Significance
Disruption of the timing of the sleep–wake cycle and circadian rhythms, such as occurs during jet lag and shift work, leads to disordered physiological rhythms, but to what extent the molecular elements of circadian rhythm generation are affected is not known. Here, we show that delaying sleep by 4 h for 3 consecutive days leads to a sixfold reduction of circadian transcripts in the human blood transcriptome to just 1%, whereas, at the same time, the centrally driven circadian rhythm of melatonin is not affected. Genes and processes affected included those at the core of circadian rhythm generation and gene expression. The data have implications for understanding the negative health outcomes of disruption of the sleep–wake cycle.
Keywords: bloodomics, chronobiology, microarray, genomics, biological rhythms
Abstract
Circadian organization of the mammalian transcriptome is achieved by rhythmic recruitment of key modifiers of chromatin structure and transcriptional and translational processes. These rhythmic processes, together with posttranslational modification, constitute circadian oscillators in the brain and peripheral tissues, which drive rhythms in physiology and behavior, including the sleep–wake cycle. In humans, sleep is normally timed to occur during the biological night, when body temperature is low and melatonin is synthesized. Desynchrony of sleep–wake timing and other circadian rhythms, such as occurs in shift work and jet lag, is associated with disruption of rhythmicity in physiology and endocrinology. However, to what extent mistimed sleep affects the molecular regulators of circadian rhythmicity remains to be established. Here, we show that mistimed sleep leads to a reduction of rhythmic transcripts in the human blood transcriptome from 6.4% at baseline to 1.0% during forced desynchrony of sleep and centrally driven circadian rhythms. Transcripts affected are key regulators of gene expression, including those associated with chromatin modification (methylases and acetylases), transcription (RNA polymerase II), translation (ribosomal proteins, initiation, and elongation factors), temperature-regulated transcription (cold inducible RNA-binding proteins), and core clock genes including CLOCK and ARNTL (BMAL1). We also estimated the separate contribution of sleep and circadian rhythmicity and found that the sleep–wake cycle coordinates the timing of transcription and translation in particular. The data show that mistimed sleep affects molecular processes at the core of circadian rhythm generation and imply that appropriate timing of sleep contributes significantly to the overall temporal organization of the human transcriptome.
Twenty-four-hour rhythms in physiology and behavior are generated through interaction between environmental cycles and endogenous self-sustained circadian oscillators (1). In mammals, circadian oscillators are present in the brain and most peripheral tissues (2). Circadian rhythmicity within cells in these tissues is generated by a molecular oscillator, which is composed of core transcriptional–translational feedback loops that can also interact with metabolic oscillators (3).
The circadian regulation of the mammalian transcriptome includes circadian transcriptional and translational regulation by proteins such as the positive transcription factors CLOCK and ARNTL (BMAL1), and the repressors PERIOD (PER) and CRYPTOCHROME (CRY) (4), chromatin modification by factors such as E1A binding protein P300 (EP300) and the methyltransferase MLL3 (4–6), RNA polymerase activity (4, 7), and posttranscriptional events such as the regulation of ribosome biogenesis and translation (8, 9). Furthermore, physiological factors such as body temperature (10) and endocrine rhythms such as cortisol (11) can modify and reinforce these regulatory processes.
The mammalian circadian system is organized in a hierarchical manner, with a central pacemaker located in the suprachiasmatic nuclei (SCN) of the hypothalamus. Synchronization of central and peripheral oscillators is achieved through direct neural connections between the SCN and target tissues, endocrine rhythms that are driven by the SCN, such as cortisol and melatonin, and behaviors such as food intake and sleep and associated changes in physiology (12–14).
The sleep–wake cycle, and associated cycles of darkness and light and fasting and feeding, interacts with the circadian system and is a major driving factor on rhythms in physiology and behavior, such that these rhythms are highly disrupted when the sleep–wake cycle is desynchronized from the central circadian clock (15). Here, we address the question of whether the molecular processes that regulate circadian gene expression are also affected when the synchrony of the sleep–wake cycle and endogenous circadian rhythmicity is disrupted, such as occurs during jet lag and shift work (16) and laboratory protocols of forced desynchrony (17, 18), which we have used in this study.
Results
Effect of the Protocol on the Melatonin Rhythm and Sleep.
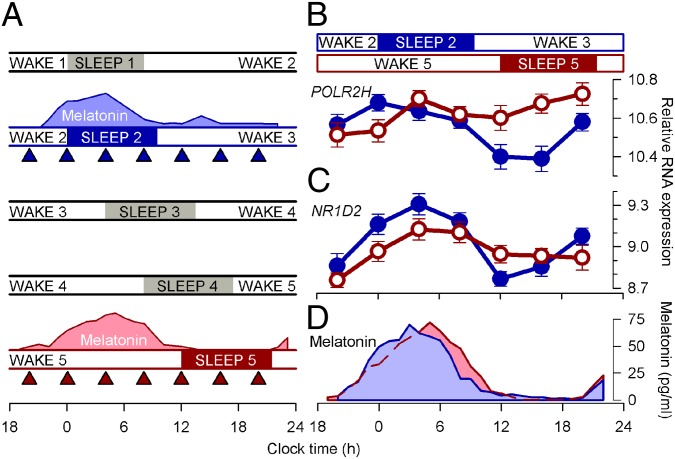
Twenty-two healthy volunteers (Table S1) participated in a forced-desynchrony protocol, in which the sleep–wake cycle and the associated fasting–feeding and dark–dim light cycles are scheduled to a 28-h day (Fig. 1A). Under these conditions of low light levels (<5 lux) during scheduled waking episodes, the rhythm of plasma melatonin, which is driven by the central circadian pacemaker in the SCN (19), oscillates at its intrinsic period (∼24.2 h) (20). Thus, the phase of the melatonin rhythm occurred at approximately the same clock time during the sleep 2/wake 3 and wake 5/sleep 5 periods, and there were no major changes in either the amplitude or the waveform of this rhythm, even though sleep was scheduled 12 h out of phase on these 2 d (Fig. 1 A and D). When sleep occurred during the biological night, in phase with plasma melatonin, polysomnographically assessed total sleep time was 450.6 ± 7.6 min (mean ± SEM). During the subsequent three sleep episodes, which were scheduled progressively later, total sleep times were 446.5 ± 11.4, 401.7 ± 78.6, and 388.3 ± 17.6 min, respectively, demonstrating the known disruptive effects of forced desynchrony on sleep (17) and the modest sleep loss. We assessed the impact of this desynchrony between the sleep–wake cycle and the melatonin rhythm on the blood transcriptome by analyzing two sets of seven RNA samples, collected both while sleeping in phase and out of phase with the circadian melatonin rhythm (Fig. 1A).
Fig. 1.
Forced-desynchrony protocol. (A) After a baseline day (wake 1, sleep 1), the sleep–wake cycle was extended to 28 h such that sleep period 2 was in phase with melatonin (blue area plot) but sleep period 5 commenced at noon, 12 h out of phase with sleep period 2 and the melatonin rhythm (pink area plot). Blood RNA was sampled during both in-phase (blue triangles) and out-of-phase (red triangles) conditions. Relative RNA expression levels (log2, mean ± SEM) of POLR2H (B) and NR1D2 (C) when sleeping in phase (blue circles) with melatonin [blue area plot; mean dim light melatonin onset (DLMO), 2159 hours ± 1 h and 13 min (±SD); n = 21)] (D) and when sleeping out of phase (red circles) with melatonin [pink area plot; DLMO, 2303 hours ± 1 h and 57 min (±SD); n = 21] (D).
Forced Desynchrony and the Transcriptome: Loss of Circadian Rhythmicity.
