Abstract
Dramatic progress has been made in our understanding of the highly heterogeneous molecular bases of sensorineural hearing loss (SNHL), demonstrating the involvement of all known forms of inheritance and a plethora of genes tangled in various molecular pathways. This progress permits the provision of prognostic information and genetic counseling for affected families, which might, nevertheless, be exceedingly challenging. Here, we describe an intricate genetic investigation that included Sanger-type sequencing, BeadArray technology, and next-generation sequencing to resolve a complex case involving one family presenting syndromic and nonsyndromic SNHL phenotypes in two consecutive generations. We demonstrate and conclude that such an effort can be completed during pregnancy.
Introduction
The wealth of genetic heterogeneity known to be causative for sensorineural hearing loss (SNHL) makes the molecular diagnosis of such cases expensive and time-consuming (Smith et al., 2005; Shearer et al., 2010), but is particularly challenging during pregnancy. Divided into syndromic (30%) and nonsyndromic (70%), genetic etiologies make up 70% of all prelingual-onset SNHL (Smith et al., 2005). The single leading cause for hereditary SNHL, in general, and for nonsyndromic prelingual SNHL, in particular, is involvement of the DFNB1 gene GJB2 (Zelante et al., 1997). The most common cause of syndromic prelingual SNHL, characterized by combined SNHL and vision loss, is the Usher syndrome (USH) (Friedman et al., 2011). Both DFNB1 and USH are recessively inherited and the numerous population-specific causative mutations identified in the respective genes make up the essence of prenatal SNHL prevention programs to date (Smith et al., 2005). The same is true for the various Jewish communities in which different founder mutations have been identified (Brownstein et al., 2009). We report the resolution of a complex case of a family of mixed Ashkenazi and Iraqi Jewish ancestry during pregnancy, illustrating the scope of clinical and molecular work-up of SNHL in the genomic era.
Materials and Methods
Subjects
A 35-year-old woman (II-2) of consanguineous (first cousins) Iraqi Jewish ancestry sought urgent genetic counseling at the beginning of her second pregnancy (Fig. 1). The woman suffers from type 2 Usher syndrome (USH2), characterized by moderate SNHL and retinal degeneration (retinitis pigmentosa), and was known to be homozygous for the founder Iraqi Jewish USH2A mutation c.239_240insGTAC (Adato et al., 2000). She reported that her husband (II-3) was of Ashkenazi Jewish origin, that her brother (II-1) suffers from congenital profound SNHL and that her 4-year-old son (III-1) suffers from congenital moderate SNHL with a normal electroretinogram (ERG) at the age of three (Fig. 1). Genetic evaluation of all family members commenced after informed consent. The study was approved by the Helsinki Committee of the Israel Ministry of Health and the Institutional Review Board Committees of the Rabin Medical Center.
