Significance
Myosin-binding protein C (MyBP-C) is a component of myosin filaments, one of the two sets of contractile elements whose relative sliding is the basis of muscle contraction. In the heart, MyBP-C modulates contractility in response to cardiac stimulation; mutations in MyBP-C lead to cardiac disease. The mechanism by which MyBP-C modulates cardiac contraction is not understood. Using electron microscopy and a light microscopic assay for filament sliding, we demonstrate that MyBP-C binds to the other set of filaments, containing actin and the regulatory component, tropomyosin. In so doing, it displaces tropomyosin from its inhibitory position to activate actin filament interaction with myosin, promoting filament sliding. These findings provide insights into the molecular basis of heart function.
Keywords: muscle regulation, muscle activation
Abstract
Myosin-binding protein C (MyBP-C) is an accessory protein of striated muscle thick filaments and a modulator of cardiac muscle contraction. Defects in the cardiac isoform, cMyBP-C, cause heart disease. cMyBP-C includes 11 Ig- and fibronectin-like domains and a cMyBP-C-specific motif. In vitro studies show that in addition to binding to the thick filament via its C-terminal region, cMyBP-C can also interact with actin via its N-terminal domains, modulating thin filament motility. Structural observations of F-actin decorated with N-terminal fragments of cMyBP-C suggest that cMyBP-C binds to actin close to the low Ca2+ binding site of tropomyosin. This suggests that cMyBP-C might modulate thin filament activity by interfering with tropomyosin regulatory movements on actin. To determine directly whether cMyBP-C binding affects tropomyosin position, we have used electron microscopy and in vitro motility assays to study the structural and functional effects of N-terminal fragments binding to thin filaments. 3D reconstructions suggest that under low Ca2+ conditions, cMyBP-C displaces tropomyosin toward its high Ca2+ position, and that this movement corresponds to thin filament activation in the motility assay. At high Ca2+, cMyBP-C had little effect on tropomyosin position and caused slowing of thin filament sliding. Unexpectedly, a shorter N-terminal fragment did not displace tropomyosin or activate the thin filament at low Ca2+ but slowed thin filament sliding as much as the larger fragments. These results suggest that cMyBP-C may both modulate thin filament activity, by physically displacing tropomyosin from its low Ca2+ position on actin, and govern contractile speed by an independent molecular mechanism.
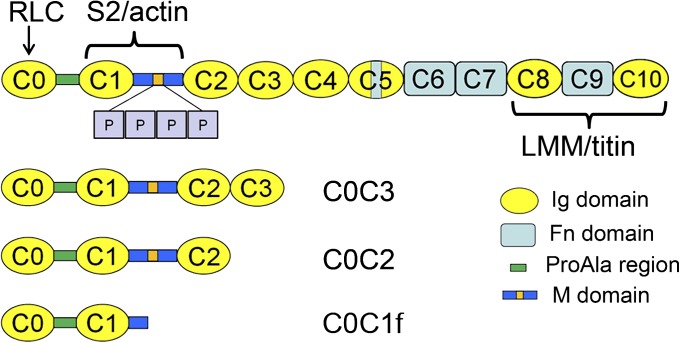
Myosin-binding protein C (MyBP-C) is an accessory protein of vertebrate striated muscle thick filaments (1) that is known to modulate cardiac muscle contraction (2). The skeletal isoform includes 10 Ig-like (Ig) and fibronectin type 3-like (Fn) domains, numbered C1 through C10 from the N terminus, together with a MyBP-C-specific motif (the M-domain) between C1 and C2 and a Pro-Ala-rich sequence at the N terminus. The cardiac isoform (cMyBP-C) has an additional N-terminal Ig domain (C0), four phosphorylation sites in the M-domain, and a 28-residue insert in the C5 domain (3) (Fig. 1). MyBP-C binds to the thick filament in the C-zone of the sarcomeric A-band (4) via its C-terminal domains (C8–C10) (5), whereas its N-terminal region contains binding sites for myosin S2 (6–10) and the myosin regulatory light chain (11).
Fig. 1.
Schematic of cMyBP-C and the expressed N-terminal fragments C0C3, C0C2, and C0C1f used in this study. cMyBP-C consists of 8 Ig and 3 Fn domains together with a cMyBP-C-specific M-domain, containing a phosphorylation region (orange) with 4 phosphorylatable serines (P), a ProAla-rich domain, and a cardiac-specific insert (blue) in the C5 domain.
In addition to binding to myosin, MyBP-C also interacts with actin (12) and with thin filaments (13) via its N-terminal region (9, 10, 14–17; cf. 18). In the in vitro motility assay, actin filament sliding over myosin is slowed by N-terminal fragments of cMyBP-C to the same extent as whole cMyBP-C (19), possibly by slowing the myosin detachment rate from actin (19) or tethering the thick to the thin filament (16, 19). In an assay closer to the in vivo situation, the sliding of F-actin over native cardiac thick filaments was slowed specifically in the C-zone, and this slowing was ablated by removal of C0C1 and the first 17 amino acids of the M-domain [known as C0C1f (16); Fig. 1] (20). On the basis of yeast 2 hybrid experiments, it was concluded that the C1 and M domains were necessary for actin binding and that replacement of endogenous cMyBP-C with actin binding-ablated cMyBP-C resulted in its abnormal sarcomeric distribution and disturbance of the sarcomeric structure (9). These in vitro demonstrations of actin binding are supported by electron tomographic observations showing MyBP-C extending from the thick to the thin filaments in the intact sarcomere, consistent with a model in which the N terminus of MyBP-C binds to the thin filament (21, 22). Together, these results suggest that actin binding is physiologically relevant and that the slowing of actin filament sliding is one possible mechanism by which cMyBP-C modulates cardiac contractility (5, 23, 24).
The structural basis of cMyBP-C’s N-terminal binding to F-actin has been studied in several ways. Neutron scattering and NMR titration analysis of F-actin decorated with the N-terminal fragment C0C2 (Fig. 1) suggested that C0C2 binds to subdomain 1 (SD1) and the DNase loop of actin (25) via key regions within C0 and C1 (10). More direct observations by negative stain electron microscopy (26) and 3D reconstruction (27) suggest that C0 and C1 bind to SD1 of actin, whereas the M domain crosses over SD2, possibly binding to the next SD1, and C2 and C3 lie above the surface of the filament (27, 28).
