Significance
Unexpected evolutionary innovations that lead to qualitatively new traits may result from complex genetic and ecological interactions that develop over long timescales. In a 25-y evolution experiment with Escherichia coli, a rare metabolic innovation arose that allowed a previously untapped resource to be exploited. By dissecting the genetics of this trait using a recursive genomewide recombination and sequencing method (REGRES), we identified a key mutation that converts a rudimentary form of the innovation into a refined trait that confers a decisive competitive advantage. The effects of this mutation demonstrate how improvement of an emergent trait can be as important to its eventual success as earlier mutations or environmental conditions that may have been necessary for it to evolve in the first place.
Keywords: experimental evolution, epistatic network, genetic basis of adaptation
Abstract
Evolutionary innovations often arise from complex genetic and ecological interactions, which can make it challenging to understand retrospectively how a novel trait arose. In a long-term experiment, Escherichia coli gained the ability to use abundant citrate (Cit+) in the growth medium after ∼31,500 generations of evolution. Exploiting this previously untapped resource was highly beneficial: later Cit+ variants achieve a much higher population density in this environment. All Cit+ individuals share a mutation that activates aerobic expression of the citT citrate transporter, but this mutation confers only an extremely weak Cit+ phenotype on its own. To determine which of the other >70 mutations in early Cit+ clones were needed to take full advantage of citrate, we developed a recursive genomewide recombination and sequencing method (REGRES) and performed genetic backcrosses to purge mutations not required for Cit+ from an evolved strain. We discovered a mutation that increased expression of the dctA C4-dicarboxylate transporter greatly enhanced the Cit+ phenotype after it evolved. Surprisingly, strains containing just the citT and dctA mutations fully use citrate, indicating that earlier mutations thought to have potentiated the initial evolution of Cit+ are not required for expression of the refined version of this trait. Instead, this metabolic innovation may be contingent on a genetic background, and possibly ecological context, that enabled citT mutants to persist among competitors long enough to obtain dctA or equivalent mutations that conferred an overwhelming advantage. More generally, refinement of an emergent trait from a rudimentary form may be crucial to its evolutionary success.
Key innovations in the history of life are often caused by the acquisition of a qualitatively new trait that is “an evolutionary novelty which allows the exploitation of new resources or habitats and thus triggers an adaptive radiation” (1). Such innovations are typically rare and difficult to predict because they result from complex nonadditive (i.e., epistatic) genetic interactions or ecological interactions, within or between species, that develop only over the course of long evolutionary trajectories (2). Evolution of a new trait can be conceptually divided into three steps: potentiation, actualization, and refinement (3). First, one or more potentiating events may be necessary to generate a genetic background or environmental conditions that make a new trait accessible to evolution. Genetic potentiation, for example, may involve a period of nonadaptive genetic drift wherein a phenotype stays constant or the accumulation of mutations that are immediately advantageous for reasons unrelated to the new trait (4, 5). Then, a keystone actualizing mutation or environmental shift may lead to expression of the new trait, possibly by coopting latent changes in a cellular network or physical structure for a new use (6–8). Finally, there may be many subsequent opportunities for further refinement mutations that improve an emergent trait so that a newly colonized niche can be fully exploited (1).
The appearance of citrate utilization in a >25-y long-term evolution experiment (LTEE) with Escherichia coli provides an opportunity to study a deep historical record leading to a key metabolic innovation (3, 9, 10). E. coli cannot ordinarily grow on citrate as a sole carbon source under aerobic conditions (11, 12), a phenotype that has been used to define it as a species (13). Just 1 of 12 replicate LTEE populations gained the ability to aerobically use citrate (Cit+), and this rare innovation happened only after ∼31,500 generations of growth in glucose-limited media, despite the presence of an excess of citrate as an untapped carbon source all along (10). The actualizing event for the Cit+ trait is known: tandem amplifications of a chromosomal region that place a copy of an aerobically active promoter upstream of a citrate transporter (citT) are present in all Cit+ isolates from this population (3).
However, the Cit+ trait was surprisingly weak when it first appeared. The earliest individuals with the citT mutation exhibit little or no growth on citrate as a sole carbon source and appear to have derived only a small benefit from this mutation under the conditions of the LTEE (3). In fact, a majority of the population remained Cit– for at least 1,500 generations (225 d) after the citT mutation evolved, and these initial Cit+ individuals were only detected retrospectively in historical samples of the population by using a sensitive indicator agar test for citrate utilization and allowing days to weeks for a positive result (10). Adding a high-copy plasmid with a module containing the new promoter configuration and citT gene to the ancestral strain of the LTEE leads to a phenotype similar to that of the early Cit+ clones, indicating that this mutation is, at least qualitatively, sufficient on its own for this rudimentary version of the new trait (3).
Shortly after ∼33,000 generations, this LTEE population experienced a massive increase in the final cell density it reached at the end of each daily growth cycle (10). This population expansion was due to the evolution of new Cit+ variants that fully use the abundant citrate in the media after glucose depletion. We call this strong phenotype Cit++, to differentiate it from the weak Cit+ phenotype of earlier isolates with just the citT mutation. Cit++ cells contain one or more additional refinement mutations that make robust growth on citrate as a sole carbon source possible. Strains with the Cit++ trait can be readily distinguished from Cit+ strains by their ability to form colonies within 48 h on minimal agar containing citrate as the only carbon source.