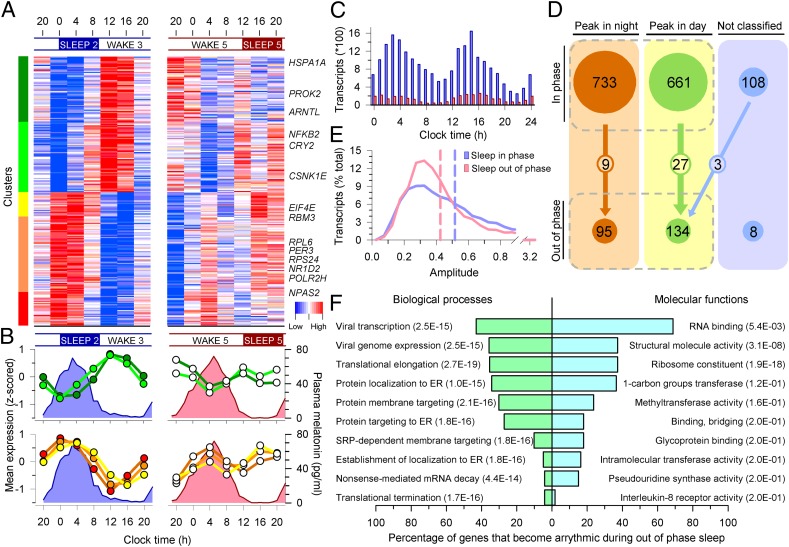
We first determined whether the time courses of transcripts contained a significant near 24-h rhythmic (circadian) component using algorithms and statistical techniques previously described (21). The expression profiles of many transcripts appeared clearly circadian when sleep occurred in phase with the melatonin rhythm (Sleep 2) but developed a nonrhythmic profile when sleep occurred out of phase with the melatonin rhythm (Sleep 5) (e.g., RNA polymerase II subunit H, POLR2H; Fig. 1B), whereas some transcripts remained rhythmic [e.g., NR1D2 (REV-ERB-β); Fig. 1C]. On a genome-wide scale, 1,502 transcripts (targeting 1,396 genes; 6.4% of all genes analyzed) were classified as having a circadian expression profile while participants were sleeping in phase with melatonin (Fig. 2A, Left). When these circadian transcripts were sorted according to the timing of their peak levels, an overall bimodal distribution was observed, with peaks during the day and night (Fig. 2 A and C). Cluster analysis further subdivided these circadian transcripts into two night- and three day-peaking clusters (Fig. 2 A and B, Left). Gene ontology (GO) analysis revealed that biological processes and molecular functions that were associated with day transcripts included response to wounding, defense response, response to stress, cytokine receptor activity, and hormone activity, whereas those associated with night transcripts included protein localization to the endoplasmic reticulum (ER), translation initiation, elongation and termination, structural constituent of the ribosome, RNA binding, and methyltransferase activity (Table S2). One small cluster of night-peaking transcripts included genes associated with lymphocyte differentiation and activation and T-cell differentiation and activation (Fig. 2 A and B, red cluster). Temporal profiles of many transcripts were markedly disrupted when sleep was timed out of phase with melatonin (Fig. 2A, Right). On a genome-wide scale, when sleeping out of phase with melatonin, only 237 transcripts (targeting 228 genes; 1.0%) were classified as circadian (Fig. 1C, Fig. S1, and Dataset S1), which represents a greater than sixfold reduction compared with the sleeping in phase condition. Of the transcripts that were classified as circadian while sleeping in phase, the majority (97%) became arrhythmic when sleeping out of phase with the melatonin rhythm (Fig. 2 A and B, Right). Known clock genes classified as circadian while sleeping in phase included ARNTL, NPAS2, PER2, PER3, CRY2, NR1D2, and the protein kinase CSKN1E. The only clock genes that remained rhythmic when sleeping out of phase were NR1D2 (Fig. 1C) and CSNK1E, whereas NR1D1 (REV-ERB-α) became rhythmic.
Fig. 2.
Mistimed sleep disrupts the circadian transcriptome. (A) Median expression profiles of circadian transcripts when sleeping in phase with melatonin (Left) and their profiles when sleeping out of phase (Right) (n = 19 paired participants). Colored bars on the left indicate clusters of day-peaking (B, Upper Left) and night-peaking (B, Lower Left) transcripts. Profiles of these transcripts were disrupted when sleep occurred out of phase with melatonin (A and B, Right). Average melatonin profiles are indicated by blue and pink area plots. (C) Peak expression phase distribution of prevalent circadian transcripts while sleeping in phase (blue bars; n = 16,253 derived from 1,502 circadian transcripts in an average of 10.84 subjects; minimum number of subjects for an FDR < 5%, 10) and out of phase (red bars; n = 2,503 derived from 237 circadian transcripts in an average of 10.56 subjects). (D) A total of 733 night and 661 day in-phase circadian transcripts reduced to 95 and 134 out-of-phase transcripts, with 9 and 27 common to both (n = 19 paired participants); 108 rhythmic transcripts were not classified as either day or night when sleeping in phase, and this category contained only eight transcripts during out of phase. (E) Out-of-phase prevalent circadian transcripts (red plot) had a lower mean amplitude (0.4264, red dashed line) than in-phase prevalent transcripts (blue plot; 0.5169, blue dashed line) (95% confidence interval, −0.1012 to −0.0797; P < 2.2 × 10−16). (F) Top 10 GO processes and functions associated with transcripts that became arrhythmic when sleeping out of phase.
Disruption of circadian rhythmicity during desynchrony was observed for both day- and night-peaking transcripts (Fig. 2 B–D) and also included a significant reduction in circadian amplitude of expression of the prevalent probes in each condition (Fig. 2E). The top 10 GO processes, that were significantly associated with transcripts that lost circadian rhythmicity when sleeping out of phase, included translation elongation and termination, targeting and localization of protein to the ER, as well as viral transcription and viral genome expression. The latter processes are likely identified by the GO analysis because these processes also use the host gene expression machinery, which was affected by desynchrony. The top 10 molecular functions included RNA binding and ribosome constituents, although it should be noted that not all of these categories remained significant after correction for multiple testing (Fig. 2F). The biological processes and molecular functions associated with transcripts that were circadian when sleeping in phase and out of phase separately are presented in Fig. S2. It should be noted that because the majority of the transcripts that were circadian while sleeping in phase became arrhythmic when sleeping out of phase, the GO analysis of transcripts that were circadian while sleeping in phase is similar to that for the transcripts that became arrhythmic. Because of the small number of transcripts that were classified as rhythmic when sleeping out of phase, the corrected P values for GO terms associated with these transcripts are not significant. However, the top-ten biological processes and molecular functions included protein glycosylation, melanocyte differentiation, B-cell receptor signaling pathway, and down-regulation of vascular permeability (Fig. S2).
Forced Desynchrony and the Transcriptome: Alterations in Expression Profiles.
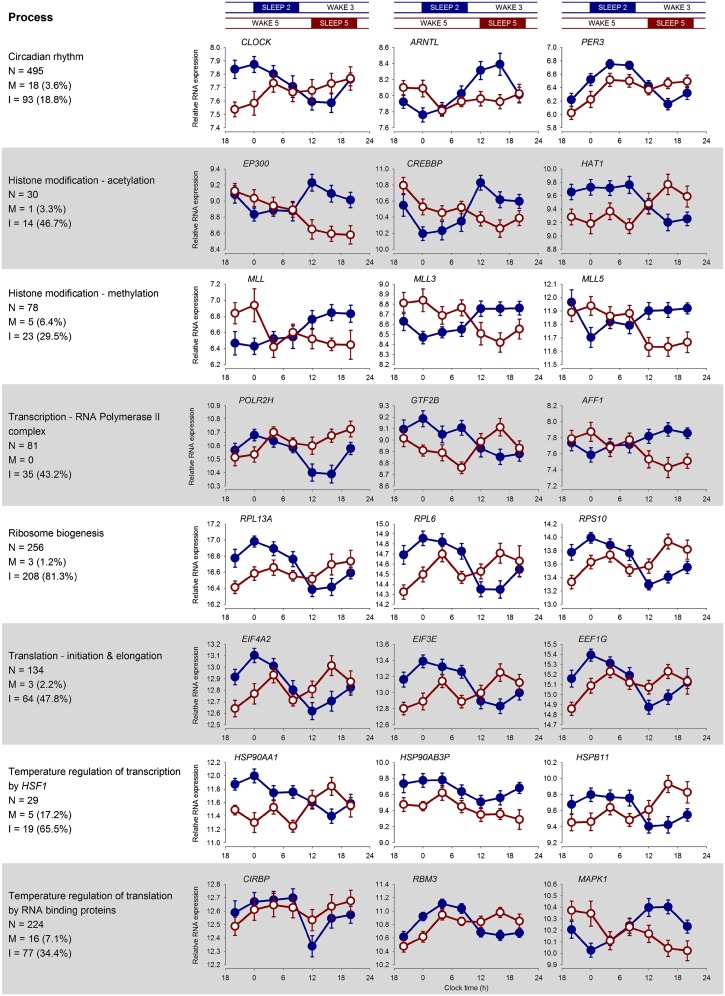
To further quantify the effects of desynchrony on the transcriptome beyond a dichotomy of rhythmic vs. nonrhythmic transcripts, we applied mixed-model ANOVA to the entire dataset. We found that the interaction between the factors “condition” (i.e., sleeping in phase vs. sleeping out of phase with melatonin) and “time point” (i.e., RNA sample numbers 1–7) was significant in 10,848 (34%) transcripts (Dataset S1). Transcripts whose temporal expression profiles according to this analysis were affected by forced desynchrony included circadian clock genes (CLOCK, ARNTL, and PER3), genes involved in chromatin modification [e.g., methyltransferases (MLL, MLL3, and MLL5), EP300, CREB-binding protein (CREBBP), histone acetyltransferase 1 (HAT1), nuclear receptor corepressor 1 (NCOR1), and SET domain-containing 2 (SETD2)], multiple transcripts for RNA polymerase II (POLR2) and its complex formation (GTF2B, AFF1), ribosome biogenesis (many transcripts for RPL and RPS proteins), and regulators of transcription and translation (TCEA, EIF, EEF, RPL, and RPS), as well as genes for heat shock proteins (HSP90AA1, HSP90AB3P, HSPB11) and temperature-sensitive RNA binding proteins (CIRBP and RBM3). Fig. 3 shows example expression profiles of transcripts from these different categories that clearly demonstrate the disruption of the time course during the forced-desynchrony protocol.
