Abstract
Alterations in lipid metabolism have been reported in many types of cancer. Lipids have been implicated in the regulation of proliferation, differentiation, apoptosis, inflammation, autophagy, motility and membrane homeostasis. It is required that their biosynthesis is tightly regulated to ensure homeostasis and to prevent unnecessary energy expenditure. This review focuses on the emerging understanding of the role of lipids and lipogenic pathway regulation in breast cancer, including parallels drawn from the study of metabolic disease models, and suggestions on how these findings can potentially be exploited to promote gains in HER2/neu-positive breast cancer research.
Introduction
Lipid biosynthesis is tightly coupled with regulatory signaling pathways to achieve appropriate growth and proliferation throughout development and homeostatic maintenance in the developed organism [1]. Therefore, it is not surprising that several lipogenic regulatory pathways are dysregulated in metabolic disorders and cancer. In metabolic disorders associated with diabetes, the best recognized pathologies are associated with pathways controlled by insulin, PI3K, Akt, and mTOR [2]. These molecules comprise a signaling network with extensive crosstalk as well as divergent upstream control and downstream functions. In type II diabetes, peripheral tissues involved in energy storage become insulin resistant, requiring more insulin production by the pancreas to achieve postprandial glucose clearance through the function of the insulin receptor (INSR) and downstream effects mediated by PI3K, Akt and mTOR [3, 4]. Importantly, the functions regulated by these molecules are among the most commonly dysregulated pathways in all cancers. In breast cancer, this includes gain-of-function mutation of PI3K, loss-of-function mutation of the PI3K negative regulator PTEN, and the amplification and hyperactivation of the receptor tyrosine kinase (RTK) HER2/neu, which shares several downstream effectors with INSR. While superficial differences in the pathological nature of these pathways in metabolic diseases and cancer would suggest they are unrelated, several important linkages exist: humans with features of metabolic disease are at increased risk of developing cancer and metabolic disease often presents with hyperlipidemia and hepatic steatosis, while many cancers exhibit aberrantly high lipogenesis, including HER2/neu-positive breast cancer [5].
The observation that HER2/neu overexpression in breast cancer confers a lipogenic phenotype has stimulated investigation into this unique metabolism of the transformed state. Our lab and others have found several potentially druggable vulnerabilities stemming from the reliance on certain aspects of this metabolic phenotype [6–9]. In this review, we begin by briefly introducing many of the lipids made by the cell, and how these lipids are acquired via exogenous sources or made through de novo biosynthesis. Next, we review recent literature investigating the mechanisms of lipogenic pathways in various experimental models, and how these findings could be relevant to breast cancer. This is followed by an introduction to endoplasmic reticulum stress and the unfolded protein response, with a focus on emerging data linking UPR/ER-stress to lipogenic hepatic steatosis and potential parallels with HER2/neu-positive breast cancer fat storage phenotypes. Finally, we summarize established links between lipid dysregulation in metabolic disease and breast cancer, highlight gaps in the research, and speculate how these gaps may be resolved by future research.
Lipids – overview of chemical species and cellular distribution
Lipids play a number of essential physiological roles in all cells. The term lipid collectively describes many different classes of compounds that mainly consist of fatty acids bound to a polyalcohol via an ester bond. Exceptions to this rule are the ether lipids or plasmalogens, in which the fatty acid that is to be incorporated into the lipid is reduced to a fatty alcohol thus forming an ether bond. While plasmalogens make up about 20 % of all phospholipids in humans, their specific function has not been fully determined. Plasmalogens have recently been reviewed [10]. The non-ether lipids are subdivided based on the alcohol that serves as the lipid “backbone” and the nature of the polar “head group”. Triacylglycerides (TAG) are comprised of three fatty acids bound to a glycerol “backbone”. These lipids are especially hydrophobic since they lack a polar head group. Substitution of one of the fatty acids in a TAG for a phosphate group forms the simplest phospholipid, phosphatidate (diacylglycerol-3-phosphate). The term phospholipid describes a large variety of lipids that differ in their polar head group. The different phosphoester moieties can be coupled to other alcohols, forming, for example, phosphatidylethanolamine, phosphatidylserine, phosphatidylcholine or phosphatidylinositol. In contrast, sphingolipids are synthesized from the amino alcohol sphingosine, which contains a long unsaturated hydrocarbon chain. A fatty acid is bound to the amino group of sphingosine in an amide linkage, whereas a phosphoester group serves as a linker to one of the aforementioned head groups. Sphingomyelin, a common component of cellular membranes and the most abundant sphingolipid [11], carries a choline head group. Glycolipids are another type of lipid derived from sphingosine, where, instead of phosphoryl choline, their primary hydroxyl group is bound to single carbohydrate (cerebroside) or to a complex chain of up to seven different carbohydrate moieties (gangliosides).
The distribution of these lipids inside the cell is tightly regulated. TAGs are predominantly found in intracellular lipid droplets whereas phospholipids are the major constituents of membranes. Even within the lipid bilayer that forms the membranes the distribution of individual lipids differs, forming an asymmetry. Glycolipids are always localized to the outer leaflet of the membranes, together with phosphatidylcholine and sphingomyelin, whereas phosphatidylinositols and phosphatidylserines are predominantly localized to the inner leaflet of the membrane [12].
Another important component of mammalian cell membranes is cholesterol, which can interact with the hydrophobic tails of other lipids as well as with their polar head groups through its hydroxyl group at C1. All of these lipids are synthesized from acetyl-CoA, but the exact pathway differs between the steroids and the fatty acid-based lipids. The latter are synthesized by the multifunctional enzyme complex fatty acid synthase (FASN), whereas the steroids are produced downstream of the mevalonate pathway. The plasmalogens are synthesized in the peroxisomes and utilize the same substrates as the phospholipid synthesis pathway, namely fatty acids and glycerol. The distribution, abundance and modification of these molecules is sensed by a large complement of signaling pathways; a complement that continues to grow. Thus lipids serve sensitive rheostats relaying information about diverse cellular states to the appropriate cellular machinery to support development and homeostasis. Corruptions of these signaling pathways and sensing mechanisms represent a significant factor in the pathophysiology of cancer and metabolic disease.
Fatty Acid Uptake
Fatty acids (FAs) can be either synthesized by the cell or be taken up from the “environment”. There are two different mechanisms of FA uptake: transporter-mediated uptake and passive permeation. In aqueous media fatty acids are readily absorbed into membranes, where they are protonated, flip-flop into the inner leaflet of the membrane and then desorbed into the cytoplasm. One of the most researched FA transporters is CD36 or fatty acid translocase, an integral membrane protein, which, when overexpressed, increases saturable FA uptake [13]. It has been comprehensively reviewed [14]. Two other families of potential FA transport proteins have been described, the fatty acid transport protein family (FATP, SLC27A1-6) and the acyl-CoA-synthetases (ACSL, 11 members). It is still unclear whether these proteins function as actual transporters or if they facilitate FA uptake by other means. In the case of the ACSL family it is conceivable that they function by trapping the passively diffused FAs in the cytoplasm through the formation of CoA-esters. Members of each family display a certain preference for FAs of various chain lengths and which are commonly denoted as short (<6 carbons, SCFA), medium (6–12 carbons, MCFA), long (12–21 carbons, LCFA) and very long chain fatty acids (22+ carbons, VLCFA). The ACSL family has been reviewed recently [15].
Most adult tissues take up circulating FAs instead of upregulating de novo FA synthesis. During malignant transformation cells have been shown to upregulate FA synthesis instead of increasing the uptake of exogenous FAs to meet increasing demand for biomass production. This is thought to be part of a general metabolic switch away from a non-proliferative catabolic phenotype to a proliferative anabolic phenotype. The metabolic requirements of cell proliferation are extensively reviewed elsewhere [16].
Fatty acid/lipid synthesis and regulation
Biosynthetic overview
Cytosolic acetyl-CoA serves as the starting point for fatty acid synthesis. In mammals this pool is derived from glucose, which is catabolized in the cytosol through glycolysis, yielding two molecules of pyruvate, 2 ATP and 2 NADH. Pyruvate is transported into the mitochondria, decarboxylated and condensed to oxaloacetate to form citrate. Citrate can be exported into the cytosol and cleaved by ATP-citrate lyase (ACLY) which generates cytosolic acetyl-CoA and oxaloacetate. Acetyl-CoA carboxylases (ACACA and ACACB) form malonyl-CoA in a biotin-dependent reaction, which is the rate-limiting step in fatty acid synthesis. The multifunctional enzyme fatty acid synthase (FASN) requires one molecule of acetyl-CoA as a chain starter and subsequently prolongs the chain by two carbons for every malonyl-CoA in a decarboxylation reaction followed by complete reduction of each two carbon unit, ultimately forming a saturated fatty acid chain 16 carbons in length (palmitate). Every elongation step requires 1 ATP, 1 malonyl-CoA and one NADPH. The final product can then be shuttled towards lipid synthesis to form either membrane phospholipids or neutral triacylglycerides (TAG). Free palmitate functions as an inhibitor of FASN in a classic example of product feedback inhibition and has been shown to induce oxidative stress in various cell lines [17]. Many enzymes work downstream of FASN to modify palmitate by elongation, desaturation or a combination of both to form a wide variety of FAs. Some of these elongases and desaturases are essential for cell survival and require the supply of exogenous, essential FAs as substrates, i.e. arachidonic acid.