Experiments that replayed the evolution of a Cit++ phenotype many times from Cit– isolates taken at different generational time points from the focal LTEE population, found evidence that one or more as-yet-unknown potentiating mutations accumulated in the lineage leading to Cit+ that made later strains more likely to access this metabolic innovation (10). Determining what evolved alleles are required for efficient citrate utilization would shed light on the evolutionary pathway that led to the appearance and refinement of this trait. However, identifying these mutations is confounded by the presence of >70 evolved alleles in the earliest strains that have the Cit++ phenotype, most of which likely improved growth on glucose by mechanisms that were not relevant for the emergence of efficient citrate utilization.
Finding the minimal set of mutations required for Cit++, as well as many other complex evolved phenotypes, is currently a daunting task in asexual microbes. Methods such as genome shuffling (14), multiplex automated genome engineering (MAGE) (15), and array-based discovery of adaptive mutations (ADAM) (16) can be used to create and test of libraries consisting of many genetic variants to dissect complex traits. However, no current technique is suitable in a bacterium like E. coli for concurrently (i) capturing the structure of epistatic networks involving multiple alleles spread across the entire chromosome, (ii) introducing point mutations without associated genetic markers, (iii) broad compatibility with different strain backgrounds, and (iv) reconstructing complex chromosomal rearrangements, mobile element insertions, or large genomic duplications, which are common features of these evolved strains (17, 18). To overcome these limitations, we developed recursive genomewide recombination and sequencing (REGRES), a method that uses conjugative chromosomal transfer, phenotypic selection, and whole-genome sequencing to identify the alleles required for an evolved trait.
Qualitative traits, including evolutionary innovations, may be the result of all-or-none epistatic interactions, where no subset of mutations is sufficient for expression of a new phenotype (5). This absolute genetic interdependence will lead to a sudden and discrete change in a trait when an actualizing or refinement mutation occurs, as observed with the two steps in the evolution of citrate utilization in the LTEE. In accordance with this model, we expected REGRES with selection for the Cit++ phenotype to preserve the citT amplification and some combination of unknown potentiation and refinement mutations. We found that a single refinement mutation affecting the dctA gene was the only evolved allele other than citT conserved across REGRES genomes. These two mutations were sufficient for qualitatively reproducing full utilization of citrate, suggesting a physiological mechanism for refinement that involved activation of a second transporter that makes citrate import by the CitT transporter independent of the production of a cosubstrate by central metabolism. Finally, we discuss an alternative to the all-or-none epistasis hypothesis for how mutations could potentiate the evolution of the strong Cit++ trait without being strictly required for its expression.
Results
REGRES Method.
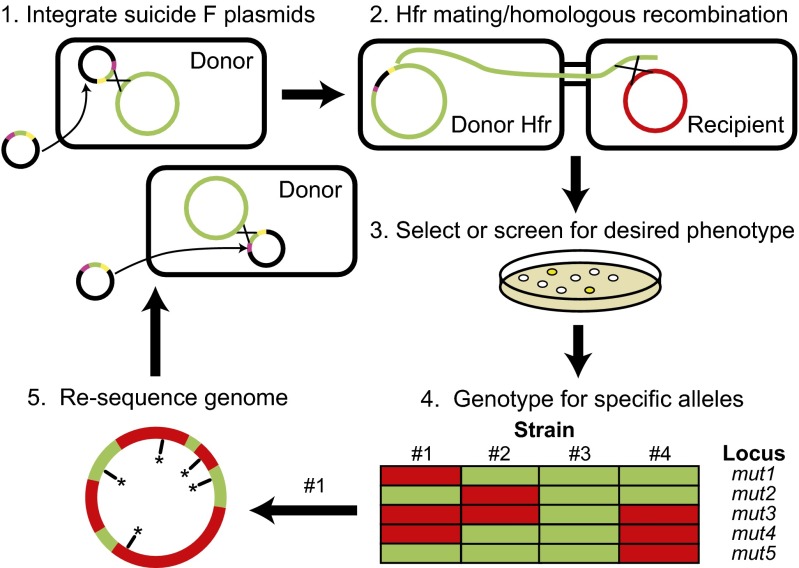
In the REGRES procedure (Fig. 1), one first constructs a collection of Hfr (high frequency of recombination) donor strains (19, 20) from one or more recA+ pir– parent strains by introducing suicide F plasmids for homology-targeted integration at different genomic locations (21). Each of these unique Hfr strains is capable of initiating genome transfer at a specific locus determined by the orientation of the oriT site in its chromosomally integrated F plasmid. The population of Hfr donor strains is then mated with an F– recipient. At this point, donor cells are eliminated to isolate transconjugants by selecting for an antibiotic resistance marker carried only by the recipient strain.
Fig. 1.
