Abstract
Accurate cell division depends on tightly regulated ubiquitylation events catalyzed by the anaphase-promoting complex. Among its many substrates, the APC/C triggers the degradation of proteins that stabilize the mitotic spindle, and loss or accumulation of such spindle assembly factors can result in aneuploidy and cancer. Although critical for cell division, it has remained poorly understood how the timing of spindle assembly factor degradation is established during mitosis. Here, we report that active spindle assembly factors are protected from APC/C-dependent degradation by microtubules. In contrast, those molecules that are not bound to microtubules are highly susceptible to proteolysis and turned over immediately after APC/C-activation. The correct timing of spindle assembly factor degradation, as achieved by this regulatory circuit, is required for accurate spindle structure and function. We propose that the localized stabilization of APC/C-substrates provides a mechanism for the selective disposal of cell cycle regulators that have fulfilled their mitotic roles.
Keywords: microtubules, spindle assembly factors, ubiquitin, Ran, APC/C
Introduction
Posttranslational modification with ubiquitin is widely used to regulate protein stability or activity, and it is essential for cell division in all eukaryotes (Komander and Rape, 2012). Substrates are selected for ubiquitylation by E3 ligases, many of which play important roles in cell cycle control (Deshaies and Joazeiro, 2009). Among the ~600 human E3s, the anaphase-promoting complex (APC/C) is of particular interest as it controls essential steps in mitosis (Peters, 2006).
The APC/C was originally identified based on its role in triggering the degradation of cyclin B1, an activating subunit of Cdk1 (reviewed in (Peters, 2006)). It is now known to ubiquitylate a large number of cell cycle regulators, thereby orchestrating progression of cells through mitosis and G1. Substrates are delivered to the APC/C by the WD40-repeat proteins Cdc20 and Cdh1, which recognize degron motifs referred to as D- or KEN-boxes (Buschhorn et al., 2011; da Fonseca et al., 2011; Tian et al., 2012; Visintin et al., 1997). Following substrate recognition, the APC/C-specific E2s Ube2C and Ube2S synthesize ubiquitin chains that are recognized by the proteasome (Garnett et al., 2009; Rape and Kirschner, 2004; Williamson et al., 2009; Wu et al., 2010; Yu et al., 1996). Depletion of Cdc20 or Ube2C and Ube2S stabilizes APC/C-substrates and arrests cells prior to anaphase (Manchado et al., 2010; Williamson et al., 2009; Wolthuis et al., 2008).
As full activation of the APC/C leads to sister chromatid separation, it has to be delayed until all chromosomes have achieved bipolar attachment to the mitotic spindle. To this end, the APC/C is inhibited during spindle formation by a signaling network referred to as the spindle checkpoint (Kim and Yu, 2011; Musacchio and Salmon, 2007). The checkpoint components Mad2, BubR1, and Bub3 associate with Cdc20 to form the mitotic checkpoint complex, which binds the APC/C and blocks recognition of most of its substrates (Chao et al., 2012; Herzog et al., 2009; Schreiber et al., 2011). Once the spindle has been built and all chromosomes have been aligned at the metaphase plate, checkpoint complexes are disassembled, APC/CCdc20 is activated, and sister chromatid separation ensues (Foster and Morgan, 2012; Reddy et al., 2007; Uzunova et al., 2012; Varetti et al., 2011). Thus, by virtue of the checkpoint, the spindle indirectly controls the stability of most APC/C-substrates.
Spindle formation is a highly dynamic process that depends on a large number of proteins referred to as spindle assembly factors. Several of these factors, such as HURP, NuSAP, or Tpx2, associate with and stabilize microtubules, the main constituents of the spindle apparatus (Gruss et al., 2001; Koffa et al., 2006; Ribbeck et al., 2006; Sillje et al., 2006; Wittmann et al., 2000; Wong and Fang, 2006). Spindle assembly factors also recruit regulators of spindle formation: Tpx2, for example, promotes the spindle association of Aurora A and Eg5, thus controlling spindle pole integrity or spindle bipolarity (Gable et al., 2012; Kufer et al., 2002). After their activation in prometaphase, spindle assembly factors continue to play important roles during ana- and telophase, when they stabilize kinetochore fibers, drive spindle elongation, or establish a microtubule-dense spindle midzone (Goshima et al., 2007; Uehara and Goshima, 2010).
Befitting their role in cell division, complete loss of spindle assembly factors leads to embryonic lethality or female infertility (Aguirre-Portoles et al., 2012; Tsai et al., 2008), whereas their overexpression can result in tumorigenesis (Aguirre-Portoles et al., 2012; Gulzar et al., 2013; Hu et al., 2012; Tsou et al., 2003). These observations imply that cells need to control the abundance of spindle assembly factors, and indeed, HURP, NuSAP, and Tpx2 are turned over during mitosis by APC/C-dependent ubiquitylation and proteasomal degradation (Song and Rape, 2010; Stewart and Fang, 2005). HURP, NuSAP, and Tpx2 are also regulated by proteins of the importin family, which restrict their association with microtubules and prevent their recognition by the APC/C (Gruss et al., 2001; Gruss et al., 2002; Koffa et al., 2006; Kufer et al., 2002; Ribbeck et al., 2006; Sillje et al., 2006; Song and Rape, 2010). A consequence of this dual regulation, the dissociation of importin-β by GTP-bound Ran not only activates spindle assembly factors, but also allows their APC/C-dependent inactivation. However, despite the importance of spindle formation for cell division, it has remained unclear how cells can sufficiently delay the degradation of HURP, NuSAP, and Tpx2 to allow these proteins to fulfill their critical roles in regulating spindle structure and function.
Here, we have discovered that spindle microtubules protect HURP, NuSAP, and Tpx2 from APC/C-dependent degradation. By contrast, those spindle assembly factor molecules that are not associated with microtubules are highly susceptible to proteolysis and degraded instantaneously after APC/C-activation in early anaphase. The proper timing of spindle assembly factor degradation, as achieved by this regulatory circuit, is critical for mitosis, and inappropriate stabilization of spindle assembly factors disrupts spindle structure and function. Our findings suggest that localized stabilization of APC/C-substrates enables the selective disposal of cell cycle regulators that have fulfilled their mitotic roles, thus coupling the activity and stability of important spindle assembly factors.
Results
Microtubules protect spindle assembly factors from degradation
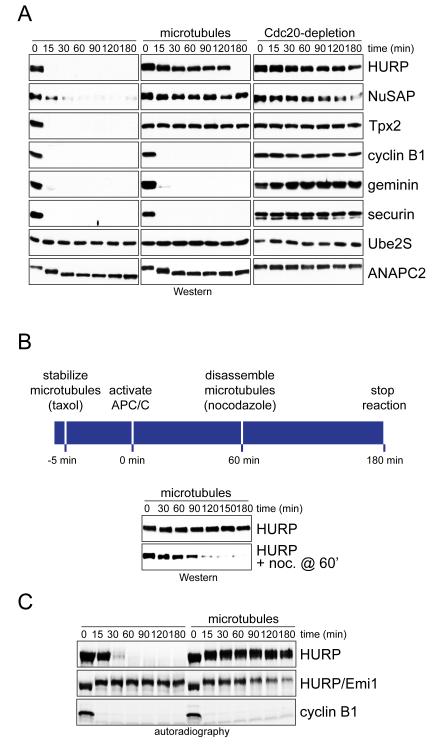
As most spindle assembly factors accumulate on the spindle during mitosis, we wished to determine whether their localization affected their stability. To this end, we tested whether microtubules, the main constituents of the spindle, influence the efficiency of spindle assembly factor degradation by the APC/C. We first generated extracts of prometaphase HeLa cells that contained inactive APC/C and soluble tubulin. We then supplemented these extracts with taxol to induce microtubule formation; GTP-charged RanQ69L to release spindle assembly factors from their inhibitors of the importin family; and p31comet to activate the APC/C. Without microtubules, these extracts supported the efficient degradation of spindle assembly factors and other known APC/C-substrates, and depletion of the essential APC/C-activator Cdc20 or addition of the APC/CCdc20-specific inhibitor Mad2 underscored the specificity of these reactions (Figure 1A; Figure S1A, B). By contrast, when microtubules were present, HURP, NuSAP and Tpx2 were protected from degradation, whereas soluble APC/C-substrates remained unstable (Figure 1A). The effects of microtubules on the stability of spindle assembly factors were reversible, as HURP was rapidly turned over once microtubules had been depolymerized with nocodazole (Figure 1B). Thus, microtubules protect multiple spindle assembly factors from degradation through APC/CCdc20 in early mitotic extracts.