Fig. 3.
Summary of the effects of desynchrony on the regulation of gene expression. Biological processes that contribute to the regulation of gene expression are shown on the left. For each process, the mean (log2 ± SEM) expression plots while sleeping in (blue) and out (red) of phase of exemplar transcripts (identified as having a significant differential expression between conditions) associated with the process are depicted. Also provided are the number of transcripts (N) in our study that have been associated with that process, together with the number and percentage of those transcripts that showed either a main effect of sleep condition (M) or an interaction between sleep condition and sample time (I) in the ANOVA. It should be noted that the custom-designed microarrays used in this study included the addition of many probes covering the length of identified transcripts for circadian clock-related genes. That is why the circadian rhythm category in this figure includes 495 probes.
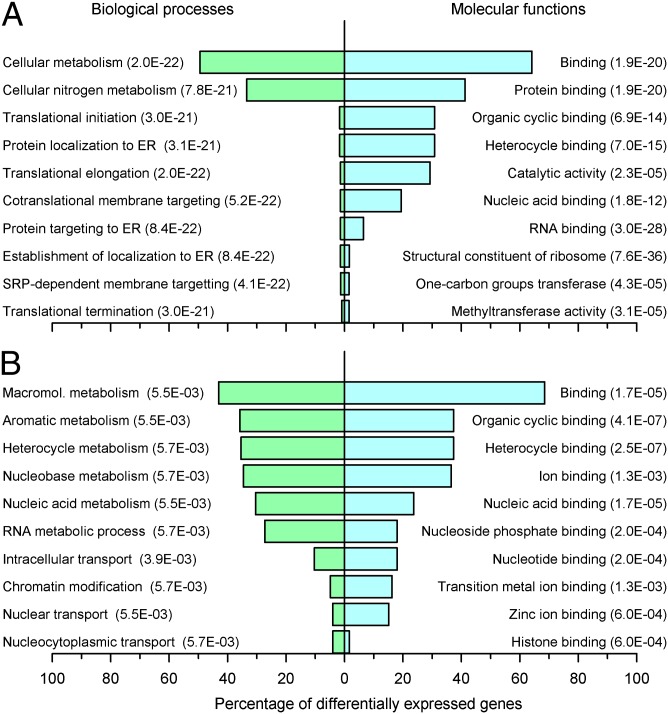
GO analysis demonstrated that transcripts whose expression showed an interaction between condition and time were associated with biological processes and molecular functions that included translation initiation, elongation and termination, protein targeting and localization to the ER, structural constituent of ribosome, RNA binding, and methyltransferase activity (Fig. 4A). In addition to large effects on ribosomal protein transcripts, we also found significant temporal disruption to many mitochondrial ribosomal proteins, which are associated with mitonuclear protein imbalance and longevity (22).
Fig. 4.
GO analysis for transcripts whose expression profiles changed between sleep conditions. Top 10 GO processes and functions associated with transcripts whose expression profile showed a significant interaction between sleep condition and sample time (A) or a main effect of sleep condition (B) (ANOVA; P < 0.05; n = 22).
ANOVA also identified a main effect (i.e., overall up or down-regulation of expression) of sleep condition on the overall expression of 1,119 transcripts, which were associated with biological processes and molecular functions that included chromatin modification, nucleic acid metabolic and cellular macromolecule metabolic processes, histone binding, and nucleic acid binding (Fig. 4B). Of these 1,119 transcripts, 913 transcripts were significantly down-regulated and 206 were up-regulated. Observed significant fold changes (log2) ranged from −0.453 (73% compared with sleeping in phase) to 0.410 (133% compared with sleeping in phase; Dataset S2). Transcripts that were down-regulated while sleeping out of phase were associated with biological processes and molecular functions that included macromolecular metabolism, gene expression, nucleic acid metabolism, RNA metabolic process, and DNA and RNA binding, whereas up-regulated transcripts were associated with hemoglobin metabolic processes, oxygen transporter activity, peroxiredoxin activity, myosin binding, and deoxyribonucleotide catabolism, among others (Fig. S3). Taken together, these data again suggest that mistimed sleep has a major suppressive effect on transcription and translation.
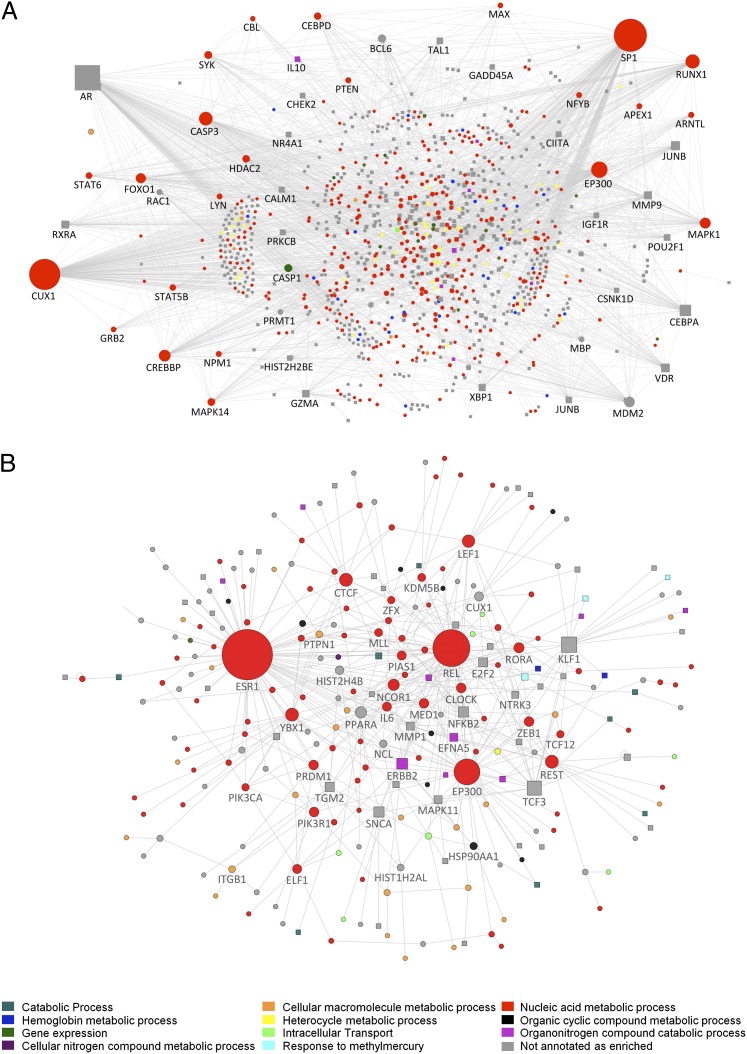
To further investigate the potential functional interactivity between transcripts that showed differential expression between sleep conditions and across time, we performed a direct interaction network analysis, which connects elements that are known to interact (at levels ranging from gene to protein). For the transcripts whose expression showed an interaction between condition and sample time, the direct interaction network revealed highly connected nodes at specificity protein 1 (SP1), EP300, CREBBP, ARNTL, and MAPK1 (Fig. 5A). The node with the most connections was SP1, which is a transcription factor that regulates the expression of a wide range of genes and also interacts with EP300 in gene expression regulation (23). EP300, together with CREBBP, regulates the circadian transcription of many genes via histone acetylation (4). Thus, SP1, together with EP300 and CREBBP, likely contributes to the regulation of the expression of thousands of genes and was itself negatively regulated by sleep (Fig. S4). Comparison with the Encyclopedia of DNA Elements cell line data shows that up to 32% of the ANOVA interaction genes contained biologically confirmed binding sites for SP1 (Table S3; see SI Methods). For those transcripts that showed a main effect of condition (i.e., were up or down-regulated), the interaction network was composed of many down-regulated nodes that are associated with nucleic acid metabolic processes (Fig. 5B). The network nodes with most direct interactions were ESR1, REL, and EP300, but other significant nodes included MLL, NCOR1, and CLOCK, genes that have also been implicated in the rhythmic control of transcription.