REGRES. (1) Suicide F plasmids are transferred to the donor strain by conjugation, and multiple F plasmid integrant Hfr strains are selected. (2) Isolated Hfr strains are mated with an F– recipient strain, permitting genome transfer and homologous recombination with the genomes of recipient cells. (3) A suitable selection or screening procedure for individual transconjugants displaying the phenotype of interest is applied. (4) Isolated strains are genotyped against a panel of known alleles. (5) Strains of interest, usually with the fewest donor alleles, are selected for whole-genome sequencing and can be used as donors for another round of REGRES, if desired.
The genomes of the resulting pool of transconjugants comprise segments from both donor and recipient parental strains, arising from (i) random crossover events during homologous recombination of the donor DNA with that of the recipient; (ii) the low efficiency of complete chromosome transfer during mating; and (iii) the possibility of transconjugants serving as recipients for multiple conjugative transfer events from different members of the population of donor Hfr strains. Finally, transconjugant clones with a phenotype of interest are isolated and screened for specific alleles to determine the degree of genetic mosaicism. Clones displaying a desired genotype may be subjected to whole-genome resequencing to fully characterize the alleles present and then used as donors or recipients in subsequent rounds of REGRES.
Cit++ REGRES Backcross.
Citrate could not be used by E. coli cells in the LTEE population before the evolution of the rnk–citG duplication that activated aerobic expression of the citT transporter (3). For this reason, the vast majority of mutations that accumulated earlier are thought to be unrelated to the Cit+ phenotype, and most probably improved growth on glucose (22). Therefore, we hypothesized that nearly all of the evolved alleles in an early Cit++ clone should be dispensable for citrate utilization, except possibly for specific mutations that potentiated the evolution of Cit+ or refined its functionality to Cit++. To remove irrelevant mutations while maintaining a Cit++ phenotype, we used REGRES with selection for growth on citrate-only media to backcross the Cit++ donor strain CZB154 with the ancestral recipient strain REL607. CZB154 is a 33,000-generation Cit++ clone that accumulated a total of 79 mutations relative to its ancestor during the evolution experiment and can fully use citrate. It contains three copies of the tandem head-to-tail amplification of 2.9-kb spanning the rnk–citG region found in all Cit+ strains, as well as a subsequent 14.9-kb dsbG–insA-9 duplication that encompasses the entire rnk–citG region (3). Together, these two mutations result in four copies of the activated rnk-citT promoter configuration.
To begin REGRES, suicide F plasmids targeting the galS and rhaM loci were introduced into CZB154 by conjugative transfer with subsequent antibiotic selection for chromosomal integration (21). The resulting library of CZB154 Hfr donor strains was then mated to REL607 cells containing a plasmid-encoded kanamycin resistance marker. REL607 transconjugants that had obtained alleles necessary for citrate utilization were selected on minimal agar plates containing citrate as the sole carbon source and kanamycin to eliminate the donor strains. After incubation for up to 4 d, several colonies were picked and genotyped by Sanger sequencing for the presence or absence of CZB154-specific alleles (Fig. S1). Strain R1 was found to contain a mixture of 50 CZB154-derived and 29 REL607-derived alleles by whole-genome sequencing, indicating successful genome transfer and recombination.
Three additional rounds of REGRES were performed in this manner, each time using REL607 as the recipient strain but alternating the use of either a kanamycin or tetracycline resistance marker to differentiate new recipients from the previous round’s recipients. After each round, resulting Cit+ clones were genotyped (Figs. S2–S4), and the genomes of selected clones were sequenced to monitor the loss of CZB154-specific alleles (Fig. 2 and Fig. S5). Interestingly, a great degree of genomic chimerism was observed after each REGRES round, likely due to multiple genome transfers and/or recombination events. We concluded the REGRES procedure with the isolation of strain R4, which was found to contain only 7 of the 79 alleles originally present in CZB154. It and all other sequenced REGRES strains still displayed high levels of citrate utilization similar to CZB154, as determined by growth curves that reached final cell densities that were significantly higher than that supported by glucose-only growth (Fig. S6).
Fig. 2.
Evolved mutations present in Cit+ REGRES strains. Whole-genome sequencing was used to determine the CZB154 alleles and other genetic changes present in seven REGRES clones chosen on the basis of genotyping results (Figs. S1–S4). Selected alleles are shown here, ordered by position in the ancestral chromosome and named for the genes they impact (e.g., yaaH), for the flanking genes if an allele is intergenic (e.g., dctA/yhjK) or for a range of genes for alleles that represent large chromosomal duplications or deletions (e.g., rbsD–yieO). Full results are shown in Fig. S5, and complete details for each allele are provided in Table S1.
dctA* and citT Mutations Are Sufficient for Cit++.