Figure 1. Microtubules stabilize spindle assembly factors.
A. Microtubules protect spindle assembly factors from APC/CCdc20-dependent degradation in extracts. Extracts of prometaphase HeLa cells were subjected to p31comet to activate APC/CCdc20, and RanQ69L to release spindle assembly factors from importin inhibitors. As indicated, extracts were treated with taxol to stabilize microtubules. Control extracts were depleted of Cdc20, and the stability of endogenous substrates was monitored by immunoblotting. B. Microtubule-dependent stabilization of spindle assembly factors is reversible. Mitotic extracts were treated with taxol and RanQ69L. As indicated, nocodazole was added after 60min to depolymerize microtubules. C. Microtubules prevent APC/CCdh1-dependent degradation of spindle assembly factors. 35S-labeled HURP or cyclin B1 was added to G1-extracts of HeLa cells with active APC/CCdh1 and treated with taxol and RanQ69L. As control, the APC/CCdh1-inhibitor Emi1 was added, and substrate stability was monitored by autoradiography. See also Figure S1.
During cytokinesis and G1, the APC/C is activated by Cdh1 (Visintin et al., 1997). To determine whether microtubules stabilize spindle assembly factors against APC/CCdh1, we generated extracts of HeLa cells that were synchronized in G1. We supplemented these extracts with taxol and RanQ69L and then measured the stability of radiolabeled APC/C-substrates. As we had seen for APC/CCdc20, microtubules protected the spindle assembly factor HURP, but not soluble cyclin B1, from APC/C-dependent degradation (Figure 1C). Soluble HURP was stabilized by the APC/CCdh1-inhibitor Emi1, which attests to the specificity of these reactions.
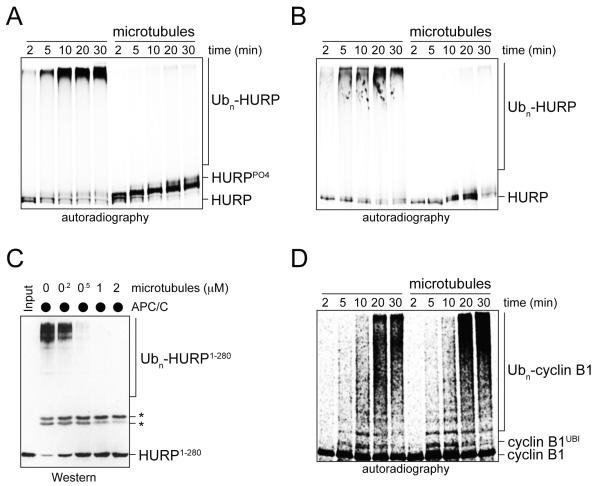
Prompted by these observations, we tested whether microtubules interfered with the ubiquitylation of spindle assembly factors by the APC/C. Indeed, the APC/CCdc20- and APC/CCdh1-dependent ubiquitylation of HURP, NuSAP, and Tpx2 were blocked by microtubules (Figure 2A, B; Figure S1C, D), with an efficiency comparable to that of Mad2, an established APC/CCdc20-inhibitor (Figure S1E). Microtubules also ablated the ubiquitylation of a bacterially purified HURP-domain that contained all APC/C-degrons and microtubule-binding domains (Figure 2C), whereas unpolymerized tubulin did not impede these reactions (Figure S1F). In contrast to their effects on spindle assembly factors, microtubules did not affect the modification of soluble APC/C-substrates, such as cyclin B1 (Figure 2D), nor did they interfere with the ubiquitylation of substrates of other E3s (Figure S1G). We conclude that microtubules protect spindle assembly factors from APC/C-dependent ubiquitylation and degradation in vitro.
Figure 2. Microtubules prevent ubiquitylation of spindle assembly factors by the APC/C.
A. Microtubules inhibit ubiquitylation of HURP by APC/CCdc20. 35S-labeled HURP was synthesized in vitro, purified, incubated with RanQ69L and subjected to ubiquitylation by APC/CCdc20. As indicated, HURP was pre-bound to microtubules. Reactions were monitored by autoradiography. B. Microtubules inhibit ubiquitylation of 35S-labeled HURP by APC/CCdh1 isolated from HeLa cells synchronized in G1. Reactions were analyzed in the presence of microtubules as described. C. Microtubules prevent the ubiquitylation of bacterially expressed HURP. HURP1-280, which contains all degrons and microtubule-binding domains, was incubated with microtubules and subjected to APC/C-dependent ubiquitylation. Reactions were analyzed by Western. The asterisks mark crossreactive bands. D. Microtubules do not affect the ubiquitylation of 35S-labeled cyclin B1 cyclin B1 by APC/CCdc20. See also Figure S1.
HURP contains two microtubule-binding domains
Multiple lines of evidence suggest that HURP, NuSAP, and Tpx2 are regulated by similar mechanisms: all proteins localize to a subset of spindle microtubules, the kinetochore fibers; their association with microtubules is inhibited by importins (Kalab and Heald, 2008); and their microtubule-regulated degradation is dependent upon the APC/C, which targets these spindle assembly factors at similar times during mitosis (Song and Rape, 2010; Stewart and Fang, 2005). Unfortunately, for none of these proteins were microtubule-binding domains sufficiently characterized to generate inactive mutants, a prerequisite for understanding how their degradation is controlled. Given the similarities in regulation, we decided to use HURP as a model substrate to define its microtubule-binding domains. We then introduced inactivating mutations into the respective domains to gain insight into the mechanism of microtubule-dependent APC/C-regulation.
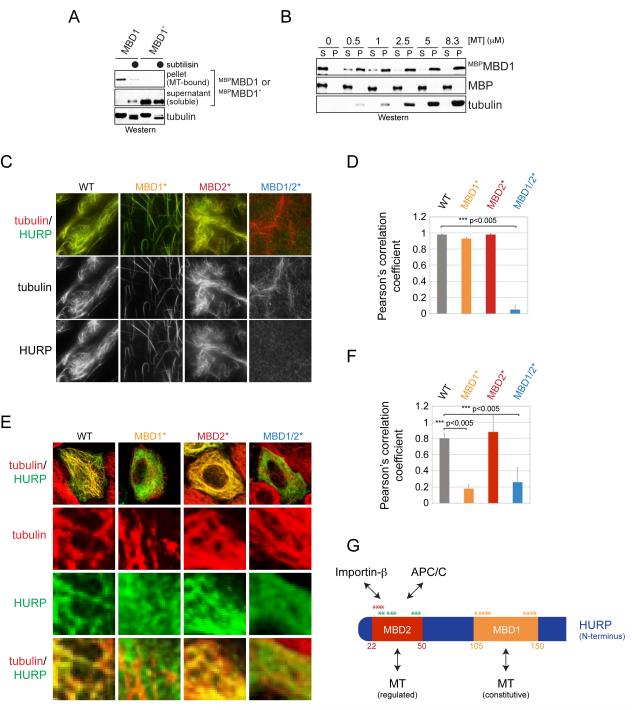
Previous work had located a microtubule-binding domain within the first 280 residues of HURP (Wong et al., 2008). By testing fragments that covered this region, we found a smaller motif, the MBD1, which associated with microtubules with similar affinity as the full-length protein (Figure 3A, B) (Wong and Fang, 2006). The interaction between microtubules and the MBD1 was disrupted by mutation of positively charged residues in the MBD1 or treatment of microtubules with subtilisin, which removes the negatively charged C-terminus of tubulin. Thus, the MBD1 is a microtubule-binding motif that likely recognizes the C-terminal tail of tubulin.
Figure 3. HURP contains two microtubule-binding motifs.