Fig. 5.
Direct interaction networks between genes for which at least one transcript had a statistically significant interaction between sleep condition and sample time (A) or a main effect of sleep condition (B). Up-regulated genes (on average) in the out-of-phase condition compared with in-phase are indicated as square nodes and down-regulated ones by circles. Node size reflects the number of direct connections a gene has within the network. Node color (see key) represents the most enriched association with a top 10 GO biological process. Downloadable, interactive versions of A and B are available at http://sleep-sysbio.fhms.surrey.ac.uk/PNAS_14/.
Separating the Contribution of Circadian Rhythmicity and Sleep to the Temporal Organization of the Transcriptome.
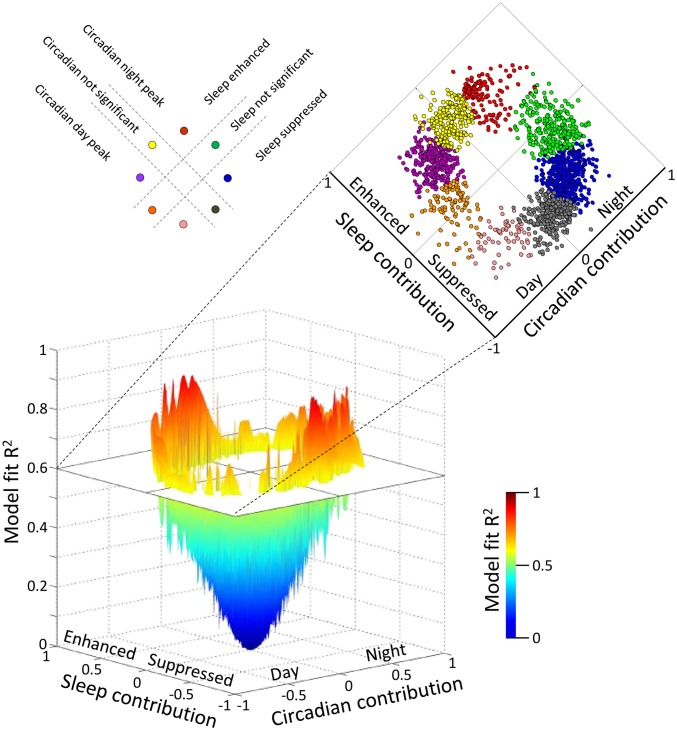
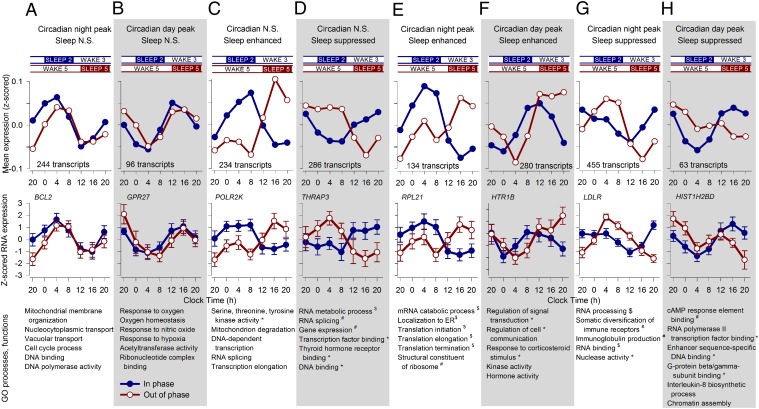
While emphasizing the overall disruptive effect of the forced-desynchrony protocol on the temporal expression of transcripts, ANOVA did not quantify the separate contribution of the sleep–wake cycle and circadian rhythmicity to the expression profile of transcripts when sleeping in or out of phase with melatonin. Some physiological variables, such as growth hormone (24) or slow wave sleep (17), are primarily driven by the sleep–wake cycle; others, such as melatonin, are almost exclusively driven by circadian rhythmicity (25), whereas variables such as core body temperature are driven by both circadian rhythmicity and the sleep–wake cycle (26, 27). We applied a simple mathematical model to estimate the contribution of circadian rhythmicity and the sleep–wake cycle to the expression profiles. In this model, the circadian influence (as indexed by the melatonin rhythm) induces peak expression levels in either the night or the day, and sleep either suppresses or enhances expression. The circadian and sleep effects combine in a linear manner (Fig. 6). This model identified two groups of transcripts that only responded to circadian rhythmicity: one group peaked during the biological night (Fig. 7A), whereas the other group had maximum expression levels during the biological day (Fig. 7B). Two other groups of transcripts only responded to sleep with an enhanced (Fig. 7C) or suppressed (Fig. 7D) drive and were not affected by circadian rhythmicity. Note that this exclusive contribution of the sleep–wake cycle to gene expression may either reflect a direct enhancing/suppressive effect of sleep on gene expression or entrainment of rhythmicity to the 28-h imposed sleep–wake cycle. Such entrainment of gene expression to temperature cycles longer than 24 h has previously been demonstrated (28). Other transcripts were enhanced by sleep and circadian rhythmicity during the biological night (Fig. 7E) or enhanced by sleep and circadian rhythmicity during the biological day (Fig. 7F), whereas others were suppressed by sleep and had either a circadian night (Fig. 7G) or day peak (Fig. 7H). The biological processes and functions that were associated with these groups were different, such that circadian-driven transcripts were associated with cellular metabolic and homeostatic processes, whereas those that were driven by sleep alone or by both circadian rhythmicity and the sleep–wake cycle were linked with the regulation of transcription and translation in particular. All GO terms for each of these categories are listed in Fig. 7. Together with the GO analyses for transcripts that lost rhythmicity (Fig. 2F) and those whose expression profile showed an interaction between sleep condition and time (Fig. 4A), the results from the modeling analysis further underline that sleep drives the expression of groups of transcripts that are specifically associated with the regulation of gene expression and protein synthesis.
Fig. 6.
Modeling the contribution of circadian and sleep–wake drive on transcript expression profiles. A model that describes the temporal profile of transcripts as a linear combination of the 28-h sleep–wake cycle and 24-h circadian rhythmicity was fitted to the median expression profile of each transcript (median of z-scored data across 19 paired participants per sample time point). In the 3D plot, the horizontal plane maps the estimated coefficient of the contribution of the sleep–wake cycle against the contribution of the circadian rhythmicity, whereas the corresponding model fit R2 value is indicated by the vertical axis (n = 41,119 transcripts). Transcripts with a model fit R2 > 0.6 (1,792 transcripts) were further classified into eight distinct categories (indicated with different colors in the vertical plane) based on the contribution of the sleep–wake cycle and the contribution of the circadian rhythmicity as follows: transcripts with no significant sleep contribution, which were primarily determined by circadian rhythmicity with a peak at night (green) or day (orange); transcripts with a significant circadian contribution with a peak at night and an enhancing effect of sleep (red) or a suppressive effect of sleep (blue); transcripts with a significant circadian contribution with a peak during the day and an enhancing effect of sleep (purple) or a suppressive effect of sleep (pink); and transcripts with no significant circadian component, which were enhanced by sleep (yellow) or suppressed (gray).
Fig. 7.