Analysis of all sequenced genomes revealed that only the nested dsbG–insA-9 and rnk–citG amplifications related to citT, and a mutation in the promoter region of the dctA C4-dicarboxylate transporter gene (dctA*) were common to all sequenced REGRES strains (Fig. 2). It has previously been established that the rnk–citG tandem duplication was the mutation that first enabled the rudimentary Cit+ phenotype (3), and duplications like the dsbG–insA-9 mutation likely serve only to further increase the copy number of the key rnk–citT promoter configuration. Based on its occurrence after the citT mutation in the lineage leading to CZB154, the evolved dctA allele was previously identified as a candidate refinement mutation, which could have played a role in strengthening an initially weak Cit+ phenotype (3). Given the evidence that a potentiated genetic background made evolution of the Cit++ phenotype more likely (3, 10), we were surprised that no mutation that occurred in the LTEE before the Cit+ trait evolved was conserved in the REGRES genomes.
To prove that the evolved dctA allele was able to support the strong Cit++ phenotype in the absence of any other mutations aside from citT, we introduced it into the genome of REL607 by allelic replacement to create strain REL607(dctA*). On its own, the dctA* mutation did not confer growth on citrate. However, when REL607(dctA*) cells were transformed with a low copy plasmid encoding the rnk-citT activated promoter arrangement (pCitT)—which was used in lieu of the evolved citT allele due to the inherent instability of chromosomal amplifications—we observed robust growth on citrate (Fig. 3A). Therefore, in the absence of any potentiating mutations or other refinement mutations, strains minimally containing the dctA* and citT mutations are able to fully use citrate in the media on glucose depletion (that is, they are qualitatively Cit++). To further investigate the contribution of the dctA* mutation to aerobic citrate utilization, we reverted the dctA* mutation in strain CZB154 to the ancestral state to create strain CZB154(dctAwt). Growth experiments with this strain showed that removal of the dctA* mutation eliminated the high level of citrate utilization on glucose depletion normally observed in strain CZB154 and instead resulted in a limited Cit+ phenotype (Fig. 3B).
Fig. 3.
Evolved dctA* mutation is sufficient for Cit++ in conjunction with CitT activation, increases dctA mRNA expression, and enables utilization of succinate. (A) Average growth curves of strains with either the ancestral (REL607) or evolved dctA allele [REL607(dctA*) and CZB154] grown in DM25 media as in the evolution experiment. Certain strains also carry a multicopy plasmid with the activated rnk–citT promoter configuration (pCitT). DM25 media contains 0.0025% (wt/vol) glucose and 0.032% (wt/vol) citrate. Error bars are the SD of at least three replicates. (B) Average growth curves in DM25 of strains containing either the ancestral or evolved dctA allele or with a reversion of the evolved allele to the ancestral state (dctAwt). Error bars are the SD of at least three replicates. (C) mRNA expression of dctA gene determined by qRT-PCR for strains containing either the ancestral or evolved dctA allele. Transcript levels are shown relative to strain REL607. Error bars are the SEM for biological triplicate samples. (D) Average growth curves of strains containing either the ancestral or evolved dctA allele in DM media with no citrate or glucose and containing 0.01% (wt/vol) succinate as the sole carbon source. Error bars are the SD of at least three replicates. Similar growth was observed for these strains on other C4-dicarboxylates such as fumarate and aspartate (Fig. S7).
dctA* Mutation Enables Transport of C4-Dicarboxylates.
The location of the dctA* mutation, 20 bases upstream of the dctA start codon, suggested that it altered regulation of this gene. To investigate this possibility, quantitative RT-PCR (qRT-PCR) was used to compare dctA transcript levels in strains with the ancestral and evolved dctA alleles (Fig. 3C). We find that the dctA* mutation increases dctA transcript levels when introduced into strain REL607 and that dctA transcript levels are also elevated in CZB154. Furthermore, reversion of the evolved dctA* mutation in strain CZB154(dctAwt) reduced dctA transcription to the level seen in the REL607 ancestral strain. Therefore, we conclude that the dctA* mutation increases expression of the messenger RNA encoding the DctA protein.
DctA is a C4-dicarboxylate transporter (23), so we next investigated whether this process had been altered in strains with the dctA* mutation by testing growth with succinate as the sole carbon source (Fig. 3D). REL607 displays severe growth defects on succinate, as noted previously in a comparison of E. coli B and K12 strains (24). CZB154 does, however, show growth on succinate, and addition of the dctA* mutation to REL607 enables it to grow on succinate. As before, we find that strain CZB154(dctAwt) loses all ability to grow on succinate due to the reverted dctA allele, ruling out the existence of any additional C4-dicarboxylate transport mechanisms in this evolved strain. Identical results were obtained in media supplemented with fumarate or aspartate, other C4-dicarboxylates known to be transported by DctA (Fig. S7).
Discussion
By using REGRES in genetic backcrosses with selection for growth on citrate as a sole carbon source, we discovered that mutations affecting the citT and dctA genes were sufficient for the refined version of the Cit+ innovation (the Cit++ phenotype) that evolved in a long-term evolution experiment with E. coli. A reconstructed strain with just these two evolved alleles is able to use citrate on glucose depletion, which greatly increases its population density in this environment, the hallmark of Cit++ clones isolated from the LTEE population after 33,000 generations. The rnk–citG mutation affecting the citT transporter had previously been identified as the mutation responsible for the emergence of the initially very weak Cit+ phenotype (3). The dctA* mutation occurred chronologically after citT during the LTEE, implying that it was important for improving this rudimentary trait to enable full citrate utilization (Fig. 4A).