The position of cMyBP-C binding on actin SD1 suggests that in addition to inhibiting actomyosin interactions, it might also affect thin filament regulation by interfering with the binding of tropomyosin/troponin (Tm/Tn) to actin. Tm and Tn regulate muscle contraction by movement of Tm in response to calcium binding by Tn (29). At low Ca2+, Tm lies on SD1, where it sterically blocks the binding of myosin to actin (the “blocked” position); the consequent inhibition of actin-myosin interaction leads to muscle relaxation. On activation, Tn binds Ca2+, causing Tm to move onto actin SD3 (the “closed” position), exposing myosin binding sites on SD1 and initiating crossbridge cycling and contraction (29–35). When a model of Tm in its low Ca2+ (blocked) state is positioned on the reconstruction of F-actin decorated with C0C3, it appears to clash with cMyBP-C’s C0 and C1 domains, suggesting that cMyBP-C and Tm might compete for binding to SD1 in the relaxed thin filament (25, 27, 28). As a consequence, cMyBP-C might be expected to activate the thin filament by physically preventing Tm from assuming its blocked position (25, 27, 28). Motility (19) and solution kinetics studies (36, 37) support this concept, showing that the N-terminal C1C2 fragment activates thin filaments in low Ca2+ similarly to rigor heads.
Here we have investigated cMyBP-C’s potential to modulate cardiac contractility by contrasting mechanisms; that is, activating the thin filament and inhibiting maximal actomyosin mechanical activity. First we used negative-staining EM and 3D reconstruction to investigate whether cMyBP-C displaces Tm at low Ca2+, by decorating regulated thin filaments (containing F-actin, Tm and Tn) with C0C2. In parallel experiments, we determined the functional consequences of such N-terminal fragments on regulated thin filament activation and sliding velocities in an in vitro motility assay. We find clear structural evidence for displacement of Tm toward the high Ca2+ (closed) position when C0C2 binds to thin filaments under low Ca2+ conditions, suggesting that N-terminal binding should activate the thin filament; this was confirmed in the motility assay. We also demonstrate that C0C2 has no effect on Tm position under high Ca2+ conditions but inhibits maximal sliding velocity in the motility assay. Interestingly, a smaller fragment (C0C1f; Fig. 1) binds to the thin filament under low Ca2+ conditions but does not displace Tm or activate thin filament sliding. However, C0C1f still inhibits thin filament sliding at high Ca2+ to the same extent as the larger N-terminal fragment. These results suggest that cMyBP-C may play two physiological roles in intact muscle, displacing Tm from the “blocked” position at low Ca2+ to modulate thin filament activation, and governing maximal sliding velocity at high Ca2+ by a potentially independent molecular mechanism.
Results
Negative Staining of Thin Filaments Decorated with C0C2 at Low and High Ca2+.
Reconstituted and native thin filaments were mixed with C0C2 (Fig. 1) at molar ratios of 1:6, 1:3, 1:1, or 7:1 (actin:C0C2) in solutions containing KAc (or NaCl) at concentrations from 100 to 180 mM (see Materials and Methods). Low Ca2+ solutions contained 0.2 mM EGTA, whereas high Ca2+ solutions were the same, with Ca2+ added to total 0.33 mM [pCa (negative log of calcium concentration), 3.9].
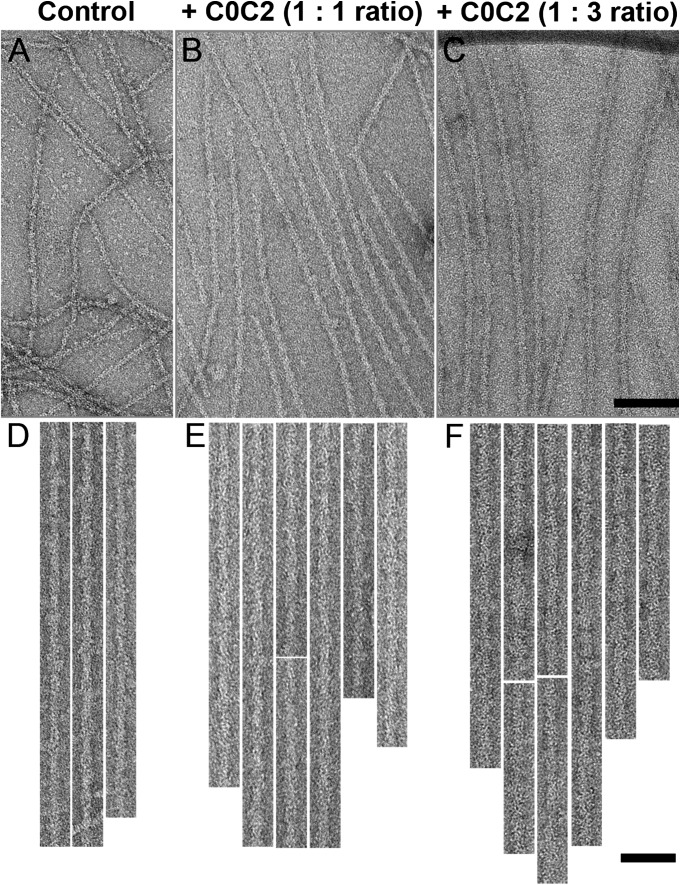
Negatively stained low-Ca2+ control (no C0C2 decoration) native (Fig. 2 A and D) and reconstituted (Figs. S1A and S2 A and D) thin filaments showed typical actin subunits and occasional resolution of Tm strands (31). C0C2 decoration caused a clear increase in filament diameter, most obviously when more C0C2 was added (1:3 and 1:6 ratios of actin:C0C2; Fig. 2 C and F; Fig. S1D; Table S1). In addition, 1:1 and 7:1 molar ratios showed a smaller, but still visible, increase (Fig. 2 B and E; Figs. S1 B and C and S2 B, C, E, and F; Table S1). Clear decoration was observed in all ionic conditions with no major difference in the level of decoration or background protein. The higher levels of decoration obscured detailed actin and tropomyosin structure. The appearance and diameter of filaments decorated under high Ca2+ conditions (Fig. S3) were similar to those at low Ca2+.
Fig. 2.
Decoration of native thin filaments with C0C2 under low Ca2+ conditions. (A, D) Undecorated control. (B, C, E, F) Filaments decorated with C0C2 at A:C0C2 molar ratios of 1:1 (B, E) and 1:3 (C, F). Filaments in D–F have been computationally straightened. [Scale bar (A–C) = 100 nm; (D–F) = 50 nm.)
Most experiments were carried out by adding C0C2 to native or preformed reconstituted thin filaments. To determine whether Tm and Tn could still bind to actin if C0C2 were bound first, we also reversed the order of mixing. The apparent level of decoration and the filament diameter appeared to be independent of the order of mixing (Fig. S2; Table S1).
3D Reconstruction of Thin Filaments Decorated with C0C2 at Low and High Ca2+.