All of these biosynthetic steps require that the FA moiety is bound to some kind of coenzyme A derivative. Leonardi et al. have written an excellent review on the biology of Coenzyme A [18]. In a state of increased lipogenesis it is conceivable that there is an increased requirement for CoA production and thus elevated pathway flux. In the case of HER2/neu positive breast cancer cells, treatment with the CoA-pathway inhibitor homopantothenate reduced the levels of stored fats and decreased cell viability. Knockdown of pantothenate kinase (pank) and coenzyme A synthase (COASY) using shRNAs also specifically decreased the viability of HER2/neu positive breast cancer cells (Baumann et al., unpublished results). Interestingly, COASY has been shown to interact with the PI3K pathway as well as with the mTOR/S6K pathway [19, 20]. The cytosolic pool of Acetyl-CoA that feeds the lipogenic phenotype is uniquely poised to serve as a rheostat for the metabolic state of the cell. Acetyl-CoA levels have been linked to histone acetylation, changes in gene expression, metabolic control and proliferation [21–23].
The importance of the fatty acid synthesis pathway in breast cancer cells has been appreciated since the discovery that FASN is induced by progestin [24]. Subsequently, FASN has been investigated as a potential target for anti-neoplastic therapy in general [8, 25]. There is evidence to suggest that in the special case of HER2/neu-positive breast cancer lipids and lipogenesis play a more important role compared to other breast cancer subtypes. HER2 has been shown to upregulate FASN transcription and increases de novo fatty acid synthesis. Both of these effects can be blocked by pharmacological inhibition of HER2/neu. Interestingly, this link between HER2/neuand FASN seems to be bi-directional. Vasquez-Martin et al. have shown that overexpresssion of FASN in MCF10A and HBL100 breast epithelial cells results in oncogenic properties, including colony formation in soft agar and upregulation and activation of endogenous HER1 (EGFR) and HER2/neu [26]. Cells that were rendered resistant to treatment with Herceptin could be resensitized by FASN inhibition [9]. In addition to the transcriptional link between HER2/neuand FASN, Jin et al. found that FASN is directly phosphorylated by HER2/neuand that this phosphorylation resulted in increased synthetic activity [27]. In agreement with earlier observations, our lab determined that HER2/neu-positive breast cancer cell lines showed increased levels of stored fats compared to other HER2/neu-normal cell lines [7]. The endproduct of fatty acid synthesis, mainly free palmitate, has been shown to be cytotoxic to cells, unless it can be shuttled towards TAG synthesis [7, 17, 28].
Regulation of lipid biosynthesis
The PPARγ binding protein (PBP) is located in the common region of HER2/neuamplification on chromosome 17q12–21 and has been shown to be frequently co-amplified along with HER2/neu [29]. Overexpression of PBP increases the transcriptional activity of PPARγ, a major regulator of cellular fat metabolism and adipogenesis [30, 31]. PPARγ itself is often upregulated in HER2/neu-positive breast cancer and pharmacological inhibition results in increased free fatty acids and apoptosis in HER2/neu-positive breast cancer cell lines [7]. While upregulation of FASN by HER2/neu has been reported to occur at the transcriptional level through the activation of the sterol-regulatory element binding protein (SREBP), mTOR-dependent translational upregulation has also been observed[32, 33]. Importantly, the PI3K/Akt/mTOR pathways are potently activated by oncogenic HER2 signaling, and abundant evidence supports a role for Akt and mTOR in the regulation of de novo lipid biosynthesis in experimental models of cancer and metabolic disease.
The lipogenic sterol response element binding protein 1 (SREBP-1) transcription factor pathway has recently been shown to be controlled by Akt/mTORC1 signaling. SREBP-1c is a global transcriptional regulator of lipid and fatty acid biosynthesis machinery that has been shown to positively regulate most genes involved in fatty acid synthesis, including acetyl-CoA carboxylase (ACC) fatty acid synthase (FASN) [34]. FOXO1 negatively regulates this pathway by repressing retinoic acid receptor alpha and liver X receptor alpha (RXRα/LXRα) mediated SREBP-1c transcription [35]. Akt inhibits FOXO transcriptional activity by mediating cytoplasmic FOXO sequestration by 14-3-3 proteins, which ultimately promotes its proteosomal degradation [36]. Therefore, Akt driven inhibition of FOXO proteins may promote SREBP1 transcription under permissive conditions, which could result in conversion of glucose to fat in many organs.
Akt has also been shown to positively regulate peroxisome proliferator activated protein gamma (PPARγ) through a similar FOXO transcriptional repression mechanism [37]. PPARγ and Akt are critical determinants of adipocyte differentiation. This is also achieved at least in part by Akt mediated repression of FOXO1 nuclear localization and transcriptional function. PPARγ transcriptional coactivator activity is repressed by FOXO1, blocking adipocyte differentiation in vitro [37]. Taken together with findings that constitutively active Akt is sufficient to induce adipocyte differentiation in vitro in the absence of hormones and Akt null mice display severe defects in adipocyte development, an Akt/FOXO1/PPARγ driven lipogenesis mechanism has solid experimental support [38, 39]. Given the abundance of TAG accumulation and high degree of Akt activity in HER2/neu-positive breast cancer cells, these pathways may contribute to the unique physiology of HER2/neu-positive breast cancer.
In addition to the Akt/FOXO1/PPARγlipogenic mechanism, Akt/mTOR signaling has also been shown to promote lipogenesis through mTOR dependent activation of SREBP1, illustrating an important mode of cooperation between Akt and mTOR in effecting metabolic programs. Under nutrient-replete conditions, SREBP1 is tethered to the inner endoplasmic reticulum membrane. Under conditions in which the cell experiences low cholesterol concentrations in the ER membrane, SREBP1 is trafficked to the Golgi apparatus, where it is proteolytically processed, ultimately resulting in localization of the n-terminal fragment to the nucleus. In the nucleus, SREBP1 activates transcription of lipogenic genes, promoting lipid and cholesterol biosynthesis [40]. Experiments in which mTOR was inhibited by rapamycin have shown that mTORC1 positively regulates this this phenomenon. Rapamycin blocks nuclear accumulation of SREBP, SREBP gene expression and SREBP target gene expression in mammalian cells, even under conditions in which Akt was constitutively activated [41, 42].
Perhaps the most direct mechanistic insight into mTOR mediated SREBP regulation was recently shown to be dependent on mTOR phosphorylation of the lipid phosphatase lipin 1. Peterson et al. established a mechanism whereby mTOR directly affects SREBP1 transcriptional activity in a rapamycin insensitive manner. Using newly available selective ATPase competitive mTOR inhibitors, lipin 1 was shown to be a rapamycin insensitive mTOR substrate. Lipin 1 negatively regulated SREBP1 nuclear localization and transcriptional activity when mTOR was inhibited, and these effects were abrogated by transfection with a constitutively active lipin 1 construct [43]. Lipin 1 is a phosphatase best characterized in catalyzing the dephosphorylation of phosphatidate in the TAG synthesis pathway in the cytoplasm. As nuclear localization and activity of lipin 1 is associated with inhibition of lipogenic phenotypes, mTOR likely regulates this pathway by modulating lipin 1 localization and activity. It is tempting to speculate that lipin 1 may transduce lipogenic repression signals when mTOR is inhibited, as inhibition of mTOR results in lipin 1 nuclear translocation and transcriptional repression of lipogenic pathways [43]. Importantly, lipin 1 expression had previously been shown to be induced by peroxisome proliferator-activated receptor gamma, coactivator 1 (PGC-1) and to subsequently cooperate with peroxisome proliferator-activated receptor alpha to suppress lipogenesis in vitro and in vivo [44]. Interestingly, cytosolic Lipin 1 phosphatidate phosphatase activity produces diacyl glycerol, which is proposed to indirectly inactivate Akt through protein kinase A activation [45]. Also of note, Lipin 1 phosphatase activity catalyzes the dephosphorylation of phosphatidate [46], which is a potent activator of mTOR activity (discussed later in this review) [47]. This suggests that Lipin 1 or the related phosphatidate phosphatases Lipin 2 or Lipin 3 could contribute to mTOR inhibition under catabolic cellular contexts. Whether this reciprocal Lipin mediated mTOR inhibition pathway exists and influences these central lipogenic pathways in breast cancer deserves further review, as deregulation of Akt, mTOR and de novo lipogenesis are hallmarks of many diseases, including breast cancer. These observations, along with activation of mTOR downstream of HER2 and the lipogenic phenotype of HER2/neu-positive breast cancer cells, suggests these mechanisms may be operative in HER2/neu-positive breast cancer cells, and should be investigated in this context.
In addition to PI3K/Akt/mTOR mediated regulation of lipogenic transcriptional programs, the enzyme responsible for the rate limiting step of FA synthesis, ACACA, has also been implicated in breast cancer pathogenesis. Notably, this occurs via cooperation with a common genomic lesion with a causal linkage to breast cancer pathogenesis. The breast cancer susceptibility gene 1 (BRCA1), mutations in which can be found in about 50 % of all breast cancer cases, binds to phosphorylated (inactive) ACACA and stabilizes this inactive state [48, 49]. Many mutations in BRCA1 can be found in the c-terminal, phosphoprotein-binding domain, which results in a hyperactivated ACACA and an upregulation of de novo fatty acid synthesis [50]. ACACA function has also been implicated breast cancer cell motility and invadopodia formation [51]. Pharmacological inhibition of ACACA induces apoptosis in breast cancer cells [37]. Both FASN and ACACA have been suggested as novel therapeutic targets for breast cancer [52, 53]. This direct repressive interaction between the BRCA1 tumor suppressor and the core fatty acid biosynthetic machinery further supports a requirement for a lipogenic phenotype in cancer cells.
