Abstract
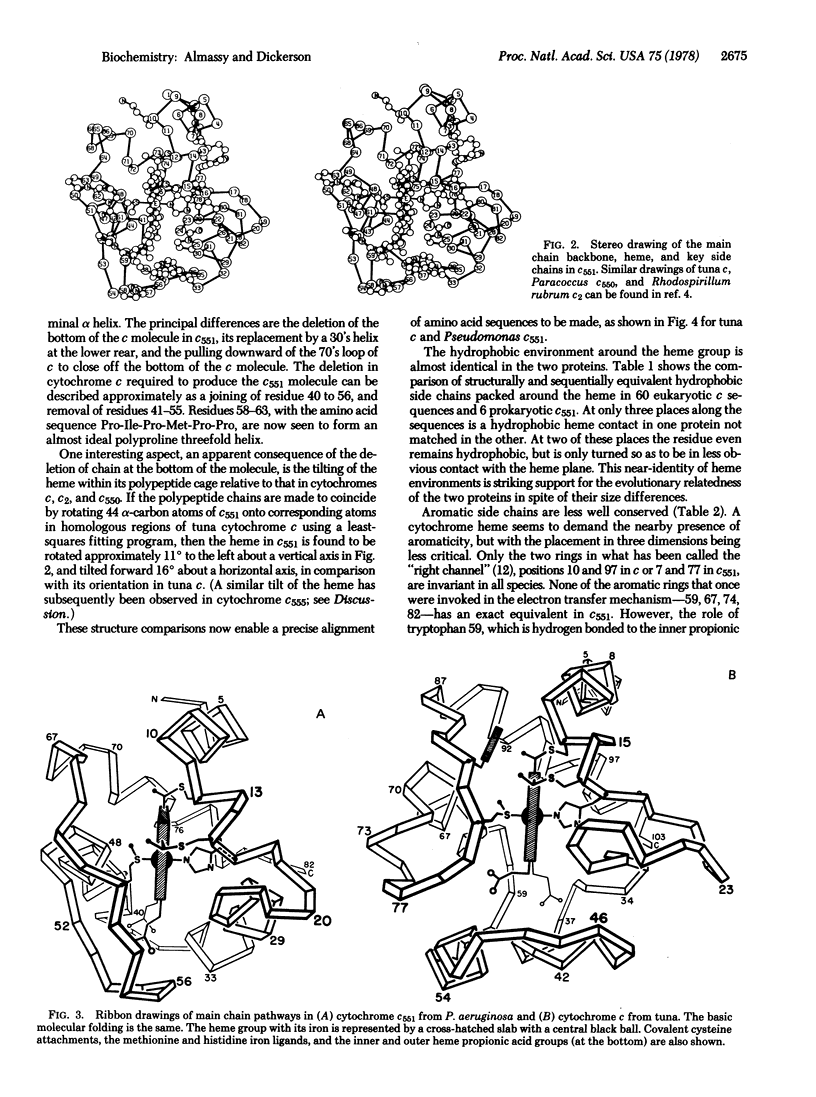
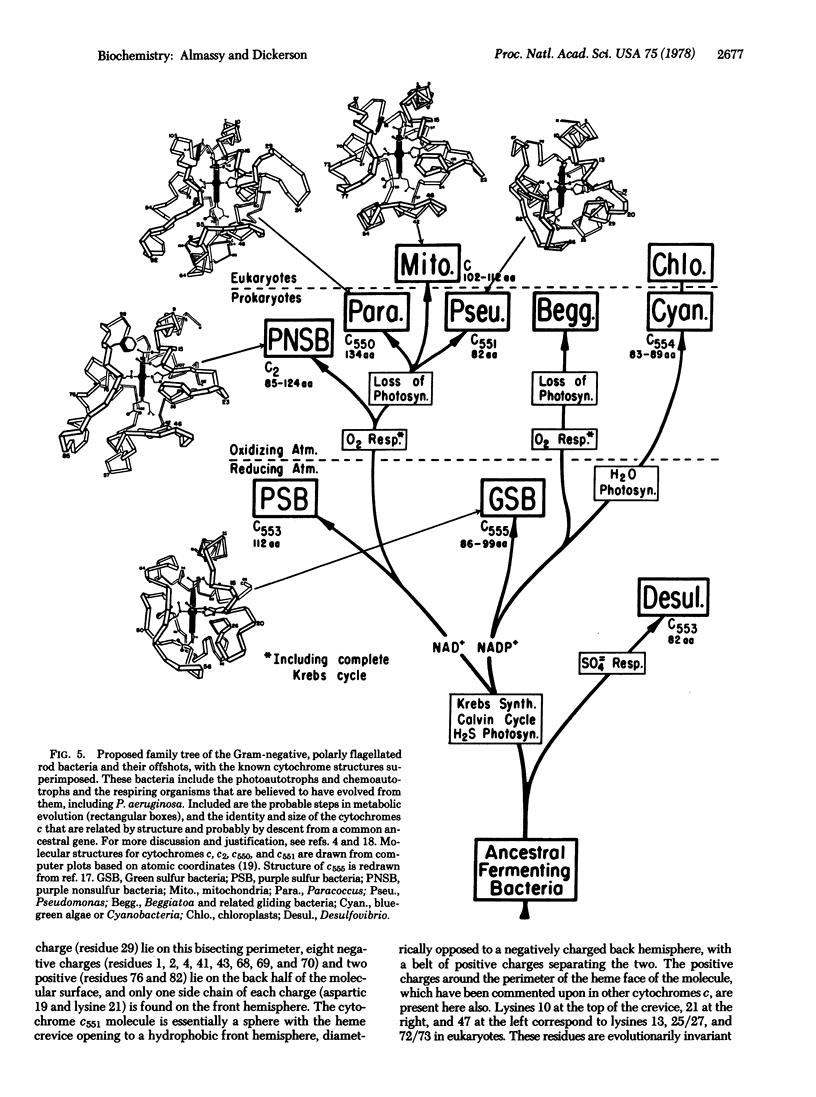
The structure of respiratory cytochrome c551 of Pseudomonas aeruginosa, with 82 amino acids, has been solved by x-ray analysis and refined to a crystallographic R factor of 16.2%. It has the same basic folding pattern and hydrophobic heme environment as cytochromes c, c2, and c550, except for a large deletion at the bottom of the heme crevice. This same "cytochrome fold" appears to be present in photosynthetic cytochromes c of green and purple sulfur bacteria, and algal cytochromes f, suggesting a common evolutionary origin for electron transport chains in photosynthesis and respiration.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- AMBLER R. P. THE AMINO ACID SEQUENCE OF PSEUDOMONAS CYTOCHROME C-551. Biochem J. 1963 Nov;89:349–378. doi: 10.1042/bj0890349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambler R. P., Meyer T. E., Kamen M. D. Primary structure determination of two cytochromes c2: close similarity to functionally unrelated mitochondrial cytochrome C. Proc Natl Acad Sci U S A. 1976 Feb;73(2):472–475. doi: 10.1073/pnas.73.2.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson R. E. Sequence and structure homologies in bacterial and mammalian-type cytochromes. J Mol Biol. 1971 Apr 14;57(1):1–15. doi: 10.1016/0022-2836(71)90116-1. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Takano T., Eisenberg D., Kallai O. B., Samson L., Cooper A., Margoliash E. Ferricytochrome c. I. General features of the horse and bonito proteins at 2.8 A resolution. J Biol Chem. 1971 Mar 10;246(5):1511–1535. [PubMed] [Google Scholar]