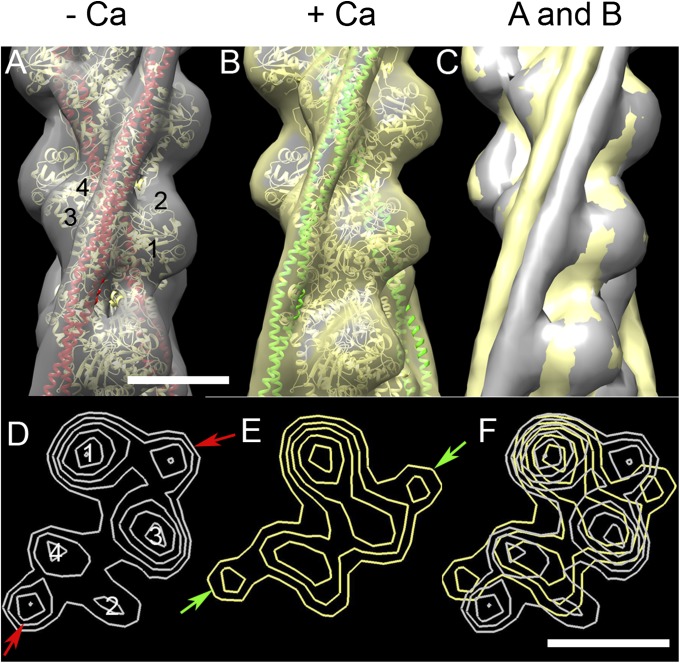
3D reconstruction of computationally straightened thin filaments was carried out by iterative helical real-space reconstruction (38). Control filaments (no decoration) in low and high Ca2+ showed clear actin subunits and elongated Tm strands running along each long-pitch actin helix (Fig. 3). At low Ca2+, Tm is seen to lie near the junction of SD1 and SD3 of actin, over the main myosin-binding site (the blocked position), as found in previous studies (31, 32) (Fig. 3 A and D). At high Ca2+, Tm moves to the “closed” position on the inner domain of actin, exposing myosin-binding sites on SD1 (Fig. 3 B and E) (31, 32). The two Tm positions were supported by fitting the reconstructions with molecular models of F-actin-Tm, where Tm is in the blocked or closed position (Fig. 3 A and B) (39). The change in Tm position induced by Ca2+ was especially clear when high and low Ca2+ reconstructions were superimposed (Fig. 3 C and F), confirming that Tm and its well-documented Ca2+-induced movement are clearly seen by our procedures. Similar results were obtained for both native and reconstituted filaments (Fig. 3 A and B; Figs. S4B and S5B).
Fig. 3.
3D reconstructions of native control thin filaments under low and high Ca2+ conditions. (A) Low Ca2+ filament (gray surface rendering) fitted with ribbon depiction of low-Ca2+ A.Tm atomic model (39) (actin monomers, yellow; Tm, red). (B) High-Ca2+ reconstruction (yellow surface rendering), fitted with high-Ca2+ A.Tm atomic model (actin monomers, yellow; Tm, green). (C) Superposition of A and B demonstrating Tm shift on to inner domain of actin at high Ca2+ (note: slight variations in actin contours in A and B cause either gray or yellow to appear on the actin surface in C). (D and E) Transverse sections of low and high Ca2+ reconstructions, respectively, showing positioning of Tm (arrows) near the junction of actin SD1 and SD3 in low Ca2+, and on SD3 in high Ca2+. (F) Superposition of D and E, demonstrating the shift of Tm. Filaments in A–C oriented with pointed end at top; actin subdomains are marked in A and D. (Scale bar = 5 nm.)
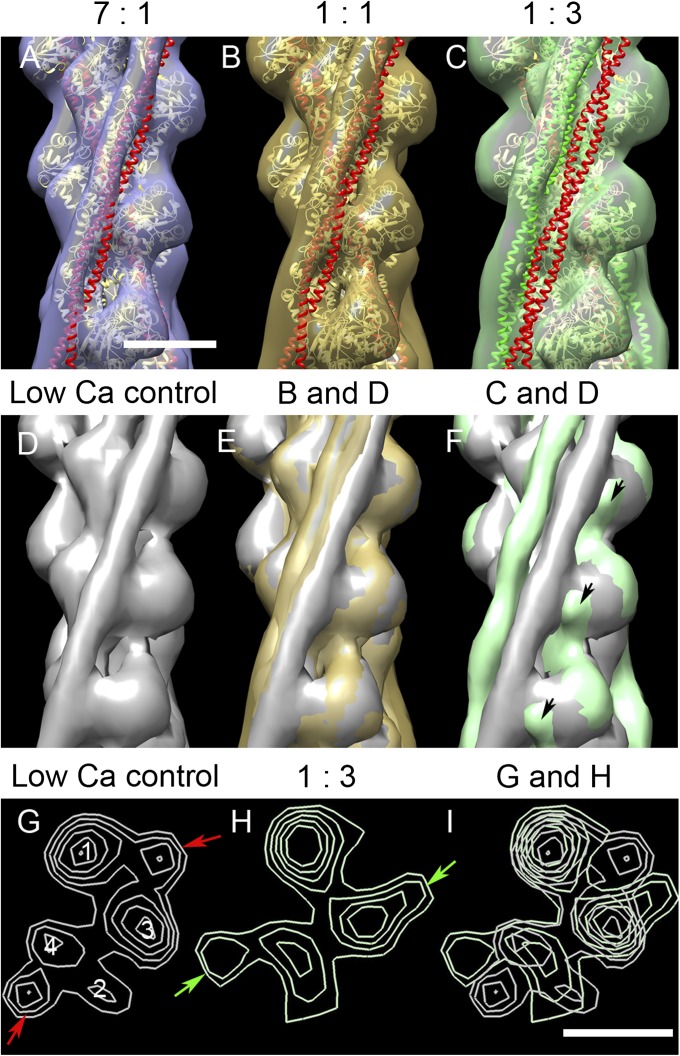
To determine the effect of C0C2 on Tm position under low Ca2+ conditions, reconstructions were computed for native filaments decorated with C0C2 at different molar ratios (Fig. 4). All reconstructions again showed clear actin subunits and Tm strands. Actin subunit shape was similar to that in control filaments, especially at the lower levels of decoration. At higher levels, the binding of C0C2 was visualized as an extra density on SD1 of actin (Fig. 4F, arrows), similar to its binding position observed previously (27, 28).
Fig. 4.