The unfolded protein response and endoplasmic reticulum stress promote lipogenesis
The unfolded protein response (UPR) pathway has recently been shown to trigger increased lipogenic capacity [54, 55], and is an emerging feature of many forms of cancer [56]. The canonical UPR/ER-stress pathway is described as a cellular stress response to unfolded proteins in the lumen of the endoplasmic reticulum, wherein the protein folding, glycosylation and quality control machinery is overwhelmed by high levels of protein synthesis or defective progression through these pathways. Eukaryotes have developed three parallel pathways that act in concert to adapt to conditions that overwhelm the ER’s protein folding capacity. The first pathway acts through protein kinase-like endoplasmic reticulum kinase (PERK) to reduce global protein synthesis by phosphorylation of eIF2α. This disrupts translation initiation by blocking the eIF2B GEF activity, trapping eIF2α in a repressed GDP bound state and decreasing the transfer of Met-tRNA to the 40S ribosomal subunit [57, 58]. The second pathway acts through endoplasmic reticulum to nucleus signaling 1 (IRE1). IRE1 is activated in response to ER-stress, which leads to a non-canonical splicing mechanism wherein the mRNA transcript of X-box binding protein 1 (XBP1) is spliced to give rise to a transcriptionally active variant that upregulates many genes involved in the UPR/ER-stress including many resident ER protein folding chaperones [59]. Lastly, activating transcription factor 6 (ATF6) activity is induced by ER-stress. ATF6 is a transmembrane ER protein that is proteolytically processed in response to ER-stress, leading to its translocation to the nucleus and activation of target gene expression. As with XBP1, ATF6 target genes include several ER chaperone proteins, promoting adaptation to increased demand on the ER protein folding and quality control machinery [60]. High intensity or prolonged duration of UPR/ER-stress can lead to apoptosis through induction of the proapoptotic regulator DNA-damage-inducible transcript 3 (DDIT3/CHOP) [61]. In addition to the canonical pathways induced byER-stress, lipogenesis has recently been described as an additional output. This could represent a mechanism to satisfy increased demands on the functional lipid components of the ER.
Lipogenic UPR/ER-stress has been implicated in the etiology of fatty liver disease and hepatic steatosis, wherein excess dietary calorie or alcohol consumption result in excessive hepatic fat accumulation. This is thought to occur through exogenous fatty acid uptake, but also through endogenous de novo lipid and fatty acid biosynthesis pathways. Importantly, several studies have shown that UPR/ER-stress leads to SREBP transcriptional activation and de novo lipogenesis [62–64]. Dissection of each arm of the UPR/ER-stress pathway yielded insights into the mechanism by which SREBP is activated in this fashion. Rutkowski et al. employed a series of transgenic mouse models deficient in each of the three pathways in the UPR that are commonly initiated by ER-stress: Atf6 −/−, Ire1 −/−, and eIF2α dominant negative (eIF2α S51A) mice enabled the study of the differential physiology of each of the canonical arms of the pathway. Each model showed impaired resolution of UPR/ER-stress after tunicamycin challenge, a drug known to induce UPR/ER-stress in experimental models. All transgenic animals developed hepatic steatosis, while the wild type mouse was able to overcome the challenge and maintained normal liver physiology. In Atf6 −/− mice, this affect was mediated by impaired β-oxidation through lower C/EBP transcriptional activity. eIF2α dominant negative mice with defective UPR/ER-stress induced Ser51 phosphorylation and Ire1 −/− mice also showed hepatic lipid accumulation, although the mechanisms driving this phenotype are not as clear as the C/EBP transcriptional defects in Atf6−/− mice [64]. These data link UPR/ER-stress to impaired β-oxidation of fats in hepatic steatosis. Recent findings have also linked the mTOR pathway with UPR/ER-stress [65].
Apart from pathophysiological states, important regulators of ER-stress have been found to be essential for the homeostasis of lipid metabolism. Experiments by Sha et al. indicate that IRE1-alpha and XBP-1 are required for adipogenesis in mouse embryonic fibroblasts [66]. Lee et al. have shown that XBP-1 expression is required for normal fat metabolism in the liver of mice on a carbohydrate-rich diet. Deletion of XBP-1 resulted in severe hypocholesterinemia and hypotriglyceridemia [67]. Similarly, overexpression of GADD34, the phosphatase that acts on ser51-phosphorylated eIF2α, resulted in decreased hepatic steatosis in mice on a high fat diet [68]. XBP-1 has also been shown to regulate ER-biogenesis which correlated with increased activities of choline cytidylyltransferase and cholinephosphotransferase as well as elevated levels of phosphatidylcholine [69]. Experiments in mice have shown that the ER-stress regulator PERK is required for the development of milk-secreting mammary gland epithelial cells. PERK was found to induce lipogenesis and loss of function resulted in markedly reduced levels of FASN, ACLY and stearoyl-CoA desaturase 1 (SCD1) as well as altered lipid content of milk secretions [70]. Pharmacological induction of UPR/ER-stress in hepatoma cells resulted in lipid droplet formation and increased expression of many pro-lipogenic proteins including PPARγ, ACACA, SCD1 and LXRα. The same pro-lipogenic signature was observed when an activated form of XBP-1 was expressed [71]. As mentioned earlier in this review, LXRα is a potent inducer of SREBP1 and is suppressed by FOXO1.
Kammoun et al. studied the effects of insulin and ER-stress induced SREBP1 activation and resultant lipogenesis phenotypes in mice. They were able to block the effects of both routes to SREBP1 activation by overexpressing a single ER chaperone protein, heat shock 70 kDa protein 5 (GRP78), in the livers of obese mice. In a series of experiments, the authors showed that ER-stress and insulin mediated SREBP1 activation could be blocked by overexpressing GRP78, resulting in decreased hepatic lipid accumulation, attenuated UPR/ER-stress marker expression and improved insulin sensitivity and glucose tolerance [72].
It is important to note that acute stress can result in catabolic effects of the UPR/ER-stress pathways, resulting in effects counter to those reviewed here. These differential effects likely reflect contextual differences in the initiation, intensity, and/or duration of UPR/ER-stress, and conform with well-established observations of UPR/ER-stress resolution through tolerance and adaptation or apoptotic pathways [73].
mTORC1 negatively regulates lipolysis
Consistent with a prolipogenic metabolic program, mTOR has also been shown to negatively regulate lipolysis. Two independent labs investigated the effect of mTOR activation and inhibition on lipid storage in the 3T3-L1 adipocyte model using orthogonal approaches. Chakrabarti et al. found that hyperactivation of mTORC1, either by ectopic overexpression of constitutively active Rheb (the small GTPase that activates mTORC1), or by genetic ablation of the mTORC1 negative regulator TSC2 enhances lipogenesis through transcriptional repression of the major lipases adipocyte triacylglycerol lipase (ATGL) and hormone sensitive lipase (HSL) [74]. In reciprocal experiments, inhibition of mTORC1 activity by rapamycin or RNAi mediated knockdown of the essential mTORC1 component RAPTOR resulted in decreases in TAG stores, with concomitant upregulation of ATGL protein levels. Consistent with these findings, in vivo analysis of white and brown adipose tissues in mice treated with rapamycin showed increased ATGL levels, confirming the mechanism in a physiologically relevant model [74]. In a parallel study of mTORC1 in 3T3-L1 adipocytes, Soliman et al. showed that rapamycin blocked insulin stimulated TAG accumulation, and enhanced β-adrenergic cAMP/PKA stimulated lipolysis, further supporting a model in which mTORC1 augments TAG accumulation by inhibiting lipolysis pathways [75]. While these studies implicate mTORC1 in a lipogenic phenotype, several gaps in the mechanistic details remain, and defining the connectivity of the intermediary machinery that affects these processes will be critical to eventually exploit these pathways therapeutically. Additionally, it remains to be seen whether these pathways are operative in the lipogenic phenotype of HER2/neu-positive breast cancer cells. If HER2/neu-positive cells use these pathways to promote TAG storage, application of combination therapies including mTOR inhibition and PKA agonists may prove useful in combating this subtype of breast cancer.
Lipid Signaling in Anabolic Metabolism
Importantly, lipids are prominent signaling molecules. Several lipid species play important roles in transducing signals that direct diverse cellular physiologies through innumerable effectors. Given the enormity of this field, we focus our discussion on lipid signaling mechanisms relevant to the biology discussed above.
Phosphatidylinositide kinases and Phospholipase D – lipid signals converge on lipogenesis through activation of mTOR
Phosphoinsitides are important constituents of cellular plasma and endomembranes and play an integral role in the instigation of numerous signaling pathways. Given the extensive body of literature reviewing the regulation of phosphoinositides and phosphatidate, we briefly review recent literature elucidating how signaling through PI3K, PI5K and PLD and the lipid products formed by these enzymes impinge on Akt and mTOR, with probable links to metabolic phenotypes discussed above. For comprehensive overviews of these pathways, see recent reviews [47, 76, 77].
