Abstract
Protein degradation constitutes a major cellular function that is responsible for maintenance of the normal cellular physiology either through the degradation of normal proteins or through the elimination of damaged proteins. The Ubiquitin–Proteasome System (UPS)1 is one of the main proteolytic systems that orchestrate protein degradation. Given that up- and down- regulation of the UPS system has been shown to occur in various normal (such as ageing) and pathological (such as neurodegenerative diseases) processes, the exogenous modulation of the UPS function and activity holds promise of (a) developing new therapeutic interventions against various diseases and (b) establishing strategies to maintain cellular homeostasis. Since the proteasome genes are evolutionarily conserved, their role can be dissected in simple model organisms, such as the nematode, Caenorhabditis elegans. In this review, we survey findings on the redox regulation of the UPS in C. elegans showing that the nematode is an instrumental tool in the identification of major players in the UPS pathway. Moreover, we specifically discuss UPS-related genes that have been modulated in the nematode and in human cells and have resulted in similar effects thus further exhibiting the value of this model in the study of the UPS.
Abbreviations: EGF, Epidermal Growth Factor; IIS, insulin/IGF-1 signaling pathway; UPS, Ubiquitin–Proteasome System
Keywords: Ubiquitin-proteasome system (UPS), Proteasome regulation, C. elegans, Protein
Graphical abstract
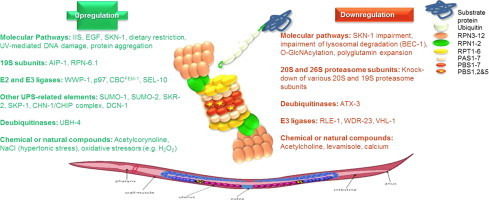
Genes, pathways and compounds that have been identified to alter the UPS function in C. elegans. Molecular pathways and natural or chemical compounds along with alterations in the various components of the UPS system (e.g. proteasome subunits, E2 and E3 ligases, DUBs) that have been revealed to affect the UPS function in C. elegans in terms of proteasome activities and/or assembly and/or expression. Positive regulators are shown in green whereas negative regulators are shown in red.
Highlights
-
•
UPS is one of the main proteolytic systems that orchestrate protein degradation.
-
•
Proteasome function can be dissected in Caenorhabditis elegans.
-
•
Nematodes can be used in the identification of major players in the UPS pathway.
Introduction
Proteolysis
Proteolysis refers to the breakdown of proteins into smaller polypeptides or amino acids. In general, this occurs through the hydrolysis of peptide bonds, and this is most commonly achieved by cellular enzymes called proteases (enzyme-mediated degradation), but may also occur through intramolecular digestion, as well as through non-enzymatic routes such as the action of mineral acids and heat.
Proteolysis has various roles on the cellular maintenance. It occurs in order to provide amino acids to the cells [153], to activate post-translationally certain proteins [22], to regulate physiological cellular processes (e.g. cell cycle regulation, apoptosis etc) and to prevent the accumulation of abnormal or misfolded proteins [26]. The two main mechanisms of cellular proteolysis that play a key role in cellular homeostasis are the lysosome-mediated intracellular protein degradation and the proteasome-mediated protein degradation.
Lysosomes are single or double membrane organelles that enclose acidic hydrolases with multiple specificities, such as peptidases, lipases and nucleases for the degradation of various biological molecules. One can distinguish different forms of autophagy mainly based on the mechanism for the delivery of cargo to the lysosomes. The main forms are the so called macroautophagy, microautophagy and chaperone-mediated autophagy. In general, autophagy is a bulk degradation process implicated in the clearance of long-lived proteins and organelles but more selective forms of autophagy have also been described for both macro- and micro-autophagy. Macroautophagy is the main pathway, responsible for the elimination of damaged cell organelles or unused proteins [96]. This involves the formation of a double-layered membrane around the organelle known as an autophagosome [16]. The autophagosome travels through the cytoplasm of the cell to get fused with a lysosome and to carry on to the endosomal pathways [110]. Within the lysosome, the contents of the autophagosome are degraded via acidic lysosomal hydrolases [60]. In microautophagy, the cargo is engulfed by single-membraned vesicles originated by invagination of the lysosomal membrane that then pinches off inside the lumen where it is rapidly degraded [112]. Finally, chaperone-mediated autophagy (CMA) is a selective protein degradation pathway by which soluble cytosolic proteins bearing in their amino acid sequences a common targeting motif (KFERQ-like pentapeptide sequence) are recognized by a chaperone complex (hsc70-containing complex) which targets them to lysosomes for degradation [105]. These proteins have to be bound to the lysosome-associated membrane protein type 2A (LAMP-2A), a CMA receptor at the lysosomal membrane, in order to be translocated into the lysosomal lumen and to get degraded by the lysosomal proteases.
Proteasome-mediated proteolysis refers to a selective cellular process that results in the specific degradation of either un-needed (but normal) or damaged/misfolded proteins. Proteasomes are part of a major mechanism by which cells regulate the concentration of particular proteins and degrade misfolded proteins, the so called Ubiquitin–Proteasome System (UPS). Proteins, that need to be degraded, are tagged with a series of a small protein called ubiquitin. The result is a poly-ubiquitin chain on the protein to be degraded that is recognized by the proteasome, allowing it to degrade the tagged protein [102]. This review will focus on the proteasome-mediated proteolysis on a model organism for ageing studies, namely the nematode Caenorhabditis elegans.
Caenorhabditis elegans
The nematode C. elegans is a roundworm. The roundworms, a phylum of smooth-skinned, unsegmented worms with a long cylindrical body shape, include free-living and parasitic forms both aquatic and terrestrial. C. elegans is a non-hazardous, non-infectious, non-pathogenic, non-parasitic organism. It is small, growing to about 1 mm in length, and lives in the soil, where it survives by feeding on microbes, primarily bacteria [183]. C. elegans genome, which is completely sequenced (approximately 100 million base pairs long), consists of five pairs of autosomes (I, II, III, IV and V) and one pair of sex chromosomes (X), so as of mitochondrial genome. C. elegans has two sexes, hermaphrodites and males. Almost all individuals are hermaphrodites, with males comprising just 0.05% on average of the total population. Sex in C. elegans is based on an X0 sex-determination system. Hermaphrodite C. elegans have a matched pair of sex chromosomes (XX) and the rare males have only one sex chromosome (X0). When self-inseminated, the wild-type worm will lay approximately 300 eggs. When inseminated by a male, the number of progeny can exceed 1000.
The basic anatomy of C. elegans includes a mouth, a pharynx, an intestine, a gonad (two bilaterally symmetric, U-shaped gonad arms that are connected to a central uterus through spermatheca) and a collagenous cuticle. The four bands of muscles that run the length of the body are connected to a neuronal system, that is thoroughly mapped, that allows the muscles to move the animal's body only in the dorsal and ventral direction.
The hermaphrodite is considered to be a specialized form of self-fertile female because the soma is female but its germ line produces male gametes firstly and then conducts the laying of eggs. Eggs are deposited externally through the uterus following internal fertilization. After hatching, juveniles pass through four stages (larval stages: L1–L4). When the population of the culture is crowded or under caloric restriction conditions, C. elegans can enter an alternative larval stage (L3 stage) called the dauer state. Dauer larvae are stress-resistant and do not age. At 20 °C, the laboratory strain of C. elegans has an average lifespan of approximately 3 weeks and a generation time of approximately 4 days.
In the three last decades, multiple studies have focused on the regulation of certain signaling pathways and cellular processes, which have been conserved through evolution in humans, such us ageing, proteolysis and response to oxidative stress. C. elegans has been shown to be a valuable model in studying these processes and in extrapolating conclusions to humans.
Proteasome biochemistry
Structure-subunits-forms (proteasome complexes)
20S Proteasome
20S proteasomes are large protein complexes of approximately 700 kDa inside all eukaryotes and archaea, as well as in some bacteria. The main catalytic function of the proteasome is to degrade normal or damaged proteins by proteolysis, a chemical reaction that breaks peptide bonds. This degradation process yields peptides of about seven to eight amino acids long, which can then be further degraded by various peptidases into amino acids and used in synthesis of new proteins. In eukaryotes, oxidized proteins or proteins with attached polyubiquitin chains are bound by the proteasome and get degraded.
The eukaryotic 20S proteasome, which is the main particle of the proteasomal mechanism also known as the 20S proteasome core, is a large cylindrical structure containing four stacked homologous rings, 2α (consisted by α-type subunits) and 2β (consisted by β-type subunits), around a central pore. Each ring is composed of seven different protein subunits (molecular mass between 20 and 30 kDa) thus giving rise to the final 20S structure, α1−7β1−7β1−7α1−7. The inner two rings are each made of seven different β-type subunits and three of those namely, β1, β2 and β5 contain three proteolytic active sites; chymotrypsin-like, trypsin-like and caspase-like activities (see below; [72]). These sites are located on the interior surface of the rings, so that the target protein must enter the central pore before it is degraded. The outer two rings maintain a “gate” through which proteins enter the cylinder. The α-type subunits are controlled by binding to “cap” structures or regulatory particles that recognize polyubiquitin tags attached to candidate protein substrates and initiate the degradation process. Table 1 summarizes the orthologues of the 20S human proteasome subunits in C. elegans.
Table 1.
The orthologues of the human 20S proteasome subunits in C. elegans.
|
α-type subunits |
β-type subunits |
||
|---|---|---|---|
| C. elegans | H. sapiens | C. elegans | H. sapiens |
| Gene name | |||
| pas-1 | PSMA6 | pbs-1 | PSMB6 |
| pas-2 | PSMA2 | pbs-2 | PSMB7 |
| pas-3 | PSMA4 | pbs-3 | PSMB3 |
| pas-4 | PSMA7 | pbs-4 | PSMB2 |
| pas-5 | PSMA5 | pbs-5 | PSMB5 |
| pas-6 | PSMA1 | pbs-6 | PSMB1 |
| pas-7 | PSMA3 | pbs-7 | PSMB4 |
α-Type subunits in C. elegans
The α-type proteasomal subunits are arranged in the two outer rings of the core proteasome and possess structural and regulatory roles. They compose the gate of the 20S core proteasome that regulates the access of the substrate towards the pore, through allosteric interactions. In C. elegans all seven α-type subunits (pas-1 to pas-7) have been mapped [24], and all of them have a statistical significant homology with the ones found in Homo sapiens and Saccharomyces cerevisiae.
The pas-1 gene encodes for a type-6 alpha subunit of the 20S core particle in humans and is required for embryonic development. By homology, it is predicted to comprise the outer rings of the proteasome and to play a role in selective degradation of ubiquitinated proteins during development. In vitro, PAS-1 interacts with VET-1, a coiled-coil domain-containing protein expressed in the early embryo and ATN-1, an alpha-actinin, in addition to other members of the proteasome [24]. The pas-2 gene encodes for a proteasome subunit with high similarity to human proteasome α2 subunit [8] and is revealed to be an orthologue of the yeast protein Sec18, member of the AAA ATPase family [24]. The pas-3 gene encodes for a type-4 alpha subunit in humans that affects embryonic viability, growth, fertility, and locomotion [38] and resembles to the human TGF-β receptor associated protein-1 [24]. The pas-4 gene encodes for a type-7 alpha subunit of the 20S core in humans. Loss of pas-4 activity via RNAi results in a wide variety of defects including embryonic and larval lethality, sterility, abnormal locomotion, slow growth, and abnormal transgene expression and subcellular localization [38,154]. Furthermore, loss of PAS-4 activity also results in activation of SKN-1, a transcription factor required in adult worms for the acute response to oxidative stress that is absolutely necessary for embryonic development [75]. The pas-5 gene encodes for a type-5 alpha subunit of the 20S core in humans. Loss of pas-5 activity via RNAi has the same results as loss of pas-4 [38,154,141,142,166]. The pas-6 gene encodes for a type-1 alpha subunit in humans. PAS-6 is required for embryonic, larval, and germline development. In vitro, PAS-6 interacts with BRP-1, a glutamine/asparagine (Q/N)-rich domain-containing protein conserved amongst nematodes, and with other proteasome subunits [28,185]. The pas-7 gene encodes for a type-3 alpha subunit in humans. Its loss via RNAi results in several defects including embryonic and larval lethality, sterility, and abnormal meiotic progression [167,141,142,34,166]. The α3 subunit plays a central role in the mammalian 20S proteasome since it regulates the stabilization of the gate and the allosteric interactions between α2, α3 and α4 by altering the conformation of its N-terminal end [49]; PAS-7 is its orthologue in C. elegans.
β-Type subunits in C. elegans
The β-type proteasomal subunits enclose the proteolytic part of the 20S core proteasome, where the degradation of polypeptides and proteins takes place. β-type subunits possess protease activities and have been conserved through evolution. 20S proteasome exhibits three different proteolytic activities; caspase-like activity that cleaves after acidic amino acids with β1 subunit being the responsible one, trypsin-like activity that cleaves after basic amino acids with β2 subunit being the responsible one and chymotrypsin-like activity that cleaves after large hydrophobic and neutral amino acids with β5 subunit being the responsible one [72]. The catalytic center of each proteolytic subunit contains an N-terminal threonine residue (Thr1) [103]. Additionally, the three-dimensional structure of the proteolytic center of each of these subunits is stabilized by a set of serines, Ser129/166/169. In C. elegans all seven β-type subunits (pbs-1 to pbs-7) have been mapped [24], bearing a statistical significant homology with the ones found in H. sapiens and S. cerevisiae.