3D reconstructions of native thin filaments decorated with C0C2 under low Ca2+ conditions. (A–C) Reconstructions with the indicated ratios of A:C0C2 fitted with A.Tm atomic models (39), as in Fig. 3, with Tm in blocked (red) or closed (green) position. With low levels of C0C2, there was a small movement of Tm from the blocked position (A and B), whereas with the highest level, Tm shifted to approximately the closed position (C). (E and F) show superposition of B and C, respectively, on the low Ca2+ control (D), demonstrating the smaller and larger shifts; black arrows indicate protrusion on SD1 surface, close to Tm, which we attribute to proximal region of C0C2. (G and H) Transverse sections of D and C, respectively, showing the shift of Tm from the blocked position in control (red arrows) to the closed position in C0C2-decorated filament (green arrows). (I) Superposition of G and H. Filaments in A–F oriented with the pointed end up. (Scale bar = 5 nm.)
The effects of C0C2 binding on Tm position were determined by superimposing decorated and control filament reconstructions (Fig. 4 D–I) and by fitting of blocked and closed position A.Tm atomic models (39) to the reconstructions (Fig. 4 A–C). Addition of C0C2 appeared to cause a small movement of Tm in the direction of the closed position when the lower amounts of C0C2 were used (Fig. 4 A, B, and E), and a larger movement, as far as the closed position or slightly further, with the highest amount of C0C2 (Fig. 4 C, F, and H) or when C0C2 was added to F-actin before the addition of Tm.Tn (Fig. S4D). These results, showing displacement of Tm from its low-Ca2+ position by C0C2, suggest direct competition between C0C2 and Tm for Tm’s low-Ca2+ binding region on actin. This displacement would be expected to activate the thin filament.
Visibility of C0C2 in the reconstructions varied with conditions. In filaments showing a large Tm shift (high ratios of C0C2 added and reverse order of addition), C0C2 density appeared stronger than in those showing a smaller shift (lower ratios, normal order; Fig. 4C and Fig. S4D; compare with Fig. 4 A and B and Fig. S4C). These observations support the view that Tm and C0C2 compete for similar sites on actin. The reconstructions show only the proximal region of C0C2, and we assume that the more distal domains are not well ordered under our experimental conditions.
In high Ca2+ conditions, the position of Tm in C0C2-decorated thin filaments was similar to that of control high Ca2+ filaments, and clear C0C2 density was visible on SD1 of actin (Fig. S5) in a similar position to that seen in F-actin decorated with C0C2 (27). Thus, C0C2 did not displace Tm from its closed position, suggesting that C0C2 and Tm occupy different binding interfaces on actin at high Ca2+.
3D Reconstruction of Thin Filaments Decorated with C0C1f at Low and High Ca2+.
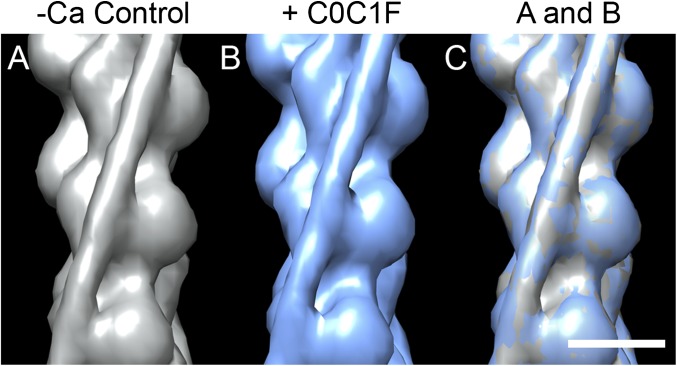
We also studied the effect on Tm position of a shorter fragment (C0C1f; Fig. 1) known to bind to F-actin (16, 27). Filaments decorated with C0C1f were wider than controls, clearly demonstrating binding of the fragment (Fig. S6 A and B; Table S1). However, 3D reconstructions showed no change in position of Tm compared with controls, either at low (Fig. 5) or high Ca2+ (Fig. S7), suggesting that C0C1f would have no activating effect. The site of binding of C0C1f was mainly on the front of actin SD1, although the density was not strong (Figs. 5 and Fig. S7).
Fig. 5.
3D reconstructions of native thin filaments decorated with C0C1f under low Ca2+ conditions. (A) Low Ca2+ control filament (gray surface rendering). (B) C0C1f-decorated filament (blue). (C) Superposition of low Ca2+ control (A) and C0C1f-decorated filament (B) showing no Tm shift. (Scale bar = 5 nm.)
Effect of N-Terminal Fragments on Native Thin Filament Sliding at Low and High Ca2+.
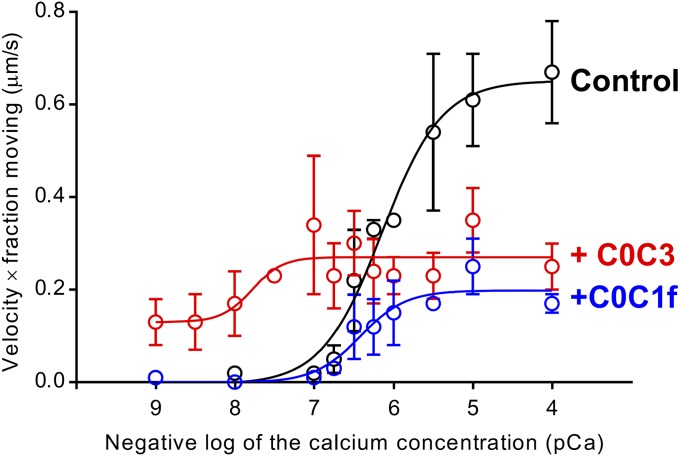
The sliding of native cardiac thin filaments on mouse cardiac myosin was observed in an in vitro motility assay over a range of Ca2+ concentrations (Fig. 6 and Fig. S8). Thin filament sliding was fully regulated by Ca2+, with little to no motion observed at low Ca2+ (pCa > 7) and sigmoidal increases in velocity and fraction of filaments moving at higher Ca2+ concentrations (Fig. S8, controls). Multiplying the thin filament velocity by the fraction of filaments moving (Fig. S8) gives an effective activation curve with a pCa50 of 6.15 ± 0.09 (Fig. 6, control). Although the shorter C0C2 fragment was used in our structural studies (to minimize interference with Tm visibility in the reconstructions), we have shown previously that C0C2 and C0C3 bind similarly to actin filaments (27) and are functionally identical in their inhibition of actin filament motility (16). On addition of 1 µM C0C3 (Fig. 1) to the assay, thin filament sliding was now observed even at the lowest Ca2+ level (Fig. S8), and there was a significant shift in pCa50 for the effective activation curve to 7.78 ± 0.33 (P < 0.05; Fig. 6). At high Ca2+, maximal sliding velocity was inhibited by 50% (Fig. S8), as reported previously for C0C2 (19). In contrast, addition of 1 µM C0C1f showed no activation of the thin filament at low calcium (Fig. S8) and no change in pCa50 for the effective activation curve (6.42 ± 0.15; P > 0.05; Fig. 6) but showed inhibition of maximal velocity by 47% at higher Ca2+ concentrations, similar to C0C3 (Fig. S8). The inability of C0C1f to activate the thin filament despite retaining its inhibitory capacity suggests that thin filament activation and inhibition of maximal velocity may be governed by two independent molecular mechanisms.