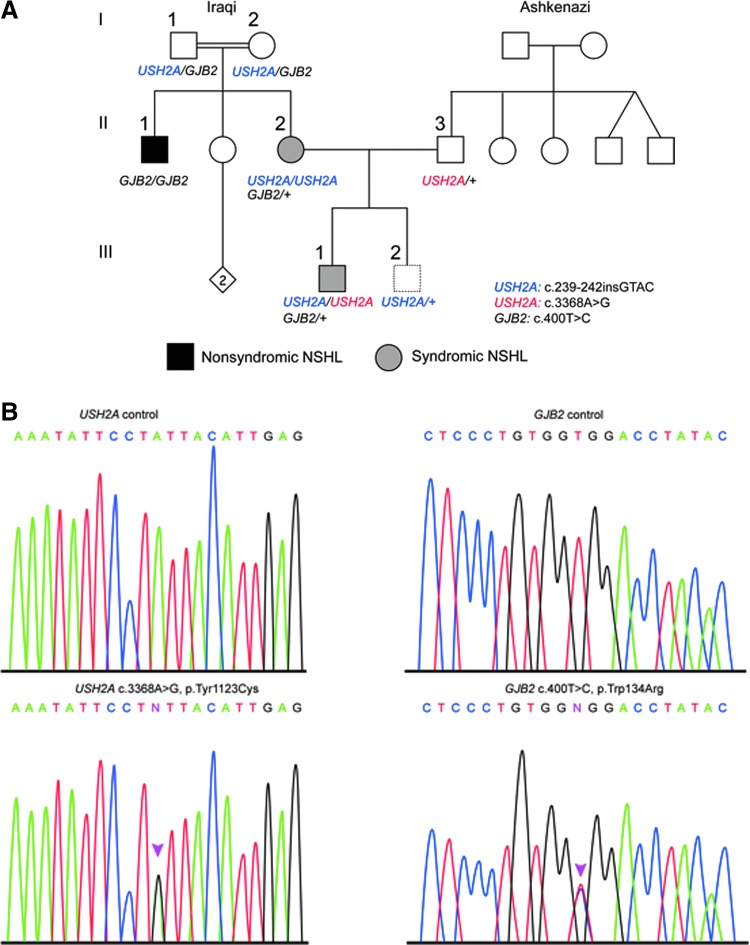
FIG. 1.
Presentation of the molecular analysis of the complex family. (A) Pedigree of the investigated family. Initial clinical presentation indicated type 2 Usher syndrome (USH2) in the mother II-2, but was suggestive of NSHL in her brother (II-1) and son (III-1). The genotypes are indicated for the ascertained individuals. (B) Sanger sequencing of the novel USH2A and GJB2 heterozygote mutations c.3368A>G, p.Tyr1123Cys, and c.400T>C, p.Trp134Arg, respectively, in a control individual and III-1. Color images available online at www.liebertpub.com/gtmb
DNA analysis
Genomic DNA was isolated from peripheral blood by using the Qiagen kit (Qiagen Ltd.) following the manufacturer's protocol. Known founder mutations were screened as previously summarized (Brownstein et al., 2009). For Sanger sequencing of the GJB2, USH2A, and SMPX genes, DNA was amplified to obtain all coding exons and their flanking regions using conventional PCR techniques. Sequencing was performed on a 3730xl DNA Analyzer (Applied Biosystems). A BeadArray-based screen for 198 common mitochondrial and autosomal NSHL mutations was completed by Asper Biotech Ltd. (www.asperbio.com). A next-generation sequencing (NGS) panel, including a total of 80 genes implicated in NSHL was carried out at Otogenetics Ltd. (www.otogenetics.com).
Results
Molecular investigation of III-1 (Fig. 1) included family screening for all known Iraqi founder DFNB1 and USH2A mutations and for all known Ashkenazi founder DFNB1, USH1F, and USH3 mutations (Brownstein et al., 2009), and yielded only the obligatory heterozygous states for the Iraqi USH2A mutation (Fig. 1 and Table 1). III-1 underwent a screen for the 198 common mitochondrial and autosomal NSHL mutations included in a diagnostic microarray (www.asperbio.com). No pathogenic mutations were found; the Iraqi USH2A c.239_240insGTAC mutation is not included in this array. II-2 requested prenatal diagnosis for SNHL. Considerable maternal stress was witnessed. As there was no evidence for retinal involvement in her son (III-1) and as no Ashkenazi USH2A founder mutations were previously reported, sequencing of the GJB2 gene in her son was performed (Brownstein et al., 2009). However, as the family's pedigree could theoretically be concordant with autosomal recessive, autosomal dominant, or X-linked inheritance modes, and as it was clear that time would not allow for hierarchical Sanger-based screening of additional genes in case of normal GJB2 sequencing, we opted to use an NGS panel, including a total of 80 genes implicated in NSHL (www.otogenetics.com) on DNA derived from III-1. In addition, Sanger sequencing of the X-linked SMPX gene was performed. A novel GJB2 c.400T>C (NM_004004); p.Trp134Arg (NP_003995) variant was detected in a heterozygous state, with a PolyPhen-2 score of 0.994 (http://genetics.bwh.harvard.edu/pph2/). Whereas it did not explain the SNHL of III-1, the segregation of the mutation clarified the diagnosis of II-1 and explained the degree of HL manifested by his audiogram (not shown), which is compatible with the profound phenotype of many GJB2 mutations. The NGS panel suggested over 100 additional alterations that were less likely to be deleterious, but the available time made it impossible to validate and segregate all variants. Fourteen days before the date of the scheduled amniocentesis, the clinical signs and symptoms were evaluated and it was concluded that the normal ERG at age three in the child III-1 did not rule out USH2 (Malm et al., 2011). Accordingly, sequencing