Among myriad other functions, the lipid second messenger phosphatidylinositol-3,4,5-trisphosphate (PIP3) instigates a singling cascade that results in activation of Akt and mTOR. Upon generation of PIP3 by activated PI3K, a subset of proteins containing PH domains which are competent to bind these lipids localize to the plasma membrane. The PH domain containing protein 3-phosphoinositide dependent protein kinase-1 (PDPK1) binds PIP3, activating its kinase activity [78]. Simultaneously, Akt isoforms are also localized to the plasma membrane by virtue the PIP3 affinity of its PH domain, and activated PDPK1 phosphorylates Akt isoforms on a key threonine residue in the kinase activation loop (Thr308) [79, 80]. Importantly, insulin and other growth factors also stimulate Akt phosphorylation on a key Akt hydrophobic motif regulatory serine (S473), resulting in full activation of Akt kinase activity. This has been shown to be regulated by the mammalian target of rapamycin kinase in the rapamycin-insensitive companion of mTOR (RICTOR) containing complex 2 (mTORC2) kinase [81]. Also notable are recent reports that mTORC2 is responsible for cotranslational phosphorylation of Akt on Thr450, which is critical to maintain stability of Akt [82]. The mechanisms by which growth factors promote these Akt directed mTORC2 activities is poorly understood, but this high degree of functional interaction between mTOR and PI3K/Akt signaling is critical to the effective transduction of insulin signals. Downstream of growth factor/PI3K/Akt/mTOR signaling, metabolic pathways affecting glycogen synthesis, lipogenesis, and protein synthesis are activated depending on the cellular and developmental context. Importantly, these molecules are activated in HER2/neu-positive breast cancer cells, and mutational activation of PI3K is among the most common genetic lesions in other forms of breast cancer [83, 84].
mTOR is activated downstream of PIP3 by an Akt dependent mechanism in a well characterized pathway involving Akt mediated phosphorylation and inhibition of the mTOR inhibitor TSC2 [85, 86]. In addition to PIP3/Akt mediated activation, mTOR requires additional inputs for maximal activation, including amino acids, permissive oxygen levels, and the additional lipid second messenger phosphatidate [47, 86, 87]. mTOR promotes global anabolic metabolism, integrating growth factor, nutrient and stress signals to regulate myriad metabolic programs, including lipogenesis. Although best studied in the context of growth factor and amino acid sensing, mTOR is also regulated by lipids. Recent work has shown mTORC1 and mTORC2 are positively regulated by phosphatidate (PA). PA binds mTOR in a manner that competes with rapamycin. Recent NMR structural analysis of the mTOR rapamycin binding domain in complex with PA provides a physical basis for this competitive binding phenomenon [88]. While PA can be generated by three distinct biochemical reactions, the most important source of PA in the activation of mTOR in cells appears to be derived by phospholipase D (PLD) [47]. Importantly, PLD derived PA was shown to regulate both mTOR complexes. Overexpression of dominant negative PLD1 or PLD2 diminished mTORC1 and mTORC2 activities as demonstrated by a potent reduction in ribosomal protein S6 kinase (p70S6K) threonine 389 phosphorylation and Akt serine 473 phosphorylation, respectively [89]. These results demonstrate that lipid signaling is necessary for mTOR signaling, and provides a mechanistic basis for the observed effects of rapamycin on both complexes.
Palmitoylation/Acylation/Prenylation
Acylation refers to the post-translational modification of a protein with a carbon chain derived from a fatty acid. While there are many different types of acylation reactions, classified by the chain length of the fatty acid, myristoylation (C14) and palmitoylation (C16) are among the most common. Myristate is usually linked to the N-terminal glycine of a protein in an amide linkage, whereas palmitate is often connected to cysteine residue via a thioester bond. Prenylation uses farnesyl- as well as geranyl- or geranylgeranyl-residues in place of fatty acids. These post-translational modifications are capable of changing the subcellular localization of the modified protein and can serve as a membrane anchor. Acylation/prenylation can also affect protein stability and it is quite common to find different types of modifications in the same protein [90]. The most prominent and probably the most researched target of acylation is the small G-protein Ras. When modified by both farnesyl and palmitate, 95% of H-ras is membrane bound and fully active. In contrast, farnesyl lipid modification of H-ras alone decreases membrane recruitment to about 10% and its biological activity is extremely decreased [91].
Many members of the src family of tyrosine kinases are post-translationally acylated, which regulates membrane recruitment and thus signaling activity [92]. Several src-family members have been implicated in breast cancer pathologies, but the exact role of acylation in these processes remains to be elucidated [93, 94]. Pharmacological blockade of src-family tyrosine kinase activity is being investigated as a potential therapy for breast cancer [95], and since membrane recruitment is required for kinase activation it is tempting to speculate that inhibition of protein acylation could result in similar effects.Other important players in breast cancer have been shown to be acylated as well. For example, ERα is palmitoylated on cys-447 and can thus be recruited to the plasma membrane where its signaling effects differ profoundly from those known to occur through nuclear ER [96, 97]. Hsp27 has been shown to promote palmitoylation of ERα and this function is required for the correct receptor trafficking to the plasma membrane [98].
Palmitoylation reactions are facilitated by palmitoyltransferases. These enzymes are characterized by a conserved aspartate-histidine-histidine-cysteine rich domain, approximately 55 amino acids in length. 25 of these DHHC proteins have been identified in humans [99]. While there is still not much known about individual DHHC proteins, their substrates and their effect on cell physiology, one study reported that overexpression of DHHC20 in NIH3T3 cells resulted in colony formation in soft agar, reduced contact inhibition as well as increased proliferation. Unfortunately, this could not yet be linked to the palmitoylation of a specific substrate [100]. Porcupine (PORCN), an O-acyltransferase which plays a role in the wnt signaling pathway, seems to be important for the proliferation of MDA-MB-231 breast cancer cells. This effect appears to be independent of the wnt signaling pathway, since siRNA mediated knockdown of PORCN regulates a different set of genes compared to wnt inhibitors [101].
Breast Cancer and Obesity
Although there is limited evidence to suggest that total dietary fat intake is a cause of breast cancer, many epidemiological studies have looked at correlations between body weight and a number of breast cancer statistics including cancer incidence and patient survival. Several meta-analyses have summarized the epidemiological data gathered over the last 3 decades, concluding that high body mass index (BMI) is significantly correlated with increased death rates for all types of cancers. Death rates that are 52% higher for men and 62% higher for women compared to the rates for men and women of normal weight have been reported [102].
In the specific case of breast cancer, which is often times dependent on the hormonal status of the patient, subdivision into pre- and post-menopausal breast cancer patients was needed to obtain reliable results. Interestingly, some studies suggest that increased body mass is correlated with a risk reduction in the case of pre-menopausal breast cancer. This phenomenon is reversed when the epidemiological data for post-menopausal breast cancer is scrutinized. In those cases increased body mass is significantly correlated with greater risk of cancer occurrence as well as with reduced survival rates [103]. The World Cancer Research Fund (WCRF) and the American Institute for Cancer Research (AICR) issued an expert report in 2007 in which a panel of experts evaluated the epidemiological and experimental data in the literature on obesity and cancer to come up with guidelines for the general public. The panel concluded that in the case of pre-menopausal breast cancer there is some evidence to suggest that weight loss would have a preventative effect. In the case of post-menopausal breast cancer there is strong evidence to suggest that excess body fat has a causative relationship with breast cancer development.
In these studies, when data are adjusted for total serum estrogen levels, the correlation with BMI is lost, suggesting a hormonal influence of obesity on cancer development [104]. Some studies have reported that a high fat diet can result in increased estrogen concentrations in the serum, potentially through increased de novo synthesis [105]. In general, we can summarize that women with breast cancer who are obese have poorer survival than women with breast cancer, who are not obese. However, no study has elucidated the causal mechanism(s) and there is currently no evidence that weight loss after diagnosis improves survival [106]. Some studies have looked at histopathological markers that are frequently associated with breast cancer, such as estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2/neu) expression. In the case of pre-menopausal women who are obese, a significantly greater risk of developing triple-negative breast cancer has been reported by at least two studies [107, 108]. One study reported significant correlation between BMI and ER positive breast cancer subtypes [107]. No significant correlation has been reported for HER2/neu expression and BMI.
Creighton et al. performed a gene expression analysis in tumor samples that were classified according to the BMI of the patient and identified a 662 gene signature that was mostly congruent with gene expression patters related to insulin-like growth factor (IGF-1) signaling [109]. IGF-1 regulates many genes associated with cell proliferation, metabolism, and DNA repair and is highly correlated with poor prognostic outcome [110]. In mice, a high fat diet correlated with higher levels of insulin, leptin and IGF-1 which results in increased signaling through PI3K as well as MAPK/p38, which have been previously linked to uncontrolled cell proliferation [111]. IGF-1 stimulates cell proliferation and inhibits apoptosis. The biologically active concentration of IGF in the blood is determined by the concentration of its main binding protein IGFBP-3. Usually more than 90% of all IGF molecules are bound to binding proteins. In the case of obesity the levels of IGF-binding proteins are decreased, resulting in a greater effective concentration of free IGF even though the total levels of IGF do not change [112]. Insulin and free IGF-1 are also able to regulate the synthesis and bioavailability of sex steroids, particularly estrogen and progesterone, which affect the development and progression of breast cancer. By downregulating the hepatic synthesis of the sex hormone-binding globulins (SHBG) this phenomenon increases the bioavailability of steroid hormones very much like the decrease in IGFBP increases the availability of IGF-1 [113]. In addition to the effects of insulin on the bioavailability of sex hormones, adipose tissue itself increases the concentration of circulating estrogens in men and postmenopausal women through the aromatization of androstenedione to estrone [114]. In postmenopausal women, ovarian production of estrogen falls to very low levels and the adipose is the primary source of circulating estrogen [115]. Whether free fatty acids or their albumin-bound counterparts have a direct effect on the signaling of tumor cells or the tumor-associated stroma remains to be elucidated.