Fig. 6.
Effect of N-terminal cMyBP-C fragments on native thin filament sliding in in vitro motility assays. The graph shows “effective activation” (thin filament velocity × fraction of filaments moving) vs. pCa. The black line shows native thin filaments demonstrating a sigmoidal response to Ca2+. The red line shows C0C3 activated the thin filaments at low Ca2+, increased Ca2+ sensitivity, and inhibited maximal velocity. The blue line shows C0C1f had no effect on activation or Ca2+ sensitivity but inhibited maximal velocity. See Fig. S8 for individual velocity and fraction-moving data.
Discussion
Elimination of MyBP-C from muscle fibers by chemical extraction or genetic ablation results in changes to the muscle’s Ca2+ sensitivity of force production (40–42), to its shortening velocity (43, 44), and to the kinetics of tension recovery after stretch (45). These alterations emphasize the physiological roles played by MyBP-C in both Ca2+-dependent muscle activation and in modulation of cardiac contractility. However, the structural basis for these distinct functions has not been determined. Using a combination of structural and molecular functional assays, we have shown that the N terminus of cMyBP-C displaces Tm to activate native thin filaments at low Ca2+ and slows thin filament sliding velocity at high Ca2+, presumably through independent molecular mechanisms.
Effect of cMyBP-C on Thin Filament Activation.
Previous structural studies of MyBP-C binding to F-actin suggested that the N-terminal region might interfere with Tm binding to actin, especially when Tm is in its low Ca2+ position (25, 27, 28). We tested this proposal directly by decorating regulated thin filaments (containing Tm and Tn) with C0C2. Our results, showing clear movement of Tm from the blocked toward the closed position under low Ca2+ conditions, directly support the hypothesis that the N terminus of cMyBP-C competes with Tm (in its blocked position) for the same binding region on actin; this competition is further suggested by the greater movement of Tm that occurs when cMyBP-C is added to F-actin before Tm and Tn. This cMyBP-C-induced shift of Tm could straightforwardly explain the activation of thin filament sliding in the in vitro motility assay by C0C3 (Fig. 6) and C0C2 (19), the reduced Ca2+-sensitivity of contraction of cardiac myocytes from cMyBP-C knockout mice (40), and the activation of myosin ATPase activity by thin filaments under low Ca2+ conditions when cMyBP-C N-terminal fragments are present (36, 37). Interestingly, solution ATPase assays found strong activation even at low levels of added C1C2 (actin:C1C2 = 7:1) (37), where Tm movement in our reconstructions was small. Variations in conditions between the ATPase and EM experiments may account for this difference. For example, thin filament activation was measured by the rate of nucleotide release from thin filament-bound myosin subfragment 1 (S1), whereas S1 is absent from our experiments. Sparse decoration of thin filaments by cMyBP-C fragments may weaken Tm binding in the blocked position without significantly changing its average position observed by EM. This could increase the freedom of movement of Tm so that single S1 binding events are no longer significantly inhibited. Physiological experiments in which N-terminal fragments of cMyBP-C are diffused into skinned myocytes (46, 47) also demonstrate activation of contraction at low (relaxing) Ca2+ concentrations. In these experiments, it was suggested that cMyBP-C affects crossbridge cycling primarily by binding to myosin, thus affecting myosin crossbridge mechanics, although mechanisms involving actin binding were also considered possible. Our results suggest that cMyBP-C increases thin filament calcium sensitivity by binding to actin and displacing Tm toward the closed position.
The cMyBP-C domains primarily involved in binding actin appear to be the C1 and M-domains (9, 15, 16, 20), although there is evidence that C0 may be important (10, 14). Previous modeling suggested that the C0 and C1 domains could clash with Tm in its blocked position (25, 27, 28), whereas experiments in which expressed N-terminal fragments were added to skinned cardiac myocytes implicated the ProAla-rich domain between C0 and C1 in modulating Ca2+ activation of crossbridge cycling (46). However, in similar skinned fiber experiments, the ProAla-rich domain was found to have no effect, whereas the C1 and M-domains were critical to the Ca2+-sensitizing and activating effects of various N-terminal domains (48). This too was the case in the present study, as only the N-terminal fragments containing the C1 and M-domains were able to displace Tm (C0C2) and activate the thin filament (C0C3). C0C1f (containing C0, the ProAla-rich domain, C1, and the first 17 amino acids of the M-domain; Fig. 1) was capable of binding to thin filaments at low Ca2+ but did not displace Tm from the blocked position and did not activate thin filament sliding. Therefore, thin filament activation by the N terminus of cMyBP-C under relaxing conditions (pCa > 7) likely involves multiple sites of interaction with actin to move Tm into the closed position. Assuming that C0C1f binds to the thin filament through the same contacts as C0C2, then additional sites of actin interaction must exist beyond the first 17 amino acids of the M-domain that are present in the C0C1f fragment. For example, arginines 279/280 (further along the M-domain) have been identified as critical to actin binding (9), and when mutated to alanines, they diminish the capacity of C1C2 fragments to activate thin filaments in skinned rat trabeculae (49).
Effect of cMyBP-C on the Inhibition of Sliding Velocity.
In addition to the thin filament activating effects of cMyBP-C at low Ca2+, cMyBP-C also governs contractility, as evidenced by enhanced unloaded cardiac muscle shortening velocities in cMyBP-C null mice (43). The inhibition of velocity in the presence of cMyBP-C occurs through cMyBP-C’s N terminus, as observed previously by ourselves and others (16, 19, 20, 50) and confirmed here (Fig. 6). Because our structural studies demonstrate that C0C2 and C0C1f bind to the thin filament at high Ca2+, inhibition of maximal sliding velocity could be explained by tethering of the thin filaments to the motility surface by the N-terminal domains, imparting a load on the actomyosin interactions (16), or by preventing myosin from interacting with the thin filament by occupying available myosin binding sites. In the intact sarcomere, MyBP-C is thought to bind by its 3 C-terminal domains to the thick filament backbone, whereas the rest of the molecule extends to adjacent thin filaments (5, 22, 51, 52). Thus, although the molar ratio of MyBP-C to actin subunits in the C-zone is only ∼1:10, the proximity of its N termini to actin may make their effective local concentration high enough to bind the thin filament without substantial occupancy of S1 binding sites. With only minimal steric interference with S1 binding, the inhibition of maximal velocity would likely arise from tethering between the thick and thin filaments. However, our structural studies cannot exclude the possibility that N-terminal domains inhibit thin filament velocity in the in vitro motility assay by binding to myosin S2 (47) or the regulatory light chain (11) to alter myosin kinetics.
