems needs to be investigated in greater detail, especially in light of recent studies of the ParM system (10).
The AlfB-parN segrosome binds preferentially to one end of AlfA filaments. This polarized association of the segrosome coupled with the destabilization of unbound ends dynamizes the system and promotes uni-directional treadmilling. Insertional polymerization can propel the DNA forward on the end of a growing comet tail or along the length of a preexisting structure through annealing of filaments. Based on these observations, we propose a simple model for AlfA-mediated plasmid movement and segregation (Fig. 4 F–K). Filament assembly is directed to plasmids by the nucleation and stabilization activity of AlfB-parN. This localized stabilization is sufficient to produce linear structures capable of finding the long axis of rod-shaped B. subtilis cells (2, 16) and is likely sufficient to place the alf-containing plasmid, pLS32, in the forespore compartment of sporulating cells. This basic behavior probably underlies the dramatic increase in the rate at which alf operon-containing plasmids survive sporulation. When two plasmid-bound filaments encounter one another, they can bundle together. If the bundling is antiparallel, which appears to be the preferred interaction, further polymerization will drive the plasmids apart. If the bundling is parallel, the distance between plasmids will be maintained as both centromeres move in the same direction until the leading filament stalls. Furthermore, the disparity in filament stability at centromere-bound and unbound ends predicts that the range of these coordinated movements will be limited by filament turnover.
Thus, the alf system provides bacterial implementations of regulatory features familiar to eukaryotic actin, such as regulated nucleation, filament destabilization driven by a cofilin-like cofactor, filament treadmilling, and motile comet tail formation. Furthermore, the suppression and rescue of inherent bundling properties of AlfA may play a role in specifying physical associations between plasmid-bound polymers. These features expand our understanding of diversity in the regulation of bacterial polymers that move cellular cargoes.
Materials and Methods
Briefly, intracellular concentrations were calculated using values from the literature (23, 24). Plasmids for B. subtilis were constructed from a fragment from pWH1520 containing an Escherichia coli origin and ampicillin resistance gene, as well as a B. subtilis tetracycline resistance gene. This fragment was ligated to repN, the origin of replication from pLS32. To this backbone, the alf operon, a lacO array, and GFP from pMutin were added. Fluorescently tagged LacI constructs were cloned under a xlyose-inducible promoter into pSG1154 from the Pogliano laboratory, which integrates into AmyE. Constructs were transformed into PY79 for integration in the AmyE locus.
Live cell imaging of B. subtilis was performed as previously described (2). Images were acquired in oblique TIRF on a Nikon TE 2000 inverted microscope controlled with MicroManager (25) as previously described (12). Plasmid foci were tracked with MicroTracker (17), and mean squared displacement plots were created using MatLab scripts.
AlfA was purified for TIRF assays, light scattering, pelleting, and negative-stain electron microscopy as described (12). AlfB was expressed with a tobacco etch virus-cleavable 6His fusion and purified by affinity. Full materials and methods (26–30) are provided in the SI Appendix.
Supplementary Material
Acknowledgments
We thank A. Derman and J. Pogliano (University of California, San Diego) for strains, plasmids, instruction on microbiological technique, and many invaluable conversations. We also thank K. C. Huang (Stanford University), A. Gopinathan (University of California, Merced), and E. C. Garner (Harvard University) for helpful discussions. This work was funded by National Institutes of Health Grants R01GM095263 and R01GM079556 (to R.D.M.) and by National Science Foundation and Genentech Graduate Research Fellowships (to J.K.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304127111/-/DCSupplemental.
References
- 1.Jones LJ, Carballido-López R, Errington J. Control of cell shape in bacteria: Helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104(6):913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 2.Derman AI, et al. Phylogenetic analysis identifies many uncharacterized actin-like proteins (Alps) in bacteria: Regulated polymerization, dynamic instability and treadmilling in Alp7A. Mol Microbiol. 2009;73(4):534–552. doi: 10.1111/j.1365-2958.2009.06771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garner EC, et al. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333(6039):222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Domínguez-Escobar J, et al. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333(6039):225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- 5.van Teeffelen S, et al. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc Natl Acad Sci USA. 2011;108(38):15822–15827. doi: 10.1073/pnas.1108999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komeili A, Li Z, Newman DK, Jensen GJ. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006;311(5758):242–245. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- 7.Ma X, Ehrhardt DW, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci USA. 1996;93(23):12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Møller-Jensen J, Jensen RB, Löwe J, Gerdes K. Prokaryotic DNA segregation by an actin-like filament. EMBO J. 2002;21(12):3119–3127. doi: 10.1093/emboj/cdf320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garner EC, Campbell CS, Weibel DB, Mullins RD. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science. 2007;315(5816):1270–1274. doi: 10.1126/science.1138527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gayathri P, et al. A bipolar spindle of antiparallel ParM filaments drives bacterial plasmid segregation. Science. 2012;338(6112):1334–1337. doi: 10.1126/science.1229091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Becker E, et al. DNA segregation by the bacterial actin AlfA during Bacillus subtilis growth and development. EMBO J. 2006;25(24):5919–5931. doi: 10.1038/sj.emboj.7601443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polka JK, Kollman JM, Agard DA, Mullins RD. 2009. The structure and assembly dynamics of plasmid actin AlfA imply a novel mechanism of DNA segregation. J Bacteriol 191(20):6219–6230.