Conclusions
Cancer cells within solid tumors are comprised of a highly dynamic microenvironment that is often characterized by stressful conditions including increased acidity, mild to severe hypoxia and fluctuating nutrient availability. It has been shown that under mild stress conditions UPR/ER-stress induction is essential for cell survival and is activated in a range of human tumors, including breast cancer [65, 116–118]. Activation of UPR/ER-stress results in the initiation of transcriptional programs that regulate glucose and lipid metabolism [67, 119]. This is especially interesting in light of the recent renaissance in the field of tumor metabolism and how it relates to Otto Warburg's initial finding of aerobic glycolysis in tumor cells [120].
UPR/ER-Stress has been shown to regulate mitochondrial metabolism through the mitochondria associated membrane (MAM) Ca2+ signaling [121]. Ca2+ levels regulate the activity of several key enzymes in the TCA cycle, including isocitrate dehydrogenase, alpha-ketoglutarate dehydrogenase and pyruvate dehydrogenase [122]. The UPR/ER-stress response may work in concert with tumorigenic genetic lesions to fine tune metabolism, sustaining the requirements for robust cellular proliferation in a hostile environment. The activation of the mTOR pathway through constitutively active HER2/PI3K/Akt signaling has the potential to promote the ER-stress response in these cancer cells [123]. It is important to note that mTOR activity has been shown to be increased in obesity, providing a potential link between tumor cell metabolism and an obesity related increase in risk of developing breast cancer [124].
The activation of the unfolded protein response in cancer has also been associated with poor clinical outcome. Nagelkerke et al. have shown that hypoxia is able to induce the PERK-ATF4 arm of the UPR, resulting in increased motility and migration. This suggests a role for UPR/ER-stress in the metastasis of breast cancer [125]. Induction of UPR/ER-stress in cancer cells may be important in determining therapeutic outcome as it has been implicated in the development of resistance to various chemotherapeutics such as doxorubicin and anti-estrogens [118].
Our current understanding of breast cancer points to the importance of lipid synthesis and lipid related signaling alterations in the cells that cause this disease. Continued efforts will explore the molecular mechanisms involved. In many cases, previous studies performed in non-cancerous cell types will provide a roadmap. For example, the acquisition of a lipogenic phenotype by HER2/neu-positive breast cancer cells that is not seen in HER2/neu-normal mammary epithelial cells, suggests that some of the same pathways regulating lipogenesis seen in adipocytes and hepatic steatosis are operative in HER2/neu-positive breast cancer cells. Fatty acid synthesis results in consumption of NADPH which indirectly leads to NAD+ regeneration which in turn supports the continued high levels of anaerobic glycolysis required by breast cancer cells for survival. The enhanced NAD+ regeneration afforded by fatty acid synthesis in HER2/neu-positive breast cancer cells is well substantiated by experiments demonstrating palmitate toxicity, increased fat storage and the reliance on the prolipogenic transcription factors, NR1D1 and PBP [6, 7, 17, 28]. That these breast cancer cells have borrowed heavily from the lipid metabolism repertoire of both liver and adipocytes for their continued survival is a situation that is likely repeated in other cancers.
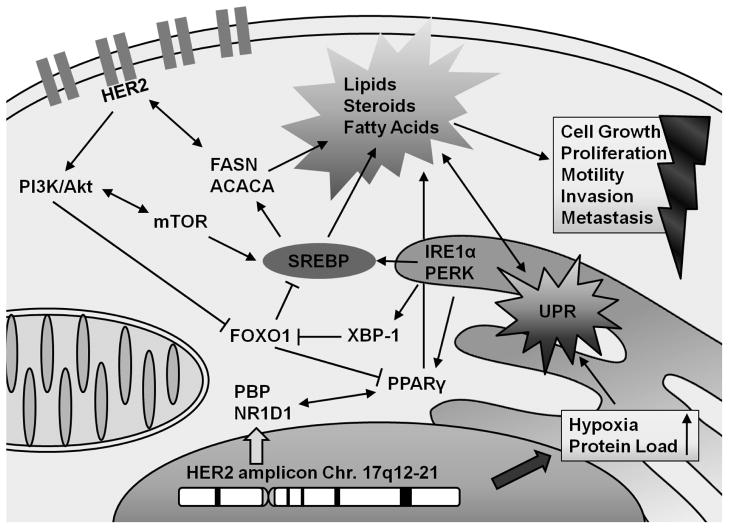
Figure 1. Integrated model of lipogenic tumor biology in HER2/neu-positive breast cancer.
Recurrent genomic amplification of chromosome 17q12-21 results in a prolipogenic signal transduction and transcriptional program. The resulting upregulation of HER2 and the transcription factors NR1D1 and PBP positively influences the production and/or function of lipogenic transcription factors SREBP and PPARγ. The concomitant activation of the unfolded protein response has the potential to further manifest and strengthen a lipogenic metabolic phenotype that is uniquely capable of sustaining cell growth, proliferation, migratory capacity and chemoresistance. This altered lipid metabolism phenotype is likely to affect tumor progression and metastasis in vivo.
Highlights.
The importance of fat metabolism to the breast cancer phenotype is reviewed.
Potential roles for the fatty acid and lipid synthesis pathways in breast cancer cells are discussed.
Fatty acid synthesis, regulation and its relevance to human HER2/neu positive breast cancer are reviewed.
The potential involvement of the unfolded protein response and endoplasmic reticulum stress in promoting lipogenesis in cancer cells is reviewed.
The central role of mTOR activation in phosphatidylinositide kinase and phospholipase D signaling during lipogenesis is discussed.
The potential relationships between dietary fat intake and lipid biology in breast cancer are reviewed.
Acknowledgments
The authors would like to thank members of the Conklin lab for helpful suggestions. Supported by U.S. Army Medical Research Acquisition Activity BC101082 to JB and NCI 1R01CA136658 to DSC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Laplante M, Sabatini DM. An emerging role of mTOR in lipid biosynthesis. Current biology : CB. 2009;19:R1046–1052. doi: 10.1016/j.cub.2009.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foukas LC, Withers DJ. Phosphoinositide signalling pathways in metabolic regulation. Curr Top Microbiol Immunol. 2010;346:115–141. doi: 10.1007/82_2010_59. [DOI] [PubMed] [Google Scholar]
- 3.Whiteman EL, Cho H, Birnbaum MJ. Role of Akt/protein kinase B in metabolism. Trends Endocrinol Metab. 2002;13:444–451. doi: 10.1016/s1043-2760(02)00662-8. [DOI] [PubMed] [Google Scholar]
- 4.Buller CL, Loberg RD, Fan MH, Zhu Q, Park JL, Vesely E, Inoki K, Guan KL, Brosius FC., 3rd A GSK-3/TSC2/mTOR pathway regulates glucose uptake and GLUT1 glucose transporter expression. Am J Physiol Cell Physiol. 2008;295:C836–843. doi: 10.1152/ajpcell.00554.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Menendez JA. Fine-tuning the lipogenic/lipolytic balance to optimize the metabolic requirements of cancer cell growth: molecular mechanisms and therapeutic perspectives. Biochim Biophys Acta. 2010;1801:381–391. doi: 10.1016/j.bbalip.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Kourtidis A, Jain R, Carkner RD, Eifert C, Brosnan MJ, Conklin DS. An RNA interference screen identifies metabolic regulators NR1D1 and PBP as novel survival factors for breast cancer cells with the ERBB2 signature. Cancer research. 2010;70:1783. doi: 10.1158/0008-5472.CAN-09-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kourtidis A, Srinivasaiah R, Carkner RD, Brosnan MJ, Conklin DS. Peroxisome proliferator-activated receptor-gamma protects ERBB2-positive breast cancer cells from palmitate toxicity. Breast Cancer Res. 2009;11:R16. doi: 10.1186/bcr2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menendez JA, Vellon L, Mehmi I, Oza BP, Ropero S, Colomer R, Lupu R. Inhibition of fatty acid synthase (FAS) suppresses HER2/neu (erbB-2) oncogene overexpression in cancer cells. Proc Natl Acad Sci U S A. 2004;101:10715–10720. doi: 10.1073/pnas.0403390101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vazquez-Martin A, Colomer R, Brunet J, Menendez JA. Pharmacological blockade of fatty acid synthase (FASN) reverses acquired autoresistance to trastuzumab (Herceptin by transcriptionally inhibiting 'HER2 super-expression' occurring in high-dose trastuzumab-conditioned SKBR3/Tzb100 breast cancer cells. International journal of oncology. 2007;31:769–776. [PubMed] [Google Scholar]
- 10.Lessig J, Fuchs B. Plasmalogens in biological systems: their role in oxidative processes in biological membranes, their contribution to pathological processes and aging and plasmalogen analysis. Current Medicinal Chemistry. 2009;16:2021–2041. doi: 10.2174/092986709788682164. [DOI] [PubMed] [Google Scholar]
- 11.Pruett ST, Bushnev A, Hagedorn K, Adiga M, Haynes Ca, Sullards MC, Liotta DC, Merrill AH. Biodiversity of sphingoid bases (“sphingosines”) and related amino alcohols. Journal of Lipid Research. 2008;49:1621–1639. doi: 10.1194/jlr.R800012-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bretscher MS. Asymmetrical lipid bilayer structure for biological membranes. Nature: New biology. 1972;236:11–12. doi: 10.1038/newbio236011a0. [DOI] [PubMed] [Google Scholar]
