Abstract
Reconstructing the biogeographic history of groups present in continuous arid landscapes is challenging due to the difficulties in defining discrete areas for analyses, and even more so when species largely overlap both in terms of geography and habitat preference. In this study, we use a novel approach to estimate ancestral areas for the small plant genus Centipeda. We apply continuous diffusion of geography by a relaxed random walk where each species is sampled from its extant distribution on an empirical distribution of time-calibrated species-trees. Using a distribution of previously published substitution rates of the internal transcribed spacer (ITS) for Asteraceae, we show how the evolution of Centipeda correlates with the temporal increase of aridity in the arid zone since the Pliocene. Geographic estimates of ancestral species show a consistent pattern of speciation of early lineages in the Lake Eyre region, with a division in more northerly and southerly groups since ∼840 ka. Summarizing the geographic slices of species-trees at the time of the latest speciation event (∼20 ka), indicates no presence of the genus in Australia west of the combined desert belt of the Nullabor Plain, the Great Victoria Desert, the Gibson Desert, and the Great Sandy Desert, or beyond the main continental shelf of Australia. The result indicates all western occurrences of the genus to be a result of recent dispersal rather than ancient vicariance. This study contributes to our understanding of the spatiotemporal processes shaping the flora of the arid zone, and offers a significant improvement in inference of ancestral areas for any organismal group distributed where it remains difficult to describe geography in terms of discrete areas.
Keywords: Australia, BEAST, biogeography, Centipeda, continuous diffusion, Pliocene, species-tree
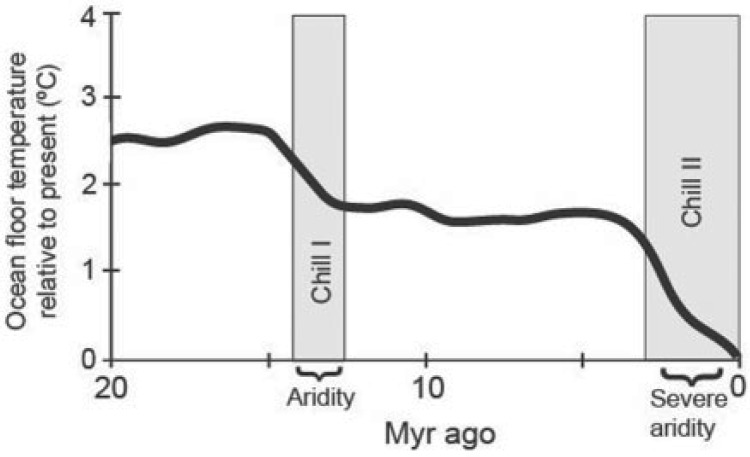
Australia hosts a large biodiversity with a flora estimated to comprise c. 21,000 species of vascular plants (Chapman 2009). Many of these are endemic to the mainland continent and exhibit various adaptations to aridity and fire, believed to have evolved as responses to climatic change from ancestors living under mesic conditions since the Miocene or during the Quaternary (Byrne et al. 2008). The modern climate regime consists of vast areas of interconnected deserts making up an arid or semi-arid zone spanning the majority of the central parts of the landscape (e.g., De Pauw et al. 2000; Fensham and Fairfax 2008), and was already established by the early Pleistocene (Martin 2006). Since the Mid-Miocene (∼15 Ma), the land has witnessed a steady progression toward increased aridity with the latest arid peak occurring soon after the timing of the last glacial maximum at 26–19 ka (Nanson et al. 1992; Clark et al. 2009) (Fig. 1).
Figure 1.
Global temperature trend measured as ocean-floor temperature relative to present from Mid-Miocene (Adapted from McGowran et al. 2004; Crisp and Cook 2007). Two major global cooling events (Chills I–II) are indicated and coincide with times of aridification in Australia.