- 13.Popp D, et al. Polymeric structures and dynamic properties of the bacterial actin AlfA. J Mol Biol. 2010;397(4):1031–1041. doi: 10.1016/j.jmb.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 14.Fuesler JA, Li H-J. Dynamic instability—A common denominator in prokaryotic and eukaryotic DNA segregation and cell division. Cell Mol Biol Lett. 2012;17(4):542–548. doi: 10.2478/s11658-012-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka T, Ogura M. A novel Bacillus natto plasmid pLS32 capable of replication in Bacillus subtilis. FEBS Lett. 1998;422(2):243–246. doi: 10.1016/s0014-5793(98)00015-5. [DOI] [PubMed] [Google Scholar]
- 16.Campbell CS, Mullins RD. In vivo visualization of type II plasmid segregation: bacterial actin filaments pushing plasmids. J Cell Biol. 2007;179(5):1059–1066. doi: 10.1083/jcb.200708206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaqaman K, et al. Robust single-particle tracking in live-cell time-lapse sequences. Nat Methods. 2008;5(8):695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weber SC, Spakowitz AJ, Theriot JA. Bacterial chromosomal loci move subdiffusively through a viscoelastic cytoplasm. Phys Rev Lett. 2010;104(23):238102. doi: 10.1103/PhysRevLett.104.238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka T. Functional analysis of the stability determinant AlfB of pBET131, a miniplasmid derivative of bacillus subtilis (natto) plasmid pLS32. J Bacteriol. 2010;192(5):1221–1230. doi: 10.1128/JB.01312-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flyvbjerg H, Jobs E, Leibler S. Kinetics of self-assembling microtubules: An “inverse problem” in biochemistry. Proc Natl Acad Sci USA. 1996;93(12):5975–5979. doi: 10.1073/pnas.93.12.5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Theriot JA. Why are bacteria different from eukaryotes? BMC Biol. 2013;11:119–135. doi: 10.1186/1741-7007-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rivera CR, Kollman JM, Polka JK, Agard DA, Mullins RD. Architecture and assembly of a divergent member of the ParM family of bacterial actin-like proteins. J Biol Chem. 2011;286(16):14282–14290. doi: 10.1074/jbc.M110.203828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fisher AJ, Rosenstiel TN, Shirk MC, Fall R. Nonradioactive assay for cellular dimethylallyl diphosphate. Anal Biochem. 2001;292(2):272–279. doi: 10.1006/abio.2001.5079. [DOI] [PubMed] [Google Scholar]
- 24.McCabe BC, Gollnick P. Cellular levels of trp RNA-binding attenuation protein in Bacillus subtilis. J Bacteriol. 2004;186(15):5157–5159. doi: 10.1128/JB.186.15.5157-5159.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van den Ent F, Löwe J. RF cloning: A restriction-free method for inserting target genes into plasmids. J Biochem Biophys Methods. 2006;67(1):67–74. doi: 10.1016/j.jbbm.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 27.Stuurman N, Edelstein AD, Amodaj N, Hoover KH, Vale RD. 2010. Computer control of microscopes using μManager. Curr Protoc Mol Biol 92(Suppl):14.20.2--14.20.17.
- 28.Rygus T, Scheler A, Allmansberger R, Hillen W. Molecular cloning, structure, promoters and regulatory elements for transcription of the Bacillus megaterium encoded regulon for xylose utilization. Arch Microbiol. 1991;155(6):535–542. doi: 10.1007/BF00245346. [DOI] [PubMed] [Google Scholar]
- 29.Vagner V, Dervyn E, Ehrlich SD. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144(Pt 11):3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 30.Ingerman E, Hsiao JY, Mullins RD. Arp2/3 complex ATP hydrolysis promotes lamellipodial actin network disassembly but is dispensable for assembly. J Cell Biol. 2013;200(5):619–633. doi: 10.1083/jcb.201211069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.