Estimating the separate contribution of circadian rhythmicity and sleep to the temporal organization of the transcriptome. A model describing the temporal profile of transcripts as a linear combination of the 28-h sleep–wake cycle and 24-h circadian rhythmicity was fitted to the median expression profiles (z-scored data across all participants per sample point) of all transcripts (Fig. 6). The model identified transcripts that were only driven by circadian rhythmicity with a positive drive during the night (A) or a positive drive during the day (B). Two groups of transcripts were only driven by sleep, which enhanced (C) or suppressed (D) the levels of these transcripts. Other transcripts were affected positively by sleep and also by circadian rhythmicity at night (E) or circadian rhythmicity during the day (F), whereas others were suppressed by sleep and had either a circadian night (G) or day peak (H). Also shown are the group time course, number of transcripts per category, example mean expression profile (log2 ± SEM) while sleeping in (blue line) and out of phase (red line), together with top 10 GO processes and functions associated with the set of transcripts (*P < 0.05; #P < 0.01; $P < 0.001).
The Effects of Mistimed Sleep Compared with Sleep Restriction.
In a previous study, we compared the transcriptome (measured in the absence of a sleep–wake cycle) after 1 wk of sufficient sleep (mean sleep duration, 8.5 h) with the transcriptome after 1 wk of sleep restriction (mean sleep duration, 5.7 h) (21). Thus, the sleep loss that accumulated during that protocol was 19.6 h and was associated with a reduction in circadian transcripts from 8.8% to 6.9%. From the mean sleep durations recorded for each sleep–wake cycle in the current protocol, the estimated cumulative sleep loss is only 1.9 h. Therefore, it seems reasonable to conclude that the much larger reduction in the number of circadian transcripts from 6.4% when sleeping in phase to just 1% when sleeping out of phase with melatonin that we observed in the current protocol is attributable largely to the mistiming of sleep and is not attributable to the modest amount of sleep loss incurred. Sleep restriction and mistimed sleep also caused loss of circadian rhythmicity in different sets of transcripts, with an overlap of only 122 genes that became arrhythmic in both protocols (Dataset S3). After 1 wk of sleep restriction, transcripts that became arrhythmic were associated with biological processes that included inositol triphosphate kinase activity, phospholipid transporter activity, transferase activity, nucleotide binding, and catalytic activity (21), whereas during mistimed sleep, transcripts that became arrhythmic were associated with biological processes and molecular functions linked with the regulation of transcription and translation (Fig. 2F) (for comparison, see Fig. S5). Mixed model ANOVA analyses showed that similar numbers of transcripts showed a main effect of sleep condition in the two studies; 711 and 1,119 transcripts were up- or down-regulated in response to sleep restriction and sleeping out of phase, respectively. Up- and down-regulated transcripts in the two studies were also associated with overlapping biological processes and molecular functions (up-regulated: catabolic processes and peroxiredoxin activity; down-regulated: macromolecular metabolism, gene expression, nucleic acid metabolism, and nucleic acid binding) (Fig. S6). However, ANOVA analysis showed that whereas only 252 transcripts (0.8%) showed an interaction between sleep condition and sample time in the previous study, 10,848 transcripts (34%) showed an interaction between sleep condition and sample time in the current study. This means that the temporal expression profiles of many more transcripts were affected by sleeping out of phase in the current study compared with the effects of sleep restriction in the previous study. However, we cannot rule out the possibility that the differential regulation of some processes that occurred in both protocols (e.g., macromolecular processes) is attributable to the sleep loss that occurs in both protocols. Taken together, we therefore conclude that in the current protocol, mistimed sleep disrupts the circadian organization of the transcriptome and that the main effects of mistimed sleep are primarily related to the change in the timing of sleep and associated changes in physiology, rather than the modest reduction in sleep time that is associated with mistimed sleep.
Discussion
We provide evidence that although the central circadian clock does remain rhythmic during the imposition of a noncircadian (i.e., 28-h) sleep–wake cycle in dim light (18, 20, 29), the majority of the blood peripheral transcriptome becomes arrhythmic, with a minority of transcripts that remain rhythmic or become rhythmic. Previously, it has been demonstrated in animal models that altering the timing of the sleep–wake cycle may also affect the transcriptome of other organs and tissues, including the brain (30), liver (31), and adipocytes (32). These human and animal transcriptome data can be interpreted within the framework of a hierarchical organized multioscillator system in which rhythmicity in local gene expression may be driven, to a varying extent, by local molecular clocks, neural or hormonal input driven by a central pacemaker, vigilance state itself, or cues and changes in endocrine and physiological variables associated with behaviors such as sleep or food intake. The varying extent of these influences on gene expression will be reflected in the extent to which their temporal profile is disrupted during forced desynchrony of, for example, the sleep–wake cycle and the centrally driven melatonin rhythm.
Circadian Transcripts That Are Robustly Rhythmic During Synchrony and Desynchrony.
The observation that 6.4% of the blood transcriptome displayed a circadian expression profile when sleep occurred in phase with the central circadian clock agrees well with the proportions of rhythmic transcripts previously reported for other tissues (33) and is comparable with our previous estimate of 8.8% in the human blood transcriptome (21). The distinct bimodal distribution of phases, with night-peaking transcripts associated with the regulation of gene expression regulation and day-peaking transcripts associated with processes linked with immunity and inflammation (Table S2), is also in accordance with our previous analyses of the circadian human transcriptome in the absence of a sleep–wake cycle (21). Although the time courses of both night- and day-peaking genes were greatly disrupted in the current study, more of the day-peaking transcripts remained robustly rhythmic. Indeed, of the 39 transcripts that were rhythmic in both conditions, 27 peaked during the day when sleeping in phase. In accordance with the identified GO terms for the day-peaking transcripts, the robustly rhythmic transcripts are linked with processes such as blood cell development and function (MAL, B4GALT5, OTX1, NELL2, ST6GALNAC2), vascular function (ADM, ANXA3), immunity (HCG27, NFAM1, TREM1), and lipid metabolism [low-density lipoprotein receptor (LDLR)]. Thus, robustly rhythmic transcripts whose expression profiles were not affected by forced desynchrony were largely those related to intrinsic blood-specific functions.
Transcripts and Associated Molecular Processes Affected by Desynchrony.
Current understanding of the molecular mechanisms underlying the circadian regulation of the mammalian transcriptome emphasizes the role of clock genes (1). Although the central circadian clock, as indexed by melatonin, remained largely unaffected, the temporal organization of expression of clock genes considered to be central to circadian rhythm generation was affected in the blood transcriptome, with only NR1D2 and CSNK1E remaining rhythmic when sleeping out of phase. Histone modification, and the control of transcription and translation (4–9), is also considered central to circadian organization of the transcriptome and the time courses of transcripts associated with all of these processes were affected in this study (Fig. 3). We also observed disruption to the time course of expression of the transcription factor SP1 (Fig. S4), which was the most connected gene node in the direct interaction network of transcripts that showed an interaction between sleep condition and time (Fig. 5A). In addition to interacting with EP300 and CREBBP in the circadian regulation of gene expression (4), SP1-binding motifs were overrepresented in the promoter regions of circadian gene sets from multiple mammalian tissues (34, 35). It has been shown in mouse liver that BMAL1 binds in a phase-specific way to several thousand DNA sites that predominantly contain two tandem E-box motifs and an adjacent SP1 site (36). The authors propose that SP1 acts with BMAL1 to coregulate circadian gene expression. We have previously shown that the promoter region of human PER3 contains two SP1 sites adjacent to two tandem noncanonical E-box motifs and that a variable number tandem-repeat polymorphism in the PER3 promoter removes one of these SP1 sites and is associated with differences in the levels of reporter gene expression (37). Thus, these data are consistent with a role for SP1 in the regulation of circadian gene expression.
Transcripts and Associated Processes Driven by the Sleep–Wake Cycle.