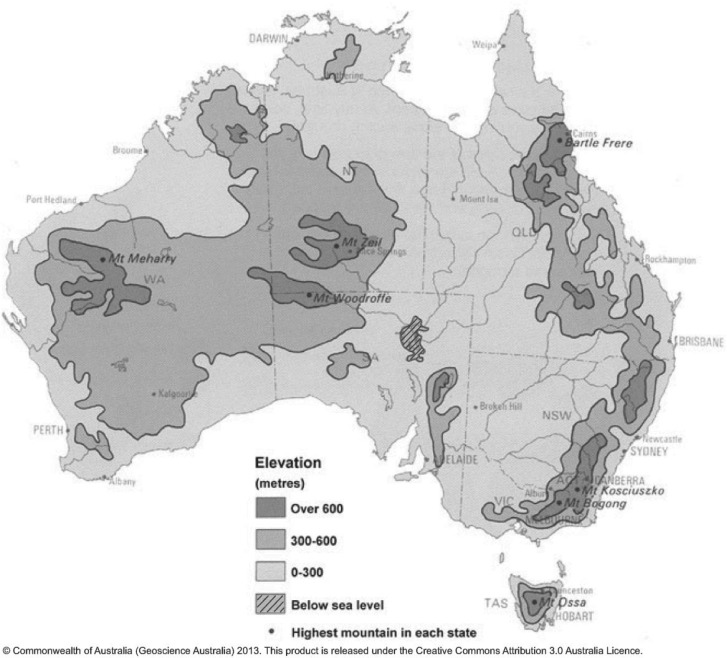
Rainfall in the Australian arid zone (∼400 mm/year) is undoubtedly the single most important factor restricting plant growth (Keast 1959). Precipitation levels are determined by the relatively flat topography (Fig. 2), which is insufficient to ensure continuous cloud formation and leads to highly seasonal rainfall (North: December–February, South: June–August). Access to water is a limiting factor for many organisms, promoting the evolution of xerophytism (i.e., long-term water storage capabilities, water preservation features, or adaptive life cycles). Over time spatial isolation in mosaic microhabitats, defined by for example level of moisture, can lead to diversification despite the lack of obvious dispersal barriers (Pianka 1972; Orr and Smith 1998; Schluter 2009), and is likely influenced by increased genetic drift due to small effective population sizes (Ne). Recent studies have shown sister lineage splits in the similar but isolated southwestern (SW) and southeastern (SE) biomes dating back to at least the Mid-Miocene (Burridge 2000; Crisp and Cook 2007), a pattern of spatial disjunction likely to have been governed by aridity, temporal incursion of the Nullabor Plain, and/or presence of calcareous soils (Mast and Givinish 2002).
Figure 2.
Elevation map of Australia with altitudinal outlines indicated in segments of 300 m. (http://www.ga.gov.au/webtemp/image_cache/GA11759.gif; last accessed December 19, 2013).
Along similar lines Australia's Central Ranges are thought to have provided long-term refuge and opportunity for speciation, particularly for mesic-adapted taxa, with some species (e.g., red cabbage palm, Livistona mariae, and the MacDonnell Ranges cycad, Macrozamia macdonnellii) considered “relicts” that have clung to survival through deep time. Recent molecular evidence however, casts doubt on these interpretations instead suggesting recent Plio-Pleistocene diversification and/or dispersal into the Central Ranges (Powney et al. 2010; Ingham et al. 2013). Thus, the gradual emergence of the arid zone does not explain all spatial patterns, and evidence of more recent splits (Close et al. 1978; Toon et al. 2007) as well as single species or groups of subspecies with cohesive distributions across multiple isolated areas in the arid zone (Brown et al. 1990; Faulks et al. 2010), indicates other or additional factors to be of equal importance for speciation, or the lack thereof.
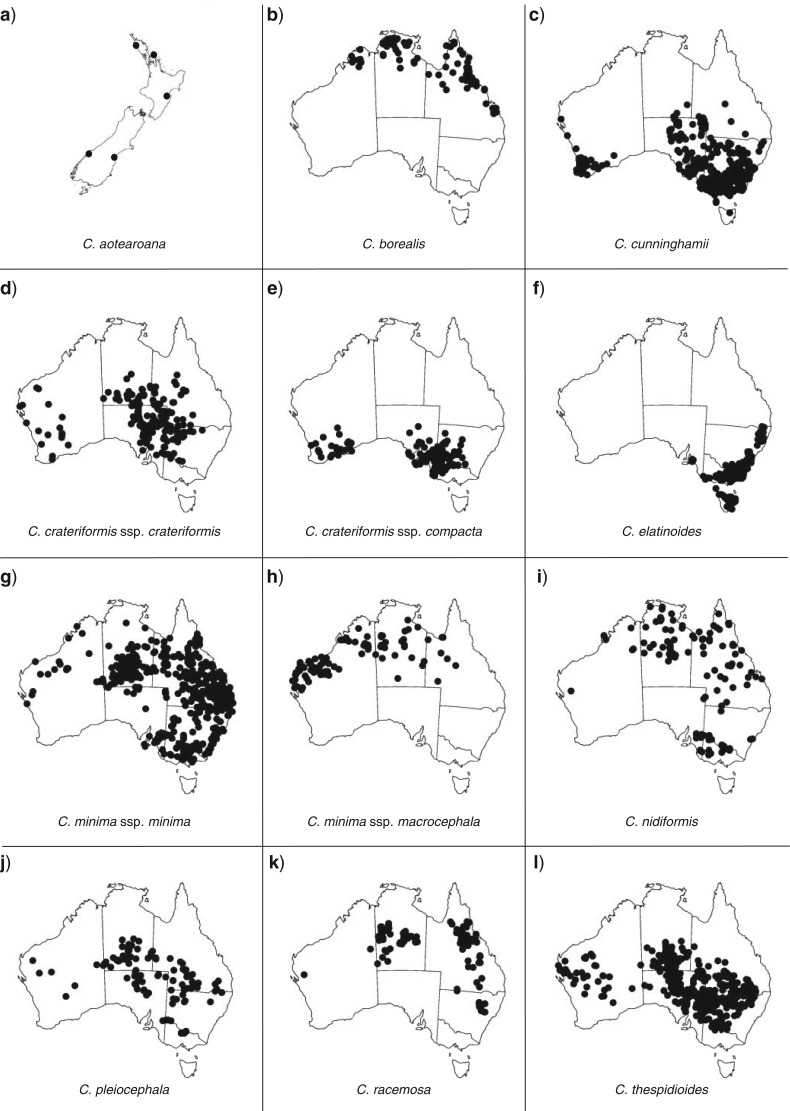
Facultative perenniality can be considered as a specialized form of xerophytism where an otherwise annual life form can continue to grow whenever access to water persists, and set seed and die when water becomes limiting. One group in which such a habit has evolved is the plant genus Centipeda, with 10 species (Walsh 2001) mainly distributed in mainland Australia (Fig. 3a–l). However, a few of its species have wider distributions and occur also in Asia, New Zealand, New Guinea, and South America. It is a group of small herbs often found in proximity to ephemeral sources of water and moisture. They are present in all parts of Australia except, generally, for the combined desert belt of the Nullabor Plain, the Great Victoria Desert, the Gibson Desert, and the Great Sandy Desert (NGGG), dividing the genus in an east–west distribution. The phylogeny and morphological character evolution of the genus was studied in detail by Nylinder et al. 2013, showing a plausible connection between species relationships and evolution of drought resistant traits. Strict annuality has evolved twice