A. Taxol-stabilized microtubules were incubated with MBPMBD1 or mutant MBPMBD1* in which positively charged residues were exchanged to alanine. As indicated, microtubules were treated with subtilisin. Binding reactions were centrifuged through a sucrose cushion, and microtubule-bound and soluble fractions were analyzed by Western. B. The MBD1 binds microtubules with similar affinity as full-length HURP. MBPMBD1 was incubated with taxol-stabilized microtubules and subjected to sucrose gradient centrifugation. C. The MBD1 and MBD2 both mediate microtubule-binding of HURP. Recombinant full-length HURP or mutants in its MBD1 or MBD2 were labeled with Oregon Green and incubated with rhodamine-labeled microtubules. Binding was analyzed by fluorescence microscopy. D. Quantification of HURP- and microtubule co-localization in vitro. Green (HURP) and red (tubulin) signals were correlated using ImageJ and JACoP, resulting in a Pearson’s correlation coefficient. E. MBD1 mediates microtubule-binding of HURP in interphase. HeLa cells were transfected with FLAGHURP or mutants in its MBDs and analyzed by immunofluorescence microscopy (green: HURP; red: β-tubulin). F. Pearson’s correlation coefficient to quantify the co-localization between HURP-mutants and microtubules in interphase cells. G. Overview of binding motifs in the N-terminal domain of HURP. The MBD1 provides a constitutive binding site for microtubules; MBD2 overlaps with APC/C-degrons and the importin-β-binding site (Song and Rape, 2010). The asterisks mark residues mutated to alanine in order to ablate the functions of the MBD1 (orange), MBD2 (red), or APC/C-degrons (green). See also Figure S2.
To test whether the MBD1 was essential for the binding of HURP to microtubules, we generated Oregon Green-labeled HURPMBD1*, in which positively charged residues of the MBD1 were replaced with alanine. We incubated HURPMBD1* with rhodamine-labeled microtubules and monitored the interaction between the two partners by fluorescence microscopy. In these assays, mutation of the MBD1 strongly reduced the capacity of HURP to bundle, but not to bind, microtubules (Figure 3C, D), suggesting that a second domain provides an additional interaction surface. Indeed, a motif that we refer to as the MBD2 and that overlaps with the importin-β-binding site in HURP was enriched in positively charged residues, a common feature of microtubule-binding domains. When these charged residues were mutated in the background of HURPMBD1* (HURPMBD1/2*), all microtubule-binding of HURP was lost (Figure 3C, D). The MBD2, but not the MBD1, also bound importin-β in vitro and in cells (Figure S2A, B), and accordingly, importin-β inhibited the capacity of the MBD2 to associate with microtubules (Figure S2C). Thus, the MBD1 provides a constitutive high-affinity binding site for microtubules, whereas the weaker interaction between the MBD2 and microtubules is regulated by importin-β.
The results of our biochemical analyses were confirmed in vivo: when expressed during interphase, HURP and HURPMBD2* co-localized with microtubules, indicative of a functional interaction (Figure 3E, F). This association was strongly reduced by mutation of the MBD1 or simultaneous inactivation of the MBD1 and MBD2. These findings show that during interphase, the MBD1 is the dominant microtubule-binding domain in HURP. As described later, the MBD2 contributes to microtubule-binding of HURP during mitosis (see below), indicating that both the MBD1 and MBD2 can mediate an interaction of the spindle assembly factor HURP to microtubules (Figure 3G).
Binding to microtubules is necessary and sufficient for HURP stabilization
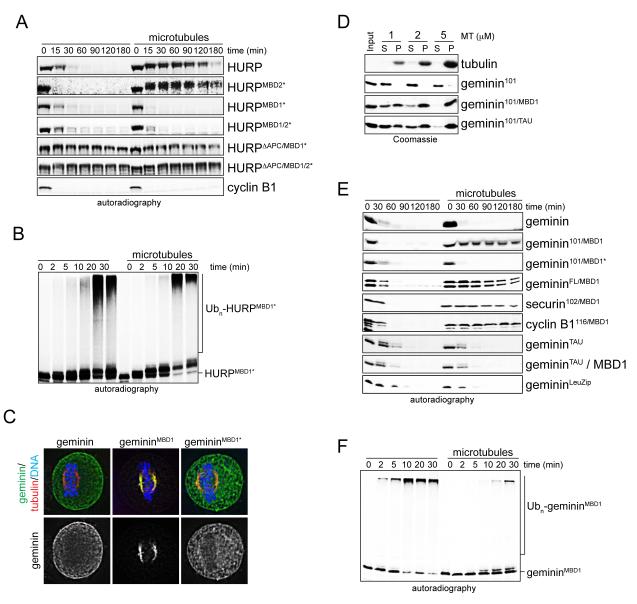
Having mapped HURP’s microtubule-binding motifs, we tested whether their integrity was required for the stabilization of APC/C-substrates by microtubules. We supplemented extracts containing APC/CCdc20, taxol, and RanQ69L with the respective HURP-mutants and monitored the degradation of these substrates over time. While microtubules stabilized HURP and HURPMBD2*, this effect was lost if the constitutive binding motif MBD1 was mutated (Figure 4A). The degradation of HURPMBD1* and HURPMBD1/2* was dependent upon the APC/C, as both proteins were stabilized by depletion of Cdc20 (Figure S3A), addition of Mad2 (Figure S3B), or mutation of APC/C-degrons (Figure 4A). Accordingly, in contrast to wild-type HURP (Figure 2A), microtubules did not inhibit the ubiquitylation of HURPMBD1* and HURPMBD1/2* by the APC/C (Figure 4B; Figure S3C). We conclude that the MBD1, but not the importin-β-regulated MBD2, is required for the stabilization of HURP by microtubules.
Figure 4. The MBD1 is required and sufficient for the regulation of HURP-stability by microtubules.
A. MBD1-mutation allows degradation of HURP in the presence of microtubules. Mitotic extracts with APC/CCdc20 and RanQ69L were supplemented with 35S-labeled HURP, HURP-mutants (MBD1*, MBD2*, ΔAPC), or cyclin B1, and degradation was monitored by autoradiography. As indicated, microtubules were stabilized by taxol. B. The MBD1 is required for the inhibition of HURP-ubiquitylation by microtubules. 35S-labeled HURPMBD1* was incubated with buffer or microtubules, before being subjected to ubiquitylation by APC/CCdc20. Reactions were followed by autoradiography. C. The MBD1 targets a soluble APC/C-substrate to the spindle. HeLa cells were transfected with geminin, gemininMBD1 (residues 1-101 of geminin fused to the MBD1 of HURP); or gemininMBD1* (charged residues in the MBD1 mutated to alanine), and localization was determined by immunofluorescence microscopy (green: geminin; red: tubulin; blue: DNA/DAPI). The bottom panel shows the localization of the geminin proteins alone. D. Fusions of HURP’s MBD1 or the N-terminal microtubule-binding domain of tau induce microtubule-binding of soluble APC/C-substrates with similar efficiency. Binding of recombinant proteins to microtubules was analyzed by sucrose gradient centrifugation (S: soluble fraction; P: microtubule-bound fraction). E. The MBD1 imposes microtubule-dependent regulation of degradation. 35S-labeled substrates were added to extracts with APC/CCdc20 and RanQ69L (MBD1: HURP’s MBD1; TAU: N-terminal microtubule-binding domain of tau; LeuZip: leucine zippers of GCN4 transcription factor). As indicated, microtubules were stabilized by taxol or the MBPMBD1 was added to induce microtubule bundling. Reactions were monitored by autoradiography. F. Microtubules inhibit the ubiquitylation of gemininMBD1. 35S-labeled gemininMBD1 was incubated with buffer or microtubules and subjected to ubiquitylation by APC/CCdc20. Reactions were monitored by autoradiography. See also Figure S3.
To test whether the MBD1 is sufficient to impose this regulatory mechanism onto APC/C-substrates, we fused it to the N-terminal half of geminin, a protein that is turned over by the APC/C in the presence of microtubules. The fusion gemininMBD1, but not WT-geminin, was targeted to microtubules in vitro and to the spindle in cells, confirming that the MBD1 mediates an interaction with microtubules during mitosis (Figure 4C, D). Importantly, degradation of gemininMBD1 in extracts and its ubiquitylation by the APC/C were strongly inhibited by microtubules (Figure 4E, F). Similar results were obtained in assays that monitored the stability of MBD1-fusions to various other APC/C-substrates. The mutation of positively charged residues within the MBD1 disrupted the spindle-binding of gemininMBD1 and ablated the regulation of its ubiquitylation and degradation by microtubules (Figures 4C, E; Figure S3D). Thus, the MBD1 is not only required, but also sufficient for the microtubule-dependent stabilization of HURP.
