Abstract
Cysteine residues, and in particular their thiolate groups, react not only with reactive oxygen species but also with electrophiles and with reactive nitrogen species. Thus, cysteine oxidation has often been linked to the toxic effects of some of these reactive molecules. However, thiol-based switches are common in protein sensors of antioxidant cascades, in both prokaryotic and eukaryotic organisms. We will describe here three redox sensors, the transcription factors OxyR, Yap1 and Pap1, which respond by disulfide bond formation to hydrogen peroxide stress, focusing specially on the differences among the three peroxide-sensing mechanisms.
Keywords: H2O2 sensor, Cys oxidation, OxyR, Pap1, Yap1, S. pombe
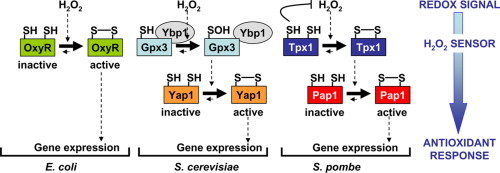
Graphical abstract
Introduction
Reactive oxygen species (ROS) are generated upon univalent reduction of oxygen from strong electron donors, such as flavoproteins or quinones, and therefore their levels rise with metabolism. ROS in general and hydrogen peroxide (H2O2) in particular harm many biological molecules. Thus, H2O2 can trigger metal loss and enzyme inactivation in iron–sulfur cluster- and iron-containing proteins, DNA damage, oxidation of cysteine (Cys) and methionine residues, as well as lipid peroxidation (for a review, see [1]). The concentration of ROS scavengers and defense activities such as DNA repair proteins is regulated in most cell types through the activation of oxidative stress pathways. The unique properties of the sulfur chemistry in Cys residues is utilized by many sensors of signaling cascades to reversibly switch from an inactive to an active state in response to H2O2.
In general, it is now well established that the reaction on most Cys residues in proteins with H2O2 is too slow and quantitatively insignificant as to be considered relevant in signaling, where the switch from an inactive to an active form is expected to be fast and to affect a major percentage of the sensor molecules in the cell. Thus, proteomic studies have reported general Cys oxidation upon H2O2 treatments, but (i) high concentrations of extracellular peroxides were applied, with several orders of magnitude over the levels known to activate signaling cascades; (ii) only a fraction of each particular protein is oxidized upon these treatments. In fact, the thioredoxin and glutaredoxin systems have been proposed to maintain thiols in their reduced state. However, genetic ablation of most cytosolic thioredoxins does not seem to trigger massive thiol oxidation [2], [3], [4], but rather induce accumulation of oxidized thioredoxin substrates such as methionine sulfoxide reductases of ribonucleotide reductases [4]. Therefore, most Cys residues in proteins, even when solvent-exposed, do not seem to easily respond to physiological H2O2 fluctuations, or to require cellular redox buffers to be kept in their thiol-reduced state under normal aerobic conditions.
However, the reactivity of some Cys residues in specific proteins can justify H2O2-driven signaling events. For instance, the structure of the active site of some H2O2 scavengers, such as peroxiredoxins and glutathione peroxidases, fits the requirements which allow a fast and complete oxidation of a thiol to sulfenic acid (SOH) and, in most cases, to a disulfide bond with another Cys of the scavenger. The rate constants of Cys oxidation in peroxiredoxins and glutathione peroxidases during peroxide scavenging, in the order of 2×103 to 4×107 M−1 s−1, fully justify that these proteins are among the first to react with H2O2. Another protein, bacterial OxyR, has been demonstrated to react efficiently with H2O2, with a rate constant of 105 M−1 s−1 [5], and to be a sensor in an antioxidant cascade. We do not pretend with this review to provide a complete repertoire of all the thiol-based sensors of H2O2 (for a recent comprehensive review see [6]). Furthermore, we are not going to describe the role of these sensors as transcriptional activators of antioxidant responses. Rather, we will focus on the eukaryotic transcription factor Pap1, which becomes activated upon moderate H2O2 stress in a peroxiredoxin-dependent manner in Schizosaccharomyces pombe, and we will compare its activation with that of other two well characterized peroxide sensors: OxyR from Escherichia coli and Yap1 from Saccharomyces cerevisiae.