Conclusions
We conclude that the N-terminal region of cMyBP-C binds to F-actin at a location that directly competes with Tm in the blocked position. This competition is sufficient to displace Tm toward the closed position and activate the thin filament, providing a means of modulating the Ca2+ sensitivity of thin filaments in cardiac muscle. Similar modulation may also occur in skeletal muscle. In addition to activating the thin filament, cMyBP-C’s N terminus can also inhibit myosin-generated thin filament sliding. However, these two distinct functions appear to occur through independent mechanisms, as emphasized by the inability of C0C1f to activate the thin filament despite retaining its inhibitory capacity, in contrast to the larger C0C2 and C0C3 fragments, which activate the thin filament (by Tm movement) while also slowing filament sliding. Although thin filament activation must be in part a result of cMyBP-C binding directly to actin, mechanical inhibition may still be through either actin and/or myosin binding. Experiments to distinguish which binding partner cMyBP-C interacts with to inhibit actomyosin interactions will be critical. We note, finally, that cMyBP-C phosphorylation in response to beta-adrenergic stimulation leads to enhanced cardiac contractility (53). Serine phosphorylation in cMyBP-C’s M-domain by a host of kinases reduces the affinity of the N terminus for actin (15) and myosin (7). Therefore, modulation of cMyBP-C’s binding capacity by phosphorylation may add a measure of tunability to cMyBP-C’s regulation of cardiac contractility in response to physiological stress.
Materials and Methods
Detailed methods are provided in SI Materials and Methods and summarized here.
Proteins.
F-actin was purified from chicken pectoralis muscle (54) and native thin filaments from porcine cardiac muscle (55, 56). Bovine cardiac tropomyosin and troponin were produced as previously described (57). cMyBP-C N-terminal fragments C0C1f (1–269), C0C2 (1–448), and C0C3 (1–539) were bacterially expressed as described previously (27). For the motility assays, myosin (58) and native thin filaments (59) (with modifications described in SI Materials and Methods) were freshly isolated from mouse hearts.
Electron Microscopy.
Native thin filaments, or F-actin preincubated with tropomyosin and troponin (60), were mixed with varying ratios of C0C1f and C0C2 under different buffer conditions used in previous thin filament studies (27, 36, 37, 60). Five-microliter aliquots were applied to EM grids coated with thin carbon and negatively stained with 1% (wt/vol) uranyl acetate. Dried grids were observed in a Philips CM120 electron microscope (FEI) and low-dose images acquired at a pixel size of 0.35 nm, using a 2K × 2K CCD camera (F224HD, TVIPS GmbH).
3D Reconstruction.
Thin filaments were unbent using ImageJ and selected regions converted to SPIDER format and cut into segments in SPIDER (v11.2; Wadsworth Center). Iterative helical real-space reconstruction was carried out using SPIDER (38, 61), with F-actin as an initial reference model. UCSF Chimera (62) was used for visualization, analysis, and atomic fitting of 3D volumes.
In Vitro Motility.
In vitro motility assays were performed on the surface of a nitrocellulose-coated flow cell, and the motion of actin filaments was observed by epifluorescence microscopy, as previously described (63).
Supplementary Material
Acknowledgments
We thank Dr. John Woodhead for discussion, Dr. Shixin Yang for help with SPIDER procedures, and Guy Kennedy from the University of Vermont Instrumentation and Model Facility for his expert microscopy design and assistance. EM work was carried out in the Core Electron Microscopy Facility at the University of Massachusetts Medical School. This work was supported by National Institutes of Health (NIH) Grants AR034711, HL007647, HL063774, P01 HL059408, and P01 HL069779. Molecular graphics images and atomic fitting were produced using UCSF Chimera from the Resource for Biocomputing, Visualization, and Informatics at UCSF (supported by NIH Grant P41 RR-01081).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1316001111/-/DCSupplemental.
References
- 1.Offer G, Moos C, Starr R. A new protein of the thick filaments of vertebrate skeletal myofibrils. Extraction, purification and characterization. J Mol Biol. 1973;74(4):653–676. doi: 10.1016/0022-2836(73)90055-7. [DOI] [PubMed] [Google Scholar]
- 2.Winegrad S. Cardiac myosin binding protein C. Circ Res. 1999;84(10):1117–1126. doi: 10.1161/01.res.84.10.1117. [DOI] [PubMed] [Google Scholar]
- 3.Barefield D, Sadayappan S. Phosphorylation and function of cardiac myosin binding protein-C in health and disease. J Mol Cell Cardiol. 2010;48(5):866–875. doi: 10.1016/j.yjmcc.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craig R, Offer G. The location of C-protein in rabbit skeletal muscle. Proc R Soc Lond B Biol Sci. 1976;192(1109):451–461. doi: 10.1098/rspb.1976.0023. [DOI] [PubMed] [Google Scholar]
- 5.Flashman E, Redwood C, Moolman-Smook J, Watkins H. Cardiac myosin binding protein C: Its role in physiology and disease. Circ Res. 2004;94(10):1279–1289. doi: 10.1161/01.RES.0000127175.21818.C2. [DOI] [PubMed] [Google Scholar]
- 6.Starr R, Offer G. The interaction of C-protein with heavy meromyosin and subfragment-2. Biochem J. 1978;171(3):813–816. doi: 10.1042/bj1710813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gruen M, Prinz H, Gautel M. cAPK-phosphorylation controls the interaction of the regulatory domain of cardiac myosin binding protein C with myosin-S2 in an on-off fashion. FEBS Lett. 1999;453(3):254–259. doi: 10.1016/s0014-5793(99)00727-9. [DOI] [PubMed] [Google Scholar]
- 8.Ababou A, et al. Myosin binding protein C positioned to play a key role in regulation of muscle contraction: Structure and interactions of domain C1. J Mol Biol. 2008;384(3):615–630. doi: 10.1016/j.jmb.2008.09.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhuiyan MS, Gulick J, Osinska H, Gupta M, Robbins J. Determination of the critical residues responsible for cardiac myosin binding protein C’s interactions. J Mol Cell Cardiol. 2012;53(6):838–847. doi: 10.1016/j.yjmcc.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu Y, Kwan AH, Trewhella J, Jeffries CM. The C0C1 fragment of human cardiac myosin binding protein C has common binding determinants for both actin and myosin. J Mol Biol. 2011;413(5):908–913. doi: 10.1016/j.jmb.2011.09.026. [DOI] [PubMed] [Google Scholar]