It is possible that any stable interaction might prevent the degradation of APC/C-substrates, or alternatively, this might be a function of the microtubule binding domains present in spindle assembly factors. To address this issue, we fused geminin to the N-terminal microtubule-binding domain of tau, a protein that interacts with microtubules in interphase, but does not act as a spindle assembly factor during mitosis (Cleveland et al., 1977). Although this domain afforded a similar affinity to microtubules as HURP’s MBD1 (Figure 4D), gemininTAU was not protected from APC/C-dependent degradation by microtubules (Figure 4E). GemininTAU was also not stabilized if the MBD1 was added in trans, showing that the MBD1 needs to be in the same peptide as the APC/C-degrons. Moreover, locking geminin into dimeric complexes by fusing it to the leucine zippers of the transcription factor GCN4 did not prevent its APC/C-dependent degradation (Figure 4E). Together, these experiments indicate that stable interactions are not sufficient to protect HURP. Instead, a specific property of the MBD1, such as its capacity to bundle microtubules, might be required for its role in stabilizing HURP on microtubules.
Microtubules stabilize HURP during mitosis
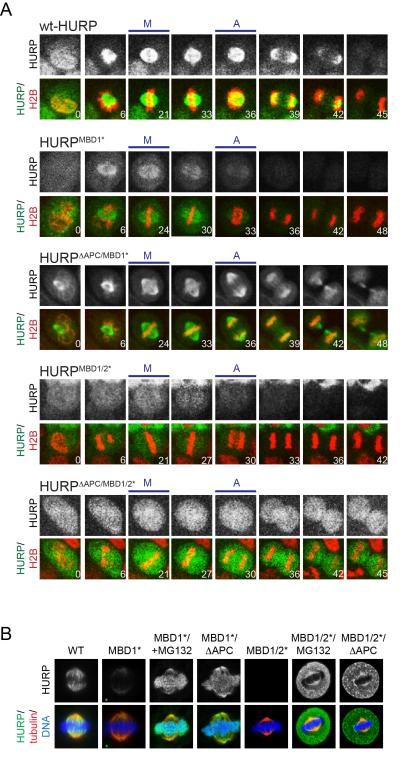
We next wished to determine whether microtubules stabilize spindle assembly factors in vivo. To this end, we expressed low levels of GFP-tagged HURP or its microtubule-binding deficient mutants using lentiviruses (Figure S4A) and monitored the abundance of these variants in HeLa cells by video microscopy. In agreement with analyses of endogenous HURP (Koffa et al., 2006; Sillje et al., 2006; Wong and Fang, 2006), GFP-tagged HURP accumulated on the spindle until telophase, and was completely turned over only after all sister chromatids had been distributed into the two daughter cells (Figure 5A). By contrast, HURPMBD1* bound the spindle with lower efficiency than WT HURP, and HURPMBD1/2* did not show significant enrichment at the spindle. Moreover, the degradation of HURPMBD1* and HURPMBD1/2* started much earlier than that of the wild-type protein, and both mutants were degraded beyond our detection limit shortly after sister chromatid separation had been initiated. Mutation of all APC/C-degrons stabilized HURPMBD1* and HURPMBD1/2* until G1, demonstrating that the premature degradation of these spindle assembly factor variants was carried out by the APC/C.
Figure 5. Microtubule-binding stabilizes HURP during mitosis.
A. HeLa cells were transduced with lentiviruses expressing mCherryhistone H2B and GFP-tagged HURP, HURPMBD1*, HURPΔAPC/MBD1/2*, HURPMBD1/2*, or HURPΔAPC/MBD1/2* (MBD1/2*: mutation of microtubule-binding motifs; ΔAPC: mutation of all degrons). Progression through mitosis was monitored by live cell imaging. The upper panels show the levels of the GFP-tagged HURP variant, whereas lower panels display GFP-HURP (green), mCherry-histone H2B (red), and the time post nuclear envelope breakdown. M: first frame with metaphase chromosome alignment; A: first frame with sister chromatid separation. B. HeLa cells were transfected with FLAGHURP, FLAGHURPMBD1* or FLAGHURPMBD1/2*; where indicated, APC/C-degrons were mutated (FLAGHURPΔAPC/MBD1*; FLAGHURPΔAPC/MBD1/2*) or cells were treated with MG132. Cells were analyzed by immunofluorescence microscopy. Upper panel: HURP-proteins; lower panel: HURP (green); tubulin (red); DNA (blue). See also Figure S4.
To independently assess whether microtubules protect spindle assembly factors from degradation, we measured the levels of microtubule-binding deficient mutants by immunofluorescence microscopy. As seen before, inactivation of the MBD1 or both the MBD1 and MBD2 caused a strong decrease in the abundance of HURP that was especially apparent shortly before anaphase (Figure 5B; Figure S4B, C). By contrast, HURP, HURPMBD1*, and HURPMBD1/2* were present at comparable levels in interphase when the APC/C was inactive (Figure S4B), or during mitosis, if the proteasome was inhibited, the APC/CCdc20-specific inhibitor Mad2 was overexpressed, or APC/C-degrons were mutated (Figure 5B; Figure S4C, D). In agreement with the premature degradation of soluble HURP, stable HURPΔAPC/MBD1*, but not HURPMBD1*, was detected on metaphase chromosomes, an observation that we also made for the GFP-tagged protein by live cell analysis (Figure 5A, B). Based on these findings, we conclude that microtubules prevent the premature degradation of HURP during mitosis. If this regulation is lost, the spindle assembly factor is highly susceptible to proteolysis and depleted from cells almost instantaneously after the APC/C has been activated.
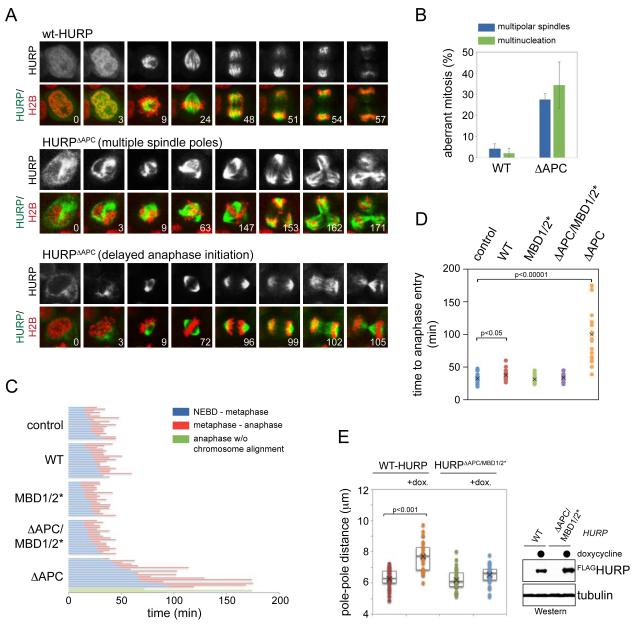
The instability of microtubule-binding deficient HURP implied that the removal of spindle assembly factors contributes to robust spindle function. To test this hypothesis, we filmed cells that expressed HURPΔAPC, a stable mutant that lacks all APC/C-degrons (Song and Rape, 2010), but retained its capacity to bind microtubules. Confirming APC/C’s role in targeting HURP, GFP-HURPΔAPC was not degraded during any stage of mitosis, allowing it to persistently bind microtubules throughout mitosis (Figure 6A). The stabilization of HURP resulted in strong mitotic defects: HURPΔAPC, but not WT-HURP or inactive HURPΔAPC/MBD1/2*, impaired the assembly of a metaphase plate with completely aligned chromosomes, induced formation of multiple spindle poles, and delayed anaphase onset (Figure 6A-D). Immunofluorescence analysis against γ-tubulin and centrin showed that expression of HURPΔAPC triggered spindle pole fragmentation, rather than centrosome amplification (Figure S5A, B). In agreement with these results, inducible overexpression of HURP, but not inactive HURPMBD1/2*, led to spindle defects, such as spindle elongation (Figure 6E). Thus, while losing its microtubule-dependent regulation caused premature degradation of HURP, the inappropriate stabilization of this spindle assembly factor interfered with accurate spindle function and, consequently, faithful cell division.
Figure 6. Stabilization of HURP interferes with spindle structure and function.