H2O2 sensing in E. coli by the transcription factor OxyR
OxyR is a transcription factor of the LysR family [7], which becomes activated upon H2O2 stress to trigger the expression of peroxide scavengers, thiol redox buffers, enzymes to repair iron–sulfur centers, and to repress iron uptake genes, among others [8]. OxyR is a tetramer which binds to the promoter of target genes before and after stress sensing. It can act as an activator but also as a repressor, as specified above. Upon H2O2 treatment, the conformation of the OxyR tetramer changes as does the DNA binding contacts, so that in most cases it becomes a potent activator of RNA polymerase [9]. Under aerobic conditions, the intracellular concentration of H2O2 is estimated to be around 20 nM, and under those conditions OxyR is inactive and in a reduced conformation. Low micromolar concentrations, in the range of 5 μM, are sufficient to fully and transiently activate OxyR through Cys oxidation to a disulfide bond (Fig. 1) [5], [10]. The active/H2O2-oxidized OxyR is then reduced back to the inactive conformation by disulfide reduction by glutaredoxin 1, with the glutathione–glutathione reductase-NADPH as the electron donor system [10].
Fig. 1.
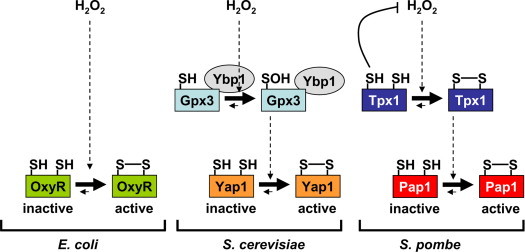
Scheme depicting the activation of OxyR, Yap1 and Pap1 upon H2O2 stress. Bacterial OxyR seems to respond directly to the oxidizing signal. The eukaryotic Yap1 and Pap1 transcription factors, however, do require the participation of upstream H2O2 sensors: the glutathione peroxidase Gpx3 and the peroxiredoxin Tpx1, respectively. See details in the text.
Even though several models have been proposed as an activating mechanism of OxyR upon agents triggering S-nitrosylation [11], [12] or S-glutathionylation [13], sensing of H2O2 by OxyR seems to occur as follows: the thiol of Cys residue 199 becomes oxidized by H2O2 to an unstable SOH, which rapidly reacts with Cys 208 to form an intramolecular disulfide bond. This oxidation triggers the conformational changes in the tetramer required to alter its interaction with RNA polymerase, enhancing its binding and/or promoting transcription initiation. No peroxiredoxin or glutathione peroxidase have been described as required to mediate the activation of OxyR by H2O2.
Oxidation of the S. cerevisiae Yap1 transcription factor by H2O2 is mediated by the glutathione peroxidase Gpx3/Orp1
Yap1 was identified in 1989 as a non-essential Jun-like transcription factor [14], which seemed to regulate the expression of antioxidant genes such as trx2 [15]. The group of Toledano described in 2000 the molecular bases of the H2O2-dependent activation of the transcription factor Yap1 in S. cerevisiae [16]. Yap1 is a b-ZIP protein of cytoplasmic localization with six Cys residues distributed in two regions, at the center and at the C-terminal domain of the polypeptide. Two of these residues, Cys 303 and Cys 598, are essential for H2O2 sensing, while two other ones (Cys 310 and Cys 629) seem to have an accessory role, since a significant decrease in total Pap1 oxidation and defective activation of the antioxidant program was reported for mutants strains carrying the yap1.C310A or yap1.C629A alleles [16]. H2O2 stress triggers the oxidation of Cys 303 and Cys 598 to a disulfide bond, with a concomitant conformational change which hinders a nuclear export signal and impairs interactions with the exportin Crm1 [16] and forces the nuclear accumulation of the transcription factor. According to transcriptome analysis, most of the genes highly induced in response to H2O2 stress are dependent on Yap1 for their induction [17]; thus, Yap1, alone or in combination with the transcription factor Skn7, controls the expression not only of classical antioxidant scavengers, but also of metabolic pathway components which regenerate reduced glutathione and NADPH [18].
After peroxide activation, a DTT-sensitive transient complex between Yap1 and an unknown protein was detected, and it was identified as the glutathione peroxidase Gpx3/Orp1 [19]. Gpx3 is supposed to scavenge peroxides using two Cys residues. However, a mutant Gpx3 containing only one of those thiols is capable of activating Yap1. Thus, oxidation of Cys 36 of Gpx3 to SOH after peroxide sensing triggers formation of an inter-molecular disulfide with Yap1 through its Cys 598. This transient intermediate is then resolved by Cys 303 of Yap1, which forms an intra-molecular disulfide with Cys 598 yielding active Yap1. The thioredoxin system was proposed to regenerate both Gpx3 (this glutathione peroxidase relies on thioredoxin and not on the glutathione pathway for recycling) and oxidized Yap1 a while after stress imposition [19]. It was soon after reported that another protein, Ybp1, was required for this redox relay, by either acting as a molecular scaffold of Gpx3 and Yap1 [20], or by avoiding Yap1 degradation [21] (Fig. 1).