In our model of the separate contribution of circadian rhythmicity and the sleep–wake cycle, many of the transcripts whose expression profiles were enhanced or suppressed by sleep were associated with the regulation of transcription and translation (Fig. 7 C–H). For those transcripts that were exclusively driven by the sleep–wake cycle, a high-amplitude rhythm was observed during both synchrony and desynchrony, but the timing of the peak was very different (Fig. 7 C and D). Within these categories, 234 transcripts were categorized as being enhanced by sleep with no circadian input and included POLR2K (Fig. 7C). A further 286 transcripts were only suppressed by sleep and included thyroid hormone receptor-associated protein 3 (THRAP3, also known as TRAP150) (Fig. 7D). THRAP3 has recently been identified as a coactivator of the CLOCK-BMAL1 complex and promotes its binding to target genes, linking it with the transcriptional machinery (38). This category also included the methyltransferase transcript MLL3, and the expression profiles of several transcripts associated with methylation are affected by mistimed sleep (Fig. 3), including METTL3, which methylates mRNA and regulates the processing of transcripts, including clock genes, thereby determining circadian period (39). The effects of desynchrony on the amplitude of expression for those transcripts that receive both a positive or negative drive from sleep, as well as a melatonin phase-linked circadian drive, obviously depends on the combination of the timing of the circadian peak and whether they are enhanced or suppressed by sleep. In the category circadian night peak and sleep enhanced, we observed a high-amplitude rhythm when sleeping in phase and a bimodal time course when sleeping out of phase (Fig. 7E). In this category, we saw a large number of transcripts for ribosomal subunits affected by sleeping out of phase (e.g., RPL21; Fig. 7E). In the category sleep-enhanced and circadian day peak, we found processes associated with hormone activity and response to corticosteroid stimulus. Note that in this category and the category circadian night peak sleep suppressed, the amplitude of expression increases during desynchrony, a phenomenon reminiscent of the effects of sleep displacement on thyroid stimulating hormone (29). The variety in combinations of sleep enhancing or suppressive effects and circadian day or night drives is consistent with the small overlap between conditions for the circadian night and day genes (Fig. 2D).
Overall, the annotation analysis suggests that when we sleep in phase, the circadian process and the sleep process combine to facilitate transcription and translation. Future analyses of the current and previously published human (21, 40) and animal data (30, 31, 41, 42) may shed further light on the role of sleep and circadian rhythmicity on the regulation of the transcriptome. A first comparison of the current study of the effects of mistimed sleep and our previous study of the effects of insufficient sleep (21) indicates that several of the observed phenomena are robust. Thus, the bimodal distribution of peak activity and the processes associated with day or night peaks were very similar in the two studies. Furthermore, both studies indicated that disruption of the sleep–wake circadian system suppresses genes associated with the regulation of gene expression and translation and enhances activity of genes associated with catabolic processes, the regulation of ATPase activity, and peroxiredoxin activity.
Mechanisms Underlying the Temporal Disruption of Gene Expression by Forced Desynchrony.
What are the mechanisms underlying persistent rhythmicity and loss of rhythmicity? The circadian rhythms of both melatonin and cortisol are known to regulate gene expression (11, 43) and are driven by the SCN. The fact that the vast majority of circadian transcripts become arrhythmic when sleeping out of phase with melatonin would indicate that the SCN neural outputs and hormones driven by the SCN (i.e., melatonin and cortisol only has a limited influence on the peripheral blood). We nevertheless found some evidence for their influence on the transcriptome. For example, BCL2 is regulated by melatonin (44) and fell within the category of genes with a circadian night peak both when sleeping in and out of phase. Thus, it was unaffected by sleep, and its time course remained linked to melatonin (Fig. 7A). In our study, NR1D1 was a transcript that became rhythmic when sleeping out of phase with melatonin, and it was classified within the category of transcripts whose expression profiles remained largely unchanged between the in-phase and out-of-phase conditions and were driven by a circadian day peak and enhanced by sleep (Fig. 7F). Transcripts within that category were associated with GO terms that include response to corticosteroid stimulus and hormone activity, and it is possible that the expression of these transcripts could be driven by cortisol (45). It is important to emphasize that effects of these endocrine signals on the transcriptome can also interact with the effects of the sleep–wake cycle. Ldlr responds to sleep deprivation in the mouse but only in the presence of a glucocorticoid signal (41). In our study, LDLR is one of the transcripts that were robustly rhythmic in both sleep conditions. However, although desynchronization did not affect the rhythmicity of this transcript, its peak of expression shifted significantly between sleep conditions (Fig. 7G). To further investigate the contribution of melatonin and cortisol on the human transcriptome, these hormones would have to be manipulated directly.
The loss of rhythmicity in many transcripts suggests that expression profiles in the periphery may depend on both local cues related to for example the sleep–wake cycle and central rhythms. When sleep occurs in phase and there is “resonance” between central and peripheral cues, rhythmicity is strong, but peripheral rhythmicity is weakened when this resonance is disrupted, whereas, at the same time, central rhythms remain robust. Robustness of SCN-driven rhythms vs. fragility of peripheral rhythms has previously been observed in mouse tissues exposed to oscillating temperature cycles; the SCN does not entrain to temperature rhythms whereas peripheral tissues do (46). It is well established that sleep and forced desynchrony affect body temperature rhythms in humans and animals (26, 47). Thus, the amplitude of the temperature rhythm is high when the temperature lowering effect of sleep coincides with the circadian phase during which melatonin is synthesized (i.e., the biological night), and the overt core body temperature amplitude is lower when sleep occurs during the biological day (48). Therefore, a potential mechanism for the observed effects of desynchrony would be that the lowered amplitude of core body temperature caused by sleeping out of phase affects the amplitude of rhythmicity in temperature-sensitive RNA-binding proteins, such as cold-inducible RNA-binding protein (CIRBP) and RNA-binding motif protein 3 (RBM3), which control the expression of key circadian regulators of transcription and translation (10, 49). CIRBP and RBM3 are both induced by lower temperature, and the observed change in the time course of expression in these transcripts when sleeping in and out of phase with melatonin (Fig. 3) is consistent with the reduced amplitude of the core body temperature rhythm when sleeping out of phase. CIRBP and RBM3 potentially regulate the circadian expression of several thousands of genes, which include core circadian clock genes and genes involved in the regulation of gene expression, such as genes for RNA polymerase II and ribosomal protein subunits (10, 49). In fact, comparison between the list of transcripts whose expression showed an interaction between sleep condition and time with published lists of gene targets for CIRBP and RBM3 (49) showed an ∼30% overlap (Fig. S7; see SI Methods). In addition, Cirbp is one of a small number of transcripts that remained rhythmic in the mouse clock-deficient liver with its expression profile entrained to temperature (50). Therefore, changes in core body temperature may be one cause of the widespread disruption of circadian rhythmicity. In addition to changes in temperature cycles, the feeding schedule and associated changes in endocrine and metabolic variables or the dim light–dark schedule, which are timed in synchrony with the imposed 28-h sleep–wake cycle (15, 18), may also contribute to the changes in the rhythmic organization of the transcriptome (14).
The classical term in circadian rhythm research for the effects of the light–dark cycle or sleep–wake would be “masking” the endogenous circadian rhythmicity, implying that these effects are separate from circadian rhythm generation (51, 52). Our current data highlight the interrelatedness of these masking effects and the genes involved in circadian rhythm generation and their combined contribution to overall temporal organization. Systemic cues are known to be important for circadian organization in other peripheral tissues. The timing of feeding, for example, is known to combine with the intrinsic clock to drive rhythms in the mouse liver, whereas restricted feeding can restore liver rhythmicity in clock-deficient mice (53). Similarly, the expression of the majority of liver transcripts became arrhythmic in mice where only the liver clock was deficient, although a small number of transcripts remained entrained by systemic cues that included temperature (50). Furthermore in the vole, which has a distinctive ultradian feeding pattern, clock gene expression has a high-amplitude circadian rhythm in the SCN but is flat in the liver (54).
Summary and Implications.
Previously, we showed that 1 wk of insufficient sleep reduced the circadian organization of the human transcriptome (from 8.8% to 6.9%), even when assessed in the absence of a sleep–wake cycle (21). Here, we show that mistimed sleep desynchronized from the central circadian clock has a much larger effect on the circadian regulation of the human transcriptome (i.e., a reduction in the number of circadian transcripts from 6.4% to 1% and changes in the overall time course of expression of 34% of transcripts). Also, although the number of genes for which the overall level of expression was up- or down-regulated was similar to the number affected by sleep restriction (21), in the present study, forced desynchrony resulted in fold changes that were greater. Overall, these data imply that outputs of the circadian timing system, such as the sleep–wake cycle and associated changes, feedback onto and reinforce processes that are at the core of the generation of circadian rhythmicity. There are two main implications of these phenomena. Firstly, the negative health outcomes of disruption of this reinforcement and resonance of the sleep–wake cycle and central circadian rhythmicity, as occurs in jet lag and shift work, may be mediated by this profound disruption of the temporal organization at the level of the transcriptome. Secondly, the powerful reinforcement of peripheral rhythmicity by the sleep–wake cycle and associated variables may be used to enhance temporal organization in those conditions in which it is weakened, such as aging. With a growing number of shift workers and insufficient sleep becoming an ever-increasing problem globally, these two implications will become increasingly relevant.