- 11.Ratti J, Rostkova E, Gautel M, Pfuhl M. Structure and interactions of myosin-binding protein C domain C0: Cardiac-specific regulation of myosin at its neck? J Biol Chem. 2011;286(14):12650–12658. doi: 10.1074/jbc.M110.156646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moos C, Mason CM, Besterman JM, Feng IN, Dubin JH. The binding of skeletal muscle C-protein to F-actin, and its relation to the interaction of actin with myosin subfragment-1. J Mol Biol. 1978;124(4):571–586. doi: 10.1016/0022-2836(78)90172-9. [DOI] [PubMed] [Google Scholar]
- 13.Yamamoto K. The binding of skeletal muscle C-protein to regulated actin. FEBS Lett. 1986;208(1):123–127. doi: 10.1016/0014-5793(86)81545-9. [DOI] [PubMed] [Google Scholar]
- 14.Kulikovskaya I, McClellan G, Flavigny J, Carrier L, Winegrad S. Effect of MyBP-C binding to actin on contractility in heart muscle. J Gen Physiol. 2003;122(6):761–774. doi: 10.1085/jgp.200308941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaffer JF, Kensler RW, Harris SP. The myosin-binding protein C motif binds to F-actin in a phosphorylation-sensitive manner. J Biol Chem. 2009;284(18):12318–12327. doi: 10.1074/jbc.M808850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weith A, et al. Unique single molecule binding of cardiac myosin binding protein-C to actin and phosphorylation-dependent inhibition of actomyosin motility requires 17 amino acids of the motif domain. J Mol Cell Cardiol. 2012;52(1):219–227. doi: 10.1016/j.yjmcc.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Squire JM, Luther PK, Knupp C. Structural evidence for the interaction of C-protein (MyBP-C) with actin and sequence identification of a possible actin-binding domain. J Mol Biol. 2003;331(3):713–724. doi: 10.1016/s0022-2836(03)00781-2. [DOI] [PubMed] [Google Scholar]
- 18.Rybakova IN, Greaser ML, Moss RL. Myosin binding protein C interaction with actin: Characterization and mapping of the binding site. J Biol Chem. 2011;286(3):2008–2016. doi: 10.1074/jbc.M110.170605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Razumova MV, et al. Effects of the N-terminal domains of myosin binding protein-C in an in vitro motility assay: Evidence for long-lived cross-bridges. J Biol Chem. 2006;281(47):35846–35854. doi: 10.1074/jbc.M606949200. [DOI] [PubMed] [Google Scholar]
- 20.Previs MJ, Beck Previs S, Gulick J, Robbins J, Warshaw DM. Molecular mechanics of cardiac myosin-binding protein C in native thick filaments. Science. 2012;337(6099):1215–1218. doi: 10.1126/science.1223602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luther PK, Craig R. Modulation of striated muscle contraction by binding of myosin binding protein C to actin. BioArchitecture. 2011;1(6):277–283. doi: 10.4161/bioa.1.6.19341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luther PK, et al. Direct visualization of myosin-binding protein C bridging myosin and actin filaments in intact muscle. Proc Natl Acad Sci USA. 2011;108(28):11423–11428. doi: 10.1073/pnas.1103216108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oakley CE, Chamoun J, Brown LJ, Hambly BD. Myosin binding protein-C: Enigmatic regulator of cardiac contraction. Int J Biochem Cell Biol. 2007;39(12):2161–2166. doi: 10.1016/j.biocel.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 24.Gautel M, Zuffardi O, Freiburg A, Labeit S. Phosphorylation switches specific for the cardiac isoform of myosin binding protein-C: A modulator of cardiac contraction? EMBO J. 1995;14(9):1952–1960. doi: 10.1002/j.1460-2075.1995.tb07187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitten AE, Jeffries CM, Harris SP, Trewhella J. Cardiac myosin-binding protein C decorates F-actin: Implications for cardiac function. Proc Natl Acad Sci USA. 2008;105(47):18360–18365. doi: 10.1073/pnas.0808903105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kensler RW, Shaffer JF, Harris SP. Binding of the N-terminal fragment C0-C2 of cardiac MyBP-C to cardiac F-actin. J Struct Biol. 2011;174(1):44–51. doi: 10.1016/j.jsb.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mun JY, et al. Electron microscopy and 3D reconstruction of F-actin decorated with cardiac myosin-binding protein C (cMyBP-C) J Mol Biol. 2011;410(2):214–225. doi: 10.1016/j.jmb.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orlova A, Galkin VE, Jeffries CM, Egelman EH, Trewhella J. The N-terminal domains of myosin binding protein C can bind polymorphically to F-actin. J Mol Biol. 2011;412(3):379–386. doi: 10.1016/j.jmb.2011.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev. 2000;80(2):853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- 30.Lehman W, Craig R. Tropomyosin and the steric mechanism of muscle regulation. Adv Exp Med Biol. 2008;644:95–109. doi: 10.1007/978-0-387-85766-4_8. [DOI] [PubMed] [Google Scholar]
- 31.Lehman W, Craig R, Vibert P. Ca2+-induced tropomyosin movement in Limulus thin filaments revealed by three-dimensional reconstruction. Nature. 1994;368(6466):65–67. doi: 10.1038/368065a0. [DOI] [PubMed] [Google Scholar]
- 32.Vibert P, Craig R, Lehman W. Steric-model for activation of muscle thin filaments. J Mol Biol. 1997;266(1):8–14. doi: 10.1006/jmbi.1996.0800. [DOI] [PubMed] [Google Scholar]
- 33.Parry DA, Squire JM. Structural role of tropomyosin in muscle regulation: Analysis of the x-ray diffraction patterns from relaxed and contracting muscles. J Mol Biol. 1973;75(1):33–55. doi: 10.1016/0022-2836(73)90527-5. [DOI] [PubMed] [Google Scholar]
- 34.Huxley HE. Structural changes in the actin- and myosin-containing filaments during contraction. Cold Spring Harb Symp Quant Biol. 1973;37:361–376. [Google Scholar]
- 35.Haselgrove JC. X-ray evidence for a conformational change in the actin-containing filaments of vertebrate striated muscle. Cold Spring Harb Symp Quant Biol. 1973;37:341–352. [Google Scholar]