In S. pombe, the Pap1 transcription factor is activated by H2O2 in a peroxiredoxin Tpx1-dependent manner
Activation of the stress-dependent S. pombe Yap1 ortholog, Pap1, was early reported to be similar but also displayed significant differences to the budding yeast system. The transcription factor Pap1 up-regulates the expression of around 40–80 genes in response to H2O2, related to both antioxidant defense and multidrug resistance [22], [23], [24], [25] (Fig. 2). Pap1 oxidizes with at least one disulfide bond [26], [27], [28] (Fig. 2), and accumulates at the nucleus only after peroxide stress imposition. This oxidation/activation only occurs at moderate doses of H2O2 [27], whereas Yap1 activation is maximal at doses of peroxide known to jeopardize cell growth [16]. Thus, Pap1 oxidation only occurs at low peroxide concentrations, whereas elevated doses strongly trigger the general stress response MAP kinase pathway driven by Sty1.
Fig. 2.
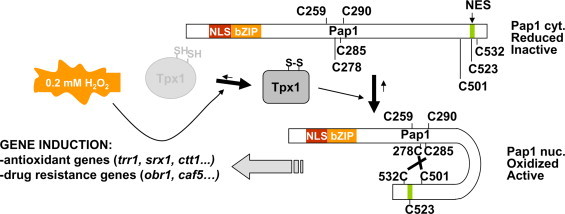
Schematic representation of Pap1 activation by H2O2. Upon moderate levels of peroxide stress (0.2 mM in the culture media), Tpx1 mediates disulfide bond formation in Pap1, which hinders the nuclear export signal (NES). Nuclear accumulation of oxidized Pap1 triggers transcription of both antioxidant and drug resistance genes. The relative positions of the seven cysteines residues (C) in Pap1 are indicated.
This intriguing property of concentration dependence in the fission yeast system was soon after unraveled thanks to the identification of Tpx1 as the upstream activator of Pap1 [29], [30]. The peroxiredoxin Tpx1 is normally scavenging peroxides during normal aerobic growth [31], [32] (Fig. 3, top panel). Only when Tpx1 accumulates on its oxidized form, with at least one disulfide within the homodimer, then the Tpx1-Pap1 redox relay occurs. At high concentrations of H2O2, Tpx1 is temporally inactivated by oxidation of its catalytic cysteine to a sulfinic acid (SO2H, Fig. 3, bottom panel). This over-oxidized form of Tpx1 can be reduced back to the SOH form by the action of the sulfiredoxin Srx1, which expression is dependent on the activation of the Sty1 pathway [29], [30]. Therefore, the use of a peroxiredoxin, which suffers substrate-dependent inactivation at high concentrations of peroxides, instead of the glutathione peroxidase Gpx3 in budding yeast explains the dose-dependent inhibition of Pap1 activation at high concentrations of H2O2.
Fig. 3.
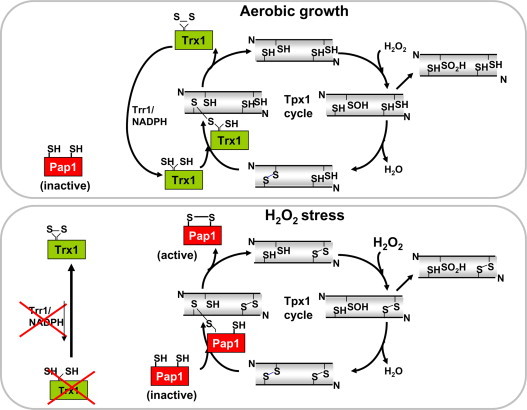
Scheme of the Tpx1 cycle. Upper panel: under aerobic growth conditions, reduced thioredoxin Trx1 breaks the intramolecular disulfide of Tpx1 by disulfide exchange leading to a transient intermediate Trx1-S-S-Tpx1 to finally release oxidized Trx1 and reduced Tpx1. Bottom panel: under mild oxidative stress, the Tpx1 cycle becomes saturated, reduced Trx1 is depleted and Pap1 acts as an alternative electron donor to Tpx1.