A. Expression of stabilized HURP in mitosis leads to persistent microtubule-binding and mitotic defects. HeLa cells transduced with lentiviruses expressing mCherryhistone H2B, and GFP-tagged HURP or HURPΔAPC, were filmed through mitosis. Upper panel: HURP; lower panel: GFP-tagged HURP (green), mCherryhistone H2B (red), and the time post nuclear envelope breakdown. B. Stabilization of HURP results in multiple spindle poles and multinucleation. Cells expressing GFP-HURP or GFP-HURPΔAPC were filmed through mitosis and analyzed for >2 spindle poles or multinucleation. Results were from three independent experiments, including at least 50 dividing cells per condition and per experiment. C. Expression of HURPΔAPC delays the assembly of a metaphase plate with completely aligned chromosomes. HeLa cells expressing GFP-tagged HURP or mutants were filmed through mitosis. The time from nuclear envelope breakdown to assembly of a metaphase plate (blue) and from metaphase to anaphase initiation (red) are shown. Expression of HURPΔAPC frequently caused anaphase initiation after a profound delay, even though a metaphase plate had never been established (green). D. Quantification of the time required for anaphase entry in the presence of HURP proteins. E. Increased levels of active HURP cause spindle defects. Stable 293T cell lines expressing HURP or HURPΔAPC/MBD1/2* under an inducible promoter were treated with doxycycline, and spindle structures were analyzed by microscopy against α- and γ-tubulin. Left panel: pole-pole distance in the presence or absence of HURP; right panel: inducible expression of HURP-variants. See also Figure S5.
The APC/C and importin-β cooperate to ensure spindle structure and function
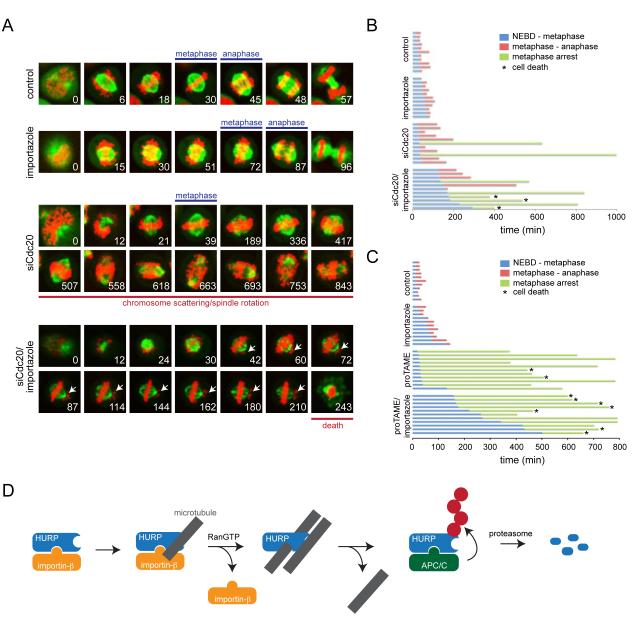
Contrary to the expression of HURPΔAPC, inhibition of the APC/C or the proteasome had little effects on spindle formation and only led to spindle breakdown after a prolonged metaphase arrest (Daum et al., 2011; Stevens et al., 2011; Zeng et al., 2010). The APC/C-dependent degradation of spindle assembly factors thus likely cooperates with another mechanism to ensure spindle function, and indeed, all microtubule-regulated APC/C-substrates studied here are also inhibited by members of the importin family. As HURPΔAPC was neither recognized by the APC/C nor importin-β (Figure S5C), the latter two regulators might cooperate in ensuring spindle function. To test this hypothesis, we inhibited the APC/C by depleting its activator Cdc20 (Figure S6A); importin-β by treating cells with importazole, a molecule that inactivates importin-β without stabilizing spindle assembly factors (Soderholm et al., 2011); or APC/C and importin-β by subjecting cells to siRNAs against Cdc20 and importazole at the same time. We then monitored cells that stably expressed mCherryhistone H2B and GFPtubulin by video microscopy.
In agreement with earlier reports (Daum et al., 2011; Stevens et al., 2011; Zeng et al., 2010), inhibition of the APC/C had no apparent effect on spindle formation, but often caused a metaphase arrest followed by cohesion fatigue and chromosome scattering, spindle elongation, and spindle rotation (Figure 7A, B). In other cases, cells depleted of Cdc20 were able to proceed into anaphase, albeit after a delay, which is consistent with reports that very low levels of Cdc20 are sufficient to support anaphase onset (Wolthuis et al., 2008). Also confirming published results (Soderholm et al., 2011), the inhibition of importin-β slightly delayed chromosome congression, yet cells eventually completed spindle assembly and initiated sister chromatid separation (Figure 7A, B). By contrast, when cells were concurrently treated with siRNAs against Cdc20 and importazole, a much larger fraction of cells arrested prior to anaphase, a metaphase plate with completely aligned chromosomes was rarely observed, and arrested cells died without visible chromosome scattering or spindle rotation (Figure 7A, B). Similar phenotypes were observed when importazole was combined with TAME (Figure 7C; Figure S6B, C), a compound that interferes with the activity of APC/CCdc20 (Zeng et al., 2010), or MG132, a molecule that blocks proteasomal degradation (Figure S6D). These findings were independent of the reporter used to monitor cell division, as we made similar observations when the APC/C and importin-β were inhibited in cells that stably expressed the kinetochore marker LAP/GFPCenpA (Figure S6E). Our experiments, therefore, confirm that neither the APC/C nor importin-β is essential for spindle formation. However, spindle function is highly compromised when the APC/C and importin-β are inhibited at the same time, a condition that simultaneously removes two players that restrict the activity of spindle assembly factors. We conclude that the rapid degradation of spindle assembly factors, a reaction that is tightly controlled by microtubules and importin-β, is a fundamental component of the control mechanisms that ensure the robustness and accuracy of metazoan cell division.
Figure 7. The APC/C and importin-β regulate spindle function.
A. Simultaneous inhibition of the APC/C and importin-β results in cooperative defects in spindle structure and function. HeLa cells stably expressing GFPtubulin and mCherryhistone H2B were treated with siRNAs against Cdc20, importazole, or siRNAs against Cdc20 and importazole at the same time. Progression of cells through mitosis was monitored by video microscopy (tubulin: green; histone: red; arrows: misaligned chromosomes). B. Quantification of the mitotic timing for HeLa cells expressing mCherryhistone H2B and GFPtubulin, after being treated with siRNAs against Cdc20 or importazole, as shown on the left. Blue: time from nuclear envelope breakdown to completion of chromosome alignment; red: time from metaphase to anaphase initiation; green: cells permanently arrested prior to anaphase. The asterisk marks cells undergoing cell death. C. The Cdc20-inhibitor proTAME displays similar functional interactions with importin-β as Cdc20-siRNAs. HeLa cells stably expressing mCherryhistone H2B and GFPtubulin were treated with proTAME, importazole, or both, and filmed through mitosis. Experiments were quantified as described. D. Model of microtubule-dependent regulation of APC/C-substrate degradation. Ran-dependent spindle assembly factors are inhibited by importin-β. As seen for HURP, importin-β does not ablate the microtubule-binding of a constitutive motif, the MBD1, indicating that spindle assembly factors might be loaded onto the spindle in their inactive state. RanGTP-dependent dissociation of HURP allows the MBD2 to engage microtubules, thereby activating the spindle assembly factor. Once HURP is not required, its release from microtubules results in rapid ubiquitylation by the APC/C and proteasomal degradation, a reaction that is required for maintaining proper spindle structure and function. See also Figure S6.
Discussion
In this study, we discovered localized stabilization as a mechanism that tightly controls the abundance of critical spindle assembly factors. At the heart of this regulatory circuit are microtubules, the main constituents of the mitotic spindle and direct binding partners of spindle assembly factors. When bound to microtubules, HURP, NuSAP, and Tpx2 are protected from APC/C-dependent degradation. By contrast, if these substrates are not associated with the spindle, they are highly susceptible to degradation and turned over shortly after the APC/C has been fully activated at the metaphase-anaphase transition. It is well established that spindle defects indirectly result in APC/C-inhibition by triggering the spindle assembly checkpoint, a signaling network that impedes the capacity of APC/CCdc20 to recognize its substrates. Our current work suggests that the spindle also directly inhibits the turnover of specific APC/C-substrates, a mechanism that is important for accurate cell division.