- Dickerson R. E., Timkovich R., Almassy R. J. The cytochrome fold and the evolution of bacterial energy metabolism. J Mol Biol. 1976 Feb 5;100(4):473–491. doi: 10.1016/s0022-2836(76)80041-1. [DOI] [PubMed] [Google Scholar]
- HORIO T., HIGASHI T., SASAGAWA M., KUSAI K., NAKAI M., OKUNUKI K. Preparation of crystalline Pseudomonas cvtochrome c-551 and its general properties. Biochem J. 1960 Oct;77:194–201. doi: 10.1042/bj0770194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korszun Z. R., Salemme F. R. Structure of cytochrome c555 of Chlorobium thiosulfatophilum: primitive low-potential cytochrome c. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5244–5247. doi: 10.1073/pnas.74.12.5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel N., Mandel G., Trus B. L., Rosenberg J., Carlson G., Dickerson R. E. Tuna cytochrome c at 2.0 A resolution. III. Coordinate optimization and comparison of structures. J Biol Chem. 1977 Jul 10;252(13):4619–4636. [PubMed] [Google Scholar]
- McLachlan A. D. Tests for comparing related amino-acid sequences. Cytochrome c and cytochrome c 551 . J Mol Biol. 1971 Oct 28;61(2):409–424. doi: 10.1016/0022-2836(71)90390-1. [DOI] [PubMed] [Google Scholar]
- Needleman S. B., Blair T. T. Homology of Pseudomonas cytochrome c-551 with eukaryotic c-cytochromes. Proc Natl Acad Sci U S A. 1969 Aug;63(4):1227–1233. doi: 10.1073/pnas.63.4.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salemme F. R., Freer S. T., Xuong N. H., Alden R. A., Kraut J. The structure of oxidized cytochrome c 2 of Rhodospirillum rubrum. J Biol Chem. 1973 Jun 10;248(11):3910–3921. doi: 10.2210/pdb1c2c/pdb. [DOI] [PubMed] [Google Scholar]
- Timkovich R., Dickerson R. E. The structure of Paracoccus denitrificans cytochrome c550. J Biol Chem. 1976 Jul 10;251(13):4033–4046. doi: 10.2210/pdb155c/pdb. [DOI] [PubMed] [Google Scholar]