Fig. 4.
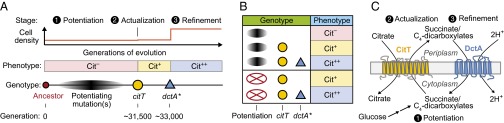
Model for the evolution of citrate utilization in the E. coli long-term evolution experiment. (A) There were three major epochs in the evolution of this metabolic innovation: potentiation, actualization, and refinement. Weakly Cit+ cells were first isolated from the population after the rnk–citG actualizing mutation that amplified and activated the CitT transporter (shown as the citT allele). There was not an appreciable increase in the final cell density at the end of each growth cycle in the evolution experiment at this point (Top). After the dctA* refinement mutation, there was a substantial increase in population size because cells were able to fully use citrate, which we distinguish from the rudimentary Cit+ phenotype as the strong Cit++ phenotype (Middle). The evolution of Cit++ is statistically more likely from certain Cit– genetic backgrounds that arose later in this population, presumably because they accumulated one or more potentiating mutations relative to the ancestor (3, 10). Key mutations are shown with their approximate timings relative to these evolutionary epochs and one another (Bottom). (B) Cit++ phenotype is not the product of all-or-none epistasis with potentiating mutations. The progression of citrate utilization phenotypes as they evolved in the LTEE in strains that contained key mutations in the context of earlier evolved alleles (Upper) and qualitative phenotypes of reconstructed strains containing only key evolved alleles in the ancestral genetic background (Bottom) are shown. The evolved citT mutation alone is sufficient for detectable but extremely limited citrate utilization, as observed in early Cit+ isolates (3). The citT and dctA* mutations together are sufficient for full citrate utilization characteristic of the Cit++ phenotype, even in the absence of potentiating mutations. (C) Mechanism of Cit++ refinement. When both the CitT and DctA transporters are expressed, due to the citT and dctA* mutations, their activities can be coupled so that the proton-motive force (H+) powers reuptake of succinate or other C4-dicarboxylate substrates for continued citrate import, yielding the Cit++ phenotype. It is possible that the unknown potentiating mutations make sufficient succinate available from glucose metabolism to power limited citrate import through CitT but that this does not result in a sustainable cycle.
There is evidence that one or more mutations in the history of this E. coli population contributed to a potentiated genetic background in which the Cit– to Cit++ evolutionary transition was more likely to occur (10). This observation was thought to result from all-or-none epistatic interactions involving potentiation mutations. According to this hypothesis, the evolution of Cit++ in replay evolution experiments, in which multiple new mutations accumulated, was made statistically more likely because one or more of the necessary mutational steps were already present in potentiated genotypes. We were surprised to find that the citT actualizing mutation and the dctA* refinement mutation are sufficient for a strong Cit++ phenotype, demonstrating that potentiating alleles are not strictly required for expression of this trait (Fig. 4B).
Further characterization of the evolved dctA* mutation revealed that it increases the mRNA expression level of the dctA C4-dicarboxylate transporter gene and enables the utilization of C4-dicarboxylates. Unlike most strains of E. coli K-12, the ancestral strain of E.coli B used in the LTEE cannot use C4-dicarboxylates (24). This difference has been attributed an inactivating mutation in the gene for DcuS (24), the sensor kinase in the DcuSR two-component system that regulates genes involved in C4-dicarboxylate transport and utilization (25, 26). The dctA* promoter mutation apparently enables this gene to be expressed in the absence of activation by DcuSR, permitting transport of C4-dicarboxylates. Interestingly, other Cit++ strains isolated from the LTEE, such as strain CZB152, have been found to contain mutations in the dcuS gene (3). It is possible that these isolates achieve some level of dctA expression by dcuS reactivation and that these variants may represent alternative mutational pathways to refinement of the Cit+ trait that transiently coexisted with the ultimately successful dctA allele.
DctA allows import of succinate to be powered by the proton-motive force. Therefore, increased expression of DctA is expected to be beneficial for aerobic citrate utilization because it makes it possible to reuptake succinate or other C4-dicarboxylates that are exported in exchange for citrate import by the CitT antiporter (27). This activity completes a cycle that permits sustained citrate transport and utilization without an unbalanced loss of intracellular C4-dicarboxylate substrates due to efflux (Fig. 4C). Strains with the dctA* mutation are able to extend their citrate-related growth phase and reach significantly higher cell densities on mixtures of glucose and citrate, like that used in the LTEE, indicating that this refinement mutation was the major driver of the population expansion that was observed after 33,000 generations (10). The dctA* and citT mutations do not completely recapitulate the improved growth rate, reduced lag phase, and higher final cell density exhibited by the evolved CZB154 clone, indicating that other evolved alleles contribute to improving the competitive fitness of these strains, even though they are not qualitatively necessary for the strong Cit++ trait.