- 13.Ibrahimi A, Sfeir Z. Expression of the CD36 homolog (FAT) in fibroblast cells: effects on fatty acid transport. Proceedings of the National Academy of Sciences. 1996;93:2646–2651. doi: 10.1073/pnas.93.7.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ge Y, Elghetany M. CD36: a multiligand molecule. Laboratory Hematology. 2005;11:31–37. doi: 10.1532/LH96.04056. [DOI] [PubMed] [Google Scholar]
- 15.Ellis J, Frahm J. Acyl-coenzyme A synthetases in metabolic control. Current opinion in cell biologyell Biology. 2010;21:212–217. doi: 10.1097/mol.0b013e32833884bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaffer JE. Lipotoxicity: when tissues overeat. Current Opinion in Lipidology. 2003;14:281–287. doi: 10.1097/00041433-200306000-00008. [DOI] [PubMed] [Google Scholar]
- 18.Leonardi R, Zhang Y-m, Rock CO, Jackowski S. Coenzyme A: back in action. Progress in lipid research. 2005;44:125–153. doi: 10.1016/j.plipres.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Breus O, Panasyuk G, Gout IT, Filonenko V, Nemazanyy I. CoA synthase is in complex with p85alphaPI3K and affects PI3K signaling pathway. Biochemical and biophysical research communications. 2009;385:581–585. doi: 10.1016/j.bbrc.2009.05.102. [DOI] [PubMed] [Google Scholar]
- 20.Nemazanyy I, Panasyuk G, Zhyvoloup A, Panayotou G, Gout IT, Filonenko V. Specific interaction between S6K1 and CoA synthase: a potential link between the mTOR/S6K pathway, CoA biosynthesis and energy metabolism. FEBS letters. 2004;578:357–362. doi: 10.1016/j.febslet.2004.10.091. [DOI] [PubMed] [Google Scholar]
- 21.Albaugh BN, Arnold KM, Denu JM. KAT(ching) metabolism by the tail: insight into the links between lysine acetyltransferases and metabolism. Chembiochem : a European journal of chemical biology. 2011;12:290–298. doi: 10.1002/cbic.201000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science (New York, NY) 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Regulation of cellular metabolism by protein lysine acetylation. Science (New York, NY) 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chalbos D, Chambon M. Fatty acid synthetase and its mRNA are induced by progestins in breast cancer cells. Journal of Biological Chemistry. 1987;262:9923–9926. [PubMed] [Google Scholar]
- 25.Kuhajda F, Jenner K. Fatty acid synthesis: a potential selective target for antineoplastic therapy. Proceedings of the National Academy of Sciences. 1994;91:6379–6383. doi: 10.1073/pnas.91.14.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vazquez-Martin A, Colomer R, Brunet J, Lupu R, Menendez JA. Overexpression of fatty acid synthase gene activates HER1/HER2 tyrosine kinase receptors in human breast epithelial cells. Cell proliferation. 2008;41:59–85. doi: 10.1111/j.1365-2184.2007.00498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin Q, Yuan LX, Boulbes D, Baek JM, Wang YN, Gomez-Cabello D, Hawke DH, Yeung SC, Lee MH, Hortobagyi GN, Hung MC, Esteva FJ. Fatty acid synthase phosphorylation: a novel therapeutic target in HER2-overexpressing breast cancer cells. Breast cancer research : BCR. 2010;12:R96. doi: 10.1186/bcr2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Listenberger LL, Han X, Lewis SE, Cases S, Farese RV, Ory DS, Schaffer JE. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3077–3082. doi: 10.1073/pnas.0630588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kauraniemi P, Bärlund M, Monni O, Kallioniemi A. New amplified and highly expressed genes discovered in the ERBB2 amplicon in breast cancer by cDNA microarrays. Cancer research. 2001;61:8235–8240. [PubMed] [Google Scholar]
- 30.Lowell BB. PPARgamma: an essential regulator of adipogenesis and modulator of fat cell function. Cell. 1999;99:239–242. doi: 10.1016/s0092-8674(00)81654-2. [DOI] [PubMed] [Google Scholar]
- 31.Zhu Y, Qi C, Jain S, Rao MS, Reddy JK. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. The Journal of biological chemistry. 1997;272:25500–25506. doi: 10.1074/jbc.272.41.25500. [DOI] [PubMed] [Google Scholar]
- 32.Yang Y, Morin PJ, Han WF, Chen T, Bornman DM, Gabrielson EW, Pizer ES. Regulation of fatty acid synthase expression in breast cancer by sterol regulatory element binding protein-1c. Experimental Cell Research. 2003;282:132–137. doi: 10.1016/s0014-4827(02)00023-x. [DOI] [PubMed] [Google Scholar]
- 33.Yoon S, Lee MY, Park SW, Moon JS, Koh YK, Ahn YH, Park BW, Kim KS. Up-regulation of acetyl-CoA carboxylase alpha and fatty acid synthase by human epidermal growth factor receptor 2 at the translational level in breast cancer cells. The Journal of biological chemistry. 2007;282:26122–26131. doi: 10.1074/jbc.M702854200. [DOI] [PubMed] [Google Scholar]
- 34.Ferre P, Foufelle F. SREBP-1c transcription factor and lipid homeostasis: clinical perspective. Horm Res. 2007;68:72–82. doi: 10.1159/000100426. [DOI] [PubMed] [Google Scholar]
- 35.Kamei Y, Miura S, Suganami T, Akaike F, Kanai S, Sugita S, Katsumata A, Aburatani H, Unterman TG, Ezaki O, Ogawa Y. Regulation of SREBP1c gene expression in skeletal muscle: role of retinoid X receptor/liver X receptor and forkhead-O1 transcription factor. Endocrinology. 2008;149:2293–2305. doi: 10.1210/en.2007-1461. [DOI] [PubMed] [Google Scholar]
- 36.Vogt PK, Jiang H, Aoki M. Triple layer control: phosphorylation, acetylation and ubiquitination of FOXO proteins. Cell Cycle. 2005;4:908–913. doi: 10.4161/cc.4.7.1796. [DOI] [PubMed] [Google Scholar]
- 37.Wang F, Tong Q. SIRT2 suppresses adipocyte differentiation by deacetylating FOXO1 and enhancing FOXO1's repressive interaction with PPARgamma. Mol Biol Cell. 2009;20:801–808. doi: 10.1091/mbc.E08-06-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kohn AD, Summers SA, Birnbaum MJ, Roth RA. Expression of a constitutively active Akt Ser/Thr kinase in 3T3-L1 adipocytes stimulates glucose uptake and glucose transporter 4 translocation. J Biol Chem. 1996;271:31372–31378. doi: 10.1074/jbc.271.49.31372. [DOI] [PubMed] [Google Scholar]
- 39.Peng XD, Xu PZ, Chen ML, Hahn-Windgassen A, Skeen J, Jacobs J, Sundararajan D, Chen WS, Crawford SE, Coleman KG, Hay N. Dwarfism, impaired skin development, skeletal muscle atrophy, delayed bone development, and impeded adipogenesis in mice lacking Akt1 and Akt2. Genes Dev. 2003;17:1352–1365. doi: 10.1101/gad.1089403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rawson RB. The SREBP pathway--insights from Insigs and insects. Nat Rev Mol Cell Biol. 2003;4:631–640. doi: 10.1038/nrm1174. [DOI] [PubMed] [Google Scholar]
- 41.Li S, Brown MS, Goldstein JL. Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc Natl Acad Sci U S A. 2010;107:3441–3446. doi: 10.1073/pnas.0914798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung YL, Schulze A. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, Sabatini DM. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Finck BN, Gropler MC, Chen Z, Leone TC, Croce MA, Harris TE, Lawrence JC, Jr, Kelly DP. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell metabolism. 2006;4:199–210. doi: 10.1016/j.cmet.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 45.Hong K, Lou L, Gupta S, Ribeiro-Neto F, Altschuler DL. A novel Epac-Rap-PP2A signaling module controls cAMP-dependent Akt regulation. The Journal of Biological Chemistry. 2008;283:23129–23138. doi: 10.1074/jbc.M800478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reue K, Brindley DN. Thematic Review Series: Glycerolipids. Multiple roles for lipins/phosphatidate phosphatase enzymes in lipid metabolism. Journal of Lipid Research. 2008;49:2493–2503. doi: 10.1194/jlr.R800019-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foster DA. Phosphatidic acid signaling to mTOR: signals for the survival of human cancer cells. Biochim Biophys Acta. 2009;1791:949–955. doi: 10.1016/j.bbalip.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brunet J, Vazquez-Martin A, Colomer R, Graña-Suarez B, Martin-Castillo B, Menendez JA. BRCA1 and acetyl-CoA carboxylase: the metabolic syndrome of breast cancer. Molecular carcinogenesis. 2008;47:157–163. doi: 10.1002/mc.20364. [DOI] [PubMed] [Google Scholar]
- 49.Magnard C, Bachelier R, Vincent A, Jaquinod M, Kieffer S, Lenoir GM, Venezia ND. BRCA1 interacts with acetyl-CoA carboxylase through its tandem of BRCT domains. Oncogene. 2002;21:6729–6739. doi: 10.1038/sj.onc.1205915. [DOI] [PubMed] [Google Scholar]
- 50.Moreau K, Dizin E, Ray H, Luquain C, Lefai E, Foufelle F, Billaud M, Lenoir GM, Venezia ND. BRCA1 affects lipid synthesis through its interaction with acetyl-CoA carboxylase. The Journal of Biological Chemistry. 2006;281:3172–3181. doi: 10.1074/jbc.M504652200. [DOI] [PubMed] [Google Scholar]