Another difference between the budding and fission yeast activation mechanisms is that Gpx3 has been reported to engage into a transient disulfide intermediate with Yap1 from a SOH form, whereas Tpx1 has been postulated to initiate the redox relay from a disulfide form (Fig. 3). The main support for both models comes from the analysis of mutants lacking the resolving Cys of either Gpx3 or Tpx1: while the Gpx3.C82S mutant is able to transduce the H2O2 redox signal to Yap1 [19], Tpx1.C169S is not capable of inducing Pap1 oxidation [29], [30]. Several reports indicate that SOH is a very unstable modification which rapidly reacts with neighboring thiols to form disulfides. The receptor thiol has to be in a very close proximity and in a locked conformation, so that the highly abundant glutathione (10 mM intracellular concentration on average) does not trap the SOH group first. An SOH in one protein reacting with the SH group in another protein may be restricted to very tightly associated proteins, which can maintain both Cys residues (donor and receiving) in a closed environmental pocket, so that glutathione does not interfere with the redox relay. A hypothesis is that Ybp1 can provide this spatial network to the Gpx3-to-Yap1 cascade, adding rigidity and favoring this unlikely redox transfer.
However, maybe the Tpx1-to-Pap1 activation mechanism is also based on an SOH-to-disulfide transfer, and not on a disulfide-to-disulfide isomerization. How to then explain the inability of Tpx1.C169S, lacking the resolving Cys, to engage the Pap1 pathway? As indicated in Fig. 3 (bottom panel), the Tpx1-to-Pap1 redox relay could occur from a Tpx1 dimer with one Cys48-SOH and an intra-dimer Cys169-Cys48 disulfide, in which case a Tpx1.C169S mutant would be unable to transfer the redox signal.
The rationale explaining the two different strategies which drive the S. pombe and S. cerevisiae responses to peroxides is unknown. We can only hypothesize that sharing the same protein, a peroxiredoxin, to link aerobic H2O2 scavenging with the activation of antioxidant signaling cascades may be an evolutionary step towards the sensing of milder fluctuations of peroxides, such as the ones suffered by pluricellular eukaryotes.
Summary
In conclusion, the three microbial transcription factors OxyR, Yap1 and Pap1 are able to sense H2O2 fluctuations and trigger antioxidant gene expression programs. Even though a comparative study using the same model system for the three proteins is missing, OxyR seems to be capable on its own to sense minor increments of peroxides [5], [10]. Yap1 and Pap1, on the contrary, rely on upstream activators, which in fact are H2O2 scavengers, to sense oxidative stress: the glutathione peroxidase Gpx3 and the peroxiredoxin Tpx1, respectively. The use of a peroxiredoxin instead of a glutathione peroxidase explains two particularities of the fission yeast system, that is, the activation only at moderate doses of peroxides and the disulfide-to-disulfide stress (see above). Furthermore, it links basal aerobic metabolism with the activation of antioxidant cascades: Tpx1 is the main H2O2 scavenger during respiration, and only when the Tpx1 cycle becomes saturated Pap1 is then activated. Interestingly enough, a very similar set up has been recently described for the budding yeast system: the commonly used S. cerevisiae strains W303 expresses a truncated form of the chaperone Ybp1, and in this strain background the activation of Yap1 relies on the peroxiredoxin Tsa1, and not on Gpx3; this is a very abundant protein, probably involved in basal H2O2 scavenging [33], and with both Cys required for signal transduction [33], [34].
Acknowledgments
This work was supported by the Spanish Ministry of Science and Innovation (BFU2009-06933 and BFU2012-32045), PLAN E and FEDER, by the Spanish Program Consolider-Ingenio 2010 Grant CSD 2007-0020, and by SGR2009-195 from Generalitat de Catalunya (Spain) to E.H. E. H. is recipient of an ICREA Academia Award (Generalitat de Catalunya).
