Significance
We report the high-resolution structure of a tension-sensing myosin-Ib. We identify a striking unique orientation of structural elements that position the motor’s lever arm. This orientation results in a cavity between the motor and lever arm that holds a 10-residue stretch of N-terminal amino acids, a region that is divergent among myosins. We show the importance of the N-terminal region of myosin in controlling the kinetics and mechanics of the motor.
Keywords: mechanochemistry, optical tweezers, structural biology, cryo-EM
Abstract
Myosins are molecular motors that power diverse cellular processes, such as rapid organelle transport, muscle contraction, and tension-sensitive anchoring. The structural adaptations in the motor that allow for this functional diversity are not known, due, in part, to the lack of high-resolution structures of highly tension-sensitive myosins. We determined a 2.3-Å resolution structure of apo-myosin-Ib (Myo1b), which is the most tension-sensitive myosin characterized. We identified a striking unique orientation of structural elements that position the motor’s lever arm. This orientation results in a cavity between the motor and lever arm that holds a 10-residue stretch of N-terminal amino acids, a region that is divergent among myosins. Single-molecule and biochemical analyses show that the N terminus plays an important role in stabilizing the post power-stroke conformation of Myo1b and in tuning the rate of the force-sensitive transition. We propose that this region plays a general role in tuning the mechanochemical properties of myosins.
The cellular functions of myosin isoforms are remarkably diverse, requiring a range of mechanical and kinetic adaptations that tailor the different isoforms to their physiological roles (1). This diversity extends to the response of myosins to mechanical loads experienced during the force-generating conformational changes that drive motility (2–5). For example, the myosin-I family includes isoforms that have motile properties appropriate for transport and power-generating processes (6), whereas other isoforms act as tension-sensitive anchors that stall and remain strongly bound to actin when they encounter forces that resist forward motility (7–9).
Several high-resolution structures of myosins that fall into the power-generating class have been determined (10–12), but there have been no high-resolution structures of myosins that act as tension-sensitive anchors. To understand the structural features that link mechanical loads to biochemical transitions better, we solved the crystal structure of myosin-Ib (Myo1b), the most tension-sensitive myosin yet identified (9). This myosin has actin-detachment kinetics that slow more than 50-fold in the presence of forces >0.5 pN, resulting in attachment lifetimes greater than 50 s. The structure reveals several striking features not yet seen in other myosins, including a unique positioning of the lever arm helix (LAH) and the presence of an N-terminal region (NTR) sandwiched between the motor domain and LAH. Single-molecule and ensemble level experiments show that this NTR is important for tuning the kinetics of the myosin and for mechanically stabilizing the post power-stroke conformation.
Results
Structure of Apo-Myo1bIQ.
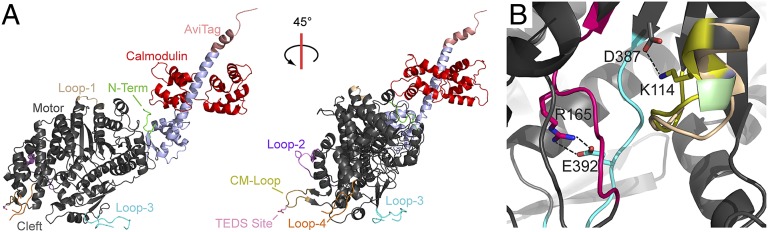
We solved a 2.3-Å resolution structure of Myo1b, the most tension-sensitive myosin yet identified (9). The crystallized protein contains the motor domain and the first IQ motif of the LAH with calmodulin bound in the nucleotide-free state (Myo1bIQ; Fig. 1A, Table S1, and Movie S1). Myo1bIQ also contains a C-terminal AviTag sequence (13) used to provide site-specific biotinylation for anchoring the protein in mechanical studies (14). Crystal contacts allow resolution of several key regions that are frequently disordered in myosin structures (Fig. 1), including loop-1, loop-2, loop-3, the cardiomyopathy loop, and most of loop-4. The TEDS site (E334), a regulatory site in some myosins that is named for the amino acids found at this position (15, 16), is also resolved. Loop-2 has a unique conformation from P555–N563 that is stabilized by hydrogen bonding between A560 and R227 from a symmetry-related molecule and by a tetragonal bipyramidal coordinated ion, whose identity could not be determined from our structural data (Fig. S1A).
Fig. 1.
Structure of apo-Myo1bIQ. (A) Overall structure of Myo1bIQ shows key structural elements and the AviTag. (B) Structural motifs from the active site of Myo1b (red box from Fig. S1B) are colored: P-loop, yellow; SW-1, blue; SW-2, red. Conserved interactions are highlighted. A113 is colored green, and G112, which is partially occluded by A113, is colored blue. The P-loop of Myo5 (PDB ID code 1OE9) is shown in tan.
The core structure of the Myo1bIQ motor domain (residues 16–620) is similar to that of apo-Myo5 [Protein Data Bank (PDB) ID code 1OE9, residues 70–686 (11)], with an rmsd between equivalent Cα atoms of 1.2 Å (Fig. S1B). The Myo1bIQ actin-binding cleft is in a closed conformation, consistent with a rigor-like state that binds tightly to actin. The central β-sheet of the motor domain, known as the transducer region because it communicates actin binding to the ATP-binding pocket (17), differs from Myo5 in that Myo1bIQ shows a twisting of β-strands 6 and 7 that is accompanied by movement of helix HH (residues G132–G153) and helix HO (residues V342–S371) (18) (Fig. S1C).