The pbs-1 gene encodes for a protease subunit with high similarity to the human proteasome type-6 beta subunit that affects fertility, embryonic viability, locomotion, and larval viability [76,154]. The pbs-2 gene encodes for a protease subunit loss of which has been found to affect apoptosis, fertility and embryonic development [48]. The pbs-2 gene shows high similarity with the human type-7 beta subunit. The pbs-3 gene encodes for a type-3 beta subunit of the 20S core. By homology, PBS-3 is predicted to function in the ATP/ubiquitin-dependent non-lysosomal protein degradation and loss of PBS-3 activity indicates that, in C. elegans, PBS-3 is required for embryonic and germline development, movement, normal body coloration, and normal body morphology [76,154]. The pbs-4 gene encodes for a type-2 beta subunit of the human 20S proteasome. PBS-4 is predicted to function in the ATP/ubiquitin-dependent non-lysosomal protein degradation, as PBS-3, and loss of PBS-4 activity reveals that PBS-4 is required for embryonic, germline, and larval development [38,167,154]. The pbs-5 gene encodes for a type-5 beta subunit, loss of which leads to reduced apoptosis, embryonic lethality and defects in locomotion [48,166,154]. The pbs-6 gene encodes for the similar to vertebrates β1 subunit of the 20S proteasome. Its loss results in embryonic lethality and defects in egg laying, larval development, morphology and movement [48,154,166]. The pbs-7 gene encodes for the similar to β4 mammalian subunit of the 20S proteasome. By homology, PBS-7 is predicted to function in the ATP/ubiquitin-dependent non-lysosomal protein degradation. Loss of PBS-7 activity indicates that PBS-7 is required for normal movement and for embryonic, germline, and larval development [38,126,154].
26S proteasome
The 26S proteasome is an over 2 MDa molecular machine that is consisted by more than 30 different subunits. This large multisubunit protease is ubiquitous in eukaryotic cells and plays a key role in cellular proteostasis. 26S proteasome consists of one 20S barrel-shaped proteolytic core complex and two 19S regulatory complexes that cap both ends of the 20S core complex, attached to the α-rings.
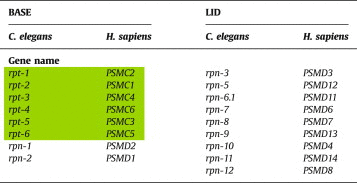
19S regulatory complex is responsible for the recognition, deubiquitination and translocation of the ubiquitinated substrates that are rapidly broken down into short peptides [44]. 19S regulatory particle is divided into a ring-like base, which consists of six different ATPases and a few non-ATPases, and a lid-like structure, which only consists of non-ATPases subunits. These two structures are considered to be linked through Rpn10 and Rpn13 subunits. Table 2 summarizes the orthologues of the 19S human proteasome subunits in C. elegans.
Table 2.
The orthologues of the human 19S proteasome subunits in C. elegans. The ATPases are shown in green background.
 |
Base complex
The ring-shaped base [116] consists of six different ATPases of the AAA+ family (ATPase Associated diverse cellular activities), namely Rpt1-6, and two non-ATPases, namely Rpn1 and Rpn2. The base complex binds in each side of the 20S core proteasome over the α-rings. Its specific role is (a) the recognition of the ubiquitinated proteins, (b) the ATP-dependent unfolding of the substrates, (c) the opening of the 20S core gate via allosteric reactions and finally, (d) the translocation of the ubiquitinated protein substrate inside the 20S catalytic particle.
In C. elegans, Rpt-1 is the orthologue of the human 26S protease regulatory subunit 2 (S7). The rpt-1 gene expression affects fertility and embryonic viability ([24,76,77,126,154]), along with the expression levels of rpn-2 [164], daf-2 (receptor of the insulin-like pathway, abnormal DAuer Formation-2) and glp-1 (abnormal Germ Line Proliferation 1) [43]. Rpt-2 is the orthologue of the human 26S protease regulatory subunit 1 (S4) and is shown to affect embryonic lethality [126,167] and lifespan expectancy via regulation of glp-1 gene expression [43]. The rpt-3 gene is required for embryonic, larval and germline development [51,167]. By homology with the human isoform 1 of the 26S protease regulatory subunit 4 (S6B), it is predicted to function in unfolding protein substrates and translocating them into the core proteasome. It has been also found to interact with daf-2 and glp-1 to regulate lifespan expectancy [43] and with fog-2 (Feminization Of Germline) to regulate oocyte maturation [46]. The rpt-4 gene encodes for another ATPase subunit that affects body morphology, embryonic viability, growth, movement and fertility [124,157,24,76,154], sharing homology with the human 26S protease regulatory subunit 6 (S10B). RPT-4 interacts with many viability and growth significant proteins such as DAF-2, GLP-1 [43], ZYG-1 (ZYGote defective 1) [124], HSP70 (Heat Shock Protein 70), GST-4 (Glutathione S-Transferase 4), PAS-6 and PBS-6 [97]. The rpt-5 gene is required for embryonic, larval and germline development development [38,126,167] and by homology with human 26S protease regulatory subunit 3 (S6A), is predicted to unfold protein substrates and to translocate them into the core particle. It has been found to regulate genes such as daf-2 and glp-1 [43] and to be regulated by PAS-5 and PBS-4 subunits and the SKN-1 transcription factor [97]. RPT-6 is the last ATPase of the 19S particle. The rpt-6 gene, the orthologue of human 26S protease regulatory subunit 5 (S8), is required for embryonic and larval development [51,167]. RPT-6 has been found to suppress the expression of daf-2 and glp-1 genes, thus leading to shortened lifespan [43].
Concerning the non-ATPase subunits, RPN-1 is a component of the 19S base subcomplex. The activity of RPN-1 is essential for embryonic, larval and germline development [167,152]. rpn-1 is the orthologue of 26S proteasome non-ATPase regulatory subunit 2 and is predicted to function in unfolding and recognition of protein substrates and/or recycling of ubiquitin moieties during protein degradation. Its expression seems to suppress daf-2 and glp-1 genes expression [43] and to regulate the expression levels of gpdh-1 (Glycerol-3-Phosphate DeHydrogenase 1) [91,92] and rpn-2 genes [164]. RPN-2, the orthologue of the isoform 2 of 26S proteasome non-ATPase regulatory subunit 1, is required for embryonic, larval and germline development [51,167,48]. It is expressed sporadically at all stages of development in the pharynx, intestine, body wall muscle, somatic gonad, neurons, and hypodermis [107,108] and regulates the expression levels of many genes such as several 26S proteasomal subunits, hsp-70, gst-10 (Glutathione S-Transferase 10), gcs-1 (gamma GlutamylCysteine Synthetase) [97], gst-4 [97,18], daf-2, glp-1 [43], gpdh-1 [91]. It is regulated by SKN-1 factor [97] and RPN-1 and RPT-1 19S subunits [164].
Lid complex
19S lid subcomplex consists only of non-ATPases subunits that play a critical role in the process of degradation of ubiquitinated substrates as well as in additional functions that are not involved in the proteasome-mediated proteolysis process. These non-ATPases share a PCI domain which serves as a scaffold for the protein–protein interaction and seem to regulate the assembly and the maintenance of the structural integrity of the proteasomal machine [65,189]. The main role of the 19S lid is to recognize ubiquitinated protein substrates and to detach the ubiquitin molecules from them. Given that these subunits have no enzymatic activity (with the exception of Rpn11), it is unlikely to participate in other functions of the lid.
In C. elegans, the rpn-3 gene, the orthologue of the human 26S proteasome regulatory non-ATPase subunit 3, plays an important role in embryonic viability [154,51], development [141,142] and growth [43]. RPN-3 has been found to suppress the expression of daf-2 and glp-1 genes and to promote a shortened lifespan [43]. The rpn-5 gene has an important role in embryonic lethality [166] sharing homology with the human 26S proteasome non-ATPase regulatory subunit 12. The rpn-6.1 gene, the orthologue of the human 26S proteasome non-ATPase subunit 11 variant, affects embryonic, larval and adult viability, fertility, and osmotic regulation [51,24,76,154]. It interacts with daf-2 and glp-1 by suppressing them [43], with gpdh-1 by modulating its expression levels [91] while it is also regulated by glp-1 [175]. The rpn-7 gene affects fertility and embryonic viability [104,126,24] and, by homology, resembles to the human 26S proteasome non-ATPase regulatory subunit 6. RPN-7 was found to suppress the expression of daf-2 and glp-1, thus leading to a shortened lifespan of the worm [43] and to regulate the expression of gst-4 [19] and pqn-47 (Prion-like-(Q/N-rich)-domain-bearing protein) [115]. RPN-8 is the product of the orthologue gene of the human 26S proteasome non-ATPase regulatory subunit 7 and is shown to suppress the expression of daf-2 and glp-1 genes [43] and to modulate the expression levels of gst-4 [19]. The rpn-9 gene, sharing homology with the human 26S proteasome non-ATPase subunit 13, affects body morphology, embryonic and larval viability, locomotion, fertility, and growth [24,76,126,154]. RPN-9 is found to alter the expression levels of gsc-1 and gst-4 [177], to suppress the expression of daf-2 and glp-1 [43] and to change the localization of SKN-1 factor [177]. RPN-10 is not completely defined whether it is a component of the base or the lid. The rpn-10 gene is the orthologue of the human isoform Rpn10A of the 26S proteasome non-ATPase regulatory subunit 4. It seems that RPN-10 acts as a linker between these two structures attached to RPN-12 of the lid and RPN-1 of the base. RPN-10 subunit has two ubiquitin interacting motifs, which recognize the polyubiquitin chains of the protein substrates through diverse conformations of the protein helices. RPN-10 has a suppressing effect on several genes, such as fem-3 (FEMinization of XX and XO animals) [152], daf-2, glp-1 [43], lin-15A (abnormal cell LINeage, vulval development), lin-15B, lin-8 [23], and alters the expression levels of tra-2 (TRAnsformer: XX animals transformed into males, transmembrane receptor of the sex determination pathway) [152] and unc-45 (UNCoordinated, muscle-specific protein) [67]. The rpn-11 gene, orthologue of the human 26S proteasome non-ATPase regulatory subunit 14, affects embryonic and adult viability, osmoregulation and movement [24,76,154]. RPN-11 is a metalloproteinase of the JAMM domain family containing a conserved metal binding site [1] thus acting as a crucial deubiquitinating enzyme (DUB) of the 26S proteasome [174,187]. RPN-11 suppresses the expression of daf-2 and glp-1 genes resulting to a shortened lifespan [43]. Li and colleagues [98] have shown that rpn-11 expression levels are regulated by several genes such as skn-1, eef-2 (Eukaryotic translation Elongation Factor), rpn-2 and pas-5. The rpn-12 gene shares homology with the human 26S proteasome non-ATPase regulatory subunit 8. RPN-12 suppresses the expression of glp-1 [43], lin-15A, lin-15B, lin-36, lin-8 [23] and gpdh-1 [91] while rpn-12 gene expression is regulated by several genes, such as skn-1, eef-1A.1, eef-2, eif-1 (Eukaryotic Initiation Factor), ifg-1 (Initiation Factor 4G (eIF4G) family), rpt-4 and pas-5 [97].
Proteasome assembly
20S assembly
The 20S core undergoes a very complex and highly regulated intracellular assembly in eukaryotic cells. The 20S proteasome assembly involves a set of proteasome-specific chaperons, called PACs (Proteasome Assembling Chaperones). Up to now, 4 different PACs, namely PAC1–PAC4, have been revealed [97,188], but their homologues in C. elegans have not been identified yet. PAC1 and PAC2 form heterodimers, as PAC3 and PAC4 do. PAC1–2 is built in order to assemble the α-ring, resulting in the activation of PAC3–4 dimer which starts the step-by-step incorporation of the β subunits to form the β-ring. More specifically, α5 and α7 subunits are the first ones that take place on the PAC1–2 complex and they are followed by α6, α1, α2, α3 and α4 subunits. After the formation of the α-ring, PAC3–4 attaches to the α2 subunit of the α-ring and initiates the assembly of the β-ring starting by the attachment of β2 and β3 subunits. Attachment of β3 is followed by disassociation of PAC3–4 complex and the binding of Ump1 (in yeast, POMP in humans) molecule (proteasome maturation factor) [97,17]. β4–β7 and β1 follow the stepwise incorporation of the rest β subunits in the β-ring [58]. After this procedure, the intermediates formed contain one complete α- and one complete β-ring as well as a PAC1–2 complex and an Ump1 factor. Hsc73 is a heat shock protein which is involved in the further dimerization of the two intermediates mentioned above and are called 16S intermediates or half-proteasomes. After this step, Hsc73 detaches and proteolytic active subunits, β1, β2 and β5, are activated through autocatalysis of their own pro-peptides. The first substrate to be degraded is the Ump1 factor that is then followed by the degradation of the two PAC1-2 complexes.
19S assembly
Base complex
As it was mentioned above, 19S regulatory particle consists of two structures, the base and the lid. The assembly of the 19S regulatory particle of the proteasome initiates with the incorporation of the base subunits. Firstly, 3 different stable intermediates are formed, namely RPN1–RPT2–RPT1–Hsm3, Nas2–RPT3–RPT6–RPN14 and Nas6–RPT5–RPT4. Hsm3, Nas2, Nas6 and RPN14 are 19S-specific assembly factors and none of them is present on the mature 26S proteasome [144,113,140]. Nas2–RPT3–RPT6–RPN14 may act as the seed of the assembly of the 19S base particle. Subsequently, the three initiative intermediates assemble into the base complex and RPN2 and RPN13 bind onto it, to join the full base complex, which will attach to the lid via RPN10. Then, the chaperones Hsm3, RPN14, Nas6 are released through the addition of RPN10.
Lid complex
The assembly of the 19S lid complex is not yet fully understood. Recent studies suggest that the lid assembly is a multistep process [66,149,40]. To date, RPN5, RPN6, RPN8 and RPN9 are the first subunits to be incorporated to a stable intermediate that are then followed by the association of RPN3, RPN7 and RPN15 subunits. These two intermediates are associated via the interaction between RPN3 and RPN5 subunits. The lid is formed after the final incorporation of RPN11 and RPN12 subunits [149,168]. Hsp90 (Heat Shock Protein 90) [64] and Yin6 (orthologue of the mammalian Int6) [147] are two of the factors that are known to be involved in the lid formation in yeast.