References
- 1.Imlay J.A. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat. Rev. Microbiol. 2013;11:443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stewart E.J., Aslund F., Beckwith J. Disulfide bond formation in the Escherichia coli cytoplasm: an in vivo role reversal for the thioredoxins. Embo J. 1998;17:5543–5550. doi: 10.1093/emboj/17.19.5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kadokura H., Katzen F., Beckwith J. Protein disulfide bond formation in prokaryotes. Annu. Rev. Biochem. 2003;72:111–135. doi: 10.1146/annurev.biochem.72.121801.161459. [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Santamarina S., Boronat S., Calvo I.A., Rodriguez-Gabriel M., Ayte J. Is oxidized thioredoxin a major trigger for cysteine oxidation? Clues from a redox proteomics approach. Antioxid. Redox Signal. 2013;18:1549–1556. doi: 10.1089/ars.2012.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aslund F., Zheng M., Beckwith J., Storz G. Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc. Natl. Acad. Sci. USA. 1999;96:6161–6165. doi: 10.1073/pnas.96.11.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antelmann H., Helmann J.D. Thiol-based redox switches and gene regulation. Antioxid. Redox Signal. 2011;14:1049–1063. doi: 10.1089/ars.2010.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christman M.F., Storz G., Ames B.N. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc. Natl. Acad. Sci. USA. 1989;86:3484–3488. doi: 10.1073/pnas.86.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng M., Wang X., Templeton L.J., Smulski D.R., LaRossa R.A. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 2001;183:4562–4570. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toledano M.B., Kullik I., Trinh F., Baird P.T., Schneider T.D. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell. 1994;78:897–909. doi: 10.1016/s0092-8674(94)90702-1. [DOI] [PubMed] [Google Scholar]
- 10.Zheng M., Aslund F., Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 11.Hausladen A., Privalle C.T., Keng T., DeAngelo J., Stamler J.S. Nitrosative stress: activation of the transcription factor OxyR. Cell. 1996;86:719–729. doi: 10.1016/s0092-8674(00)80147-6. [DOI] [PubMed] [Google Scholar]
- 12.Seth D., Hausladen A., Wang Y.J., Stamler J.S. Endogenous protein S-Nitrosylation in E. coli: regulation by OxyR. Science. 2012;336:470–473. doi: 10.1126/science.1215643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S.O., Merchant K., Nudelman R., Beyer W.F., Jr., Keng T. OxyR: a molecular code for redox-related signaling. Cell. 2002;109:383–396. doi: 10.1016/s0092-8674(02)00723-7. [DOI] [PubMed] [Google Scholar]
- 14.Harshman K.D., Moye-Rowley W.S., Parker C.S. Transcriptional activation by the SV40 AP-1 recognition element in yeast is mediated by a factor similar to AP-1 that is distinct from GCN4. Cell. 1988;53:321–330. doi: 10.1016/0092-8674(88)90393-5. [DOI] [PubMed] [Google Scholar]
- 15.Kuge S., Jones N. YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J. 1994;13:655–664. doi: 10.1002/j.1460-2075.1994.tb06304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delaunay A., Isnard A.D., Toledano M.B. H(2)O(2) sensing through oxidation of the Yap1 transcription factor. EMBO J. 2000;19:5157–5166. doi: 10.1093/emboj/19.19.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasch A.P., Spellman P.T., Kao C.M., Carmel-Harel O., Eisen M.B. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J., Godon C., Lagniel G., Spector D., Garin J. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 1999;274:16040–16046. doi: 10.1074/jbc.274.23.16040. [DOI] [PubMed] [Google Scholar]
- 19.Delaunay A., Pflieger D., Barrault M.B., Vinh J., Toledano M.B. A thiol peroxidase is an h(2)o(2) receptor and redox-transducer in gene activation. Cell. 2002;111:471–481. doi: 10.1016/s0092-8674(02)01048-6. [DOI] [PubMed] [Google Scholar]
- 20.Veal E.A., Ross S.J., Malakasi P., Peacock E., Morgan B.A. Ybp1 is required for the hydrogen peroxide-induced oxidation of the Yap1 transcription factor. J. Biol. Chem. 2003;278:30896–30904. doi: 10.1074/jbc.M303542200. [DOI] [PubMed] [Google Scholar]