Methods
Ethics and Participants.
The protocol received a favorable opinion from the University of Surrey Ethics Committee and was conducted in accordance with the principles of the Declaration of Helsinki. Participants were recruited as reported in ref. 20. Transcriptome data were obtained from 22 participants (mean ± SD of age, 26.3 ± 3.4 y; 11 males and 11 females) and are presented in this report (Table S1). Participants were all white and homozygous for the PER3 VNTR polymorphism (rs57875989), with 11 participants carrying the shorter allele. Participants were in good health, as assessed by medical history, physical examination, and standard biochemistry and hematology. None suffered from sleep disorders, as assessed by self-report questionnaires [Pittsburgh Sleep Quality Index ≤5 (55)] and a clinical polysomnographic recording. Habitual sleep duration was 7 h and 57 min ± 52 min (SD).
Study Protocol.
The forced-desynchrony protocol, during which participants were resident in a clinical research center, was modified from previous protocols (17, 56), and this version has been described previously (20, 57). Briefly, following a baseline 8-h sleep episode at habitual bedtime (assessed from 1 wk of field actigraphy and sleep diaries) and a 16-h wake period, the sleep–wake cycle, the dark–dim light cycle, and meals were all scheduled to a 28-h period such that the sleep episode began 4 h later in each cycle.
Melatonin Assay and Assessment of Circadian Phase.
Plasma melatonin levels were measured and analyzed as previously described (20).
Polysomnography.
Polysomnography was performed as previously described (21, 57).
RNA Extraction, Labeling, and Hybridization.
Extracted and labeled cRNA was hybridized to Whole Human Genome 4 × 44K custom oligonucleotide microarrays, as previously described (21). Microarray data were deposited in the Gene Expression Omnibus database (accession no. GSE48113).
Microarray Statistical Analysis.
Quality-control preprocessing of the microarray data were performed as described previously (21).
Time series analysis.
Log2 values were quantile-normalized using the R Bioconductor package limma (58). Non–control-replicated probes, along with their corresponding flags, were averaged. We assumed that transcripts whose expression levels have a circadian component will show one full oscillation every ∼24 h. To identify the set of transcripts with circadian profiles, we followed a time–domain analysis described previously (21). Transcripts targeted by probes identified as overt circadian in a minimum number of subjects (ns) were defined as having a prevalent circadian expression in the associated test condition. We used ns to keep the false-discovery rate (FDR) to a maximum of 5%, as previously described (21).
ANOVA.
For the analyses aimed at identifying effects of forced desynchrony on the transcriptome, independent of classifications of rhythmic vs. nonrhythmic patterns, we used a mixed-model ANOVA approach (Procedure Mixed in SAS version 9.2). In this model, class variables were participant, sleep condition (in phase, out of phase), time point (sample numbers 1–7), and genotype (PER35/5 vs. PER34/4). The analysis investigated whether the expression level was affected by sleep condition, time point, genotype, and their simple interactions, taking into account the repeated measurement design. Few effects of genotype were found and these results are not discussed. P values were corrected for multiple testing using the Benjamini and Hochberg approach (59). The significant effects (corrected P value, <0.05) were investigated using differences in least-square means.
Clustering analysis of prevalent circadian transcripts.
Transcript median profiles (median across all paired participants, per sampling time point, of z-scored time series) were entered in the clustering analysis. The coexpression coefficient-based circular self-organizing map (60) was used to partition the data into distinct groups, and the number of clusters was established using the Bayesian index criterion (61).
Gene Enrichment and Functional Annotation Analyses.
Gene enrichment and functional annotation analyses were performed as described previously (21).
Direct Interaction Networks.
For details of how the direct interaction networks were constructed and visualized, see SI Methods.
Contribution of the Sleep–Wake Cycle and Circadian Rhythmicity to the Time Course of Transcripts.
For details of the model used to define the contribution of the sleep-wake cycle and circadian rhythmicity to the expression profiles of transcripts, see SI Methods.
Supplementary Material
Acknowledgments
We thank John Groeger for helpful discussions; Patrick McGabe for help with statistical analyses; Sibah Hasan and June Lo for help with data acquisition; and the nurses, research officers, program managers, and study physicians of the Surrey Clinical Research Centre for clinical conduct of the study. We also thank Raphaelle Winsky-Sommerer and Jonathan Johnston for helpful comments on the manuscript. The primary support for this research was provided by Biotechnology and Biological Sciences Research Council Grant BB/F022883, with additional support from Air Force Office of Scientific Research Grant FA9550-08-1-0080 and the Royal Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE48113).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1316335111/-/DCSupplemental.
References
- 1.Buhr ED, Takahashi JS. Molecular components of the mammalian circadian clock. Handbook Exp Pharmacol. 2013;217(217):3–27. doi: 10.1007/978-3-642-25950-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bass J. Circadian topology of metabolism. Nature. 2012;491(7424):348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- 4.Koike N, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338(6105):349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421(6919):177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 6.Valekunja UK, et al. Histone methyltransferase MLL3 contributes to genome-scale circadian transcription. Proc Natl Acad Sci USA. 2013;110(4):1554–1559. doi: 10.1073/pnas.1214168110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Martelot G, et al. CycliX Consortium Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles. PLoS Biol. 2012;10(11):e1001442. doi: 10.1371/journal.pbio.1001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jouffe C, et al. The circadian clock coordinates ribosome biogenesis. PLoS Biol. 2013;11(1):e1001455. doi: 10.1371/journal.pbio.1001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kojima S, Sher-Chen EL, Green CB. Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev. 2012;26(24):2724–2736. doi: 10.1101/gad.208306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morf J, et al. Cold-inducible RNA-binding protein modulates circadian gene expression posttranscriptionally. Science. 2012;338(6105):379–383. doi: 10.1126/science.1217726. [DOI] [PubMed] [Google Scholar]
- 11.Reddy TE, Gertz J, Crawford GE, Garabedian MJ, Myers RM. The hypersensitive glucocorticoid response specifically regulates period 1 and expression of circadian genes. Mol Cell Biol. 2012;32(18):3756–3767. doi: 10.1128/MCB.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hastings M, O’Neill JS, Maywood ES. Circadian clocks: Regulators of endocrine and metabolic rhythms. J Endocrinol. 2007;195(2):187–198. doi: 10.1677/JOE-07-0378. [DOI] [PubMed] [Google Scholar]
- 13.Albrecht U. Timing to perfection: The biology of central and peripheral circadian clocks. Neuron. 2012;74(2):246–260. doi: 10.1016/j.neuron.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 14.Saini C, Suter DM, Liani A, Gos P, Schibler U. The mammalian circadian timing system: Synchronization of peripheral clocks. Cold Spring Harb Symp Quant Biol. 2011;76:39–47. doi: 10.1101/sqb.2011.76.010918. [DOI] [PubMed] [Google Scholar]
- 15.Morris CJ, Aeschbach D, Scheer FA. Circadian system, sleep and endocrinology. Mol Cell Endocrinol. 2012;349(1):91–104. doi: 10.1016/j.mce.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zee PC, Goldstein CA. Treatment of shift work disorder and jet lag. Curr Treat Options Neurol. 2010;12(5):396–411. doi: 10.1007/s11940-010-0090-9. [DOI] [PubMed] [Google Scholar]