As far as the association of 19S regulatory particle and 20S core particle is concerned, there has been found a so-called modulator complex that facilitates it. This modulator complex consists of three factors: RPT-4, RPT-5 and p27 (Cyclin-dependent kinase inhibitor 1B) [35].
Protein degradation
The UPS system consists of the ubiquitin system that is responsible for the recognition of the candidate for degradation protein and for its “tagging” with ubiquitin, and the proteasome system that performs the degradation of the protein. There are several proteins and enzymes that are recruited in UPS in order to perform the recognition, the tagging and the degradation, namely ubiquitin, E1–E3 ligases and deubiquitinases. Ubiquitin is covalently attached to the protein to be degraded in a multistep process coordinated by the sequential action of E1–E3 ligases [59]. The ubiquitin-activating enzyme (E1) is the first one to act by adenylating the C-terminal glycine of ubiquitin and forming a thioester bond between the activated glycine residue and a cysteine residue on the E1 catalytic site, through an ATP-dependent mechanism. The product of this reaction is a reactive E1–ubiquitin thioester intermediate. The activated ubiquitin is then passed to a conserved E2 active-site cysteine of a ubiquitin-conjugating enzyme (E2) by transthiolation. Meanwhile, a specific ubiquitin-protein ligase (E3) has already recruited the target protein and mediates the transfer of the activated ubiquitin from the E2 enzyme to the substrate. In most cases, an ε-NH2 group of a lysine residue on the tagged protein attacks the thioester bond between the ubiquitin and E2, and an isopeptide bond is formed, linking the activated C-terminal glycine of ubiquitin to the amino group in the attacking lysine of the target substrate [129]. If however, the substrate does not have any accessible lysine residues, ubiquitin can be conjugated to the α-amino group of the N-terminus of the target protein [135]. This ubiquitination cycle is successively repeated, thus producing polyubiquitin chains on the target protein (according to the sequential addition model). It has been shown that at least 4 ubiquitin residues are needed for efficient recognition and processing by the 26S proteasome [127,169].
Ubiquitin
Ubiquitin (Ub) is a small protein composed of 76 amino acids that is highly conserved in eukaryotes [170]. In C. elegans there are two loci, namely ubq-1 and ubq-2. ubq-1 is a polyubiquitin locus [47] which produces a polyubiquitin protein and is post-translationally cleaved into 11 Ub molecules by ubiquitin C-terminal hydrolases [69,56,57]. ubq-2 is a conserved locus in eukaryotes with an intact canonical Ub sequence fused to the ribosomal large subunit protein (L40) [70]. When the protein product is cleaved, Ub molecule is functional to be used for protein targeting [120]. Ubiquitin folds up into a globular structure and is found ubiquitously throughout the cell, but it can exist either as a free monomer or as part of a complex (polyubiquitin chains) attached to other proteins.
E1 enzymes
As far as the polyubiquitin chains are concerned, Ub molecule is conjugated to proteins through the bond between the glycine at its C-terminus and a lysine of the target protein. Conjugation process depends on the hydrolysis of ATP [128,130] and on the activation of the Ub molecule by an ubiquitin-activating enzyme (E1). In C. elegans, there is only one such enzyme, UBA-1 [89], disruption of which results in complete inactivation of the UPS proteolysis [126,51,104,76,149].
E2 enzymes
As a kind of an intermediate step, UBA-1 transfers the activated Ub molecule to an ubiquitin-conjugating enzyme (E2 ligases or UBCs). In C. elegans there are 22 proteins that act as E2 enzymes which are numbered as UBC-1–3, UBC-6–9 and UBC-12–26, but their numbering is not corresponding to the UBCs of S. cerevisiae and humans [71,104,31,146,76]. UBCs are attached to the C-terminal glycine 76 of the Ub molecule via a thioester bond and each of these can activate a special set of E3 enzymes in order to produce a lot of different E2–E3 combinations for each target protein. Table 3 summarizes the UBCs found in C. elegans.
Table 3.
Ubiquitin-conjugating enzymes (E2), UBCs, their orthologues in humans and the observed phenotype in C. elegans upon RNAi treatment or in mutants.
|
Gene orthologous grouping |
Phenotype following RNAi treatment or in mutant strains | References | |
|---|---|---|---|
| C. elegans | H. sapiens | ||
| ubc-1 | UBE2A;UBE2B | WT | [93,71,76] |
| ubc-2/let-70 | UBE2D1/UBCH5A; UBE2D2/UBCH5B; UBE2D3/UBCH5C | Embryonic and larval lethality with defects in sarcomere assembly | [192,71]; Stevens and Candido, 1998; [61,15] |
| ubc-3 | CDC34;FLJ20419 | WT | [71,76] |
| ubc-6 | NCUBE1 | WT | [71,85] |
| ubc-7 | UBE2G1 | WT | [104,62,67] |
| ubc-8 | UBE2H | WT | [80] |
| ubc-9 | UBE2I | Deficiencies in embryogenesis, larval development, vulval development, posterior morphogenesis and DNA damage response | [9,71]; Leight and Kornfeld, 2002 |
| ubc-12 | UBE2M | Deficiencies in embryogenesis and terminal hypodermal differentiation, embryonic arrest, vulval eversion at L4 stage, defective alae | [71] |
| ubc-13 | UBE2N; BAA93711 | Embryonic lethality and reduced brood size | [71,141,142,87] |
| ubc-14 | UBE2G2 | Embryonic lethality at a stage after gastrulation but prior to muscle twitching, and abnormal larval tail morphology | [71] |
| ubc-15 | NCUBE1 | WT | [71,76] |
| ubc-16 | BAA91954 | WT | [71] |
| ubc-17 | BAB14320;BAB14724 | WT | [71] |
| ubc-18 | UBE2L1;UBE2L3/UBCH7;UBE2L6 | Reduced growth rate and brood size | [71,31] |
| ubc-19 | - | Unhealthy larvae | [104] |
| ubc-20 | HIP2 | Late larval arrest and reduced brood size | [71,95] |
| ubc-21 | HIP2 | WT | [71] |
| ubc-22 | UBE2L1;UBE2L3/UBCH7;UBE2L6 | WT | [80,85] |
| ubc-23 | HIP2 | WT | [80] |
| ubc-24 | - | Reduced fat content-lipid metabolism | [80,3] |
| ubc-25 | UBE2Q1; UBE2Q2 | Defective postembryonic neuromuscular function | [146] |
| ubc-26 | NCUBE1 | – | [85] |
E3 enzymes
These proteins are ubiquitin-protein ligases which act as a bridge between an Ub-loaded E2 enzyme and the protein substrate. There are mainly four classes of ubiquitin-protein ligases: HECT-domain proteins, U-box proteins, monomeric RING finger proteins and multisubunit complexes that contain a RING finger protein [123].
HECT-domain proteins
Up to now, 9 HECT-domain E3 ligases have been identified in C. elegans that however are not widely studied. In HECT-domain proteins, the Ub molecule is transferred to a conserved cysteine residue of the E3 ligase through a thiolester linkage, and then the E3 transfers the Ub to the protein substrate. Table 4 summarizes the known HECT-domain E3 ligases in C. elegans.
Table 4.
HECT-domain ubiquitin ligases in C. elegans.
| Gene name | Phenotype upon RNAi treatment | References |
|---|---|---|
| hecd-1 | WT | [23] |
| D2085.4 | Deficiencies in germline development | [48] |
| F36A2.13 | WT | [71] |
| hecw-1 | WT | [76] |
| oxi-1 | WT | [76,52,166] |
| herc-1 | WT | [76,38,166] |
| wwp-1 | Embryonic lethality | [4,166,155,34] |
| eel-1 | Low range embryonic lethality | [104,191] |
| Y92H12A.2 | WT | [38,104,76,166] |
U-box domain proteins
U-box domain proteins are encoded by four genes in C. elegans and act as E3 or E4 (ubiquitin-chain elongation factor) enzymes. The chn-1 gene encodes for a protein orthologous to the human CHIP. RNAi treatment in worms showed that CHN-1 is essential for the larval development and viability [81]. Another gene that encodes for a U-box domain protein is cyn-4 or cyp-4 or mog-6, a divergent member of the cyclophilin family orthologous to human hCyP-60. CYN-4 activity is required for the sperm to oocyte switch, robust germline proliferation and for the mitosis or meiosis decision. Loss of cyn-4 via RNAi results to morphological defects at early larval stages [121,5,125,79]. The ufd-2 gene encodes for an E4 enzyme orthologous to S. cerevisiae Ufd2p and to human UBE4B, which both catalyze multiubiquitin chain assembly. RNAi treatment of ufd-2 results in increased protein expression and feminization of the germline [152]. The last of the U-box domain genes, prp-19, encodes for a yeast PRP (splicing factor) related protein, loss of which leads to embryonic lethality [76,141,142,166,55] and sterility [48,55].
Monomeric RING finger proteins
With regard to the monomeric RING finger proteins, there have been found 15 in the C. elegans genome [85], but it is not clear if all of them act in vivo as E3 ligases. The RING finger motif consists of eight histidine or cysteine residues that bind two Zn2+ ions in a cross-like structure [30]. The monomeric RING finger proteins bind both to the E2 enzyme and the substrate protein in order to catalyze the transaction of the ubiquitin molecule to the substrate. There are two classes of monomeric RING finger proteins: the ones that share the motif H2 and those that share the motif HC [30]. RNAi treatment of the majority of these proteins present a phenotype without abnormalities (WT), while the rest are involved in embryonic and larval development, so as in sterility [85,76,104,71,166,51,111,48].
Multisubunit RING finger complexes
The final category of E3 ligases consists of the multisubunit complexes that contain a RING finger protein, and are subcategorized into 4 classes based on the participating cullin protein: CUL-1, CUL-2, CUL-3 and CUL-4. Six different cullin genes have been found in the C. elegans genome (cul-1 to cul-6; [86]).
The CUL-1-based complexes are known as SCF complexes because they consist of an adaptor which acts as a ligand between an F-box domain protein, Skp-1, a cullin, CUL-1, and an F-box domain protein which is the substrate recognition subunit and is placed between Skp-1 and the substrate protein. Among the other components of the SCF complex, there is the RBX-1 protein which bridges the CUL-1 and the Ub-bound E2 enzyme. There are more than 326 F-box domain proteins in C. elegans, in comparison with humans and S. cerevisiae and they form numerous combinations with SKRs (Skp-1-Related proteins, 21 different proteins in C. elegans) which are different in every complex, so as more proteins can get recognized for ubiquitination. The most known and well-studied SCF complexes are SCFLIN-23 [83,114,124,43], SCFSEL-10 [62,124,25,184] and SCFFSN-1 ([99,63,109]).
The CUL-2 based complexes are very similar in structure to SCFs. They consist of a CUL-2, an Skp-1 related elongin C (ELC-1), an elongin B (ELB-1) and an RBX-1 protein. ELC-1 binds to the substrate protein via a BC-box motif [78] and to ELB-1 which contains an ubiquitin-like domain [84]. During development, CUL-2 regulates diverse biological processes via the ubiquitin-mediated proteolysis, such as the initiation of meiotic anaphase II, cytokinesis and mitotic chromosome segregation, embryonic cell fate specification and sex determination. CUL-2 is expressed in diverse cell types during embryonic and larval development and is also expressed in the adult germ line [32,28,158,161].
The CUL-3-based complex does not share the exact same structure with CUL-1 and CUL-2-base complexes. A single protein, namely BTB/POZ-domain protein, acts as both the adaptor of the CUL-3 to the substrate-protein and the substrate recognition subunit. There are over 100 BTB-domain genes in C. elegans, resulting in many combinations for CUL-3-based complexes [172,41,162,185]. The cul-3 gene encodes for one of the six C. elegans cullins and in the early embryo, maternal CUL-3 is essential for degradation of MEI-1, a meiosis-specific subunit of the microtubule-severing katanin complex, via MEL-26, a substrate-specific adaptor protein that also binds MEI-1. CUL-3 activity is regulated by cycles of neddylation and deneddylation [132,185,90,180,68].
The CUL-4-based complex structure is not yet fully studied. The cul-4 gene encodes for a cullin protein whose activity is essential for negative regulation of DNA-replication licensing and thus, for maintenance of genome stability. CUL-4 functions as part of a CUL-4/DDB-1 ubiquitin ligase complex that degrades the replication licensing factor CDT-1 during S phase. It also promotes nuclear export of the replication licensing factor CDC-6 by negatively regulating the levels of the CKI-1 CDK inhibitor, in order to regulate DNA replication [19,193,83]. DDB-1 protein acts as an adaptor and substrate recognition subunit of the CUL-4-based complex [82].
CUL-5 and CUL-6 are not part of these 4 classes of E3 ligases. CUL-5 is not involved in the degradation process [186,76] and little is known about CUL-6 [114].
Apart from the multisubunit RING finger cullin-based complexes, there are multisubunit RING finger APC/C complexes. APC/C complexes consist of nine core and two assistant components in C. elegans. The two assistant components are FZY-1 and FZR-1. APC/C complexes have been shown to have an important role in the chromosome separation during meiosis I [42,148], in the anterior–posterior polarity in the one-cell embryo [134], in the regulation of cyclins during G1 phase [31] and in the regulation of the amount of GLR-1 glutamate receptors in the nerve cells of the ventral cord [73].