The question remains: how did the potentiating mutations make the highly advantageous Cit++ phenotype more accessible to evolution in this particular LTEE population? In light of our results that argue against the all-or-none epistasis hypothesis, we propose that the potentiating mutations have a quantitative epistatic effect that makes the two-step mutational path required for evolution of the complex Cit++ phenotype from Cit– cells more likely to be realized in the context of an evolving population. Both the evolved citT and dctA alleles may have deleterious or nearly neutral effects on fitness when they are added alone to the ancestral strain or most genetic backgrounds that existed in the LTEE. In this case, weakly Cit+ variants with the citT mutation would rapidly be lost from the population after they arose, due to competition with other alleles that are beneficial for glucose utilization (22). In fact, however, genotypes with the citT mutation persisted in the population for >1,500 generations (3), and a small, but significant, fitness benefit was found when the citT allele was added to a Cit– isolate believed to have the potentiating mutations (10).
Therefore, on the basis of our results with dctA*, we hypothesize that potentiation may have altered central metabolism in some way that was directly beneficial for growth on glucose but fortuitously increased the supply of C4-dicarboxylates available to power citrate import by the CitT transporter, such that the activating citT mutation became slightly beneficial, rather than neutral or deleterious. This change could have enabled weakly Cit+ lineages to persist in the population long enough to pick up dctA* or other refinement mutations that yielded the highly beneficial, self-sufficient Cit++ phenotype. Assessment of the fitness effect of the citT mutation at various points along the lineage that achieved Cit++ in the future may provide insight into the identity of the potentiating mutations. Finally, it is also possible that the initially very subtle benefit of the Cit+ phenotype depended to some extent on the ecological interactions involving differential uptake and secretion of nutrients by cells in this genetically diverse population (3).
In addition to backcrossing to determine the genetic basis of complex traits, the REGRES procedure may also be useful for a variety of other bacterial strain improvement applications, including purging deleterious mutations introduced during mutagenesis and combining desirable traits between multiple, separately evolved strains. Alternative species-specific or broad host-range conjugative systems, such as those from IncP plasmids (28), could also be used to generalize the REGRES method so that an equivalent procedure could be used in other bacterial species. Overall, our discovery of the mechanism of a key refinement step in the evolution of a metabolic innovation highlights the degree to which interactions between alleles shape the evolution of complex traits and emphasizes the need for novel whole-genome methods to explore such relationships.
Materials and Methods
Strains and Culture Conditions.
E. coli strain REL607 is an Ara+ derivative of strain REL606 (29). E. coli strain CZB154 is a 33,000 generation clone from the Ara-3 LTEE population that was initiated from the Ara– strain REL606 (3). Strains CAG60452 and CAG60453 are BW38029 rrnB3 φ (lacZp4105(UV5)-lacY)638 ΔlacZ hsdR514Δ(araBAD)567 rph+, ΔdapA::pir, containing pMNT3 (21) suicide F plasmids CIP8 (pMNT3:galS) and CIP16 (pMNT3:rhaM), respectively. When appropriate, antibiotics were added as follows: kanamycin, 30 μg/mL; carbenicillin, 100 μg/mL; tetracycline, 10 μg/mL; spectinomycin, 30 μg/mL; chloramphenicol, 34 μg/mL; gentamicin, 15 μg/mL.
Hfr Strain Generation.
To create Cit+ Hfr strains, Cit+ strains were grown overnight in Lysogeny Broth (LB) media plus appropriate antibiotic to select for strains harboring plasmids pBR322 (carbenicillin), pET30 (Novagen) (kanamycin), pROlar.A122 (Clontech) (kanamycin), or pEQ367 (tetracycline). Additionally, CAG60452 and CAG60453 were grown in LB broth supplemented with spectinomycin and 0.3 mM 2,6-diaminopimelic acid (DAP). Saturated overnight cultures were diluted 1:100 into LB broth plus 0.3 mM DAP and grown to an OD600 ∼0.6. Equal volumes of the Cit+ strain and either CAG60452 or CAG60453 were mixed and incubated without shaking at 37 °C for 4 h. The conjugation mixture was spun down, and the cell pellet was washed in an equal volume of LB broth and then plated onto LB agar plates containing spectinomycin plus the appropriate antibiotic to select for the Cit+ strain. Single colonies were picked and regrown in selective media to confirm the identity of the integrant.
Hfr Matings.
For round 1 matings, Cit+ Hfr donor strains CZB154:CIP8 and CZB154:CIP16, each containing plasmid pBR322, were grown overnight to saturation in LB broth supplemented carbenicillin and spectinomycin. The Cit− recipient strain REL607, containing plasmid pET30 was grown overnight in LB broth plus kanamycin. These cultures were diluted 1:100 into LB broth without antibiotics and grown to an OD600∼0.6. One hundred microliters of each of the donor and recipient strains was added to 40 mL of fresh LB broth. The conjugation mixture was subjected to orbital shaking at 140 rpm over a diameter of one inch overnight at 37 °C. One milliliter of the overnight culture was spun down and washed twice in Davis Minimal (DM) salts (30) and plated onto minimal citrate (MC; DM salts + 4.5 g/L sodium citrate) agar plates supplemented with kanamycin. Plates were incubated aerobically at 37 °C for 4 d. Additionally, a 1:100 dilution of the conjugation mixture was transferred into fresh LB media, and the procedure was repeated for 4 consecutive d. After 4 d, candidate Cit+ clones were picked and restreaked to single colonies on MC agar plates plus kanamycin for confirmation of the phenotype.