- 21.Patterson M.J., McKenzie C.G., Smith D.A., da Silva Dantas A., Sherston S. Ybp1 and Gpx3 signaling in Candida albicans govern hydrogen peroxide-induced oxidation of the Cap1 transcription factor and macrophage escape. Antioxid. Redox Signal. 2003;19:2244–2260. doi: 10.1089/ars.2013.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toda T., Shimanuki M., Yanagida M. Fission yeast genes that confer resistance to staurosporine encode an AP-1-like transcription factor and a protein kinase related to the mammalian ERK1/MAP2 and budding yeast FUS3 and KSS1 kinases. Genes Dev. 1991;5:60–73. doi: 10.1101/gad.5.1.60. [DOI] [PubMed] [Google Scholar]
- 23.Toone W.M., Kuge S., Samuels M., Morgan B.A., Toda T. Regulation of the fission yeast transcription factor Pap1 by oxidative stress: requirement for the nuclear export factor Crm1 (Exportin) and the stress-activated MAP kinase Sty1/Spc1 [published erratum appears in Genes Dev. 1998 Aug 15;12(16):2650] [see comments] Genes Dev. 1998;12:1453–1463. doi: 10.1101/gad.12.10.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen D., Wilkinson C.R., Watt S., Penkett C.J., Toone W.M. Multiple pathways differentially regulate global oxidative stress responses in fission yeast. Mol. Biol. Cell. 2008;19:308–317. doi: 10.1091/mbc.E07-08-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calvo I.A., Garcia P., Ayte J., Hidalgo E. The transcription factors Pap1 and Prr1 collaborate to activate antioxidant, but not drug tolerance, genes in response to H2O2. Nucleic Acids Res. 2012;40:4816–4824. doi: 10.1093/nar/gks141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castillo E.A., Ayte J., Chiva C., Moldon A., Carrascal M. Diethylmaleate activates the transcription factor Pap1 by covalent modification of critical cysteine residues. Mol. Microbiol. 2002;45:243–254. doi: 10.1046/j.1365-2958.2002.03020.x. [DOI] [PubMed] [Google Scholar]
- 27.Vivancos A.P., Castillo E.A., Jones N., Ayte J., Hidalgo E. Activation of the redox sensor Pap1 by hydrogen peroxide requires modulation of the intracellular oxidant concentration. Mol. Microbiol. 2004;52:1427–1435. doi: 10.1111/j.1365-2958.2004.04065.x. [DOI] [PubMed] [Google Scholar]
- 28.Calvo I.A., Ayte J., Hidalgo E. Reversible thiol oxidation in the H2O2-dependent activation of the transcription factor Pap1. J. Cell Sci. 2013;126:2279–2284. doi: 10.1242/jcs.124370. [DOI] [PubMed] [Google Scholar]
- 29.Vivancos A.P., Castillo E.A., Biteau B., Nicot C., Ayte J. A cysteine-sulfinic acid in peroxiredoxin regulates H2O2-sensing by the antioxidant Pap1 pathway. Proc. Natl. Acad. Sci. USA. 2005;102:8875–8880. doi: 10.1073/pnas.0503251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bozonet S.M., Findlay V.J., Day A.M., Cameron J., Veal E.A. Oxidation of a eukaryotic 2-Cys peroxiredoxin is a molecular switch controlling the transcriptional response to increasing levels of hydrogen peroxide. J. Biol. Chem. 2005;280:23319–23327. doi: 10.1074/jbc.M502757200. [DOI] [PubMed] [Google Scholar]
- 31.Jara M., Vivancos A.P., Calvo I.A., Moldon A., Sanso M. The peroxiredoxin Tpx1 is essential as a H2O2 scavenger during aerobic growth in fission yeast. Mol. Biol. Cell. 2007;18:2288–2295. doi: 10.1091/mbc.E06-11-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calvo I.A., Boronat S., Domenech A., Garcia-Santamarina S., Ayte J. Dissection of a redox relay: H2O2-dependent activation of the transcription factor Pap1 through the peroxidatic Tpx1-thioredoxin cycle. Cell Rep. 2013;5:1413–1424. doi: 10.1016/j.celrep.2013.11.027. [DOI] [PubMed] [Google Scholar]
- 33.Tachibana T., Okazaki S., Murayama A., Naganuma A., Nomoto A. A major peroxiredoxin-induced activation of Yap1 transcription factor is mediated by reduction-sensitive disulfide bonds and reveals a low level of transcriptional activation. J. Biol. Chem. 2009;284:4464–4472. doi: 10.1074/jbc.M807583200. [DOI] [PubMed] [Google Scholar]
- 34.Okazaki S., Naganuma A., Kuge S. Peroxiredoxin-mediated redox regulation of the nuclear localization of Yap1, a transcription factor in budding yeast. Antioxid. Redox Signal. 2005;7:327–334. doi: 10.1089/ars.2005.7.327. [DOI] [PubMed] [Google Scholar]