Deubiquitinases
Ubiquitination is a reversible post-translational modification with key roles in various signal transduction cascades and a well-documented role in determining protein stability. Deubiquitinating enzymes (DUBs) are a large group of proteases [179] that cleave ubiquitin from protein substrates and other molecules [136,137]. DUBs are also known as deubiquitinases, deubiquitinating peptidases, ubiquitin isopeptidases, deubiquitinating isopeptidases, ubiquitin proteases and ubiquitin hydrolases. DUBs have been implicated in several important pathways including cell growth and differentiation, development, neuronal disease and transcriptional regulation. DUBs also play several roles in the ubiquitin pathway. One of the best functional characteristic of DUBs is the removal of monoubiqutin and polyubiquitin chains from proteins since they can cleave the peptide or isopeptide bond between ubiquitin and the protein substrate where ubiquitin is attached. Therefore, DUBs possess an antagonistic role in the axis of ubiquitination through the reversal of the fate of the proteins [136,137]. Free polyubiquitin chains are also cleaved by DUBs to produce monoubiquitin. The chains may be produced by the E1–E2–E3 machinery in the cell, free from any substrate protein. More specifically, DUBs function has major effects on the regulation of protein half-life and quality. DUBs activities include the cleavage of ubiquitin precursors, the editing and rescuing of ubiquitin conjugates, the coupling of protein deubiquitination and proteasome-mediated degradation, the disassembly of ubiquitin oligomers and the deubiquitination and membrane protein trafficking [2].
In humans there are nearly 100 genes and in C. elegans 45 genes that encode for deubiquitinating enzymes, which can be classified into two main classes: (a) cysteine proteases and, (b) metalloproteases. Both of these classes are subcategorized; cysteine proteases comprise ubiquitin-specific proteases (USPs), Machado–Josephin domain proteases (MJDs), ubiquitin C-terminal hydrolases (UCHs), ovarian tumour proteases (OTU) and metalloproteases consist of JAMM domain proteases. USPs undergo a variety of sizes (50–300 kDa) usually produced by different N-terminal extensions which may function in substrate recognition, subcellular localization and protein–protein interactions. USPs can process ubiquitin precursors, remove ubiquitin from protein conjugates and disassemble ubiquitin chains. In contrast, UCHs are relatively small enzymes (20–30 kDa). They catalyze the removal of peptides and small molecules from the C-terminus of ubiquitin. Most UCHs cannot generate monomeric ubiquitin from protein conjugates or disassemble poly-ubiquitin chains. The JAMM isopeptidases (which bind zinc), otubains and ataxin-3/josephin have been identified as ubiquitin-specific proteases [133]. There are also enzymes that specifically hydrolyze C-terminal isopeptide bonds for ubiquitin-like proteins: SUMO-specific proteases (SENPs), NEDD8-specific proteases (COP9 signalosome) and ISG15-specific proteases (UBP43) [182]. Table 5 summarizes the deubiquitinases identified in C. elegans.
Table 5.
DUBs genes and their catalytic cores in C. elegans.
|
Deubiquitinating enzymes (DUBs) |
||||||
|---|---|---|---|---|---|---|
| Gene category | OTU | JAMM | USP | UCH | MJD | |
| Catalytic core | Cys–His–Asp | His–Asp–Zn2+ | Cys–His boxes | Cys–His boxes | Cys–Gln–His–Asp | |
| Gene name | otub-1 | csn-5 | math33 | C04E6.5 | ubh-1 | atx-3 |
| otub-2 | eif-3.H | H34C03.2 | usp-14 | ubh-2 | Y67D2.2 | |
| otub-3 | prp-8 | K02C4.3 | C34F6.9 | ubh-3 | ||
| otub-4 | eif-3.F | K08B4.5 | F07A11.4 | ubh-4 | ||
| Y50C1A.1 | F37A4.5 | usp-33 | usp-39 | |||
| rpn-11 | usp-46 | E01B7.1 | ||||
| rpn-8 | T05H10.1 | duo-2 | ||||
| csn-6 | T22F3.2 | usp-48 | ||||
| T24B8.7 | F35B3.1 | |||||
| T27A3.2 | duo-1 | |||||
| duo-3 | cyld-1 | |||||
| Y67D2.2 | F59E12.6 | |||||
| usp-3 | H12I13.2 | |||||
| cyk-3 | ||||||
| References | [117,2]; Hutchins et al., 2013; [150]; www.dude-db.org | |||||
UPS regulation
The subunits and pathways that have been shown to either up- or down-regulate the UPS function upon their modulation are discussed below. Fig. 1 summarizes these factors.
Fig. 1.
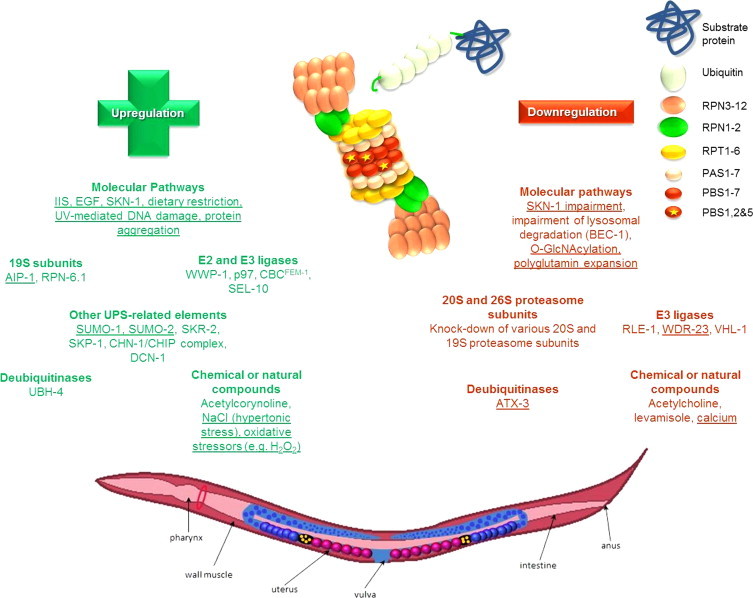
Genes, pathways and compounds that have been identified to alter the UPS function in C. elegans. Factors that have been revealed to alter the UPS function in C. elegans in terms of proteasome activities and/or assembly and/or expression. These factors include different molecular pathways and natural or chemical compounds along with alterations in the various components of the UPS system (e.g. proteasome subunits, E2 and E3 ligases, DUBs). Positive regulators are shown in green whereas negative regulators are shown in red. The regulators that have been shown to be redox sensitive are underlined.
Upregulation of proteasomal function in C. elegans
There are several chemical and genetic factors that can promote the upregulation of the proteasome function. Studies have focused on these mechanisms in order to reveal proteasome activators that can be used in putative anti-ageing or homeostasis maintaining strategies. Several genes are responsible for the regulation of the proteasome and as far as C. elegans is concerned, a number of studies have focused on the mechanisms that are involved in the degradation of muscle proteins, nervous system proteins, pathologically aggregated and oxidized proteins, so as to pathways that are involved in the worm development and sex determination. The various UPS-related factors that have been shown to result in upregulation of the proteasome function in C. elegans are summarized below.
19S proteasome subunits
AIP-1 is an arsenite-inducible protein and component of the proteasome 19S regulatory cap, which is encoded by an early-induced gene and has been found to protect against Aβ induced paralysis caused by accumulation of the toxic peptide in a C. elegans strain that serves as a model for Alzheimer's disease. AIP-1 is the homologue of the mammalian AIRAP and it was reported that its expression increases the accessibility of the protein substrate to the proteasome, assists to the adaptation of the worms under proteotoxic stress [190] and facilitates the degradation of damaged proteins in stress response conditions e.g. following arsenic treatment or fumarylacetoacetate or maleylacetoacetate treatment [33,54]. In contrast, animals maintained in aip-1 RNAi exhibit a shorter median lifespan [190]. In accordance with these results, AIRAP, the mammalian homologue of AIP-1, has been shown to exert a protective role in mammalian cells following arsenite treatment [156]. More recently, it was revealed that overexpression of AIRAP modulates the 20S core proteasome to counteract proteotoxicity induced by an environmental toxin like arsenite [160]. More specifically, association of AIRAP with the 19S cap leads to enhanced cleavage of peptide substrates thus enabling cells to cope with the arsenite-induced proteotoxicity [160].
Modulation of the proteasome activity has been recently achieved through overexpression of a 19S subunit in C. elegans. More specifically, Vilchez and colleagues [175] have revealed that glp-1 mutants, which lack their germ line, exhibit markedly increased proteasome activity in addition to decreased levels of polyubiquitinated proteins. A 19S proteasome subunit, namely rpn-6.1, has been identified to be necessary for this phenotype. Overexpression of this subunit leads to increased proteasome activities, increased survival to various stresses, and ameliorated response to proteotoxicity and lifespan extension under conditions of mild heat stress at 25 °C but not at 20 °C. Accordingly, proteasome activity in human embryonic stem cells has been shown to be correlated with increased levels of the 19S subunit, PSMD11, the orthologue of RPN-6 in humans [176].
In total, the similarities between the results in C. elegans and in mouse and human cells dictate that worms constitute a valuable model to study alterations of different constituents of the UPS.
E2 and E3 ligases
Overexpression of wwp-1, a HECT E3 ligase, has been shown to promote a 20% lifespan extension under conditions of ad libitum feeding while it is absolutely required for the extension of lifespan following dietary restriction (DR). Moreover, the specific E2 ligase that is responsible for the regulation of DR induced longevity, namely UBC-18 has been also identified [13]. The authors suggested that these two enzymes coordinate the ubiquitination of substrates that regulate DR-induced longevity. In accordance, it was recently revealed that overexpression of the human WWP1 delays cellular senescence in human fibroblasts, while its knock-down promotes irreversible premature senescence. The authors revealed a reverse correlation between the protein levels of p27(Kip1) and WWP1, thus suggesting that WWP1 promotes the ubiquitin-mediated degradation of the endogenous p27 (Kip1) [12]. Nevertheless, WWP1 E3 ligase was not shown to have any impact on the long lifespan of animals with reduced IIS pathway. In contrast, Ghazi and colleagues [43] have revealed components of the Skp1-Cul1-F-Box E3 ligase that play a pivotal role in the extended lifespan of IIS mutants by affecting DAF-16/FOXO activity.
Sex determination in C. elegans is characterized by numerous procedures including regulated degradation of specific proteins. TRA-1 plays a major role in sex determination of C. elegans. It has been found that p97 (CDC-48 in C. elegans), an ubiquitin-selective AAA chaperone, interacts with CUL-2/NPL-4 complex in order to upregulate the proteasome-mediated degradation of TRA-1 and thus to result to sex determination [145]. CBCFEM-1 is also a complex that targets TRA-1 proteasome-dependent degradation and has the same effect on proteasome-mediated degradation as p97 [161]. Previous studies have also revealed that SEL-10, an F-Box protein which regulates LIN-12 Notch signaling and promotes female development, increases the levels of proteasome-mediated degradation of FEM-1 and FEM-3, proteins which are responsible for the primary sex-determining signal [74].
Deubiquitinases
It was firstly revealed in yeast that upon ubiquitin stress (when ubiquitin levels are low), the stress is sensed thus leading to the induction of the proteasome-associated DUB Ubp6. As a result of this induction, more proteasomes are loaded with Ubp6, thereby altering proteasome function and consequently maintaining the ubiquitin pool [53]. Lately, the role of specific DUBs in the modulation of the UPS and the downstream effects in stress/proteotoxicity resistance and longevity has been also examined in C. elegans. UBH-4 is a proteasome-associated DUB in C. elegans required for protein homeostasis maintenance. RNAi of ubh-4 positively regulates the proteasome activity without however affecting its abundance. Moreover, UBH-4 was shown to slightly affect lifespan and brood size while ubh-4 levels were found to differ in wt and daf-2 mutants [106]. Most importantly, it was shown that loss of uchl5, the human orthologue of ubh-4, also increases UPS activity and the degradation of proteotoxic proteins in mammalian cells [106], thus further verifying the value of C. elegans as a model organism to evaluate the consequences of UPS modulation.
Other UPS-related elements
Ubiquitin-mediated proteasome function involves many proteins, enzymes and large complexes. SUMOs are small ubiquitin-related modifiers, with SMO-1 being the orthologue in C. elegans. Recent studies have shown that in transgenic animals that overexpress human SUMO-1 and SUMO-2 there is an upregulation of a cluster of genes encoding components of the UPS, such as ubc-9, ulp-5, smo-1 and rfp-1 [143]. Moreover, SKR-1 and SKR-2 are SKP-1 mammalian homologues that are parts of the SCF complex which mediates ubiquitination. These two proteins have been found to act normally as upregulators of the proteasomal degradation of ZYG-1 protein which is responsible for cell division regulation [124]. Another complex that plays a key role in substrate ubiquitination is the CHN-1/CHIP, which acts as an E3/E4 complex with chaperone-like function and is expressed in both the nervous system and the musculature. CHN-1/CHIP has been shown to target substrates for ubiquitination and proteasome-mediated degradation [118]. Furthermore, DCN-1, a protein with an UBA-like ubiquitin-binding domain required for cullin neddylation, has been also found to upregulate the 26S proteasome activity [90].
Various redox conditions and molecular pathways
IIS and EGF signaling pathway are two main pathways that play a key role in C. elegans growth and differentiation. DAF-2 is the main receptor of the IIS pathway in C. elegans that has been shown to upregulate the proteasome activities [163]. More specifically, using mutants that do not express the functional DAF-2 receptor, Stout and colleagues [163] have revealed that in daf-2 mutants where daf-16 expression is increased, worms exhibit a lower proteasomal activity. Moreover, they have shown that in these mutants the whole mRNA transcription and translation levels were reduced, thus leading to lower levels of misfolded proteins and indicating that the normal function of DAF-2 receptor confers enhancement of proteasomal activity. Consistent with these findings Bowerman and colleagues [10] showed that RLE-1, a transposon-tagged allele, regulates the longevity of C. elegans by promoting DAF-16 (transcription factor of the FOXO family that is the downstream regulator of the IIS pathway in C. elegans) polyubiquitination and proteasome-dependent degradation through its direct binding onto DAF-16. Recent studies have also revealed the positive role of EGF signaling via Ras–MAPK pathway on the UPS activity which not only increases the proteasomal function but also regulates the timing of the UPS activation through SKR-5, a UPS component, and exerts regulation of the organismal lifespan [100,101]. Finally, Vilchez and colleagues [175] have suggested that in glp-1 mutants chymotrypsin-like proteasome activity is increased through DAF-16 activation.