- 17.Dijk DJ, Czeisler CA. Contribution of the circadian pacemaker and the sleep homeostat to sleep propensity, sleep structure, electroencephalographic slow waves, and sleep spindle activity in humans. J Neurosci. 1995;15(5 Pt 1):3526–3538. doi: 10.1523/JNEUROSCI.15-05-03526.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheer FA, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci USA. 2009;106(11):4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pevet P, Challet E. Melatonin: Both master clock output and internal time-giver in the circadian clocks network. J Physiol Paris. 2011;105(4-6):170–182. doi: 10.1016/j.jphysparis.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 20.Hasan S, et al. Assessment of circadian rhythms in humans: Comparison of real-time fibroblast reporter imaging with plasma melatonin. FASEB J. 2012;26(6):2414–2423. doi: 10.1096/fj.11-201699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Möller-Levet CS, et al. Effects of insufficient sleep on circadian rhythmicity and expression amplitude of the human blood transcriptome. Proc Natl Acad Sci USA. 2013;110(12):E1132–E1141. doi: 10.1073/pnas.1217154110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houtkooper RH, et al. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497(7450):451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki T, Kimura A, Nagai R, Horikoshi M. Regulation of interaction of the acetyltransferase region of p300 and the DNA-binding domain of Sp1 on and through DNA binding. Genes Cells. 2000;5(1):29–41. doi: 10.1046/j.1365-2443.2000.00302.x. [DOI] [PubMed] [Google Scholar]
- 24.Van Cauter E, Spiegel K, Tasali E, Leproult R. Metabolic consequences of sleep and sleep loss. Sleep Med. 2008;9(Suppl 1):S23–S28. doi: 10.1016/S1389-9457(08)70013-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wyatt JK, Ritz-De Cecco A, Czeisler CA, Dijk DJ. Circadian temperature and melatonin rhythms, sleep, and neurobehavioral function in humans living on a 20-h day. Am J Physiol. 1999;277(4 Pt 2):R1152–R1163. doi: 10.1152/ajpregu.1999.277.4.r1152. [DOI] [PubMed] [Google Scholar]
- 26.Dijk DJ, Duffy JF, Riel E, Shanahan TL, Czeisler CA. Ageing and the circadian and homeostatic regulation of human sleep during forced desynchrony of rest, melatonin and temperature rhythms. J Physiol. 1999;516(Pt 2):611–627. doi: 10.1111/j.1469-7793.1999.0611v.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleitman N, Jackson DP. Body temperature and performance under different routines. J Appl Physiol. 1950;3(6):309–328. doi: 10.1152/jappl.1950.3.6.309. [DOI] [PubMed] [Google Scholar]
- 28.Saini C, Morf J, Stratmann M, Gos P, Schibler U. Simulated body temperature rhythms reveal the phase-shifting behavior and plasticity of mammalian circadian oscillators. Genes Dev. 2012;26(6):567–580. doi: 10.1101/gad.183251.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Czeisler CA, Klerman EB. Circadian and sleep-dependent regulation of hormone release in humans. Recent Prog Horm Res. 1999;54:97–130. [PubMed] [Google Scholar]
- 30.Maret S, et al. Homer1a is a core brain molecular correlate of sleep loss. Proc Natl Acad Sci USA. 2007;104(50):20090–20095. doi: 10.1073/pnas.0710131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barclay JL, et al. Circadian desynchrony promotes metabolic disruption in a mouse model of shiftwork. PLoS ONE. 2012;7(5):e37150. doi: 10.1371/journal.pone.0037150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Husse J, Hintze SC, Eichele G, Lehnert H, Oster H. Circadian clock genes Per1 and Per2 regulate the response of metabolism-associated transcripts to sleep disruption. PLoS ONE. 2012;7(12):e52983. doi: 10.1371/journal.pone.0052983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duffield GE. DNA microarray analyses of circadian timing: The genomic basis of biological time. J Neuroendocrinol. 2003;15(10):991–1002. doi: 10.1046/j.1365-2826.2003.01082.x. [DOI] [PubMed] [Google Scholar]
- 34.Yan J, Wang H, Liu Y, Shao C. Analysis of gene regulatory networks in the mammalian circadian rhythm. PLOS Comput Biol. 2008;4(10):e1000193. doi: 10.1371/journal.pcbi.1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bozek K, et al. Regulation of clock-controlled genes in mammals. PLoS ONE. 2009;4(3):e4882. doi: 10.1371/journal.pone.0004882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rey G, et al. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9(2):e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Archer SN, et al. Polymorphism in the PER3 promoter associates with diurnal preference and delayed sleep phase disorder. Sleep. 2010;33(5):695–701. doi: 10.1093/sleep/33.5.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lande-Diner L, Boyault C, Kim JY, Weitz CJ. A positive feedback loop links circadian clock factor CLOCK-BMAL1 to the basic transcriptional machinery. Proc Natl Acad Sci USA. 2013;110(40):16021–16026. doi: 10.1073/pnas.1305980110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fustin J-M, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155(4):793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 40.Pellegrino R, et al. Whole blood genome-wide gene expression profile in males after prolonged wakefulness and sleep recovery. Physiol Genomics. 2012;44(21):1003–1012. doi: 10.1152/physiolgenomics.00058.2012. [DOI] [PubMed] [Google Scholar]
- 41.Mongrain V, et al. Separating the contribution of glucocorticoids and wakefulness to the molecular and electrophysiological correlates of sleep homeostasis. Sleep. 2010;33(9):1147–1157. doi: 10.1093/sleep/33.9.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anafi RC, et al. Sleep is not just for the brain: Transcriptional responses to sleep in peripheral tissues. BMC Genomics. 2013;14:362. doi: 10.1186/1471-2164-14-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Unfried C, Burbach G, Korf HW, von Gall C. Melatonin receptor 1-dependent gene expression in the mouse pars tuberalis as revealed by cDNA microarray analysis and in situ hybridization. J Pineal Res. 2010;48(2):148–156. doi: 10.1111/j.1600-079X.2009.00738.x. [DOI] [PubMed] [Google Scholar]
- 44.Luchetti F, et al. Melatonin signaling and cell protection function. FASEB J. 2010;24(10):3603–3624. doi: 10.1096/fj.10-154450. [DOI] [PubMed] [Google Scholar]
- 45.Pezük P, Mohawk JA, Wang LA, Menaker M. Glucocorticoids as entraining signals for peripheral circadian oscillators. Endocrinology. 2012;153(10):4775–4783. doi: 10.1210/en.2012-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buhr ED, Yoo SH, Takahashi JS. Temperature as a universal resetting cue for mammalian circadian oscillators. Science. 2010;330(6002):379–385. doi: 10.1126/science.1195262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Franken P, Tobler I, Borbély AA. Sleep and waking have a major effect on the 24-hr rhythm of cortical temperature in the rat. J Biol Rhythms. 1992;7(4):341–352. doi: 10.1177/074873049200700407. [DOI] [PubMed] [Google Scholar]
- 48.Dijk DJ, Duffy JF, Czeisler CA. Contribution of circadian physiology and sleep homeostasis to age-related changes in human sleep. Chronobiol Int. 2000;17(3):285–311. doi: 10.1081/cbi-100101049. [DOI] [PubMed] [Google Scholar]
- 49.Liu Y, et al. Cold-induced RNA-binding proteins regulate circadian gene expression by controlling alternative polyadenylation. Sci Rep. 2013;3:2054. doi: 10.1038/srep02054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5(2):e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rietveld WJ, Minors DS, Waterhouse JM. Circadian rhythms and masking: An overview. Chronobiol Int. 1993;10(4):306–312. doi: 10.1080/07420529309059713. [DOI] [PubMed] [Google Scholar]
- 52.Mrosovsky N. Masking: History, definitions, and measurement. Chronobiol Int. 1999;16(4):415–429. doi: 10.3109/07420529908998717. [DOI] [PubMed] [Google Scholar]
- 53.Vollmers C, et al. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci USA. 2009;106(50):21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Veen DR, et al. Impact of behavior on central and peripheral circadian clocks in the common vole Microtus arvalis, a mammal with ultradian rhythms. Proc Natl Acad Sci USA. 2006;103(9):3393–3398. doi: 10.1073/pnas.0507825103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buysse DJ, Reynolds CF, 3rd, Monk TH, Berman SR, Kupfer DJ. The Pittsburgh Sleep Quality Index: A new instrument for psychiatric practice and research. Psychiatry Res. 1989;28(2):193–213. doi: 10.1016/0165-1781(89)90047-4. [DOI] [PubMed] [Google Scholar]
- 56.Czeisler CA, et al. Stability, precision, and near-24-hour period of the human circadian pacemaker. Science. 1999;284(5423):2177–2181. doi: 10.1126/science.284.5423.2177. [DOI] [PubMed] [Google Scholar]
- 57.Lazar AS, et al. Circadian period and the timing of melatonin onset in men and women: Predictors of sleep during the weekend and in the laboratory. J Sleep Res. 2013;22(2):155–159. doi: 10.1111/jsr.12001. [DOI] [PubMed] [Google Scholar]
- 58.Gentleman RC, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B. 1995;57(1):289–300. [Google Scholar]
- 60.Moller-Levet CS, Yin H. Circular SOM for temporal characterisation of modelled gene expressions. Lect Notes Comput Sci. 2005;3578:319–326. [Google Scholar]
- 61.Schwarz G. Estimating the dimension of a model. Ann Stat. 1978;6(2):461–464. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.