- 51.Scott KEN, Wheeler FB, Davis AL, Thomas MJ, Ntambi JM, Seals DF, Kridel SJ. Metabolic regulation of invadopodia and invasion by acetyl-CoA carboxylase 1 and de novo lipogenesis. PloS one. 2012;7:e29761. doi: 10.1371/journal.pone.0029761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chajès V, Cambot M, Moreau K, Lenoir GM, Joulin V. Acetyl-CoA carboxylase alpha is essential to breast cancer cell survival. Cancer Research. 2006;66:5287–5294. doi: 10.1158/0008-5472.CAN-05-1489. [DOI] [PubMed] [Google Scholar]
- 53.Flavin R, Peluso S, Nguyen PL, Loda M. Fatty acid synthase as a potential therapeutic target in cancer. Future Oncology. 2010;6:551–562. doi: 10.2217/fon.10.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malhi H, Kaufman RJ. Endoplasmic reticulum stress in liver disease. J Hepatol. 2011;54:795–809. doi: 10.1016/j.jhep.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol. 2004;167:35–41. doi: 10.1083/jcb.200406136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Healy SJ, Gorman AM, Mousavi-Shafaei P, Gupta S, Samali A. Targeting the endoplasmic reticulum-stress response as an anticancer strategy. Eur J Pharmacol. 2009;625:234–246. doi: 10.1016/j.ejphar.2009.06.064. [DOI] [PubMed] [Google Scholar]
- 57.de Haro C, Mendez R, Santoyo J. The eIF-2alpha kinases and the control of protein synthesis. FASEB J. 1996;10:1378–1387. doi: 10.1096/fasebj.10.12.8903508. [DOI] [PubMed] [Google Scholar]
- 58.Kimball SR. Eukaryotic initiation factor eIF2. Int J Biochem Cell Biol. 1999;31:25–29. doi: 10.1016/s1357-2725(98)00128-9. [DOI] [PubMed] [Google Scholar]
- 59.Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, Yoshida H, Mori K, Kaufman RJ. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol. 2000;20:6755–6767. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot RT, Remotti H, Stevens JL, Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Colgan SM, Tang D, Werstuck GH, Austin RC. Endoplasmic reticulum stress causes the activation of sterol regulatory element binding protein-2. Int J Biochem Cell Biol. 2007;39:1843–1851. doi: 10.1016/j.biocel.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 63.Lee JN, Ye J. Proteolytic activation of sterol regulatory element-binding protein induced by cellular stress through depletion of Insig-1. J Biol Chem. 2004;279:45257–45265. doi: 10.1074/jbc.M408235200. [DOI] [PubMed] [Google Scholar]
- 64.Rutkowski DT, Wu J, Back SH, Callaghan MU, Ferris SP, Iqbal J, Clark R, Miao H, Hassler JR, Fornek J, Katze MG, Hussain MM, Song B, Swathirajan J, Wang J, Yau GD, Kaufman RJ. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell. 2008;15:829–840. doi: 10.1016/j.devcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ozcan U, Ozcan L, Yilmaz E, Duvel K, Sahin M, Manning BD, Hotamisligil GS. Loss of the tuberous sclerosis complex tumor suppressors triggers the unfolded protein response to regulate insulin signaling and apoptosis. Molecular cell. 2008;29:541–551. doi: 10.1016/j.molcel.2007.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sha H, He Y, Chen H, Wang C, Zenno A, Shi H, Yang X, Zhang X, Qi L. The IRE1alpha-XBP1 pathway of the unfolded protein response is required for adipogenesis. Cell metabolism. 2009;9:556–564. doi: 10.1016/j.cmet.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science (New York, NY) 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oyadomari S, Harding HP, Zhang Y, Oyadomari M, Ron D. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell metabolism. 2008;7:520–532. doi: 10.1016/j.cmet.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sriburi R, Bommiasamy H, Buldak GL, Robbins GR, Frank M, Jackowski S, Brewer JW. Coordinate regulation of phospholipid biosynthesis and secretory pathway gene expression in XBP-1(S)-induced endoplasmic reticulum biogenesis. The Journal of biological chemistry. 2007;282:7024–7034. doi: 10.1074/jbc.M609490200. [DOI] [PubMed] [Google Scholar]
- 70.Bobrovnikova-Marjon E, Hatzivassiliou G, Grigoriadou C, Romero M, Cavener DR, Thompson CB, Diehl JA. PERK-dependent regulation of lipogenesis during mouse mammary gland development and adipocyte differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16314–16319. doi: 10.1073/pnas.0808517105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee J, Mendez R, Heng H. Pharmacological ER stress promotes hepatic lipogenesis and lipid droplet formation. American journal of …. 2012;4:102–113. [PMC free article] [PubMed] [Google Scholar]
- 72.Kammoun HL, Chabanon H, Hainault I, Luquet S, Magnan C, Koike T, Ferre P, Foufelle F. GRP78 expression inhibits insulin and ER stress-induced SREBP-1c activation and reduces hepatic steatosis in mice. J Clin Invest. 2009;119:1201–1215. doi: 10.1172/JCI37007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wouters BG, Koritzinsky M. Hypoxia signalling through mTOR and the unfolded protein response in cancer. Nat Rev Cancer. 2008;8:851–864. doi: 10.1038/nrc2501. [DOI] [PubMed] [Google Scholar]
- 74.Chakrabarti P, English T, Shi J, Smas CM, Kandror KV. Mammalian target of rapamycin complex 1 suppresses lipolysis, stimulates lipogenesis, and promotes fat storage. Diabetes. 2010;59:775–781. doi: 10.2337/db09-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Soliman GA, Acosta-Jaquez HA, Fingar DC. mTORC1 inhibition via rapamycin promotes triacylglycerol lipolysis and release of free fatty acids in 3T3-L1 adipocytes. Lipids. 2010;45:1089–1100. doi: 10.1007/s11745-010-3488-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 77.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McManus EJ, Collins BJ, Ashby PR, Prescott AR, Murray-Tait V, Armit LJ, Arthur JS, Alessi DR. The in vivo role of PtdIns(3,4,5)P3 binding to PDK1 PH domain defined by knockin mutation. EMBO J. 2004;23:2071–2082. doi: 10.1038/sj.emboj.7600218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings BA. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 80.Thomas CC, Deak M, Alessi DR, van Aalten DM. High-resolution structure of the pleckstrin homology domain of protein kinase b/akt bound to phosphatidylinositol (3,4,5)-trisphosphate. Curr Biol. 2002;12:1256–1262. doi: 10.1016/s0960-9822(02)00972-7. [DOI] [PubMed] [Google Scholar]
- 81.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 82.Oh WJ, Wu CC, Kim SJ, Facchinetti V, Julien LA, Finlan M, Roux PP, Su B, Jacinto E. mTORC2 can associate with ribosomes to promote cotranslational phosphorylation and stability of nascent Akt polypeptide. EMBO J. 2010;29:3939–3951. doi: 10.1038/emboj.2010.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Holbro T, Beerli RR, Maurer F, Koziczak M, Barbas CF, 3rd, Hynes NE. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc Natl Acad Sci U S A. 2003;100:8933–8938. doi: 10.1073/pnas.1537685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tee AR, Anjum R, Blenis J. Inactivation of the tuberous sclerosis complex-1 and -2 gene products occurs by phosphoinositide 3-kinase/Akt-dependent and -independent phosphorylation of tuberin. The Journal of Biological Chemistry. 2003;278:37288–37296. doi: 10.1074/jbc.M303257200. [DOI] [PubMed] [Google Scholar]
- 86.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nature reviews Molecular cell biology. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sun Y, Fang Y, Yoon MS, Zhang C, Roccio M, Zwartkruis FJ, Armstrong M, Brown HA, Chen J. Phospholipase D1 is an effector of Rheb in the mTOR pathway. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:8286–8291. doi: 10.1073/pnas.0712268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Veverka V, Crabbe T, Bird I, Lennie G, Muskett FW, Taylor RJ, Carr MD. Structural characterization of the interaction of mTOR with phosphatidic acid and a novel class of inhibitor: compelling evidence for a central role of the FRB domain in small molecule-mediated regulation of mTOR. Oncogene. 2008;27:585–595. doi: 10.1038/sj.onc.1210693. [DOI] [PubMed] [Google Scholar]
- 89.Toschi A, Lee E, Xu L, Garcia A, Gadir N, Foster DA. Regulation of mTORC1 and mTORC2 complex assembly by phosphatidic acid: competition with rapamycin. Molecular and Cellular Biology. 2009;29:1411–1420. doi: 10.1128/MCB.00782-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aicart-Ramos C, Valero RA, Rodriguez-Crespo I. Protein palmitoylation and subcellular trafficking. Biochim Biophys Acta. 2011;1808:2981–2994. doi: 10.1016/j.bbamem.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 91.Cadwallader KA, Paterson H, Macdonald SG, Hancock JF. N-terminally myristoylated Ras proteins require palmitoylation or a polybasic domain for plasma membrane localization. Molecular and Cellular Biology. 1994;14:4722–4730. doi: 10.1128/mcb.14.7.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sato I, Obata Y, Kasahara K, Nakayama Y, Fukumoto Y, Yamasaki T, Yokoyama KK, Saito T, Yamaguchi N. Differential trafficking of Src, Lyn, Yes and Fyn is specified by the state of palmitoylation in the SH4 domain. Journal of Cell Science. 2009;122:965–975. doi: 10.1242/jcs.034843. [DOI] [PubMed] [Google Scholar]