SKN-1 pathway is another significant pathway in C. elegans that has been shown to regulate the proteasome genes depending on the redox conditions. SKN-1 transcription factor is the orthologue of the mammalian Nrf2 that is responsible for the response to oxidative stress as well as for detoxification and defense against free radicals and toxic small molecules. SKN-1 pathway has been also linked to longevity [6,7,171] and to oxidative stress resistance [75] in worms while it has been shown that it responds to proteasome perturbations [19,75,119,122]. Upon proteasome perturbation either following RNAi of proteasome subunits or proteasome inhibition, SKN-1 is activated to produce a selective oxidative-stress response that includes the upregulation of various proteasome genes. Oxidative stress, caused by H2O2 pre-treatment of worms, was also found to increase 20S proteasome activity and simultaneously the expression levels of pas-7 subunit through SKN-1 pathway, similarly to the effect of Nrf2 in mammalian cell cultures [131]. In mature oocytes, there is a significant elevation of Reactive Oxygen Species (ROS) that cause cell cycle arrest and results in an increase in the oxidatively damaged proteins. Goudeau and colleagues [45] showed that when the yolk is endocytosized, proteasome-mediated proteolysis is activated and lowers the levels of oxidatively damaged proteins.
Treatment with ultraviolet radiation that causes DNA damage in C. elegans cells has been shown to activate the innate immune system of the worms and subsequently to positively modulate the UPS to confer stress resistance [29].
Dietary restriction in worms through feeding with a low cell density OP50 culture was also shown to increase the protein levels of several α- and β-type subunits, components of the 20S core proteasome, along with the increased expression of various subunits of the 19S regulatory complex [27].
Protein aggregation has a specific effect on proteasome activation. Vartiainen and colleagues [173] used transgenic worms that overexpress A53T human synuclein, a protein responsible for the formation of cellular inclusions in Parkinson Disease, Lewy Body Dementia and Multiple System Atrophy. Increased RNA expression levels of key ubiquitin-proteasome-relevant genes, such as pdr-1, ubc-7, pas-5, pbs-4, rpt-2 and psmd9 were detected in these transgenic animals.
Chemical or natural compounds
There are several compounds that have been shown to act as proteasome activators in cell culture [20]. Nevertheless, up to now very few compounds have been examined as proteasome activators in C. elegans. Fu and colleagues [39] recently showed that acetylcorynoline, the major alkaloid component derived from Corydalis bungeana, a herb that is used in traditional Chinese medicine, increases the expression levels of rpn-5, thus leading to an enhanced activity of the proteasome in C. elegans. Under hypertonic stress, caused by NaCl treatment, proteasome degradation is increased in order to protect from the accumulation of damaged proteins induced by the stress [11].
Downregulation of proteasomal function in C. elegans
Several genes and mechanisms have been revealed to negatively regulate the proteasome function in order to protect the cell from an un-programmed or un-needed proteolysis. These components are summarized below.
20S and 19S proteasome subunits
RNAi of various proteasome subunits has been shown to induce proteasome dysfunction that in turn activates the SKN-1-mediated stress response [75] but also the heat shock response [50]. RNAi of proteasome subunits has been shown to confer a substantial lifespan shortening in WT animals but also in various long-lived mutants like the IIS mutants and the glp-1 mutants ([43,163]). Additionally, in glp-1 mutants, knock-down of a 19S proteasome subunit, namely rpn-6.1, has been shown to markedly decrease the proteasome activity [175].
E3 ligases
RLE-1 is an E3 ligase that was identified to regulate C. elegans ageing through polyubiquitination of DAF-16. Elimination of RLE-1 decreases the levels of DAF-16 polyubiquitination thus promoting increased DAF-16 transcriptional activation that ultimately leads to increased lifespan ([97]). WDR-23, a WD40 Repeat Protein, is another protein that interacts with the CUL4/DDB1 ubiquitin ligase and probably functions as a substrate recognition substrate. Loss of wdr-23 has been shown to increase SKN-1 protein levels thus revealing the regulatory role of WDR-23 on SKN-1 protein abundance. As expected, loss of wdr-23 significantly enhanced stress tolerance and extended nematodes lifespan [19]. The C. elegans von Hippel-Lindau tumor suppressor homologue of VHL-1 is another cullin E3 ligase that negatively regulates the hypoxic response by promoting polyubiquitination and degradation of HIF-1 transcription factor. Consequently, loss of vhl-1 significantly increased resistance to proteotoxicity and lifespan [109].
Deubiquitinases
The Machado-Joseph disease deubiquitinase ataxin-3 (ATX-3) was the first DUB in C. elegans [139] to be shown to exert its influence on longevity and proteostasis [88]. More specifically, ATX-3 has been shown to interact with the chaperone-like AAA ATPase CDC-48 (Cdc-48 in yeast, CDC-48 in C. elegans and p97 in mammals) and thereby to hydrolyse polyubiquitinated proteins [159,181]. Kuhlbrodt and colleagues [88] showed that the deubiquitinase activity of ATX-3 is trivial for normal lifespan in nematodes. Worms deficient in both cdc-48.1 and atx-3 showed elevated substrate stabilization (thus decreased degradation of the substrate) and a 50% increased longevity that was dependent on the IIS.
Various redox and molecular pathways
SKN-1 pathway has been linked with downregulation of proteasome function. More specifically, under normal conditions, RNAi of skn-1 in worms slightly decreases the expression of certain proteasome genes such as rpn-2, pas-4 and pbs-6. These results show the influence of SKN-1 on proteasome expression that however is limited under normal conditions. In bright contrast, as discussed in UPS regulation section, SKN-1 is activated upon proteasome perturbation to cope for the lost expression of various proteasome genes [19,75,119,122]. Li and colleagues [97] have revealed that SKN-1 activity is also induced upon inhibition of translation initiation or elongation. More specifically, they showed that impairment of translation elongation results to reduced UPS activity in the intestine, thus linking protein synthesis and degradation. A previous work had already identified several genes involved in protein folding or degradation, translation elongation (TEFs) and initiation (TIFs) factors that upon RNAi, a SKN-1-dependent transcriptional response was initiated, thus promoting stress resistance and lifespan [177].
The lysosome-mediated proteolysis is the other major proteolytic pathway of the cell. BEC-1 is a protein responsible for the formation of autophagosomes. Impairement of bec-1 through RNAi in C. elegans leads to proteasome activation. Additionally, in mev-1 mutants (mev-1 is a gene that encodes for a component of complex II; the succinate dehydrogenase cytochrome b), that have been subjected to bec-1 RNAi, no further reduction in their lifespan was reported, in contrast to mev-1 mutants that have been subjected to RNAi for both bec-1 and ubq-1 (a major protein of the UPS pathway) and have been found to have a reduced lifespan. These results indicate that BEC-1 negatively regulates the proteasome activity [37].
O-GlcNAcylation is a post-translational modification that occurs in the brain tissue cells and is involved in several neurodegenerative diseases. Wang and colleagues [178] used mutant strains of C. elegans that expressed null alleles of the enzymes of O-GlcNAc cycling and concluded that O-GlcNAcylation downregulates proteasome function through a DAF-16-dependent mechanism.
Polyglutamine expansion (polyQs) plays a key role in many age-related neurodegenerative diseases. Polyglutamine expansion has been detected in proteins such ataxin-3 and huntingtin. These proteins form inclusions in the nucleus or the cytoplasm in neuronal cells and as they aggregate, they induce neuronal toxicity, thus leading to the activation of protective mechanisms. Expanded polyQs cause the impairment of the ubiquitin-mediated proteasomal function [80]. More specifically, ataxin-3 (ΑΤΧ-3) is a polyQ protein that as mentioned above is involved in Machado-Joseph disease. AΤΧ-3 binds tο ubiquitin molecules, NEDD8 (an ubiquitin-like molecule) and ubiquitinated proteins and acts as a deubiquitylating enzyme (DUB) cleaving off the chains of ubiquitin from the protein substrates. Mutant worms that overexpress atx-3 exhibit lower rates of proteasome-mediated degradation through a DAF-16-dependent mechanism [138,36].
Chemical or natural compounds
Acetylcholine (Ach) is an organic molecule that acts as a neurotransmitter through synapses and levamisole is a synthetic agonist of acetylcholine. Treatment of worms with one of these molecules causes inhibition of proteasome-mediated proteolysis [94] and negatively regulates the proteasome-mediated degradation of various muscle proteins [165]. Calcium is another molecule that has been implicated to proteasome regulation. More specifically, RNAi screening for several genes that are responsible for the regulation of muscle protein degradation in C. elegans revealed that calcium is a negative regulator of the proteasome and a positive regulator of autophagy [151].
Conclusions
Protein degradation is implicated in normal function/physiology of cells and consequently in organismal homeostasis. Defective function results in increased pathophysiology that can lead to various diseases and even death. Since life is tightly linked to redox regulation, it is not bizarre that the UPS system is equally subjected to redox-mediated regulation. It is of immense importance to identify all the possible negative or positive regulators of the UPS system since proteolysis modulation is a promising area in the battle against different conditions/diseases where up- or down-regulation of the system occurs. For example, proteolysis activation is desirable during the progression of ageing [20,21] or the progression of neurodegenerative diseases ([14]) where UPS is found downregulated or inhibited. On the opposite side, proteolysis inhibition is also needed in situations of hyperactivation of the UPS like in various cancers. The model organism C. elegans seems to be a nice tool in the identification of UPS modulators. Several UPS-related proteins that have been identified so far to alter the proteasome function in the worms have also been shown to exert a similar role in human cells (e.g. UBH-4, AIP-1 etc; see above). Moreover, given that genetic manipulation is rather easy in this organism while there are several strains that can serve as models for various diseases (e.g. neurodegenerative diseases), the value of this model to study redox regulation of the UPS is undeniable.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Ambroggio X., Rees D., Deshaies R. JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2004;2:E2. doi: 10.1371/journal.pbio.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amerik A., Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Ashrafi K., Chang F., Watts J., Fraser A., Kamath R., Ahringer J. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- 4.Astin J., O'Neil N., Kuwabara P. Nucleotide excision repair and the degradation of RNA pol II by the Caenorhabditis elegans XPA and Rsp5 orthologues, RAD-3 and WWP-1. DNA Repair. 2008;7:267–280. doi: 10.1016/j.dnarep.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Belfiore M., Pugnale P., Saudan Z., Puoti A. Roles of the C. elegans cyclophilin-like protein MOG-6 in MEP-1 binding and germline fates. Development (Cambridge, England) 2004;131:2935–2945. doi: 10.1242/dev.01154. [DOI] [PubMed] [Google Scholar]
- 6.Bishop N., Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- 7.Bishop N., Guarente L. Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nat. Rev. Genet. 2007;8:835–844. doi: 10.1038/nrg2188. [DOI] [PubMed] [Google Scholar]
- 8.Blumenthal T., Evans D., Link C., Guffanti A., Lawson D., Thierry-Mieg J. A global analysis of Caenorhabditis elegans operons. Nature. 2002;417:851–854. doi: 10.1038/nature00831. [DOI] [PubMed] [Google Scholar]
- 9.Boulton S., Gartner A., Reboul J., Vaglio P., Dyson N., Hill D. Combined functional genomic maps of the C. elegans DNA damage response. Science (New York, N.Y.) 2002;295:127–131. doi: 10.1126/science.1065986. [DOI] [PubMed] [Google Scholar]
- 10.Bowerman B., Kurz T. Degrade to create: developmental requirements for ubiquitin-mediated proteolysis during early C. elegans embryogenesis. Development (Cambridge, England) 2006;133:773–784. doi: 10.1242/dev.02276. [DOI] [PubMed] [Google Scholar]
- 11.Burkewitz K., Choe K., Lee E., Deonarine A., Strange K. Characterization of the proteostasis roles of glycerol accumulation, protein degradation and protein synthesis during osmotic stress in C. elegans. PloS One. 2012;7:e34153. doi: 10.1371/journal.pone.0034153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao X., Xue L., Han L., Ma L., Chen T., Tong T. WW domain-containing E3 ubiquitin protein ligase 1 (WWP1) delays cellular senescence by promoting p27(Kip1) degradation in human diploid fibroblasts. J. Biol. Chem. 2011;286:33447–33456. doi: 10.1074/jbc.M111.225565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrano A., Liu Z., Dillin A., Hunter T. A conserved ubiquitination pathway determines longevity in response to diet restriction. Nature. 2009;460:396–399. doi: 10.1038/nature08130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.B. Catalgol, T. Grune, Proteasome and neurodegeneratıve diseases., Progress in Molecular Biology and Translational Science, 109, 2012, 397–414. [DOI] [PubMed]