The nucleotide-binding site of Myo1bIQ, composed of switch-1 (SW1), switch-2 (SW2), and the P-loop, would require a conformational change to allow binding to Mg+2 and nucleotide (Fig. 1B). The apo-conformation of SW2 is stabilized by side-chain interactions similar to those described for the apo-structure of Myo5 (11). In particular, R165 interacts with E392 and K114 interacts with D387. Three residues of the P-loop (A112, G113, and K114) form an additional turn of the adjacent helix, occluding the nucleotide-binding site (Fig. 1B). This P-loop structure has not been seen in other myosin structures, and it may be stabilized by the presence of a solvent molecule that was a component of the crystallization solution.
Unique Position of the Myo1b Lever Arm.
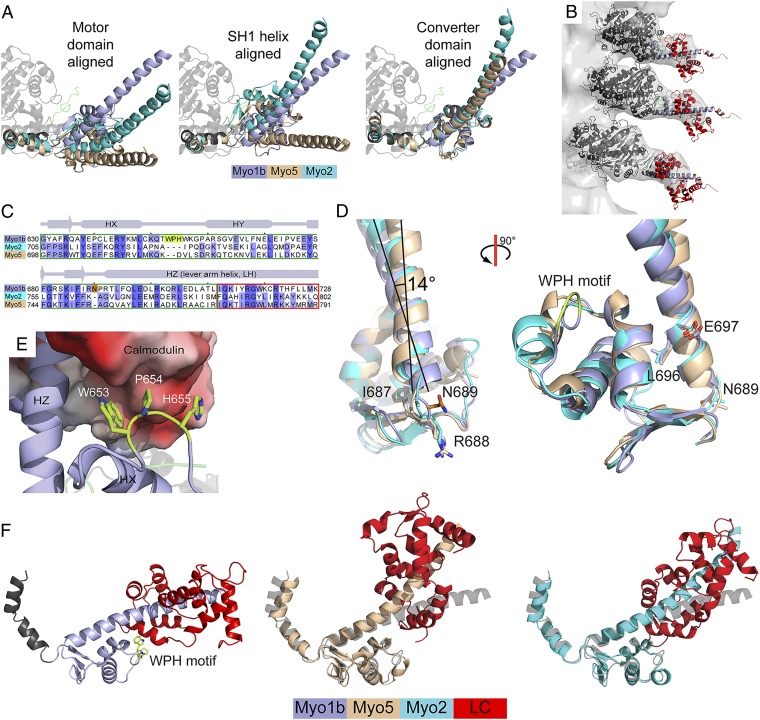
In contrast to the rigor Myo5 structure and other characterized power-generating myosins, there is a striking reorientation of the structural elements adjacent to the region that acts as the myosin lever arm, leading to a reorientation of the lever arm position. The SH1 helix (residues 621–629), converter domain (residues 634–701), LAH, and calmodulin light chain are substantially repositioned compared with the motor domains of apo-Myo5 and near-rigor Myo2 (Fig. 2A and Fig. S1B), such that the LAH orientation is ∼90° with respect to the long axis of the actin filament when docked in a rigor conformation (Fig. S1 D and E). This orientation results in a novel cavity that holds the NTR of the motor domain (see below). Cryo-EM images of nucleotide-free Myo1bIQ bound to actin confirm this conformation exists when bound to actin to 27-nm resolution (Fig. 2B), as do EM images of actin-bound vertebrate Myo1A and Myo1C isoforms (19, 20). A recent 8-Ǻ EM model of actin-bound myosin-IE from Dictyostelium is also consistent with this position of the converter domain (21), but the comparison is limited, due to the absence of the LAH and light chain.
Fig. 2.
(A) Comparison of the LAH of Myo1bIQ, Myo5, and Myo2. The structures are aligned using the core motor domain (Myo1bIQ residues 26–607, Left), the SH1 helix (residues 621–629, Center), and the converter domain (residues 634–700, Right). (B) Helical reconstruction of cryoelectron micrographs shows that the long axis of the LAH of Myo1bIQ in the nucleotide-free state is nearly perpendicular to the long axis of the actin filament. Three duplicate models of the Myo1bIQ structure are docked into the EM map. The low signal-to-noise ratio of the density from the N-terminal lobe of the calmodulin likely resulted from conformational flexibility and the greater disorder that occurs at the higher radius of the decorated filament. The flexibility of the calmodulin N-terminal lobe is consistent with the higher b-factors for this region in the crystal structure. (C) Structure-based alignment of the converter domains and IQ motifs of Myo1bIQ, Myo5 (PDB ID code 1OE9), and Myo2 (PDB ID code 2MYS). The two myosin-I–specific insertions are highlighted in lime and orange. (D) N689 insertion shifts the base of the LAH between conserved residues immediately N-terminal (R688) and C-terminal (L696), which are structurally equivalent Myo1b, Myo5, and Myo2. The insertion buckles the loop in which it resides, causing a 3.9-Å shift in the N terminus of the LAH. (E) Myo1b WPH motif interacts with a hydrophobic and acidic surface on the C-terminal lobe of the calmodulin bound to the IQ motif. (F) Distinct LAH positions of Myo1bIQ, Myo5, and Myo2 result in distinct positions of the bound light chain (LC). Structures were aligned using converter domains (residues 634–700 in Myo1bIQ). Light chains are shown in red, and Myo1bIQ is shown in gray.