independently in the genus and is observed in species restricted to arid (Fig. 3j,l), or present in arid, temperate, and/or tropical (Fig. 3i), zones. A majority of the facultative perennials, that is C. minima, C. elatinoides, C. crateriformis, and C. cunninghamii (Fig. 3c–h) are also present outside the arid zone, and many species in the genus overlap both in terms of geographic distributions and/or habitat preference. Centipeda borealis (Fig. 3b) and C. aotearoana (Fig. 3a) are the only two species not represented in the arid zone, the former being found in tropical and monsoonal climates, and the latter restricted to New Zealand. All species in the genus lack a pappus (tufts of hair assisting wind dispersal), and are likely dispersed by adhesion to animals frequenting moist sites where the species are present, or by transport along streams (Walsh 2001).
Figure 3.
Extant species distributions in Centipeda (a–l) in mainland Australia presented as specimen plots based on 5035 records of reported observations derived from the Atlas of Living Australia (ALA) database (www.ala.org.au—snapshot as of 22 February 2013), with additional specimen information from Walsh (2001). Dots in figures correspond to singular specimens. Collections listed by Walsh 2001) but not indicated here include specimens for C. borealis from Papua New Guinea, C. cunninghamii from New Zealand, C. elatinoides from New Zealand and Chile, and C. minima ssp. minima from New Zealand, New Caledonia, Japan, and multiple locations throughout S, E, and SE Asia. For a more complete view on species distributions and mapping of locations for specimens sampled in this study (Supplementary Figs. S1–S5, available at http://dx.doi.org/10.5061/dryad.69329).
Recent studies of vertebrates in the arid zone have largely failed to establish a correlation between phylogenetic structure and the biotic processes structuring their communities (e.g., Nguyen et al. 2004; Kuch et al. 2005; Faulks et al. 2010; Lanier et al. 2013), while similar studies relating to plant communities are few (but see review by Byrne et al. 2008). Therefore investigating the timing and patterns of diversification in Centipeda in relation to climate fluctuations and the aridification process of Australia can contribute significant knowledge on how groups respond to and speciate in a changing climate (Fried and Smith 1992; Hughes and Hillyer 2006). To understand the effects of such processes on Centipeda we need to estimate the species relationships and accommodate for the underlying genetic diversity of individual species, while modeling the spatiotemporal shifts in geography by means of a non-discrete process.
In the past decade or so, a range of methods for biogeographic inference based on likelihood estimates has been developed (for overview see, e.g., Ronquist and Sanmartín 2011). Many of these methods optimize area relationships as discrete traits on a fixed tree or distribution of trees (e.g., Ronquist 1997; Pagel 1999; Ree et al. 2005; Nylander et al. 2008), or co-estimate phylogeographic model uncertainty and tree topology (Lemey et al. 2009). However, such approaches are likely to be of less use when it remains difficult to make abstractions of geography in terms of discrete areas or habitats, or when the evolution of such areas is not well understood from a spatial and/or temporal perspective. In a species-tree framework where the terminals are representative of the entire species the relationship between each terminal and the individual species' extant distributions are less clear and call for an alternative approach.