Round 2 was performed as in round 1 except that E. coli strain R1 was used as the Cit+ donor and E. coli strain REL607 containing plasmid pEQ367 was used as the Cit− recipient. Round 3 was performed as in round 1 except that E. coli strains R2.1, R2.2, R2.3, and R2.4 were equally mixed and used as Cit+ donors and REL607 containing plasmid pROlar.A122 was used as the Cit− recipient. Round 4 was performed as in round 1 except that E. coli strain R3 was used as the Cit+ donor and REL607 containing plasmid pEQ367 was used as the Cit− recipient.
Mutation Analysis.
Cultures of selected Cit+ REGRES clones were used as templates for PCR and Sanger sequencing. The results of these assays are shown in Figs. S1–S4. All primers used in this study are listed in Table S2.
Whole-Genome Sequencing and Mutation Detection.
Genomic DNA was purified using the GenElute Bacterial Genomic DNA kit (Sigma) and sequenced on Illumina HiSeq and MiSeq instruments by the University of Texas at Austin Genome Sequencing and Analysis Facility (UT GSAF). Read data have been deposited in the National Center for Biotechnology Information Sequence Read Archive (SRP018688). Reads were mapped to the reference genome of REL606 (31), the Ara– progenitor of strain REL607 that was used in the REGRES experiments, and mutations were predicted using the breseq computational pipeline (version 0.22) available from http://code.google.com/p/breseq.
Strain and Plasmid Construction.
Isogenic strains containing dctA alleles were constructed using the pKO3 allelic replacement method (32). Plasmid pCitT was constructed by PCR amplification and cloning of the rnk–citT allele from strain CZB154 into the pSB3K3 vector (33). Details of strain and plasmid construction are described in SI Materials and Methods.
Growth Trajectories.
Strains of interest were revived from frozen stocks and grown in LB broth overnight followed by acclimation to DM25 media, plus antibiotic when appropriate, over two successive 24-h culture cycles. Culture densities were normalized, diluted 1:100 into the assay media, and aliquoted into a 96-well culture plate. The plate was incubated at 37 °C in a continuously shaking plate reader. Optical density was monitored at 420 nm. Full details are provided in SI Materials and Methods.
RNA Extraction and qRT-PCR.
Standard strain growth, RNA extraction, cDNA synthesis, and qRT-PCR data collection and analysis procedures were used (SI Materials and Methods.).
Supplementary Material
Acknowledgments
We thank A. Typas and C. Gross for strains CAG60452 and CAG60453, R. Lenski for strains CZB154 and REL607, Z. Blount for helpful advice, C. Barnhart, and N. Hajela for technical assistance. This project was supported by Defense Advanced Research Projects Agency Grant HR-0011-10-1-0052 (to G.G.), National Institutes of Health Grant R00-GM087550 (to J.E.B.), Welch Foundation Grant F-1654 (to A.D.E.), and a Cancer Prevention Research Institute of Texas research traineeship (to D.E.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the National Center for Biotechnology Information Sequence Read Archive, www.ncbi.nlm.nih.gov/sra (accession no. SRP018688).
See Commentary on page 2056.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314561111/-/DCSupplemental.