- 93.Bilal E, Alexe G, Yao M, Cong L, Kulkarni A, Ginjala V, Toppmeyer D, Ganesan S, Bhanot G. Identification of the YES1 Kinase as a Therapeutic Target in Basal-Like Breast Cancers. Genes & Cancer. 2010;1:1063–1073. doi: 10.1177/1947601910395583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yadav V, Denning MF. Fyn is induced by Ras/PI3K/Akt signaling and is required for enhanced invasion/migration. Molecular Carcinogenesis. 2011;50:346–352. doi: 10.1002/mc.20716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Montero JC, Seoane S, Ocaña A, Pandiella A. Inhibition of SRC family kinases and receptor tyrosine kinases by dasatinib: possible combinations in solid tumors. Clinical Cancer Research. 2011;17:5546–5552. doi: 10.1158/1078-0432.CCR-10-2616. [DOI] [PubMed] [Google Scholar]
- 96.Byrne C, Khemtémourian L, Pelekanou V, Kampa M, Leclercq G, Sagan S, Castanas E, Burlina F, Jacquot Y. ERα17p, a peptide reproducing the hinge region of the estrogen receptor α associates to biological membranes: A biophysical approach. Steroids. 2012;77:979–987. doi: 10.1016/j.steroids.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 97.Rai D, Frolova A, Frasor J, Carpenter AE, Katzenellenbogen BS. Distinctive actions of membrane-targeted versus nuclear localized estrogen receptors in breast cancer cells. Molecular Endocrinology. 2005;19:1606–1617. doi: 10.1210/me.2004-0468. [DOI] [PubMed] [Google Scholar]
- 98.Razandi M, Pedram A, Levin ER. Heat shock protein 27 is required for sex steroid receptor trafficking to and functioning at the plasma membrane. Molecular and Cellular Biology. 2010;30:3249–3261. doi: 10.1128/MCB.01354-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rocks O, Gerauer M, Vartak N, Koch S, Huang ZP, Pechlivanis M, Kuhlmann J, Brunsveld L, Chandra A, Ellinger B, Waldmann H, Bastiaens PIH. The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell. 2010;141:458–471. doi: 10.1016/j.cell.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 100.Draper JM, Smith CD. DHHC20: a human palmitoyl acyltransferase that causes cellular transformation. Molecular Membrane Biology. 2010;27:123–136. doi: 10.3109/09687681003616854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Covey TM, Kaur S, Tan Ong T, Proffitt KD, Wu Y, Tan P, Virshup DM. PORCN moonlights in a Wnt-independent pathway that regulates cancer cell proliferation. PloS One. 2012;7:e34532. doi: 10.1371/journal.pone.0034532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Calle E. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of US adults. New England Journal of Medicine. 2003;348:1625–1638. doi: 10.1056/NEJMoa021423. [DOI] [PubMed] [Google Scholar]
- 103.Rosato V, Bosetti C, Talamini R, Levi F, Montella M, Giacosa A, Negri E, La Vecchia C. Metabolic syndrome and the risk of breast cancer in postmenopausal women. Ann Oncol. 2011;22:2687–2692. doi: 10.1093/annonc/mdr025. [DOI] [PubMed] [Google Scholar]
- 104.Marmot M, Atinmo T, Byers T. J Chen, Food, nutrition, physical activity, and the prevention of cancer: a global perspective. 2007 [Google Scholar]
- 105.Goldin BR, Woods MN, Spiegelman DL, Longcope C, Morrill-LaBrode A, Dwyer JT, Gualtieri LJ, Hertzmark E, Gorbach SL. The effect of dietary fat and fiber on serum estrogen concentrations in premenopausal women under controlled dietary conditions. Cancer. 1994;74:1125–1131. doi: 10.1002/1097-0142(19940801)74:3+<1125::aid-cncr2820741521>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 106.Protani M, Coory M, Martin JH. Effect of obesity on survival of women with breast cancer: systematic review and meta-analysis. Breast Cancer Research and Treatment. 2010;123:627–635. doi: 10.1007/s10549-010-0990-0. [DOI] [PubMed] [Google Scholar]
- 107.Phipps AI, Buist DSM, Malone KE, Barlow WE, Porter PL, Kerlikowske K, O'Meara ES, Li CI. Breast density, body mass index, and risk of tumor marker-defined subtypes of breast cancer. Annals of Epidemiology. 2012;22:340–348. doi: 10.1016/j.annepidem.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pierobon M, Frankenfeld CL. Obesity as a risk factor for triple-negative breast cancers: a systematic review and meta-analysis. Breast Cancer Research and Treatment. 2012 doi: 10.1007/s10549-012-2339-3. [DOI] [PubMed] [Google Scholar]
- 109.Creighton CCJ, Sada YHY, Zhang Y, Tsimelzon A, Wong H, Dave B, Landis MD, Bear HD, Rodriguez A, Chang JC. A gene transcription signature of obesity in breast cancer. Breast cancer research and treatment. 2012;132:993–1000. doi: 10.1007/s10549-011-1595-y. [DOI] [PubMed] [Google Scholar]
- 110.Creighton CJ, Casa A, Lazard Z, Huang S, Tsimelzon A, Hilsenbeck SG, Osborne CK, Lee AV. Insulin-like growth factor-I activates gene transcription programs strongly associated with poor breast cancer prognosis. Journal of Clinical Oncology. 2008;26:4078–4085. doi: 10.1200/JCO.2007.13.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sung MK, Yeon JY, Park SY, Park JHY, Choi MS. Obesity-induced metabolic stresses in breast and colon cancer. Annals of the New York Academy of Sciences. 2011;1229:61–68. doi: 10.1111/j.1749-6632.2011.06094.x. [DOI] [PubMed] [Google Scholar]
- 112.Calle EE, Thun MJ. Obesity and cancer. Oncogene. 2004;23:6365–6378. doi: 10.1038/sj.onc.1207751. [DOI] [PubMed] [Google Scholar]
- 113.Kaaks R. Nutrition, hormones, and breast cancer: is insulin the missing link? Cancer Causes & Control : CCC. 1996;7:605–625. doi: 10.1007/BF00051703. [DOI] [PubMed] [Google Scholar]
- 114.Madigan MP, Troisi R, Potischman N, Dorgan JF, Brinton LA, Hoover RN. Serum hormone levels in relation to reproductive and lifestyle factors in postmenopausal women (United States) Cancer Causes & Control : CCC. 1998;9:199–207. doi: 10.1023/a:1008838412423. [DOI] [PubMed] [Google Scholar]
- 115.Cleary MP, Grossmann ME, Ray A. Effect of obesity on breast cancer development. Veterinary Pathology. 2010;47:202–213. doi: 10.1177/0300985809357753. [DOI] [PubMed] [Google Scholar]
- 116.Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N, Harding H, Novoa I, Varia M, Raleigh J, Scheuner D, Kaufman RJ, Bell J, Ron D, Wouters BG, Koumenis C. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. The EMBO journal. 2005;24:3470–3481. doi: 10.1038/sj.emboj.7600777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Koumenis C, Wouters BG. “Translating” tumor hypoxia: unfolded protein response (UPR)-dependent and UPR-independent pathways. Molecular cancer research : MCR. 2006;4:423–436. doi: 10.1158/1541-7786.MCR-06-0150. [DOI] [PubMed] [Google Scholar]
- 118.Scriven P, Coulson S, Haines R, Balasubramanian S, Cross S, Wyld L. Activation and clinical significance of the unfolded protein response in breast cancer. British journal of cancer. 2009;101:1692–1698. doi: 10.1038/sj.bjc.6605365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang LM, Myers MG, Jr, Sun XJ, Aaronson SA, White M, Pierce JH. IRS-1: essential for insulin- and IL-4-stimulated mitogenesis in hematopoietic cells. Science. 1993;261:1591–1594. doi: 10.1126/science.8372354. [DOI] [PubMed] [Google Scholar]
- 120.Warburg O, Wind F, Negelein E. The Metabolism of Tumors in the Body. J Gen Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bravo R, Gutierrez T, Paredes F, Gatica D, Rodriguez AE, Pedrozo Z, Chiong M, Parra V, Quest AF, Rothermel BA, Lavandero S. Endoplasmic reticulum: ER stress regulates mitochondrial bioenergetics. The international journal of biochemistry & cell biology. 2012;44:16–20. doi: 10.1016/j.biocel.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hayashi T, Rizzuto R, Hajnoczky G, Su TP. MAM: more than just a housekeeper. Trends in cell biology. 2009;19:81–88. doi: 10.1016/j.tcb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Polak P, Hall MN. mTOR and the control of whole body metabolism. Curr Opin Cell Biol. 2009;21:209–218. doi: 10.1016/j.ceb.2009.01.024. [DOI] [PubMed] [Google Scholar]
- 124.Khamzina L, Veilleux A, Bergeron S, Marette A. Increased activation of the mammalian target of rapamycin pathway in liver and skeletal muscle of obese rats: possible involvement in obesity-linked insulin resistance. Endocrinology. 2005;146:1473–1481. doi: 10.1210/en.2004-0921. [DOI] [PubMed] [Google Scholar]
- 125.Nagelkerke A, Bussink J, Mujcic H, Wouters BG, Lehmann S, Sweep FC, Span PN. Hypoxia stimulates migration of breast cancer cells via the PERK/ATF4/LAMP3-arm of the unfolded protein response. Breast cancer research : BCR. 2013;15:R2. doi: 10.1186/bcr3373. [DOI] [PMC free article] [PubMed] [Google Scholar]