Based on our findings, we propose a framework for the temporal and spatial regulation of spindle assembly factors (Figure 7D): HURP, NuSAP, or Tpx2 are initially sequestered by importins, which inhibit and stabilize Ran-dependent spindle assembly factors (Kalab and Heald, 2008; Song and Rape, 2010). As importin-β does not block HURP’s high-affinity microtubule-binding domain, spindle assembly factors could be loaded onto the spindle in their inactive, importin-bound states. In contrast to surgical mutations in microtubule- and importin-β binding sites, expression of dominant negative importin- or Ran-variants displaces HURP from the spindle (Sillje et al., 2006), suggesting that the global interference with the Ran-gradient also has secondary effects upon spindle assembly factor targeting. In proximity to chromatin, RanGTP dissociates HURP from importin-β, thereby activating the spindle assembly factor by allowing its MBD2 to engage microtubules. Importantly, as the MBD1 is already bound to microtubules, the active spindle assembly factor remains protected from recognition by the APC/C. When HURP is no longer required, it will be released from the spindle, and -as suggested by the instantaneous degradation of soluble HURP upon APC/C-activation - rapidly degraded. In this model, only spindle assembly factors that have fulfilled their mitotic role are turned over: inactive spindle assembly factors are stabilized by importin-β, whereas those molecules that are engaged in spindle formation are protected by microtubules. The microtubule-dependent regulation of APC/C-substrate degradation, therefore, effectively couples the activity and stability of critical cell cycle proteins.
How microtubules stabilize specific APC/C-substrates requires further analyses, but our results indicate that a direct interaction with spindle assembly factors plays an important role: whereas microtubules stabilized HURP, NuSAP and Tpx2, they had no effects on soluble APC/C-substrates. Moreover, the mutation of HURP’s MBD1 allowed the degradation of this spindle assembly factor in the presence of microtubules, while the transfer of the MBD1 to soluble APC/C-substrates was sufficient to impose this control mechanism. By associating with spindle assembly factors, microtubules could impede the recognition of substrates by the APC/C or interfere with rate-limiting steps of the ubiquitylation reaction, such as chain initiation (Williamson et al., 2011). Potentially pointing towards the latter mechanism, the MBD1 does not overlap with the D-box, KEN-box or initiation motif in HURP (Song and Rape, 2010), and its transfer to other substrates allowed their stabilization even though these proteins had their degrons at distinct positions than HURP. These findings make it seem unlikely that microtubules simply shield a critical degron from recognition by the APC/C. Instead, we favor the idea that the MBD1 confers a property onto microtubules, such as bundling, that is not conducive to the build-up of ubiquitin chains by the APC/C. Indeed, all spindle assembly factors subject to this regulatory mechanism bundle microtubules, and HURP requires the MBD1 for this function. Moreover, a microtubule-binding domain that does not bundle microtubules failed to impose microtubule-dependent stabilization, even though it could target APC/C-substrates to microtubules with similar efficiency as the MBD1.
As microtubule-bundlers often accumulate on kinetochore fibers, but not astral microtubules, this mechanism of stabilization might be of particular relevance for the critical subset of spindle microtubules that mediate chromosome attachment and sister chromatid separation. Supporting this notion, the stabilization of HURP impaired the assembly of a metaphase plate with completely aligned chromosomes. This result is consistent with the finding that overexpression of spindle assembly factors can impair cell division and lead to tumorigenesis, and that it has been inversely correlated with therapeutic outcome (Aguirre-Portoles et al., 2012; Gulzar et al., 2013; Perez de Castro and Malumbres, 2012; Tsou et al., 2003). Without proper APC/C-activity, spindle assembly factors might accumulate over multiple cell cycles and exert their effects due to a gradual increase in their abundance. It is also possible that a fraction of spindle assembly factor molecules are degraded immediately after the last kinetochore has been attached to the spindle, a hypothesis that is consistent with the high processivity of HURP ubiquitylation (Rape et al., 2006; Song and Rape, 2010). Alternatively, spindle assembly factor molecules that were released from microtubules might need to be turned over even during spindle formation and checkpoint signaling, potentially to ensure microtubule dynamics. In agreement with this hypothesis, proteasome inhibition stabilized the microtubule-binding deficient HURP in mitotic cells that had retained high levels of kinetochore-bound Mad1 and BubR1 (data not shown), a condition that usually signals an active spindle checkpoint (Kim and Yu, 2011; Musacchio and Salmon, 2007).
Our work underscores the notion that two essential cell cycle regulators, the APC/C and Ran, cooperate in establishing a robust spindle that can effectively separate sister chromatids during mitosis. Similar to the misregulation of spindle assembly factors, aberrant activity of the APC/C or Ran has been linked to tumorigenesis (Garcia-Higuera et al., 2008; Jung et al., 2006; Kalab and Heald, 2008; Manchado et al., 2010; van Ree et al., 2010; Wagner et al., 2004). Understanding how ubiquitin-dependent proteolysis is integrated with other mitotic regulators, such as Ran, to allow faithful cell division and how this interplay between multiple levels of cell cycle control is disrupted in disease will be an important avenue for future work.
Materials and Methods
Plasmids, siRNAs, antibodies and proteins
cDNAs encoding APC/C-substrates were cloned into pCS2 for IVT/T and into pCS2-HA and pcDNA5/FRT/TO (Invitrogen) for expression in cells and stable cell line generation. A N-terminal Flag-tag was introduced into pcDNA5/FRT/TO by PCR. MBD1 consisted of residues 65-174 of HURP, and MBD2 consisted of the first 69 residues. HURPMBD1* & HURPMBD2* were generated by replacing the following residues with Ala: MBD1*: K105, K107, R110, K112, K114, R115; MBD2*: R20, K22, R26, K27. HURPΔAPC was prepared by mutating D-, KEN-, and TEK-boxes in HURP as described (Song and Rape, 2010). The gemininMBD1 fusion was generated by fusing the first 101 residues of geminin to the MBD1 by hybrid PCR. For recombinant proteins, cDNAs were cloned into pMAL and pET28 for expression in E.coli, and the purification procedures for MBP-tagged and 6xHis-tagged proteins were as described (Song and Rape, 2010).
ON-TARGETplus Human Cdc20 siRNA-SMARTpool was purchased from Dharmacon.
The following antibodies were used: αHURP & αimportin β (polyclonal; Bethyl Laboratories); αNuSAP (polyclonal; ProteinTech Group); αTpx2 (polyclonal; Novus); αCdc27 [AF3.1], αCdc20/p55CDC [E-7], αC-Myc [9E10] (monoclonal; Santa Cruz Biotechnology); αcyclin B1, αgeminin, αsecurin, αGFP & αANAPC2 (polyclonal; Santa Cruz Biotechnology); αUbe2S (polyclonal; Novus); αMBP (monoclonal; NEB Biolab); αtubulin [DM1A] (monoclonal; Calbiochem); αFlag M2 (monoclonal), αFlag (polyclonal) & α γtubulin (monoclonal) (Sigma); αHA [C29F4] (monoclonal; Cell Signaling); αcentrin [20H5] (monoclonal; Millipore); αMad2 (monoclonal; BD Biosciences); αβ-tubulin (Developmental Studies Hybridoma Bank); Goat anti-rabbit Alexa488 (Invitrogen); Donkey anti-mouse DyLight 549, Goat anti-mouse DyLight 549 (Jackson Laboratories).
In Vitro Degradation Assays
In vitro degradation assays were performed as described (Song and Rape, 2010). To test microtubule-dependent stabilization of substrates, 20 μM paclitaxel/taxol (Sigma) was added to HeLa S3 extracts (pre-warmed to 30 °C) to promote microtubule polymerization; where indicated, 140 μM nocodazole (Sigma) was added to destabilize microtubules. To deplete Cdc20, 400μl mitotic extract was depleted twice at 4°C for 1h μg monoclonal αCdc20 antibody.
In Vitro Ubiquitylation Reactions
APC/C- or SCFβTrCP-dependent ubiquitylation of 35S-labeled substrates was performed as described (Song and Rape, 2010; Wickliffe et al., 2011). As indicated, 1μg microtubules were pre-incubated with purified substrates (Williamson et al., 2011) at 25°C for 5min before being added to ubiquitylation reactions. For ubiquitylation of recombinant HURP, ~0.25μM Flag-tagged HURP1-280 was used.