References
- 1.Heard S, Hauser D. Key evolutionary innovations and their ecological mechanisms. Hist Biol. 1995;10(2):151–173. [Google Scholar]
- 2.Phillips PC. Epistasis—The essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet. 2008;9(11):855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blount ZD, Barrick JE, Davidson CJ, Lenski RE. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature. 2012;489(7417):513–518. doi: 10.1038/nature11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagner A. The Origins of Evolutionary Innovations. New York: Oxford Univ Press; 2011. [Google Scholar]
- 5.Meyer JR, et al. Repeatability and contingency in the evolution of a key innovation in phage lambda. Science. 2012;335(80):428–432. doi: 10.1126/science.1214449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gould S, Vrba E. Exaptation—A missing term in the science of form. Paleobiology. 1982;8(1):4–15. [Google Scholar]
- 7.True JR, Carroll SB. Gene co-option in physiological and morphological evolution. Annu Rev Cell Dev Biol. 2002;18:53–80. doi: 10.1146/annurev.cellbio.18.020402.140619. [DOI] [PubMed] [Google Scholar]
- 8.Barve A, Wagner A. A latent capacity for evolutionary innovation through exaptation in metabolic systems. Nature. 2013;500(7461):203–206. doi: 10.1038/nature12301. [DOI] [PubMed] [Google Scholar]
- 9.Lenski RE. Phenotypic and genomic evolution during a 20,000-generation experiment with the bacterium Escherichia coli. Plant Breed Rev. 2004;24:225–265. [Google Scholar]
- 10.Blount ZD, Borland CZ, Lenski RE. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc Natl Acad Sci USA. 2008;105(23):7899–7906. doi: 10.1073/pnas.0803151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koser SA. Correlation of citrate utilization by members of the Colon-Aerogenes Group with other differential characteristics and with habitat. J Bacteriol. 1924;9(1):59–77. doi: 10.1128/jb.9.1.59-77.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lütgens M, Gottschalk G. Why a co-substrate is required for anaerobic growth of Escherichia coli on citrate. J Gen Microbiol. 1980;119(1):63–70. doi: 10.1099/00221287-119-1-63. [DOI] [PubMed] [Google Scholar]
- 13.Scheutz F, Strockbine NA. In: Bergey’s Manual of Systematic Bacteriology, Volume Two. Garrity GM, Brenner DJ, Kreig NR, Staley JR, editors. New York: Springer; 2005. pp. 607–624. [Google Scholar]
- 14.Zhang Y-X, et al. Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature. 2002;415(6872):644–646. doi: 10.1038/415644a. [DOI] [PubMed] [Google Scholar]
- 15.Wang HH, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460(7257):894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodarzi H, Hottes AK, Tavazoie S. Global discovery of adaptive mutations. Nat Methods. 2009;6(8):581–583. doi: 10.1038/nmeth.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider D, Duperchy E, Coursange E, Lenski RE, Blot M. Long-term experimental evolution in Escherichia coli. IX. Characterization of insertion sequence-mediated mutations and rearrangements. Genetics. 2000;156(2):477–488. doi: 10.1093/genetics/156.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneider D, Lenski RE. Dynamics of insertion sequence elements during experimental evolution of bacteria. Res Microbiol. 2004;155(5):319–327. doi: 10.1016/j.resmic.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 19.Curtiss R., 3rd Bacterial conjugation. Annu Rev Microbiol. 1969;23:69–136. doi: 10.1146/annurev.mi.23.100169.000441. [DOI] [PubMed] [Google Scholar]
- 20.Smith GR. Conjugational recombination in E. coli: Myths and mechanisms. Cell. 1991;64(1):19–27. doi: 10.1016/0092-8674(91)90205-d. [DOI] [PubMed] [Google Scholar]
- 21.Typas A, et al. High-throughput, quantitative analyses of genetic interactions in E. coli. Nat Methods. 2008;5(9):781–787. doi: 10.1038/nmeth.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrick JE, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461(7268):1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- 23.Davies SJ, et al. Inactivation and regulation of the aerobic C(4)-dicarboxylate transport (dctA) gene of Escherichia coli. J Bacteriol. 1999;181(18):5624–5635. doi: 10.1128/jb.181.18.5624-5635.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoon SH, et al. Comparative multi-omics systems analysis of Escherichia coli strains B and K-12. Genome Biol. 2012;13(5):R37. doi: 10.1186/gb-2012-13-5-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zientz E, Bongaerts J, Unden G. Fumarate regulation of gene expression in Escherichia coli by the DcuSR (dcuSR genes) two-component regulatory system. J Bacteriol. 1998;180(20):5421–5425. doi: 10.1128/jb.180.20.5421-5425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Golby P, Davies S, Kelly DJ, Guest JR, Andrews SC. Identification and characterization of a two-component sensor-kinase and response-regulator system (DcuS-DcuR) controlling gene expression in response to C4-dicarboxylates in Escherichia coli. J Bacteriol. 1999;181(4):1238–1248. doi: 10.1128/jb.181.4.1238-1248.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pos KM, Dimroth P, Bott M. The Escherichia coli citrate carrier CitT: A member of a novel eubacterial transporter family related to the 2-oxoglutarate/malate translocator from spinach chloroplasts. J Bacteriol. 1998;180(16):4160–4165. doi: 10.1128/jb.180.16.4160-4165.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas CM, Smith CA. Incompatibility group P plasmids: Genetics, evolution, and use in genetic manipulation. Annu Rev Microbiol. 1987;41:77–101. doi: 10.1146/annurev.mi.41.100187.000453. [DOI] [PubMed] [Google Scholar]
- 29.Lenski RE, Rose MR, Simpson SC, Tadler SC. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat. 1991;138(6):1315–1341. [Google Scholar]
- 30.Carlton BC, Brown BJ. In: Manual of Methods for General Bacteriology. Gerhardt P, editor. Washington, DC: American Society for Microbiology; 1981. pp. 222–242. [Google Scholar]
- 31.Jeong H, et al. Genome sequences of Escherichia coli B strains REL606 and BL21(DE3) J Mol Biol. 2009;394(4):644–652. doi: 10.1016/j.jmb.2009.09.052. [DOI] [PubMed] [Google Scholar]
- 32.Link AJ, Phillips D, Church GM. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: Application to open reading frame characterization. J Bacteriol. 1997;179(20):6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shetty RP, Endy D, Knight TF., Jr Engineering BioBrick vectors from BioBrick parts. J Biol Eng. 2008;2:5. doi: 10.1186/1754-1611-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.