The repositioning of the Myo1bIQ converter domain, LAH, and calmodulin results in an ∼15° rotation of the long axis of the SH1 helix about a fulcrum at the SH1-SH2 helix junction compared with Myo5 and Myo2 (Fig. 2A, Left). The structure is stabilized by a hydrophobic cluster of residues at the interface between the motor domain, converter, and LAH (see below), but this reorientation is not simply the result of a rigid rotation of the converter domain and LAH relative to the motor. Rather, two insertions in the Myo1b converter domain (Fig. 2C) contribute to this unique confirmation, which is best seen when Myo5 and Myo2 are aligned to the Myo1bIQ SH1 helix (residues 621–629 in Myo1b) (Fig. 2A, Center) or converter domain (residues 634–700 in Myo1b) (Fig. 2A, Right). An asparagine residue (N689) at the base of the LAH prevents the loop in which it resides from adopting a conformation similar to that in Myo5 and Myo2, shifting the position of the N terminus of the LAH by ∼3.9 Å relative to the converter domain (Fig. 2 C and D). This shift reorients the long axis of the LAH by ∼14° about a pivot point formed by two conserved residues (L696 and E697) that occupy identical positions (relative to the converter) in all three structures. The position of the LAH is further altered by an insertion in the converter domain (W653, P654, and H655), which we call the WPH motif (Fig. 2 C and E). This motif alters the conformation of the loop in which it resides, allowing it to interact with the calmodulin bound to the IQ motif, producing a distinct bend in the LAH and concomitant repositioning of the calmodulin (Fig. 2F).
NTR of Myo1bIQ.
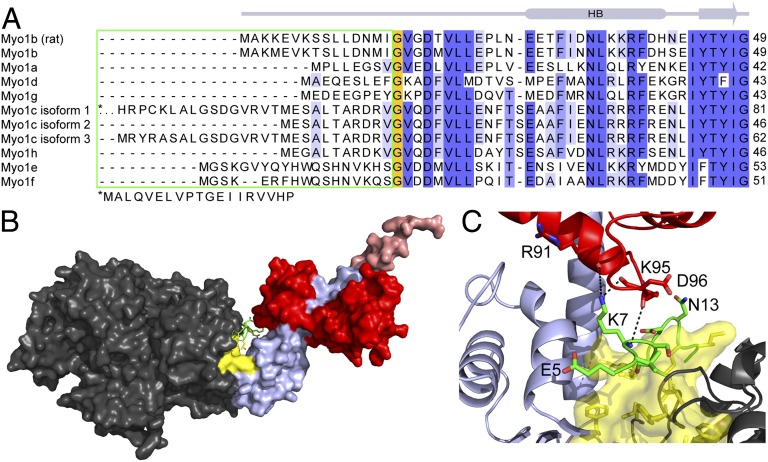
The NTR of Myo1bIQ, which we define as the region that precedes the highly conserved G16 (Fig. 3A), is in a position that has not been observed in other myosin isoforms (Figs. 1A and 3B and Movie S1). Myosin-I isoforms do not have the Src-homology 3 (SH3)-like domain that resides at the N terminus of other myosin families (10, 22) (Fig. 3A). Rather, we find that the NTR is sandwiched into the unique pocket created by the unique orientations of the converter, LAH, and calmodulin (Fig. 3 B and C). The side chain of residue K7 forms hydrogen bonds with the backbone carbonyls of R91 and D96 of the calmodulin, whereas the backbone amide forms a hydrogen bond with amide nitrogen of K95 of calmodulin (Fig. 3C). Several NTR residues (L10, L11, M14, and I15) form a hydrophobic cluster with side chains from both the motor domain (V17, V21, L22, L23, Y47, S50, F77, Y78, and P82) and the LAH (F634 and F694; Fig. 3C and Fig. S2).
Fig. 3.
Myo1bIQ NTR is located between the motor domain and LAH. (A) Sequence alignment of the N termini of human myosin-I isoforms and the rat Myo1b isoform used in this study. The green box shows the divergent NTR. High sequence conservation is present after the invariant glycine residue (orange box). For clarity, the first 18 residues of Myo1c isoform-1 are shown below the alignment. (B) NTR (green) is sandwiched between the motor (gray) and LAH (blue). A hydrophobic core (yellow), composed of residues from the motor, NTR, and LAH, is at the base of the LAH. (C) Magnification of the base of the LAH shows the concentration of hydrophobic residues. Interactions between K7 of the NTR and the C-terminal lobe of calmodulin are shown. The molecule is rotated ∼180° about the y axis, compared with B.
NTR Affects Myo1b Biochemistry and Mechanics.