HURP-Microtubule interaction assays
Taxol-stabilized microtubules were prepared by incubating pre-warmed porcine brain tubulin with 80μM taxol at 37°C for 10min. Polymerized microtubules were stabilized by bringing the final concentration of taxol to 160μM. Subtilisin-treated microtubules were prepared by incubating taxol-stabilized microtubules with 200μg/ml subtilisin (gift from Eva Nogales) at 37°C for 30min. The reaction was stopped by 2mM PMSF. Microtubules were spun down at room temperature (RT) at 14K rpm for 20min, and pellets were resuspended in binding buffer consisting of 1xBRB80 (80mM PIPES, 1mM MgCl2, 1mM EGTA, pH 6.8), 1mM DTT, 5% sucrose and 20μM taxol.
For co-pelleting experiments, 250nM MBPMBD1 was incubated with control or subtilisin-treated microtubules in a 20μl reaction for 10min at RT. Reactions were spun at 14K rpm for 20min at RT. The supernatant and pellet fractions were solubilized in 2x Laemmli buffer. 10% of supernatant and pellet were subjected to αMBP immunoblot or Coomassie.
For monitoring the HURP-microtubule interaction by microscopy, MBPHURP was labeled with Oregon Green 488 iodoacetamide (Invitrogen), cleaned by PD SpinTrap G-25 columns (GE Healthcare), and mixed with rhodamine-labeled microtubules. As indicated, 6xHisimportin-β was pre-incubated with labeled HURP at equal molar ratio for 5min. Images were taken using 60x magnification on an Olympus IX81 microscope, and processed using ImageJ.
Cell culture, transfection, and immunofluorescence
HeLa cells were cultured in DMEM containing 10% FBS. For plasmid transfection, cells were plated on coverslips and transfected using TransIT-LT1 (Mirus). For siRNA, cells were transfected with 50nM oligos using RNAiMAX (Invitrogen). Cells were fixed 48h post transfection for immunofluorescence or subjected to live cell imaging 24h post transfection. For drug treatment, cells were treated with 20μM MG132, 20μM proTAME (Boston Biochem; gift from Randy King) or 20μM importazole (gift from Rebecca Heald) for 6~8h before fixation. Cells were fixed with cold methanol (-20 °C) for 3min or 3.7% formaldehyde for 20min. Images were taken using Zeiss LSM 710 confocal microscope or Olympus IX81 microscope, deconvolved using Metamorph, and processed using ImageJ.
Flp-In T-REx 293 cells to stably express Flag-tagged HURP were generated following manufacturer’s manual (Invitrogen). For immunofluorescence, cells were plated on coverslips coated with poly-D-lysine and induced with 0.5μg/ml doxycycline for 48h before fixation.
Production of Lentiviruses and Transduction
eGFP-tagged HURP and mCherry-tagged histone H2B were cloned into pEF-1α/pENTR vector and recombined into pLenti X1 DEST by LR recombination to generate lentiviral expression constructs. Lentiviruses were produced in 293T cells by co-transfection of lentiviral constructs with packaging plasmids (Addgene) for 48~72 hours. Transduction was carried out by infecting ~50% confluent HeLa cells with lentiviruses in the presence of 6 μg/ml Polybrene (Sigma).
Live Cell Imaging
HeLa cells that stably express mCherry-H2B and GFP-tubulin (gift from Rebecca Heald) or LAP-tagged CenpA (gift from Iain Cheeseman) were maintained in DMEM-phenol red media containing 10% FBS at 37 °C with 5% CO2, and imaged at a single focal plane every 3min for the indicated time periods beginning 4h post drug treatment. Images were taken with a 0.95 NA 40x objective on an Olympus Revolution XD spinning disk confocal microscope equipped with a charged-couple device camera (Andor technology) and a Yokogawa spinning disk. Movies were assembled using Metamorph and analyzed with Photoshop.
Cell synchronization, immunoprecipitation and pulldown assays
Cell synchronization, immunoprecipitation and MBP pull-down assays were performed as previously described (Song and Rape, 2010).
Supplementary Material
Highlights.
Several Ran-dependent spindle assembly factors are stabilized by microtubules.
Soluble spindle assembly factors are rapidly degraded dependent upon the APC/C.
Loss of spindle assembly factor degradation by the APC/C disrupts spindle function.
The APC/C and Ran cooperate to ensure robust spindle formation.
Acknowledgements
We are grateful to Randy King for sending us proTAME, Rebecca Heald for importazole and stable cell lines, Gabriel Lander and Eva Nogales for providing subtilisin, Iain Cheeseman for the GFPCenpA cell line, and Priscilla Cooper for help with lentivirus production. We thank Julia Schaletzky, Rebecca Heald, and Karsten Weis for comments on the manuscript, and all members of our lab for discussions and support. This work was funded by grants of the NIH and March of Dimes to MR. AC is an NSF predoctoral fellow. MR is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguirre-Portoles C, Bird AW, Hyman A, Canamero M, Perez de Castro I, Malumbres M. Tpx2 controls spindle integrity, genome stability, and tumor development. Cancer Res. 2012;72:1518–1528. doi: 10.1158/0008-5472.CAN-11-1971. [DOI] [PubMed] [Google Scholar]
- Buschhorn BA, Petzold G, Galova M, Dube P, Kraft C, Herzog F, Stark H, Peters JM. Substrate binding on the APC/C occurs between the coactivator Cdh1 and the processivity factor Doc1. Nature structural & molecular biology. 2011;18:6–13. doi: 10.1038/nsmb.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao WC, Kulkarni K, Zhang Z, Kong EH, Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484:208–213. doi: 10.1038/nature10896. [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Hwo SY, Kirschner MW. Purification of tau, a microtubule-associated protein that induces assembly of microtubules from purified tubulin. Journal of molecular biology. 1977;116:207–225. doi: 10.1016/0022-2836(77)90213-3. [DOI] [PubMed] [Google Scholar]
- da Fonseca PC, Kong EH, Zhang Z, Schreiber A, Williams MA, Morris EP, Barford D. Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature. 2011;470:274–278. doi: 10.1038/nature09625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum JR, Potapova TA, Sivakumar S, Daniel JJ, Flynn JN, Rankin S, Gorbsky GJ. Cohesion fatigue induces chromatid separation in cells delayed at metaphase. Curr Biol. 2011;21:1018–1024. doi: 10.1016/j.cub.2011.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Foster SA, Morgan DO. The APC/C subunit Mnd2/Apc15 promotes Cdc20 autoubiquitination and spindle assembly checkpoint inactivation. Molecular cell. 2012;47:921–932. doi: 10.1016/j.molcel.2012.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gable A, Qiu M, Titus J, Balchand S, Ferenz NP, Ma N, Collins ES, Fagerstrom C, Ross JL, Yang G, et al. Dynamic reorganization of Eg5 in the mammalian spindle throughout mitosis requires dynein and TPX2. Molecular biology of the cell. 2012;23:1254–1266. doi: 10.1091/mbc.E11-09-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I, Manchado E, Dubus P, Canamero M, Mendez J, Moreno S, Malumbres M. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat Cell Biol. 2008;10:802–811. doi: 10.1038/ncb1742. [DOI] [PubMed] [Google Scholar]
- Garnett MJ, Mansfeld J, Godwin C, Matsusaka T, Wu J, Russell P, Pines J, Venkitaraman AR. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat Cell Biol. 2009;11:1363–1369. doi: 10.1038/ncb1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, Mattaj IW. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell. 2001;104:83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- Gruss OJ, Wittmann M, Yokoyama H, Pepperkok R, Kufer T, Sillje H, Karsenti E, Mattaj IW, Vernos I. Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nature cell biology. 2002;4:871–879. doi: 10.1038/ncb870. [DOI] [PubMed] [Google Scholar]
- Gulzar ZG, McKenney JK, Brooks JD. Increased expression of NuSAP in recurrent prostate cancer is mediated by E2F1. Oncogene. 2013;32:70–77. doi: 10.1038/onc.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog F, Primorac I, Dube P, Lenart P, Sander B, Mechtler K, Stark H, Peters JM. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science. 2009;323:1477–1481. doi: 10.1126/science.1163300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Wu G, Rusch M, Lukes L, Buetow KH, Zhang J, Hunter KW. Integrated cross-species transcriptional network analysis of metastatic susceptibility. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3184–3189. doi: 10.1073/pnas.1117872109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CR, Hwang KS, Yoo J, Cho WK, Kim JM, Kim WH, Im DS. E2-EPF UCP targets pVHL for degradation and associates with tumor growth and metastasis. Nat Med. 2006;12:809–816. doi: 10.1038/nm1440. [DOI] [PubMed] [Google Scholar]