of the USH2A gene, which was not included in the NGS panel, was performed. Partial results obtained a day before the amniocentesis yielded the novel USH2A variant c.3368A>G (NM_206933); p.Tyr1123Cys (NP_ 996816), scoring 0.999 by PolyPhen-2, in a heterozygous state in both the husband, II-3, and son, III-1 (Fig. 1). Consequently, we concluded that two different phenotypes and three different genotypes affect the family. The mother II-2 is affected by USH2, due to a homozygous USH2A Iraqi founder mutation (Adato et al., 2000), her brother II-1 has a homozygous novel DFNB1-related nonsyndromic SNHL resulting from parental consanguinity, and her son III-1 suffers from USH2A due to a compound heterozygous state of the known Iraqi mutation (Auslender et al., 2008) and a second novel mutation. Amniocentesis was carried out at week 20 and the embryo was found to carry only the Iraqi USH2A mutation in a heterozygous state. At 1 year of age (III-2), there were no indications of SNHL, including a normal auditory brainstem response test.
Table 1.
Clinical Characteristics of the Family Members
| Patient | Phenotype | USH2A genotype | GJB2 genotype |
|---|---|---|---|
| I-1 | Normal hearing | 239–242insGTAC/+ | 400T>C/+ |
| I-2 | Normal hearing | 239–242insGTAC/+ | 400T>C/+ |
| II-1 | Profound SNHL | 400T>C/400T>C | |
| II-2 | Moderate SNHL | 239–242insGTAC/239–242insGTAC | 400T>C/+ |
| II-3 | Normal hearing | 3368A>G/+ | |
| III-1 | Moderate SNHL; normal ERG at age 3 | 239–242insGTAC/3368A>G | 400T>C/+ |
| III-2 | Normal ABR at age 1 | 239–242insGTAC/+ |
SNHL, sensorineural hearing loss; ERG, electroretinogram; ABR, auditory brainstem response.
Discussion
The presented case is a compelling example of the complexity and extreme heterogeneity of hereditary hearing loss in the genomic era (Smith et al., 2005; Brownstein et al., 2009; Shearer et al., 2010), illustrating the need to consider various modes of inheritance, syndromic versus nonsyndromic phenotypes, and the use of various genomic technologies to resolve, during pregnancy, the multiple etiologies affecting one family. Clinically, it should be emphasized that normal ERG at the age of 3 years should not rule out the diagnosis of USH (Malm et al., 2011). In this report, the different parental ancestries, the common practice of primarily relying on the identification of founder deleterious mutations among Ashkenazi Jews (Brownstein et al., 2009), the normal ERG, and the hope for a favorable nonsyndromic versus syndromic prognosis for the elder son, were misleading. Hence, while the presented case supports the recommendation to sequence the GJB2 gene first in each SNHL individual (Brownstein et al., 2009), the sequence of any gene known to affect a family member should be considered even if the odds for its involvement appear to be extremely low. The molecular diagnostics was carried out under a very constricted schedule of ∼20 weeks. This required relying solely on prediction algorithms such as PolyPhen-2, rather than functional tests to infer mutagenicity for the detected variants, thus highlighting the need to complete genetic investigations well ahead of the actual pregnancy. It should be reiterated that preconception planning might be particularly important in similar cases because genetic testing for SNHL has raised some unique ethical issues stemming from cultural differences in attitudes toward hearing loss (Dagan et al., 2002) and the fact that treatments such as cochlear implantation are now commonly practiced. Accordingly, genetic counseling should