- 15.Ceron J., Rual J.-F., Chandra A., Dupuy D., Vidal M., van den Heuvel S. Large-scale RNAi screens identify novel genes that interact with the C. elegans retinoblastoma pathway as well as splicing-related components with synMuv B activity. BMC Dev. Biol. 2007;7:30. doi: 10.1186/1471-213X-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Česen M., Pegan K., Spes A., Turk B. Lysosomal pathways to cell death and their therapeutic applications. Exp. Cell Res. 2012;318:1245–1251. doi: 10.1016/j.yexcr.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Chen Q., Thorpe J., Dohmen J., Li F., Keller J. Ump1 extends yeast lifespan and enhances viability during oxidative stress: central role for the proteasome? Free Radic. Biol. Med. 2006;40:120–126. doi: 10.1016/j.freeradbiomed.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 18.Choe K., Strange K. Genome-wide RNAi screen and in vivo protein aggregation reporters identify degradation of damaged proteins as an essential hypertonic stress response. Am. J. Physiol. Cell Physiol. 2008;295:C1488–C1498. doi: 10.1152/ajpcell.00450.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choe K., Przybysz A., Strange K. The WD40 repeat protein WDR-23 functions with the CUL4/DDB1 ubiquitin ligase to regulate nuclear abundance and activity of SKN-1 in Caenorhabditis elegans. Mol. Cell. Biol. 2009;29:2704–2715. doi: 10.1128/MCB.01811-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chondrogianni N., Petropoulos I., Grimm S., Georgila K., Catalgol B., Friguet B. Protein damage, repair and proteolysis. Mol. Asp. Med. 2014;35:1–71. doi: 10.1016/j.mam.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Chondrogianni N., Gonos E. Structure and function of the ubiquitin-proteasome system: modulation of components. Prog. Mol. Biol. Trans. Sci. 2012;109:41–74. doi: 10.1016/B978-0-12-397863-9.00002-X. [DOI] [PubMed] [Google Scholar]
- 22.Creighton T., Bagley C., Cooper L., Darby N., Freedman R., Kemmink J. On the biosynthesis of bovine pancreatic trypsin inhibitor (BPTI), structure, processing, folding and disulphide bond formation of the precursor in vitro and in microsomes. J. Mol. Biol. 1993;232:1176–1196. doi: 10.1006/jmbi.1993.1470. [DOI] [PubMed] [Google Scholar]
- 23.Cui M., Chen J., Myers T., Hwang B., Sternberg P., Greenwald I. SynMuv genes redundantly inhibit lin-3/EGF expression to prevent inappropriate vulval induction in C. elegans. Dev. Cell. 2006;10:667–672. doi: 10.1016/j.devcel.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Davy A., Bello P., Thierry-Mieg N., Vaglio P., Hitti J., Doucette-Stamm L. A protein-protein interaction map of the Caenorhabditis elegans 26S proteasome. EMBO Rep. 2001;2:821–828. doi: 10.1093/embo-reports/kve184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de la Cova C., Greenwald I. SEL-10/Fbw7-dependent negative feedback regulation of LIN-45/Braf signaling in C. elegans via a conserved phosphodegron. Genes Dev. 2012;26:2524–2535. doi: 10.1101/gad.203703.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Strooper B. Proteases and proteolysis in Alzheimer disease: a multifactorial view on the disease process. Physiol. Rev. 2010;90:465–494. doi: 10.1152/physrev.00023.2009. [DOI] [PubMed] [Google Scholar]
- 27.G. Depuydt, F. Xie, V. Petyuk, N. Shanmugam, A. Smolders, I. Dhondt, et al., Reduced insulin/IGF-1 signaling and dietary restriction inhibit translation but preserve muscle mass in Caenorhabditis elegans., Mol. Cell. Proteomics: MCP, 12, 2013, 3624–3639 [DOI] [PMC free article] [PubMed]
- 28.DeRenzo C., Reese K., Seydoux G. Exclusion of germ plasm proteins from somatic lineages by cullin-dependent degradation. Nature. 2003;424:685–689. doi: 10.1038/nature01887.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ermolaeva M., Segref A., Dakhovnik A., Ou H.-L., Schneider J., Utermöhlen O. DNA damage in germ cells induces an innate immune response that triggers systemic stress resistance. Nature. 2013;501:416–420. doi: 10.1038/nature12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang S., Lorick K., Jensen J., Weissman A. RING finger ubiquitin protein ligases: implications for tumorigenesis, metastasis and for molecular targets in cancer. Semin. Cancer Biol. 2003;13:5–14. doi: 10.1016/s1044-579x(02)00095-0. [DOI] [PubMed] [Google Scholar]
- 31.Fay D., Large E., Han M., Darland M. lin-35/Rb and ubc-18, an E2 ubiquitin-conjugating enzyme, function redundantly to control pharyngeal morphogenesis in C. elegans. Development (Cambridge, England). 2003;130:3319–3330. doi: 10.1242/dev.00561. [DOI] [PubMed] [Google Scholar]
- 32.Feng H., Zhong W., Punkosdy G., Gu S., Zhou L., Seabolt E. CUL-2 is required for the G1-to-S-phase transition and mitotic chromosome condensation in Caenorhabditis elegans. Nat. Cell Biol. 1999;1:486–492. doi: 10.1038/70272. [DOI] [PubMed] [Google Scholar]
- 33.Ferguson A., Springer M., Fisher A. skn-1-Dependent and -independent regulation of aip-1 expression following metabolic stress in Caenorhabditis elegans. Mol. Cell. Biol. 2010;30:2651–2667. doi: 10.1128/MCB.01340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernandez A., Gunsalus K., Huang J., Chuang L.-S., Ying N., Liang H.-L. New genes with roles in the C. elegans embryo revealed using RNAi of ovary-enriched ORFeome clones. Genome Res. 2005;15:250–259. doi: 10.1101/gr.3194805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.K. Ferrell, C. Wilkinson, W. Dubiel, C. Gordon, Regulatory subunit interactions of the 26S proteasome, a complex problem, Trends in Biochemical Sciences, 25, 2000, 83–8 [DOI] [PubMed]
- 36.Ferro A., Carvalho A., Teixeira-Castro A., Almeida C., Tomé R., Cortes L. NEDD8: a new ataxin-3 interactor. Biochim. Biophys. Acta. 2007;1773:1619–1627. doi: 10.1016/j.bbamcr.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 37.Fitzenberger E., Boll M., Wenzel U. Impairment of the proteasome is crucial for glucose-induced lifespan reduction in the mev-1 mutant of Caenorhabditis elegans. Biochim. Biophys. Acta. 2013;1832:565–573. doi: 10.1016/j.bbadis.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 38.Fraser A., Kamath R., Zipperlen P., Martinez-Campos M., Sohrmann M., Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 39.Fu R.-H., Wang Y.-C., Chen C.-S., Tsai R.-T., Liu S.-P., Chang W.-L. Acetylcorynoline attenuates dopaminergic neuron degeneration and α-synuclein aggregation in animal models of Parkinson's disease. Neuropharmacology. 2013 doi: 10.1016/j.neuropharm.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 40.Fukunaga K., Kudo T., Toh-e A., Tanaka K., Saeki Y. Dissection of the assembly pathway of the proteasome lid in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2010;396:1048–1053. doi: 10.1016/j.bbrc.2010.05.061. [DOI] [PubMed] [Google Scholar]
- 41.Furukawa M., He Y., Borchers C., Xiong Y. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat. Cell Biol. 2003;5:1001–1007. doi: 10.1038/ncb1056. [DOI] [PubMed] [Google Scholar]
- 42.Furuta T., Tuck S., Kirchner J., Koch B., Auty R., Kitagawa R. EMB-30: an APC4 homologue required for metaphase-to-anaphase transitions during meiosis and mitosis in Caenorhabditis elegans. Mol. Biol. Cell. 2000;11:1401–1419. doi: 10.1091/mbc.11.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghazi A., Henis-Korenblit S. Regulation of Caenorhabditis elegans lifespan by a proteasomal E3 ligase complex. Proc. Natl. Acad. Sci. USA. 2007;104:5947–5952. doi: 10.1073/pnas.0700638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glickman M., Adir N. The proteasome and the delicate balance between destruction and rescue. PLoS Biol. 2004;2:E13. doi: 10.1371/journal.pbio.0020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goudeau J., Aguilaniu H. Carbonylated proteins are eliminated during reproduction in C. elegans. Aging Cell. 2010;9:991–1003. doi: 10.1111/j.1474-9726.2010.00625.x. [DOI] [PubMed] [Google Scholar]
- 46.Govindan J., Cheng H., Harris J., Greenstein D. Galphao/i and Galphas signaling function in parallel with the MSP/Eph receptor to control meiotic diapause in C. elegans. Curr. Biol.: CB. 2006;16:1257–1268. doi: 10.1016/j.cub.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 47.Graham R., Jones D., Candido E. UbiA, the major polyubiquitin locus in Caenorhabditis elegans, has unusual structural features and is constitutively expressed. Mol. Cell. Biol. 1989;9:268–277. doi: 10.1128/mcb.9.1.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Green R., Kao H.-L., Audhya A., Arur S., Mayers J., Fridolfsson H. A high-resolution C. elegans essential gene network based on phenotypic profiling of a complex tissue. Cell. 2011;145:470–482. doi: 10.1016/j.cell.2011.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Groll M., Bajorek M., Köhler A., Moroder L., Rubin D., Huber R. A gated channel into the proteasome core particle. Nat. Struct. Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 50.Guisbert E., Czyz D., Richter K., McMullen P., Morimoto R. Identification of a tissue-selective heat shock response regulatory network. PLoS Genet. 2013;9:e1003466. doi: 10.1371/journal.pgen.1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gönczy P., Echeverri C., Oegema K., Coulson A., Jones S., Copley R. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- 52.Hamamichi S., Rivas R., Knight A., Cao S., Caldwell K., Caldwell G. Hypothesis-based RNAi screening identifies neuroprotective genes in a Parkinson's disease model. Proc. Natl. Acad. Sci. USA. 2008;105:728–733. doi: 10.1073/pnas.0711018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hanna J., Meides A., Zhang D., Finley D. A ubiquitin stress response induces altered proteasome composition. Cell. 2007;129:747–759. doi: 10.1016/j.cell.2007.03.042. [DOI] [PubMed] [Google Scholar]
- 54.Hassan W., Merin D., Fonte V., Link C. AIP-1 ameliorates beta-amyloid peptide toxicity in a Caenorhabditis elegans Alzheimer's disease model. Hum. Mol. Genet. 2009;18:2739–2747. doi: 10.1093/hmg/ddp209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hebeisen M., Drysdale J., Roy R. Suppressors of the cdc-25.1(gf)-associated intestinal hyperplasia reveal important maternal roles for prp-8 and a subset of splicing factors in C. elegans. RNA (New York, N.Y.) 2008;14:2618–2633. doi: 10.1261/rna.1168408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hicke L. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 57.Hicke L. A new ticket for entry into budding vesicles-ubiquitin. Cell. 2001;106:527–530. doi: 10.1016/s0092-8674(01)00485-8. [DOI] [PubMed] [Google Scholar]
- 58.Hirano Y., Kaneko T., Okamoto K., Bai M., Yashiroda H., Furuyama K. Dissecting beta-ring assembly pathway of the mammalian 20S proteasome. EMBO J. 2008;27:2204–2213. doi: 10.1038/emboj.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hochstrasser M. Lingering mysteries of ubiquitin-chain assembly. Cell. 2006;124:27–34. doi: 10.1016/j.cell.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 60.Homma K., Suzuki K., Sugawara H. The Autophagy Database: an all-inclusive information resource on autophagy that provides nourishment for research. Nucl. Acids Res. 2011;39:D986–D990. doi: 10.1093/nar/gkq995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoppe T., Cassata G., Barral J., Springer W., Hutagalung A., Epstein H. Vol. 118. 2004. Regulation of the myosin-directed chaperone UNC-45 by a novel E3/E4-multiubiquitylation complex in C. elegans; pp. 337–349. (Cell). [DOI] [PubMed] [Google Scholar]
- 62.Hubbard E., Wu G., Kitajewski J., Greenwald I. sel-10, a negative regulator of lin-12 activity in Caenorhabditis elegans, encodes a member of the CDC4 family of proteins. Genes Dev. 1997;11:3182–3193. doi: 10.1101/gad.11.23.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.W. Hung, C. Hwang, S. Gao, E. Liao, J. Chitturi, Y. Wang, et al., Attenuation of insulin signalling contributes to FSN-1-mediated regulation of synapse development., The EMBO J. 32, 2013, 1745–60. [DOI] [PMC free article] [PubMed]
- 64.Imai J., Maruya M., Yashiroda H., Yahara I., Tanaka K. The molecular chaperone Hsp90 plays a role in the assembly and maintenance of the 26S proteasome. EMBO J. 2003;22:3557–3567. doi: 10.1093/emboj/cdg349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Isono E., Saeki Y., Yokosawa H., Toh-e A. Rpn7 Is required for the structural integrity of the 26S proteasome of Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:27168–27176. doi: 10.1074/jbc.M314231200. [DOI] [PubMed] [Google Scholar]
- 66.Isono E., Saito N., Kamata N., Saeki Y., Toh-E A. Functional analysis of Rpn6p, a lid component of the 26S proteasome, using temperature-sensitive rpn6 mutants of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:6537–6547. doi: 10.1074/jbc.M409364200. [DOI] [PubMed] [Google Scholar]
- 67.Janiesch P., Kim J., Mouysset J., Barikbin R., Lochmüller H., Cassata G. The ubiquitin-selective chaperone CDC-48/p97 links myosin assembly to human myopathy. Nat. Cell Biol. 2007;9:379–390. doi: 10.1038/ncb1554. [DOI] [PubMed] [Google Scholar]
- 68.Johnson T. Caenorhabditis elegans 2007: the premier model for the study of aging. Exp. Gerontol. 2008;43:1–4. doi: 10.1016/j.exger.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Johnston S., Riddle S., Cohen R., Hill C. Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J. 1999;18:3877–3887. doi: 10.1093/emboj/18.14.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jones D., Candido E. Novel ubiquitin-like ribosomal protein fusion genes from the nematodes Caenorhabditis elegans and Caenorhabditis briggsae. J. Biol. Chem. 1993;268:19545–19551. [PubMed] [Google Scholar]