- Kalab P, Heald R. The RanGTP gradient - a GPS for the mitotic spindle. J Cell Sci. 2008;121:1577–1586. doi: 10.1242/jcs.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Yu H. Mutual regulation between the spindle checkpoint and APC/C. Semin Cell Dev Biol. 2011 doi: 10.1016/j.semcdb.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koffa MD, Casanova CM, Santarella R, Kocher T, Wilm M, Mattaj IW. HURP is part of a Ran-dependent complex involved in spindle formation. Curr Biol. 2006;16:743–754. doi: 10.1016/j.cub.2006.03.056. [DOI] [PubMed] [Google Scholar]
- Komander D, Rape M. The ubiquitin code. Annual review of biochemistry. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Kufer TA, Sillje HH, Korner R, Gruss OJ, Meraldi P, Nigg EA. Human TPX2 is required for targeting Aurora-A kinase to the spindle. The Journal of cell biology. 2002;158:617–623. doi: 10.1083/jcb.200204155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchado E, Guillamot M, de Carcer G, Eguren M, Trickey M, Garcia-Higuera I, Moreno S, Yamano H, Canamero M, Malumbres M. Targeting mitotic exit leads to tumor regression in vivo: Modulation by Cdk1, Mastl, and the PP2A/B55alpha, delta phosphatase. Cancer Cell. 2010;18:641–654. doi: 10.1016/j.ccr.2010.10.028. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Perez de Castro I, Malumbres M. Mitotic Stress and Chromosomal Instability in Cancer: The Case for TPX2. Genes Cancer. 2012;3:721–730. doi: 10.1177/1947601912473306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Rape M, Kirschner MW. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature. 2004;432:588–595. doi: 10.1038/nature03023. [DOI] [PubMed] [Google Scholar]
- Rape M, Reddy SK, Kirschner MW. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell. 2006;124:89–103. doi: 10.1016/j.cell.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Reddy SK, Rape M, Margansky WA, Kirschner MW. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446:921–925. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- Ribbeck K, Groen AC, Santarella R, Bohnsack MT, Raemaekers T, Kocher T, Gentzel M, Gorlich D, Wilm M, Carmeliet G, et al. NuSAP, a mitotic RanGTP target that stabilizes and cross-links microtubules. Mol Biol Cell. 2006;17:2646–2660. doi: 10.1091/mbc.E05-12-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber A, Stengel F, Zhang Z, Enchev RI, Kong EH, Morris EP, Robinson CV, da Fonseca PC, Barford D. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature. 2011;470:227–232. doi: 10.1038/nature09756. [DOI] [PubMed] [Google Scholar]
- Sillje HH, Nagel S, Korner R, Nigg EA. HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr Biol. 2006;16:731–742. doi: 10.1016/j.cub.2006.02.070. [DOI] [PubMed] [Google Scholar]
- Soderholm JF, Bird SL, Kalab P, Sampathkumar Y, Hasegawa K, Uehara-Bingen M, Weis K, Heald R. Importazole, a small molecule inhibitor of the transport receptor importin-beta. ACS Chem Biol. 2011;6:700–708. doi: 10.1021/cb2000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Rape M. Regulated degradation of spindle assembly factors by the anaphase-promoting complex. Mol Cell. 2010;38:369–382. doi: 10.1016/j.molcel.2010.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens D, Gassmann R, Oegema K, Desai A. Uncoordinated loss of chromatid cohesion is a common outcome of extended metaphase arrest. PLoS One. 2011;6:e22969. doi: 10.1371/journal.pone.0022969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S, Fang G. Anaphase-promoting complex/cyclosome controls the stability of TPX2 during mitotic exit. Mol Cell Biol. 2005;25:10516–10527. doi: 10.1128/MCB.25.23.10516-10527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W, Li B, Warrington R, Tomchick DR, Yu H, Luo X. Structural analysis of human Cdc20 supports multisite degron recognition by APC/C. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:18419–18424. doi: 10.1073/pnas.1213438109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CY, Chou CK, Yang CW, Lai YC, Liang CC, Chen CM, Tsai TF. Hurp deficiency in mice leads to female infertility caused by an implantation defect. J Biol Chem. 2008;283:26302–26306. doi: 10.1074/jbc.C800117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou AP, Yang CW, Huang CY, Yu RC, Lee YC, Chang CW, Chen BR, Chung YF, Fann MJ, Chi CW, et al. Identification of a novel cell cycle regulated gene, HURP, overexpressed in human hepatocellular carcinoma. Oncogene. 2003;22:298–307. doi: 10.1038/sj.onc.1206129. [DOI] [PubMed] [Google Scholar]
- Uehara R, Goshima G. Functional central spindle assembly requires de novo microtubule generation in the interchromosomal region during anaphase. J Cell Biol. 2010;191:259–267. doi: 10.1083/jcb.201004150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzunova K, Dye BT, Schutz H, Ladurner R, Petzold G, Toyoda Y, Jarvis MA, Brown NG, Poser I, Novatchkova M, et al. APC15 mediates CDC20 autoubiquitylation by APC/C(MCC) and disassembly of the mitotic checkpoint complex. Nature structural & molecular biology. 2012;19:1116–1123. doi: 10.1038/nsmb.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ree JH, Jeganathan KB, Malureanu L, van Deursen JM. Overexpression of the E2 ubiquitin-conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. J Cell Biol. 2010;188:83–100. doi: 10.1083/jcb.200906147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varetti G, Guida C, Santaguida S, Chiroli E, Musacchio A. Homeostatic control of mitotic arrest. Molecular cell. 2011;44:710–720. doi: 10.1016/j.molcel.2011.11.014. [DOI] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Wagner KW, Sapinoso LM, El-Rifai W, Frierson HF, Butz N, Mestan J, Hofmann F, Deveraux QL, Hampton GM. Overexpression, genomic amplification and therapeutic potential of inhibiting the UbcH10 ubiquitin conjugase in human carcinomas of diverse anatomic origin. Oncogene. 2004;23:6621–6629. doi: 10.1038/sj.onc.1207861. [DOI] [PubMed] [Google Scholar]
- Williamson A, Banerjee S, Zhu X, Philipp I, Iavarone AT, Rape M. Regulation of ubiquitin chain initiation to control the timing of substrate degradation. Molecular cell. 2011;42:744–757. doi: 10.1016/j.molcel.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson A, Wickliffe KE, Mellone BG, Song L, Karpen GH, Rape M. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc Natl Acad Sci U S A. 2009;106:18213–18218. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Wilm M, Karsenti E, Vernos I. TPX2, A novel xenopus MAP involved in spindle pole organization. The Journal of cell biology. 2000;149:1405–1418. doi: 10.1083/jcb.149.7.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolthuis R, Clay-Farrace L, van Zon W, Yekezare M, Koop L, Ogink J, Medema R, Pines J. Cdc20 and Cks direct the spindle checkpoint-independent destruction of cyclin A. Mol Cell. 2008;30:290–302. doi: 10.1016/j.molcel.2008.02.027. [DOI] [PubMed] [Google Scholar]
- Wong J, Fang G. HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J Cell Biol. 2006;173:879–891. doi: 10.1083/jcb.200511132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J, Lerrigo R, Jang CY, Fang G. Aurora A regulates the activity of HURP by controlling the accessibility of its microtubule-binding domain. Molecular biology of the cell. 2008;19:2083–2091. doi: 10.1091/mbc.E07-10-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Merbl Y, Huo Y, Gallop JL, Tzur A, Kirschner MW. UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proc Natl Acad Sci U S A. 2010;107:1355–1360. doi: 10.1073/pnas.0912802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, King RW, Peters JM, Kirschner MW. Identification of a novel ubiquitin-conjugating enzyme involved in mitotic cyclin degradation. Curr Biol. 1996;6:455–466. doi: 10.1016/s0960-9822(02)00513-4. [DOI] [PubMed] [Google Scholar]
- Zeng X, Sigoillot F, Gaur S, Choi S, Pfaff KL, Oh DC, Hathaway N, Dimova N, Cuny GD, King RW. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell. 2010;18:382–395. doi: 10.1016/j.ccr.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.