The NTR is located in a highly active region of the molecule that undergoes large structural rearrangements during the repriming and working stroke steps. In addition to its structural role, we hypothesize that the NTR has a role in communicating the position of the LAH to the motor domain, because it makes interactions with the motor domain, LAH, and bound calmodulin. Specifically, the Myo1b NTR interacts with a loop–helix–loop (LHL) motif (residues L132–H154 in Myo2 or equivalent residues S60–H82 in Myo1b) that is adjacent to the transducer β-sheet (Fig. S3). We investigated the extent to which the NTR plays a role in modulating myosin’s biochemical and mechanical properties by characterizing a Myo1b protein construct in which residues 1–13 were removed (Myo1b∆N). The construct contained the LAH of the Myo1bb splice isoform, which can bind five calmodulins to facilitate mechanical experiments; however, we have previously shown that the size of the lever arm has little effect on Myo1b’s unloaded kinetics (7, 14, 23).
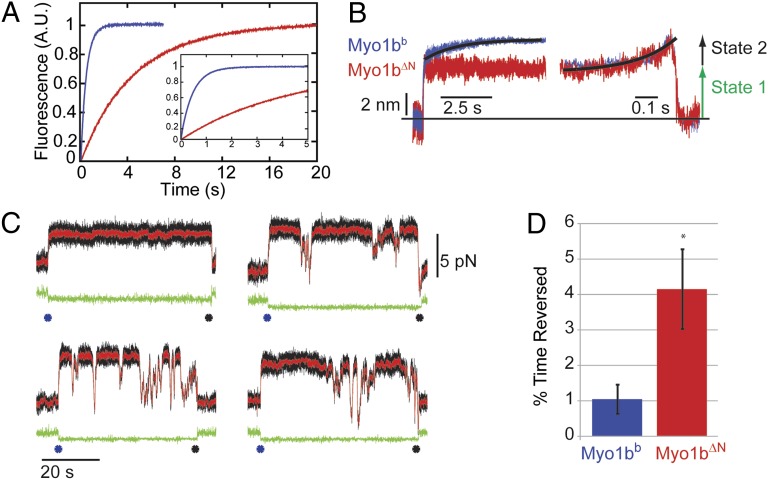
Strikingly, stopped-flow kinetic measurements show that the rate of ADP release from actoMyo1b∆N (0.22 ± 0.0010 s−1) is 12-fold slower than from actoMyo1bb (Fig. 4A) and actoMyo1bIQ (2.6 ± 0.48 s−1; P < 0.001; Table 1). The maximum rate of ATP binding to actoMyo1b∆N (k2′) is 3.6-fold faster than to actoMyo1bIQ (P < 0.001), but the apparent affinity for ATP (K1′) is decreased 3.3-fold (P < 0.001; Fig. S4 and Table 1). The probability of nucleotide-free actoMyo1b entering a state that does not bind nucleotide (Kα; Scheme 1) is 2.5-fold reduced in the mutant (P < 0.001; Table 1). The rates into and out of this state are also slowed (Table 1 and Scheme 1).
Fig. 4.
Transient kinetic and optical trapping measurements of Myo1bb (blue) and Myo1bΔN (red). (A) Rate of ADP release, as measured using stopped-flow transient kinetics (Materials and Methods), is slowed ∼10-fold by deletion of the NTR. Shown are the normalized fluorescence transients as a function of time. (Inset) Transient is shown at a shorter time scale. Fluorescence is plotted in arbitrary units (A.U.). (B) Ensemble averages of the Myo1bb (n = 561) and Myo1bΔN (n = 230) working stroke were constructed as described in Materials and Methods. The time-forward ensemble averages with a single-exponential fit to the Myo1bb data (Left) and the time-reversed averages with a single-exponential fit to the Myo1bΔN data (Right) are shown. Both constructs show a two-step working stroke with similarly sized substeps. The measured values for the step and substep sizes can be found in Table 2. (C) Sample data collected with the Myo1bΔN construct under a load imposed by the isometric optical clamp. Each panel is a single actomyosin-binding event that begins when the covariance of the two trapped beads, shown in green, becomes low (blue asterisk) and ends when the covariance becomes high (black asterisk). Raw data are shown in black, and the Savitzky–Golay-filtered data are shown in red. The first actomyosin-binding event shows no reversals, whereas the other binding events show transient decreases in force without actomyosin detachment (as seen by the constant covariance over the course of the binding event). (D) Quantification of the percentage of time spent in the reversed state as described in Materials and Methods. Myo1bΔN spends fourfold the time reversed compared with Myo1bb.
Table 1.
Rate and equilibrium constants for key steps of the actomyo1b ATPase cycle at 20 °C
| Myo1bIQ* | Myo1bΔN | |
| ATP binding | ||
| 1/K1′, μM†,‡ | 290 ± 35 | 970 ± 110 |
| k+2′, s−1†,‡ | 33 ± 1.2 | 120 ± 5.4 |
| K1′k+2′, μM−1⋅s−1§ | 0.12 ± 0.015 | 0.12 ± 0.016 |
| ADP release | ||
| k+5′, s−1† | 2.6 ± 0.48 | 0.22 ± 0.0010 |
| Nucleotide-free isomerization step | ||
| Kα†,‡ | 2.6 ± 0.48 | 6.5 ± 0.46 |
| k+α, s−1†,‡ | 3.1 ± 0.26 | 0.15 ± 0.0061 |
| k−α, s−1§ | 1.2 ± 0.24 | 0.023 ± 0.0019 |
All experiments were performed in KMg25 buffer at 20 °C.
Values are from Lewis et al. (7).