- 71.Jones D., Crowe E., Stevens T., Candido E. Functional and phylogenetic analysis of the ubiquitylation system in Caenorhabditis elegans: ubiquitin-conjugating enzymes, ubiquitin-activating enzymes, and ubiquitin-like proteins. Genome Biol. 2002;3 doi: 10.1186/gb-2001-3-1-research0002. (RESEARCH0002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jung T., Catalgol B., Grune T. The proteasomal system. Mol. Asp. Med. 2009;30:191–296. doi: 10.1016/j.mam.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 73.Juo P., Kaplan J. The anaphase-promoting complex regulates the abundance of GLR-1 glutamate receptors in the ventral nerve cord of C. elegans. Curr. Biol.: CB. 2004;14:2057–2062. doi: 10.1016/j.cub.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 74.Jäger S., Schwartz H., Horvitz H. The Caenorhabditis elegans F-box protein SEL-10 promotes female development and may target FEM-1 and FEM-3 for degradation by the proteasome. Proc. Natl. Acad. Sci. USA. 2004 doi: 10.1073/pnas.0405087101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kahn N., Rea S., Moyle S., Kell A., Johnson T. Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidative-stress response in Caenorhabditis elegans. Biochem. J. 2008;409:205–213. doi: 10.1042/BJ20070521. [DOI] [PubMed] [Google Scholar]
- 76.Kamath R., Fraser A., Dong Y., Poulin G., Durbin R., Gotta M. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 77.Kamath R., Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods (San Diego, Calif.) 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 78.Kamura T., Maenaka K., Kotoshiba S., Matsumoto M., Kohda D., Conaway R. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 2004;18:3055–3065. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kerins J., Hanazawa M., Dorsett M., Schedl T. PRP-17 and the pre-mRNA splicing pathway are preferentially required for the proliferation versus meiotic development decision and germline sex determination in Caenorhabditis elegans. Dev. Dyn.: Off. Publ. Am. Assoc. Anatom. 2010;239:1555–1572. doi: 10.1002/dvdy.22274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Khan L., Bauer P., Miyazaki H., Lindenberg K., Landwehrmeyer B., Nukina N. Expanded polyglutamines impair synaptic transmission and ubiquitin-proteasome system in Caenorhabditis elegans. J. Neurochem. 2006;98:576–587. doi: 10.1111/j.1471-4159.2006.03895.x. [DOI] [PubMed] [Google Scholar]
- 81.Khan L., Nukina N. Molecular and functional analysis of Caenorhabditis elegans CHIP, a homologue of Mammalian CHIP. FEBS Lett. 2004;565:11–18. doi: 10.1016/j.febslet.2004.03.084. [DOI] [PubMed] [Google Scholar]
- 82.Kim S.-H., Michael W. Regulated proteolysis of DNA polymerase eta during the DNA-damage response in C. elegans. Mol. Cell. 2008;32:757–766. doi: 10.1016/j.molcel.2008.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim Y., Kipreos E. The Caenorhabditis elegans replication licensing factor CDT-1 is targeted for degradation by the CUL-4/DDB-1 complex. Mol. Cell. Biol. 2007;27:1394–1406. doi: 10.1128/MCB.00736-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim W., Kaelin W. The von Hippel-Lindau tumor suppressor protein: new insights into oxygen sensing and cancer. Curr. Opin. Genet. Dev. 2003;13:55–60. doi: 10.1016/s0959-437x(02)00010-2. [DOI] [PubMed] [Google Scholar]
- 85.Kipreos E. Ubiquitin-mediated pathways in C. elegans. WormBook: Online Rev. C. Elegans Biol. 2005:1–24. doi: 10.1895/wormbook.1.36.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kipreos E., Lander L., Wing J., He W., Hedgecock E. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- 87.Kramer L., Shim J., Previtera M., Isack N., Lee M.-C., Firestein B. UEV-1 is an ubiquitin-conjugating enzyme variant that regulates glutamate receptor trafficking in C. elegans neurons. PloS One. 2010;5:e14291. doi: 10.1371/journal.pone.0014291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kuhlbrodt K., Janiesch P., Kevei É., Segref A., Barikbin R., Hoppe T. The Machado-Joseph disease deubiquitylase ATX-3 couples longevity and proteostasis. Nat. Cell Biol. 2011;13:273–281. doi: 10.1038/ncb2200. [DOI] [PubMed] [Google Scholar]
- 89.Kulkarni M., Smith H. E1 ubiquitin-activating enzyme UBA-1 plays multiple roles throughout C. elegans development. PLoS Genet. 2008;4:e1000131. doi: 10.1371/journal.pgen.1000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kurz T., Ozlü N., Rudolf F., O'Rourke S., Luke B., Hofmann K. The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature. 2005;435:1257–1261. doi: 10.1038/nature03662. [DOI] [PubMed] [Google Scholar]
- 91.Lamitina T., Huang C., Strange K. Genome-wide RNAi screening identifies protein damage as a regulator of osmoprotective gene expression. Proc. Natl. Acad. Sci. USA. 2006;103:12173–12178. doi: 10.1073/pnas.0602987103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lamitina T. Functional genomic approaches in C. elegans. Methods Mol. Biol. (Clifton, N.J.) 2006;351:127–138. doi: 10.1385/1-59745-151-7:127. [DOI] [PubMed] [Google Scholar]
- 93.Leggett D., Candido P. Biochemical characterization of Caenorhabditis elegans UBC-1: self-association and auto-ubiquitination of a RAD6-like ubiquitin-conjugating enzyme in vitro. Biochem. J. 1997;327(Pt 2):357–361. doi: 10.1042/bj3270357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lehmann S., Shephard F., Jacobson L., Szewczyk N. Integrated control of protein degradation in C. elegans muscle. Worm. 2012;1:141–150. doi: 10.4161/worm.20465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lehner B., Tischler J., Fraser A. RNAi screens in Caenorhabditis elegans in a 96-well liquid format and their application to the systematic identification of genetic interactions. Nat. Protocols. 2006;1:1617–1620. doi: 10.1038/nprot.2006.245. [DOI] [PubMed] [Google Scholar]
- 96.Levine B., Mizushima N., Virgin H. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.W. Li, B. Gao, S. Lee, K. Bennett, D. Fang, RLE-1, an E3 Ubiquitin Ligase, Regulates C. elegans Aging by Catalyzing DAF-16 Polyubiquitination, Developmental Cell 12, 2007, 235-246. [98] [DOI] [PubMed]
- 98.Li X., Matilainen O., Jin C., Glover-Cutter K., Holmberg C., Blackwell T. Specific SKN-1/Nrf stress responses to perturbations in translation elongation and proteasome activity. PLoS Genet. 2011;7:e1002119. doi: 10.1371/journal.pgen.1002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liao E., Hung W., Abrams B., Zhen M. An SCF-like ubiquitin ligase complex that controls presynaptic differentiation. Nature. 2004;430:345–350. doi: 10.1038/nature02647. [DOI] [PubMed] [Google Scholar]
- 100.Liu G., Rogers J., Murphy C., Rongo C. EGF signalling activates the ubiquitin proteasome system to modulate C. elegans lifespan. EMBO J. 2011;30:2990–3003. doi: 10.1038/emboj.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu G. In vivo study of the ubiquitin-proteasome system in C. elegans. Genetics. 2011 [Google Scholar]
- 102.Lodish H, Berk A, Matsudaira P, Kaiser CA, Krieger M, Scott MP, Zipursky SL, Darnell J, Molecular cell biology (5th ed.), 2004, 66–72
- 103.Löwe J., Stock D., Jap B., Zwickl P., Baumeister W., Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science (New York, N.Y.) 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 104.Maeda I., Kohara Y., Yamamoto M., Sugimoto A. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr. Biol.: CB. 2001;11:171–176. doi: 10.1016/s0960-9822(01)00052-5. [DOI] [PubMed] [Google Scholar]
- 105.Massey A., Kaushik S., Cuervo A. Lysosomal chat maintains the balance. Autophagy. 2006;2:325–327. doi: 10.4161/auto.3090. [DOI] [PubMed] [Google Scholar]
- 106.Matilainen O., Arpalahti L., Rantanen V., Hautaniemi S., Holmberg C. Insulin/IGF-1 signaling regulates proteasome activity through the deubiquitinating enzyme UBH-4. Cell Rep. 2013;3:1980–1995. doi: 10.1016/j.celrep.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 107.McKay S., Johnsen R., Khattra J., Asano J., Baillie D., Chan S. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harbor Symp. Quant. Biol. 2003;68:159–169. doi: 10.1101/sqb.2003.68.159. [DOI] [PubMed] [Google Scholar]
- 108.McKay R., McKay J., Avery L., Graff J. C elegans: a model for exploring the genetics of fat storage. Dev. Cell. 2003;4:131–142. doi: 10.1016/s1534-5807(02)00411-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mehta R., Steinkraus K., Sutphin G., Ramos F., Shamieh L., Huh A. Proteasomal regulation of the hypoxic response modulates aging in C. elegans. Science (New York, N.Y.) 2009;324:1196–1198. doi: 10.1126/science.1173507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mizushima N., Ohsumi Y., Yoshimori T. Autophagosome formation in mammalian cells. Cell Struct. Funct. 2002;27:421–429. doi: 10.1247/csf.27.421. [DOI] [PubMed] [Google Scholar]
- 111.Moore R., Boyd L. Analysis of RING finger genes required for embryogenesis in C. elegans. Genesis (New York, N.Y.: 2000. 2004;38:1–12. doi: 10.1002/gene.10243. [DOI] [PubMed] [Google Scholar]
- 112.Mortimore G., Pösö A. Amino acid control of intracellular protein degradation. Methods Enzymol. 1988;166:461–476. doi: 10.1016/s0076-6879(88)66060-5. [DOI] [PubMed] [Google Scholar]
- 113.Nakamura Y., Umehara T., Tanaka A., Horikoshi M., Padmanabhan B., Yokoyama S. Structural basis for the recognition between the regulatory particles Nas6 and Rpt3 of the yeast 26S proteasome. Biochem. Biophys. Res. Commun. 2007;359:503–509. doi: 10.1016/j.bbrc.2007.05.138. [DOI] [PubMed] [Google Scholar]
- 114.Nayak S., Santiago F., Jin H., Lin D., Schedl T., Kipreos E. The Caenorhabditis elegans Skp1-related gene family: diverse functions in cell proliferation, morphogenesis, and meiosis. Curr. Biol.: CB. 2002;12:277–287. doi: 10.1016/s0960-9822(02)00682-6. [DOI] [PubMed] [Google Scholar]
- 115.Neri C. Value of Invertebrate Genetics and Biology to Develop Neuroprotective and Preventive Medicine in Huntington’s Disease, Neurobiology of Huntington's Disease: Applications to Drug Discovery, CRC Press, 2011,Chapter 6. [PubMed]
- 116.Nickell S., Beck F., Scheres S., Korinek A., Förster F., Lasker K. Insights into the molecular architecture of the 26S proteasome. Proc. Natl. Acad. Sci. USA. 2009;106:11943–11947. doi: 10.1073/pnas.0905081106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nijman S., Luna-Vargas M., Velds A., Brummelkamp T., Dirac A., Sixma T. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 118.Nyamsuren O., Faggionato D., Loch W., Schulze E., Baumeister R. A mutation in CHN-1/CHIP suppresses muscle degeneration in Caenorhabditis elegans. Dev. Biol. 2007;312:193–202. doi: 10.1016/j.ydbio.2007.09.033. [DOI] [PubMed] [Google Scholar]
- 119.Oliveira R., Porter Abate J., Dilks K., Landis J., Ashraf J., Murphy C. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell. 2009;8:524–541. doi: 10.1111/j.1474-9726.2009.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ozkaynak E., Finley D., Solomon M., Varshavsky A. The yeast ubiquitin genes: a family of natural gene fusions. EMBO J. 1987;6:1429–1439. doi: 10.1002/j.1460-2075.1987.tb02384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Page A., Winter A. Enzymes involved in the biogenesis of the nematode cuticle. Adv. Parasitol. 2003;53:85–148. doi: 10.1016/s0065-308x(03)53003-2. [DOI] [PubMed] [Google Scholar]
- 122.Park S.-K., Tedesco P., Johnson T. Oxidative stress and longevity in Caenorhabditis elegans as mediated by SKN-1. Aging Cell. 2009;8:258–269. doi: 10.1111/j.1474-9726.2009.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Passmore L., Barford D. Getting into position: the catalytic mechanisms of protein ubiquitylation. Biochem. J. 2004;379:513–525. doi: 10.1042/BJ20040198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Peel N., Dougherty M., Goeres J., Liu Y., O'Connell K. The C. elegans F-box proteins LIN-23 and SEL-10 antagonize centrosome duplication by regulating ZYG-1 levels. J. Cell Sci. 2012;125:3535–3544. doi: 10.1242/jcs.097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pemberton T., Kay J. Identification and comparative analysis of the peptidyl-prolyl cis/trans isomerase repertoires of H. sapiens, D. melanogaster, C. elegans, S. cerevisiae and Sz. pombe. Comp. Funct. Genomics. 2005;6:277–300. doi: 10.1002/cfg.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Piano F., Schetter A., Mangone M., Stein L., Kemphues K. RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr. Biol.: CB. 2000;10:1619–1622. doi: 10.1016/s0960-9822(00)00869-1. [DOI] [PubMed] [Google Scholar]
- 127.Pickart C., Fushman D. Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 128.Pickart C. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 129.Pickart C., Eddins M. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 130.Pickart C. Ubiquitin enters the new millennium. Mol. Cell. 2001;8:499–504. doi: 10.1016/s1097-2765(01)00347-1. [DOI] [PubMed] [Google Scholar]
- 131.Pickering A., Staab T., Tower J., Sieburth D., Davies K. A conserved role for the 20S proteasome and Nrf2 transcription factor in oxidative stress adaptation in mammals, Caenorhabditis elegans and Drosophila melanogaster. J. Exp.Biol. 2013;216:543–553. doi: 10.1242/jeb.074757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pintard L., Kurz T., Glaser S., Willis J., Peter M., Bowerman B. Neddylation and deneddylation of CUL-3 is required to target MEI-1/Katanin for degradation at the meiosis-to-mitosis transition in C. elegans. Curr. Biol.: CB. 2003;13:911–921. doi: 10.1016/s0960-9822(03)00336-1. [DOI] [PubMed] [Google Scholar]
- 133.Quesada V., Díaz-Perales A., Gutiérrez-Fernández A., Garabaya C., Cal S., López-Otín C. Cloning and enzymatic analysis of 22 novel human ubiquitin-specific proteases. Biochem. Biophys. Res. Commun. 2004;314:54–62. doi: 10.1016/j.bbrc.2003.12.050. [DOI] [PubMed] [Google Scholar]