- 36.White HD, Harris S. Activation of cardiac thin filaments by N-terminal domains of cardiac myosin binding protein C. Biophys J. 2012;102(3):435a. [Google Scholar]
- 37.White HD, Belknap B, Harris SP. Activation and inhibition of F-actin and cardiac thin filaments by the N-terminal domains of cardiac myosin binding protein C. Biophys J. 2013;104(2):158a–159a. [Google Scholar]
- 38.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85(4):225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 39.Poole KJ, et al. A comparison of muscle thin filament models obtained from electron microscopy reconstructions and low-angle X-ray fibre diagrams from non-overlap muscle. J Struct Biol. 2006;155(2):273–284. doi: 10.1016/j.jsb.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 40.Harris SP, et al. Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ Res. 2002;90(5):594–601. doi: 10.1161/01.res.0000012222.70819.64. [DOI] [PubMed] [Google Scholar]
- 41.Korte FS, McDonald KS, Harris SP, Moss RL. Loaded shortening, power output, and rate of force redevelopment are increased with knockout of cardiac myosin binding protein-C. Circ Res. 2003;93(8):752–758. doi: 10.1161/01.RES.0000096363.85588.9A. [DOI] [PubMed] [Google Scholar]
- 42.Hofmann PA, Hartzell HC, Moss RL. Alterations in Ca2+ sensitive tension due to partial extraction of C-protein from rat skinned cardiac myocytes and rabbit skeletal muscle fibers. J Gen Physiol. 1991;97(6):1141–1163. doi: 10.1085/jgp.97.6.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palmer BM, et al. Role of cardiac myosin binding protein C in sustaining left ventricular systolic stiffening. Circ Res. 2004;94(9):1249–1255. doi: 10.1161/01.RES.0000126898.95550.31. [DOI] [PubMed] [Google Scholar]
- 44.Hofmann PA, Greaser ML, Moss RL. C-protein limits shortening velocity of rabbit skeletal muscle fibres at low levels of Ca2+ activation. J Physiol. 1991;439:701–715. doi: 10.1113/jphysiol.1991.sp018689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stelzer JE, Dunning SB, Moss RL. Ablation of cardiac myosin-binding protein-C accelerates stretch activation in murine skinned myocardium. Circ Res. 2006;98(9):1212–1218. doi: 10.1161/01.RES.0000219863.94390.ce. [DOI] [PubMed] [Google Scholar]
- 46.Herron TJ, et al. Activation of myocardial contraction by the N-terminal domains of myosin binding protein-C. Circ Res. 2006;98(10):1290–1298. doi: 10.1161/01.RES.0000222059.54917.ef. [DOI] [PubMed] [Google Scholar]
- 47.Kunst G, et al. Myosin binding protein C, a phosphorylation-dependent force regulator in muscle that controls the attachment of myosin heads by its interaction with myosin S2. Circ Res. 2000;86(1):51–58. doi: 10.1161/01.res.86.1.51. [DOI] [PubMed] [Google Scholar]
- 48.Razumova MV, Bezold KL, Tu AY, Regnier M, Harris SP. Contribution of the myosin binding protein C motif to functional effects in permeabilized rat trabeculae. J Gen Physiol. 2008;132(5):575–585. doi: 10.1085/jgp.200810013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bezold KL, Shaffer JF, Khosa JK, Hoye ER, Harris SP. A gain-of-function mutation in the M-domain of cardiac myosin-binding protein-C increases binding to actin. J Biol Chem. 2013;288(30):21496–21505. doi: 10.1074/jbc.M113.474346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shaffer JF, Razumova MV, Tu AY, Regnier M, Harris SP. Myosin S2 is not required for effects of myosin binding protein-C on motility. FEBS Lett. 2007;581(7):1501–1504. doi: 10.1016/j.febslet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 51.Zoghbi ME, Woodhead JL, Moss RL, Craig R. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc Natl Acad Sci USA. 2008;105(7):2386–2390. doi: 10.1073/pnas.0708912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee K, Sadayappan S, Harris S, Craig R. Determination of MyBP-C orientation in the cardiac sarcomere by immuno-EM. Biophys J. 2013;104(2):309a. [Google Scholar]
- 53.Tong CW, Stelzer JE, Greaser ML, Powers PA, Moss RL. Acceleration of crossbridge kinetics by protein kinase A phosphorylation of cardiac myosin binding protein C modulates cardiac function. Circ Res. 2008;103(9):974–982. doi: 10.1161/CIRCRESAHA.108.177683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85(Pt B):164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- 55.Spiess M, et al. Isolation, electron microscopic imaging, and 3-D visualization of native cardiac thin myofilaments. J Struct Biol. 1999;126(2):98–104. doi: 10.1006/jsbi.1999.4111. [DOI] [PubMed] [Google Scholar]
- 56.Matsumoto F, et al. Conformational changes of troponin C within the thin filaments detected by neutron scattering. J Mol Biol. 2004;342(4):1209–1221. doi: 10.1016/j.jmb.2004.07.086. [DOI] [PubMed] [Google Scholar]
- 57.Tobacman LS, Adelstein RS. Mechanism of regulation of cardiac actin-myosin subfragment 1 by troponin-tropomyosin. Biochemistry. 1986;25(4):798–802. doi: 10.1021/bi00352a010. [DOI] [PubMed] [Google Scholar]
- 58.Debold EP, et al. Hypertrophic and dilated cardiomyopathy mutations differentially affect the molecular force generation of mouse alpha-cardiac myosin in the laser trap assay. Am J Physiol Heart Circ Physiol. 2007;293(1):H284–H291. doi: 10.1152/ajpheart.00128.2007. [DOI] [PubMed] [Google Scholar]
- 59.Lehman W, Vibert P, Uman P, Craig R. Steric-blocking by tropomyosin visualized in relaxed vertebrate muscle thin filaments. J Mol Biol. 1995;251(2):191–196. doi: 10.1006/jmbi.1995.0425. [DOI] [PubMed] [Google Scholar]
- 60.Pirani A, et al. Single particle analysis of relaxed and activated muscle thin filaments. J Mol Biol. 2005;346(3):761–772. doi: 10.1016/j.jmb.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 61.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116(1):190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 62.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 63.Palmiter KA, et al. R403Q and L908V mutant beta-cardiac myosin from patients with familial hypertrophic cardiomyopathy exhibit enhanced mechanical performance at the single molecule level. J Muscle Res Cell Motil. 2000;21(7):609–620. doi: 10.1023/a:1005678905119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.