be provided in an appropriate setting after the pathogenicity of the identified mutation is well established and when the parents can fully comprehend and weigh the risks, benefits, and limitations of genetic testing. Finally, it has already been suggested that comprehensive NGS platforms are critical for the molecular analysis of NSHL cases, since the number of currently known genes involved in this phenotype makes it difficult, if not impossible, to offer clinically useful screening based on Sanger-type sequencing (Shearer et al., 2010; Brownstein et al., 2011). In this study, we implemented NGS and identified a novel GJB2 mutation. The development of comprehensive SNHL gene panels using NGS, including robust analytical tools appropriate for rapid clinical management, are a major challenge. Whereas a number of these platforms consisting of target gene capture and NGS have been developed for deafness and other diseases (Shearer et al., 2010; Brownstein et al., 2011), a timely analysis of the multiple variants is difficult and usually precludes providing a definitive answer within the time frame of a pregnancy. This continues to be a challenge that should be overcome in the coming years.
Acknowledgments
We thank Shaked Shivatzki for preparation of the figure. The research was partially supported by NIH (National Institute on Deafness and Other Communication Disorders) (R01DC011835), I-CORE Gene Regulation in Complex Human Disease, Center No. 41/11. The Slava Smolokowski Research Fund at Rambam Medical Center has contributed to this research and is gratefully acknowledged.
Author Disclosure Statement
No competing financial interests exist.
References
- Adato A, Weston MD, Berry A, et al. (2000) Three novel mutations and twelve polymorphisms identified in the USH2A gene in Israeli USH2 families. Hum Mutat 15:388. [DOI] [PubMed] [Google Scholar]
- Auslender N, Bandah D, Rizel L, et al. (2008) Four USH2A founder mutations underlie the majority of Usher syndrome type 2 cases among non-Ashkenazi Jews. Genet Test 12:289–294 [DOI] [PubMed] [Google Scholar]
- Brownstein Z, Avraham KB. (2009) Deafness genes in Israel: implications for diagnostics in the clinic. Pediatr Res 66:128–134 [DOI] [PubMed] [Google Scholar]
- Brownstein Z, Friedman LM, Shahin H, et al. (2011) Targeted genomic capture and massively parallel sequencing to identify genes for hereditary hearing loss in Middle Eastern families. Genome Biol 12:R89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan O, Hochner H, Levi H, et al. (2002) Genetic testing for hearing loss: different motivations for the same outcome. Am J Med Genet 113:137–143 [DOI] [PubMed] [Google Scholar]
- Friedman TB, Schultz JM, Ahmed ZM, et al. (2011) Usher syndrome: hearing loss with vision loss. Adv Otorhinolaryngol 70:56–65 [DOI] [PubMed] [Google Scholar]
- Malm E, Ponjavic V, Moller C, et al. (2011) Alteration of rod and cone function in children with Usher syndrome. Eur J Ophthalmol 21:30–38 [DOI] [PubMed] [Google Scholar]
- Shearer AE, DeLuca AP, Hildebrand MS, et al. (2010) Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci USA 107:21104–21109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RJ, Bale JF, Jr., White KR. (2005) Sensorineural hearing loss in children. Lancet 365:879–890 [DOI] [PubMed] [Google Scholar]
- Zelante L, Gasparini P, Estivill X, et al. (1997) Connexin26 mutations associated with the most common form of non-syndromic neurosensory autosomal recessive deafness (DFNB1) in Mediterraneans. Hum Mol Genet 6:1605–1609 [DOI] [PubMed] [Google Scholar]