Pyrene-actin fluorescence.
All solutions also included 100 U/mL pyruvate kinase and 0.5 mM phospho(enol)pyruvate.
Calculated.
Scheme 1.
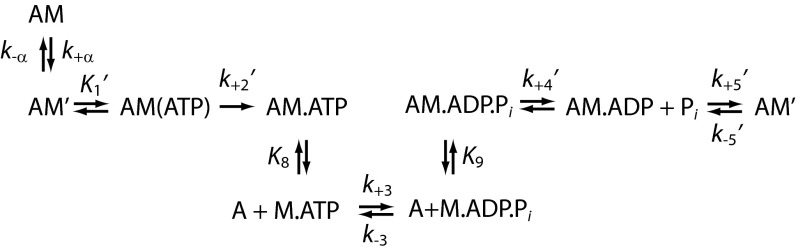
Actomyosin ATPase Cycle.
To determine the effect of the NTR on the Myo1b working stroke, we measured the unitary displacement of Myo1b∆N using an optical trapping assay utilizing the three-bead geometry, where a single actin filament, suspended between two beads held by separate optical traps, is brought close to the surface of a pedestal bead that is sparsely coated with myosin (24). Myo1bb has a two-step working stroke, where the first displacement correlates with phosphate release and the population of a structural state that correlates with the AM.ADP state, and the second displacement correlates with ADP release and the population of a structural state that correlates with the AM state (9, 25) (Scheme 1). Using ensemble averaging techniques (9, 25, 26), we resolve two substeps in the Myo1b∆N working stroke that have similar displacements to the WT protein (Fig. 4B and Table 2). The rate of the rise in displacement of the time-forward ensemble averages (Fig. 4B, Left) reports the rate of transition from the first structural state to the second structural state. The rate of the rise in displacement of the time-reversed ensemble averages (Fig. 4B, Right) reports the rate of exit from the second structural state. The rate for exiting the structural state after the second substep for Myo1b∆N, 6.9 ± 0.94 s−1, is consistent with the expected rate of ATP-induced dissociation from the AM biochemical state in the presence of 50 μM ATP (6.1 s−1). As can be seen from the time-forward ensemble averages (Fig. 4B, Left), the rate of transition from the first structural state to the second structural state is so slow for Myo1b∆N that it is not resolved compared with the WT protein (Fig. 4B). The rate of this transition for Myo1bb was 0.63 ± 0.10 s−1, consistent with our previous measurements (8, 9) showing that this rate was limited by ADP release. We were not able to determine this rate for Myo1b∆N because experimental noise during the long-lived attachments prevented the reliable averaging needed for kinetic analysis. Nevertheless, the long lifetime of this mechanical state indicates that it correlates with the long-lived AM.ADP biochemical state observed in the stopped-flow experiments.
Table 2.
Displacements and force-dependent actin detachment kinetics for Myo1bb and Myo1bΔN
| Myo1bb (n = 561) | Myo1bΔN (n = 230) | |
| Working stroke displacements | ||
| Total, nm | 8.8 ± 0.12 | 8.5 ± 0.21 |
| Substep 1, nm | 5.9 ± 0.012 | 5.2 ± 0.013 |
| Substep 2, nm | 2.9 ± 0.12 | 3.3 ± 0.21 |
| Force sensitivity | ||
| k0, s−1† | 1.6 (+0.50/−0.35)* | 0.098 (+0.020/−0.028) |
| d, nm† | 12 (+1.6/−3.0)* | 12 (+2.9/−2.9) |
Performed in the optical trap in KMg25 buffer in the presence of 50 μM ATP at 20 °C.
Values are from Laakso et al. (9).
Fit to a probability distribution function and optimized with a maximum likelihood estimation routine (90% confidence interval, n = 1,000 simulations; Materials and Methods).
The detachment of Myo1bb from actin in the presence of ATP is highly force-sensitive (8, 9). Small forces (>0.5 pN) that resist the direction of the working stroke decrease the rate of ADP release >50-fold, resulting in slow actin-detachment kinetics (Scheme 1). The mechanism of this force-dependent slowing of Myo1b detachment kinetics appears to be a direct inhibition of the LAH rotation that accompanies ADP release (9). Because the NTR interacts with the LAH and motor domain, and it affects the rate of ADP release, we examined the force-dependent behavior of Myo1b∆N (Fig. S4D). Increasing loads substantially slow the actin-detachment kinetics of Myo1b∆N with a force dependence similar to Myo1bb (Fig. S4D). The force dependence of the detachment rate, kdet(F), was modeled as follows (27):
where k0 is the rate of actin detachment in the absence of force, F is force, k is the Boltzmann constant, T is temperature, and d is the distance parameter, which reports the distance to the transition state of the primary force-sensitive step and gives a measure of force sensitivity. The value of d for Myo1b∆N (12 ± 2.9 nm) is the same as reported for Myo1bb [12 (+1.6/−0.3) nm] (9). The rate of k0 is ∼10-fold slower for Myo1b∆N (Fig. S4D), consistent with the observed decrease in the rate of ADP release in Myo1b∆N measured in stopped-flow experiments (Table 1). Importantly, these results suggest that the NTR activates ADP release by decreasing the energetic barrier to the nucleotide-free state as the LAH rotates to the post power-stroke state, but the position of the transition state along the reaction coordinate is unaffected by the NTR.