- 134.Rappleye C., Tagawa A., Lyczak R., Bowerman B., Aroian R. The anaphase-promoting complex and separin are required for embryonic anterior–posterior axis formation. Dev. Cell. 2002;2:195–206. doi: 10.1016/s1534-5807(02)00114-4. [DOI] [PubMed] [Google Scholar]
- 135.Ravid T., Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell. Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Reyes-Turcu F., Wilkinson K. Polyubiquitin binding and disassembly by deubiquitinating enzymes. Chem. Rev. 2009;109:1495–1508. doi: 10.1021/cr800470j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Reyes-Turcu F., Ventii K., Wilkinson K. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rodrigues A., Neves-Carvalho A., Teixeira-Castro A., Rokka A., Corthals G., Logarinho E. Absence of ataxin-3 leads to enhanced stress response in C. elegans. PloS One. 2011;6:e18512. doi: 10.1371/journal.pone.0018512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rodrigues A.-J., Coppola G., Santos C., Costa M., Ailion M., Sequeiros J. Functional genomics and biochemical characterization of the C. elegans orthologue of the Machado-Joseph disease protein ataxin-3. FASEB J.: Off. Publ. Fed. Am. Soc. Exp. Biol. 2007;21:1126–1136. doi: 10.1096/fj.06-7002com. [DOI] [PubMed] [Google Scholar]
- 140.Roelofs J., Park S., Haas W., Tian G., McAllister F., Huo Y. Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature. 2009;459:861–865. doi: 10.1038/nature08063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rual J.-F., Ceron J., Koreth J., Hao T., Nicot A.-S., Hirozane-Kishikawa T. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rual J.-F., Hill D., Vidal M. ORFeome projects: gateway between genomics and omics. Curr. Opin. Chem. Biol. 2004;8:20–25. doi: 10.1016/j.cbpa.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 143.Rytinki M., Lakso M., Pehkonen P., Aarnio V., Reisner K., Peräkylä M. Overexpression of SUMO perturbs the growth and development of Caenorhabditis elegans. Cell. Mol. Life Sci.: CMLS. 2011;68:3219–3232. doi: 10.1007/s00018-011-0627-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Saeki Y., Toh-E A., Kudo T., Kawamura H., Tanaka K. Multiple proteasome-interacting proteins assist the assembly of the yeast 19S regulatory particle. Cell. 2009;137:900–913. doi: 10.1016/j.cell.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 145.Sasagawa Y., Otani M., Higashitani N., Higashitani A., Sato K., Ogura T. Caenorhabditis elegans p97 controls germline-specific sex determination by controlling the TRA-1 level in a CUL-2-dependent manner. J. Cell Sci. 2009;122:3663–3672. doi: 10.1242/jcs.052415. [DOI] [PubMed] [Google Scholar]
- 146.Schulze W., Reinders A., Ward J., Lalonde S., Frommer W. Interactions between co-expressed Arabidopsis sucrose transporters in the split-ubiquitin system. BMC Biochem. 2003;4:3. doi: 10.1186/1471-2091-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sha Z., Yen H.-C.S., Scheel H., Suo J., Hofmann K., Chang E. Isolation of the Schizosaccharomyces pombe proteasome subunit Rpn7 and a structure-function study of the proteasome-COP9-initiation factor domain. J. Biol. Chem. 2007;282:32414–32423. doi: 10.1074/jbc.M706276200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shakes D., Sadler P., Schumacher J., Abdolrasulnia M., Golden A. Developmental defects observed in hypomorphic anaphase-promoting complex mutants are linked to cell cycle abnormalities. Development (Cambridge, England) 2003;130:1605–1620. doi: 10.1242/dev.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sharon M., Taverner T., Ambroggio X., Deshaies R., Robinson C. Structural organization of the 19S proteasome lid: insights from MS of intact complexes. PLoS Biol. 2006;4:e267. doi: 10.1371/journal.pbio.0040267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Shaye D., Greenwald I. OrthoList: a compendium of C. elegans genes with human orthologs. PloS One. 2011;6:e20085. doi: 10.1371/journal.pone.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shephard F., Adenle A., Jacobson L., Szewczyk N. Identification and functional clustering of genes regulating muscle protein degradation from amongst the known C. elegans muscle mutants. PloS One. 2011;6:e24686. doi: 10.1371/journal.pone.0024686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Shimada M., Kanematsu K., Tanaka K., Yokosawa H., Kawahara H. Proteasomal ubiquitin receptor RPN-10 controls sex determination in Caenorhabditis elegans. Mol. Biol. Cell. 2006;17:5356–5371. doi: 10.1091/mbc.E06-05-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Silk D. Progress report. Peptide absorption in man. Gut. 1974;15:494–501. doi: 10.1136/gut.15.6.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Simmer F., Moorman C., van der Linden A., Kuijk E., van den Berghe P., Kamath R. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1:E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Skop A., Liu H., Yates J., Meyer B., Heald R. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science (New York, N.Y.) 2004;305:61–66. doi: 10.1126/science.1097931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sok J., Calfon M., Lu J., Lichtlen P., Clark S., Ron D. Arsenite-inducible RNA-associated protein (AIRAP) protects cells from arsenite toxicity. Cell Stress Chaperones. 2001;6:6–15. doi: 10.1379/1466-1268(2001)006<0006:airapa>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Song M., Liu Y., Anderson D., Jahng W., O'Connell K. Protein phosphatase 2A-SUR-6/B55 regulates centriole duplication in C. elegans by controlling the levels of centriole assembly factors. Dev. Cell. 2011;20:563–571. doi: 10.1016/j.devcel.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sonneville R., Gönczy P. Zyg-11 and cul-2 regulate progression through meiosis II and polarity establishment in C. elegans. Development (Cambridge, England) 2004;131:3527–3543. doi: 10.1242/dev.01244. [DOI] [PubMed] [Google Scholar]
- 159.Sowa M., Bennett E., Gygi S., Harper J. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Stanhill A., Haynes C., Zhang Y., Min G., Steele M., Kalinina J. An arsenite-inducible 19S regulatory particle-associated protein adapts proteasomes to proteotoxicity. Mol. Cell. 2006;23:875–885. doi: 10.1016/j.molcel.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 161.Starostina N., Lim J., Schvarzstein M., Wells L., Spence A., Kipreos E. A CUL-2 ubiquitin ligase containing three FEM proteins degrades TRA-1 to regulate C. elegans sex determination. Dev. Cell. 2007;13:127–139. doi: 10.1016/j.devcel.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Stogios P., Downs G., Jauhal J., Nandra S., Privé G. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Stout G., Stigter E., Essers P., Mulder K., Kolkman A., Snijders D. Insulin/IGF-1-mediated longevity is marked by reduced protein metabolism. Mol. Syst. Biol. 2013;9:679. doi: 10.1038/msb.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sugiyama Y., Nishimura A., Ohno S. Symmetrically dividing cell specific division axes alteration observed in proteasome depleted C. elegans embryo. Mech. Dev. 2008;125:743–755. doi: 10.1016/j.mod.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 165.Szewczyk N., Hartman J., Barmada S. Genetic defects in acetylcholine signalling promote protein degradation in muscle cells of Caenorhabditis elegans. J. Cell Sci. 2000 doi: 10.1242/jcs.113.11.2003. [DOI] [PubMed] [Google Scholar]
- 166.Sönnichsen B., Koski L., Walsh A., Marschall P., Neumann B., Brehm M. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- 167.Takahashi M., Iwasaki H., Inoue H., Takahashi K. Reverse genetic analysis of the Caenorhabditis elegans 26S proteasome subunits by RNA interference. Biol. Chem. 2002;383:1263–1266. doi: 10.1515/BC.2002.140. [DOI] [PubMed] [Google Scholar]
- 168.Taverner T., Hernández H., Sharon M., Ruotolo B., Matak-Vinković D., Devos D. Subunit architecture of intact protein complexes from mass spectrometry and homology modeling. Acc. Chem. Res. 2008;41:617–627. doi: 10.1021/ar700218q. [DOI] [PubMed] [Google Scholar]
- 169.Thrower J., Hoffman L., Rechsteiner M., Pickart C. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tomohiro M., Mizuno A. Vol. 39. 1995. Alteration of lens sulfhydryl groups induced by oxidative stress: Raman spectroscopic study of hydrogen peroxide-treated rat lens; pp. 130–136. (Jpn. J. Ophthalmol.). [PubMed] [Google Scholar]
- 171.Tullet J., Hertweck M., An J., Baker J., Hwang J., Liu S. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.van den Heuvel S. Protein degradation: CUL-3 and BTB – partners in proteolysis. Curr. Biol.: CB. 2004;14:R59–R61. [PubMed] [Google Scholar]
- 173.Vartiainen S., Pehkonen P., Lakso M., Nass R., Wong G. Identification of gene expression changes in transgenic C. elegans overexpressing human alpha-synuclein. Neurobiol. Dis. 2006;22:477–486. doi: 10.1016/j.nbd.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 174.Verma R., Aravind L., Oania R., McDonald W., Yates J., Koonin E. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science (New York, N.Y.) 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 175.Vilchez D., Boyer L., Morantte I., Lutz M., Merkwirth C., Joyce D. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature. 2012;489:304–308. doi: 10.1038/nature11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Vilchez D., Morantte I., Liu Z., Douglas P., Merkwirth C., Rodrigues A. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012;489:263–268. doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- 177.Wang J., Robida-Stubbs S., Tullet J., Rual J.-F., Vidal M., Blackwell T. RNAi screening implicates a SKN-1-dependent transcriptional response in stress resistance and longevity deriving from translation inhibition. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wang P., Lazarus B., Forsythe M., Love D., Krause M., Hanover J. O-GlcNAc cycling mutants modulate proteotoxicity in Caenorhabditis elegans models of human neurodegenerative diseases. Proc. Natl. Acad. Sci. USA. 2012;109:17669–17674. doi: 10.1073/pnas.1205748109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wilkinson K. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J.: Off. Publ. Fed. Am. Soc. Exp. Biol. 1997;11:1245–1256. doi: 10.1096/fasebj.11.14.9409543. [DOI] [PubMed] [Google Scholar]
- 180.Wilson K., Qadota H., Mains P., Benian G. UNC-89 (obscurin) binds to MEL-26, a BTB-domain protein, and affects the function of MEI-1 (katanin) in striated muscle of Caenorhabditis elegans. Mol. Biol. Cell. 2012;23:2623–2634. doi: 10.1091/mbc.E12-01-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Winborn B., Travis S., Todi S., Scaglione K., Xu P., Williams A. The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits Lys63 linkages in mixed linkage ubiquitin chains. J. Biol. Chem. 2008;283:26436–26443. doi: 10.1074/jbc.M803692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wing S. Deubiquitinating enzymes – the importance of driving in reverse along the ubiquitin-proteasome pathway. Int. J. Biochem. Cell Biol. 2003;35:590–605. doi: 10.1016/s1357-2725(02)00392-8. [DOI] [PubMed] [Google Scholar]
- 183.Wood W. Determination of pattern and fate in early embryos of Caenorhabditis elegans. Developmental Biology (New York, N.Y. : 1985) 1988;5:57–78. [DOI] [PubMed]
- 184.Wu G., Lyapina S., Das I., Li J., Gurney M., Pauley A. SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Mol. Cell. Biol. 2001;21:7403–7415. doi: 10.1128/MCB.21.21.7403-7415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Xu L., Wei Y., Reboul J., Vaglio P., Shin T.-H., Vidal M. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature. 2003;425:316–321. doi: 10.1038/nature01985. [DOI] [PubMed] [Google Scholar]
- 186.Yamanaka A., Yada M., Imaki H., Koga M., Ohshima Y., Nakayama K.-I. Multiple Skp1-related proteins in Caenorhabditis elegans: diverse patterns of interaction with Cullins and F-box proteins. Curr. Biol.: CB. 2002;12:267–275. doi: 10.1016/s0960-9822(02)00657-7. [DOI] [PubMed] [Google Scholar]
- 187.Yao T., Cohen R. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 188.Yashiroda H., Mizushima T., Okamoto K., Kameyama T., Hayashi H., Kishimoto T. Crystal structure of a chaperone complex that contributes to the assembly of yeast 20S proteasomes. Nat. Struct. Mol. Biol. 2008;15:228–236. doi: 10.1038/nsmb.1386. [DOI] [PubMed] [Google Scholar]
- 189.Yen H.-C.S., Espiritu C., Chang E. Rpn5 is a conserved proteasome subunit and required for proper proteasome localization and assembly. J. Biol. Chem. 2003;278:30669–30676. doi: 10.1074/jbc.M302093200. [DOI] [PubMed] [Google Scholar]
- 190.Yun C., Stanhill A., Yang Y., Zhang Y., Haynes C., Xu C.-F. Proteasomal adaptation to environmental stress links resistance to proteotoxicity with longevity in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2008;105:7094–7099. doi: 10.1073/pnas.0707025105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zahreddine H., Zhang H., Diogon M., Nagamatsu Y., Labouesse M. CRT-1/calreticulin and the E3 ligase EEL-1/HUWE1 control hemidesmosome maturation in C. elegans development. Curr. Biol.: CB. 2010;20:322–327. doi: 10.1016/j.cub.2009.12.061. [DOI] [PubMed] [Google Scholar]
- 192.Zhen M., Schein J., Baillie D., Candido E. An essential ubiquitin-conjugating enzyme with tissue and developmental specificity in th nematode Caenorhabditis elegans. EMBO J. 1996;15:3229–3237. [PMC free article] [PubMed] [Google Scholar]
- 193.Zhong W., Feng H., Santiago F., Kipreos E. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature. 2003;423:885–889. doi: 10.1038/nature01747. [DOI] [PubMed] [Google Scholar]