We observed transient drops in force when single Myo1b∆N molecules were held under load (Fig. 4C and Materials and Methods). Quantification of this behavior shows that when Myo1b∆N is placed under load, it spends 4.2 ± 1.2% of its attached time in a reversed, low-force state, whereas Myo1bb only spends 1.0 ± 0.4% of its attached time in this state (P < 0.001; Fig. 4D). The increase in the frequency of these drops in force when the NTR is deleted suggests that the NTR plays a role in stabilizing the post power-stroke state. It is possible that these drops in force represent reversal of the power stroke; however, we are unable to resolve clearly transitions that occur faster than the time constant of the isometric optical clamp feedback (50 ms). Interestingly, reversal of the power stroke under load without actomyosin detachment has previously been seen in Myo5 (28). Whether or not drops in force represent genuine reversals of the power stroke to a prepower stroke state or some other destabilization of the state is not clear.
Discussion
The conformation of the core of apo-Myo1bIQ is very similar to that of apo-Myo5 (Fig. S1B). It has been proposed that the apo-Myo5 structure closely resembles its actin-bound rigor conformation (11, 29), and this is also likely to be the case for the Myo1b structure determined here. However, despite these clear structural similarities, there are differences between Myo1bIQ and Myo5 that may have functional consequences for the biochemical and mechanical activities of the motors. There is a twisting of the central β-sheet of apo-Myo1bIQ that is not seen in apo-Myo5 (Fig. S1C). Similar twisting is seen in several other structures, including the apo-Myo2 structure (PDB ID code 2MYS) (10) and the prepower stroke of Dictyostelium myosin-IE (PDB ID code 1LKX) (30). There is a major reorientation of the Myo1b LAH that results in a lever arm position that is nearly perpendicular to the long axis of the actin helix (Fig. 2B and Fig. S1D). This position of the LAH in the apo-state is different from other characterized myosins, including Myo2 (31), Myo5 (11), and Acanthamoeba myosin-IB (32), which have LAHs that are oriented toward the barbed end of the actin in the rigor state. Interestingly, this perpendicular orientation of the LAH relative to the actin filament has been observed in helical reconstructions of EM images of all characterized rigor vertebrate myosin-I isoforms [Myo1a (19, 33), Myo1b (Fig. 2B), and Myo1c (20)].
Taken together, the identified structural differences in the motor domain and LAH positioning suggest that the observed nucleotide-free and actin-bound conformations of Myo1bIQ may not be the same rigor state seen in other myosins, despite actually being in an equilibrium rigor position. It is possible that during ATPase cycling, the mechanical state that precedes detachment of actoMyo1b is a short-lived state that has the LAH rotated beyond the longer lived mechanical state observed in optical trapping experiments (7–9, 25) and in the nucleotide-free structure in the absence of actin that we have reported here. The presence of such a conformation may explain why the distance to the transition state for Myo1b force-dependent detachment is substantially larger than the size of the resolved individual substeps of the working stroke (Fig. 4B).
The identification of the NTR as an important structural feature is a key finding of this study. The NTR is located in a region of the molecule that undergoes large structural rearrangements during the repriming and working stroke steps, and our data suggest that it is an important structural element that modulates myosin’s functional properties. Removal of the NTR from Myo1b slows the rate of ADP release 10-fold (Table 1), but it does not change the force dependence of this transition (Fig. S4D and Table 2). Thus, removal of the NTR increases the free energy of the AM.ADP-to-AM transition state without changing the distance from the AM.ADP state to the transition state in the dimension of the working stroke. This result is consistent with the model that force slows the rate of ADP release in Myo1b by inhibiting the rotation of the lever arm that accompanies ADP release (8, 9, 34). The distance to the transition state is likely unaffected by the removal of the NTR because the size of the working stroke (i.e., the rotation of the lever arm) is unaffected by the deletion (Fig. 4 and Table 2).
Based on our kinetic and optical trapping studies, we propose that one of the roles of the NTR in Myo1b is to accelerate the rate of ADP release (i.e., lower the energy of the transition state) while slowing the maximal rate of ATP binding. It is interesting that removal of the NTR differentially affects the rates of ATP binding and ADP release, consistent with different structural transitions regulating these biochemical transitions.
In addition to its role in modulation of Myo1b kinetics, we propose that the NTR plays an important structural role in stabilizing the post power-stroke state (Fig. 4D). Interestingly, reversal of the power stroke under load without actomyosin detachment has previously been seen in Myo5 (28). The N terminus of Myo5 consists of an SH3-like domain that lies on the motor domain (11); however, it does not contain an NTR extension that resides between the LHL motif and LAH as does the Myo1b NTR (Fig. S3). We speculate that NTR-mediated stabilization of the post power-stroke state may be a more general feature of myosins; however, future studies are necessary to address this possibility.
Alignment of vertebrate myosin-I sequences shows that there is substantial sequence diversity in the NTR, with a high level of conservation beyond G16 (Fig. 3A). Notably, Myo1c is alternatively spliced within this region. Thus, given its position at the interface between the LHL motif of the motor domain and LAH, it is likely that this sequence diversity within the NTR tunes the mechanochemical relationship between lever arm movement and ATPase activity. The NTR is also quite divergent across the myosin superfamily. Myosin-I isoforms differ from other myosins in that they lack an N-terminal SH3-like domain structure that connects to the point homologous to G16 in Myo1bIQ. Myo2, a low-duty ratio motor, has a sequence before the start of the SH3-like domain (residues 1–33; PDB ID code 2MYS) in its NTR that interacts with the LHL motif of the motor domain, but without interacting with the LAH or bound light chain (10, 12) (Fig. S3). Deletion of this region in Dictyostelium Myo2 results in a slowing of the rate of ADP release in the absence of actin. This result led to the suggestion that deletion of the Myo2 N terminus results in a high-duty ratio motor (22). High-resolution structures of the high-duty ratio motors Myo5 and Myo6 show that their NTRs do not interact with the LHL motif (11, 35). Interestingly, these high-duty ratio motors also have ADP release as their rate-limiting step (36–38).
Given our structural data and our observation that the NTR accelerates the rate of ADP release in Myo1b, we speculate that the position of the NTR tunes the rate of ADP release in myosin, such that motors with N termini that come across the motor domain and interact with the LHL motif have low-duty ratios, whereas those that do not have high-duty ratios (Fig. S3). Interestingly, this loop region lies immediately adjacent to the transducer β-sheet, and changes in the transducer conformation could be relayed through the loop to the NTR. Future experiments are necessary to understand the role of NTR in the functional adaptation of all myosins.
Materials and Methods
Details of experiments and solution conditions are described in SI Materials and Methods. X-ray data collection and structure determination methods are provided in SI Materials and Methods. Cryo-EM sample preparation, data collection, and image processing (Fig. S5) were performed as discussed in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank V. Stojanoff, J. Jakoncic, and Edwin E. Lazo for help with synchrotron data collection. Data collection at beamline X6A of the National Synchrotron Light Source was supported by National Institutes of Health (NIH) Grant GM-0080 and Department of Energy Contract DE-AC02-98CH10886. This work was also supported by NIH Grants GM057247 (to E.M.O.), GM073791 (to R.D.), and AR053461 and GM097889 (to M.J.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.M.W. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4L79).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321022111/-/DCSupplemental.
References
- 1.De La Cruz EM, Ostap EM. Relating biochemistry and function in the myosin superfamily. Curr Opin Cell Biol. 2004;16(1):61–67. doi: 10.1016/j.ceb.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 2.Greenberg MJ, Ostap EM. Regulation and control of myosin-I by the motor and light chain-binding domains. Trends Cell Biol. 2013;23(2):81–89. doi: 10.1016/j.tcb.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Veigel C, Schmitz S, Wang F, Sellers JR. Load-dependent kinetics of myosin-V can explain its high processivity. Nat Cell Biol. 2005;7(9):861–869. doi: 10.1038/ncb1287. [DOI] [PubMed] [Google Scholar]
- 4.Takagi Y, Homsher EE, Goldman YE, Shuman H. Force generation in single conventional actomyosin complexes under high dynamic load. Biophys J. 2006;90(4):1295–1307. doi: 10.1529/biophysj.105.068429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Capitanio M, et al. Ultrafast force-clamp spectroscopy of single molecules reveals load dependence of myosin working stroke. Nat Methods. 2012;9(10):1013–1019. doi: 10.1038/nmeth.2152. [DOI] [PubMed] [Google Scholar]
- 6.Greenberg MJ, Lin T, Goldman YE, Shuman H, Ostap EM. Myosin IC generates power over a range of loads via a new tension-sensing mechanism. Proc Natl Acad Sci USA. 2012;109(37):E2433–E2440. doi: 10.1073/pnas.1207811109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis JH, Greenberg MJ, Laakso JM, Shuman H, Ostap EM. Calcium regulation of myosin-I tension sensing. Biophys J. 2012;102(12):2799–2807. doi: 10.1016/j.bpj.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laakso JM, Lewis JH, Shuman H, Ostap EM. Control of myosin-I force sensing by alternative splicing. Proc Natl Acad Sci USA. 2010;107(2):698–702. doi: 10.1073/pnas.0911426107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laakso JM, Lewis JH, Shuman H, Ostap EM. Myosin I can act as a molecular force sensor. Science. 2008;321(5885):133–136. doi: 10.1126/science.1159419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rayment I, et al. Three-dimensional structure of myosin subfragment-1: A molecular motor. Science. 1993;261(5117):50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 11.Coureux PD, et al. A structural state of the myosin V motor without bound nucleotide. Nature. 2003;425(6956):419–423. doi: 10.1038/nature01927. [DOI] [PubMed] [Google Scholar]
- 12.Dominguez R, Freyzon Y, Trybus KM, Cohen C. Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: Visualization of the pre-power stroke state. Cell. 1998;94(5):559–571. doi: 10.1016/s0092-8674(00)81598-6. [DOI] [PubMed] [Google Scholar]
- 13.Schatz PJ. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: A 13 residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology (N Y) 1993;11(10):1138–1143. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- 14.Lin T, Tang N, Ostap EM. Biochemical and motile properties of Myo1b splice isoforms. J Biol Chem. 2005;280(50):41562–41567. doi: 10.1074/jbc.M508653200. [DOI] [PubMed] [Google Scholar]
- 15.Bement WM, Mooseker MS. TEDS rule: A molecular rationale for differential regulation of myosins by phosphorylation of the heavy chain head. Cell Motil Cytoskeleton. 1995;31(2):87–92. doi: 10.1002/cm.970310202. [DOI] [PubMed] [Google Scholar]
- 16.Ostap EM, Lin T, Rosenfeld SS, Tang N. Mechanism of regulation of Acanthamoeba myosin-IC by heavy-chain phosphorylation. Biochemistry. 2002;41(41):12450–12456. doi: 10.1021/bi0262193. [DOI] [PubMed] [Google Scholar]
- 17.Coureux PD, Sweeney HL, Houdusse A. Three myosin V structures delineate essential features of chemo-mechanical transduction. EMBO J. 2004;23(23):4527–4537. doi: 10.1038/sj.emboj.7600458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cecchini M, Houdusse A, Karplus M. Allosteric communication in myosin V: From small conformational changes to large directed movements. PLoS Computational Biology. 2008;4(8):e1000129. doi: 10.1371/journal.pcbi.1000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jontes JD, Milligan RA. Brush border myosin-I structure and ADP-dependent conformational changes revealed by cryoelectron microscopy and image analysis. J Cell Biol. 1997;139(3):683–693. doi: 10.1083/jcb.139.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Batters C, et al. Myo1c is designed for the adaptation response in the inner ear. EMBO J. 2004;23(7):1433–1440. doi: 10.1038/sj.emboj.7600169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Behrmann E, et al. Structure of the rigor actin-tropomyosin-myosin complex. Cell. 2012;150(2):327–338. doi: 10.1016/j.cell.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujita-Becker S, et al. Functional characterization of the N-terminal region of myosin-2. J Biol Chem. 2006;281(47):36102–36109. doi: 10.1074/jbc.M605171200. [DOI] [PubMed] [Google Scholar]
- 23.Lewis JH, Lin T, Hokanson DE, Ostap EM. Temperature dependence of nucleotide association and kinetic characterization of myo1b. Biochemistry. 2006;45(38):11589–11597. doi: 10.1021/bi0611917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finer JT, Simmons RM, Spudich JA. Single myosin molecule mechanics: Piconewton forces and nanometre steps. Nature. 1994;368(6467):113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 25.Veigel C, et al. The motor protein myosin-I produces its working stroke in two steps. Nature. 1999;398(6727):530–533. doi: 10.1038/19104. [DOI] [PubMed] [Google Scholar]
- 26.Chen C, et al. Kinetic schemes for post-synchronized single molecule dynamics. Biophys J. 2012;102(6):L23–L25. doi: 10.1016/j.bpj.2012.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200(4342):618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 28.Sellers JR, Veigel C. Direct observation of the myosin-Va power stroke and its reversal. Nat Struct Mol Biol. 2010;17(5):590–595. doi: 10.1038/nsmb.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holmes KC, Schröder RR, Sweeney HL, Houdusse A. The structure of the rigor complex and its implications for the power stroke. Philos Trans R Soc Lond B Biol Sci. 2004;359(1452):1819–1828. doi: 10.1098/rstb.2004.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kollmar M, Dürrwang U, Kliche W, Manstein DJ, Kull FJ. Crystal structure of the motor domain of a class-I myosin. EMBO J. 2002;21(11):2517–2525. doi: 10.1093/emboj/21.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whittaker M, et al. A 35-A movement of smooth muscle myosin on ADP release. Nature. 1995;378(6558):748–751. doi: 10.1038/378748a0. [DOI] [PubMed] [Google Scholar]
- 32.Jontes JD, Ostap EM, Pollard TD, Milligan RA. Three-dimensional structure of Acanthamoeba castellanii myosin-IB (MIB) determined by cryoelectron microscopy of decorated actin filaments. J Cell Biol. 1998;141(1):155–162. doi: 10.1083/jcb.141.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jontes JD, Wilson-Kubalek EM, Milligan RA. A 32 degree tail swing in brush border myosin I on ADP release. Nature. 1995;378(6558):751–753. doi: 10.1038/378751a0. [DOI] [PubMed] [Google Scholar]
- 34.Nyitrai M, Geeves MA. Adenosine diphosphate and strain sensitivity in myosin motors. Philos Trans R Soc Lond B Biol Sci. 2004;359(1452):1867–1877. doi: 10.1098/rstb.2004.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ménétrey J, et al. The structure of the myosin VI motor reveals the mechanism of directionality reversal. Nature. 2005;435(7043):779–785. doi: 10.1038/nature03592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De La Cruz EM, Ostap EM, Sweeney HL. Kinetic mechanism and regulation of myosin VI. J Biol Chem. 2001;276(34):32373–32381. doi: 10.1074/jbc.M104136200. [DOI] [PubMed] [Google Scholar]
- 37.De La Cruz EM, Sweeney HL, Ostap EM. ADP inhibition of myosin V ATPase activity. Biophys J. 2000;79(3):1524–1529. doi: 10.1016/S0006-3495(00)76403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De La Cruz EM, Wells AL, Rosenfeld SS, Ostap EM, Sweeney HL. The kinetic mechanism of myosin V. Proc Natl Acad Sci USA. 1999;96(24):13726–13731. doi: 10.1073/pnas.96.24.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.