The overlap of different taxa in many areas, as is the case in Centipeda, represents another challenge for approaches using discretely coded areas. Widespread taxa have always been a challenge for biogeography (e.g., Sanmartín and Ronquist 2002), because they dilute the historical signal from other more narrowly distributed taxa (in ancestral area inference), or require additional assumptions (to infer an area cladogram). However, this highlights the underlying challenges of coding suitable areas, a procedure that is often undertaken for every group independently and is subjective and often difficult to justify in all but the simplest of cases. One solution is to estimate the ancestral node areas by continuous diffusion (Lemmon and Lemmon 2008; Lemey et al. 2010), but such approaches are founded on a gene tree basis where the sampled specimens represent geographic point estimates. Further, identifying temporal changes in diffusion rates across a species-tree rather than a gene tree can provide a valuable tool for understanding the evolutionary history of a group in relation to climate or other biological factors.
In this study, we aim to test whether the onset of Centipeda radiation correlates to the onset of the latest dry spell in Australia (26–19 ka), if the observed east–west disjunction is a result of a vicariance initiated by an emerging arid zone, and/or whether speciation events in the genus correlate to a gradual emergence of suitable habitats. We do this by (i) time-calibrating the Centipeda species-trees, as inferred in the study by Nylinder et al. (2013), by applying two separate probability distributions for substitution rates in the nuclear ITS region, and (ii) by applying a model for relaxed continuous geographic diffusion on the posterior distribution of time-calibrated species-trees to estimate the geographic distribution at ancestral nodes at times of speciation.
Materials and Methods
Time-Calibration of the Species-Tree
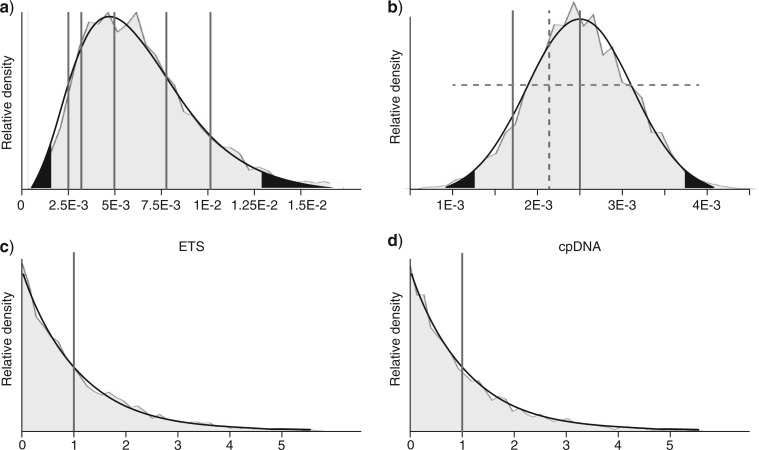
A well resolved species-tree for Centipeda was inferred in the study by Nylinder et al. (2013), based on 129 specimens sampled from all 10 putative species and multiple loci (two nuclear and three chloroplast, see online Appendix, http://www.sysbio.oxfordjournals.org/), but without estimating node ages. To estimate divergence times for Centipeda, we adapted the analysis by Nylinder et al. (2013) using two prior distributions of ITS substitution rates derived from the studies by Tremetsberger et al. (2012) on the Dandelion tribe (Asteraceae-Cichorieae), and two summaries of estimated ITS rates across Angiosperms (Kay et al. 2006; Lancaster 2010). Of the six rates relating to Asteraceae representatives in Kay et al. (2006) and Lancaster (2010), three were derived from narrow island endemics (Hawaiian Silverswords, Dendroseris D. Don, and Robinsonia DC.), which could be subject to elevated substitution rates when the effective population size (Ne) is small, a phenomenon observed in various organisms (e.g., Drosophila, Kliman et al. 2000, and Bacteria/Fungi, Woolfit and Bromham 2003, but see also Woolfit and Bromham 2005). A median rate was calculated for the five most rapidly evolving Asteraceae representatives (Table 1), and a gamma prior distribution was fitted such that all five rates fall well within the 95% credible interval of the prior distribution (Fig. 4a). This fitted curve also overlaps with the distribution of average rates for herbaceous annual/perennial species across Angiosperms (Kay et al. 2006). A second calibration based on the three most slowly evolving substitution rates for ITS (Artemisia L., Ericameria Nutt., and Eupatorium L.) (Table 1) was modeled by a normal distribution with parameters set such that the upper and lower 95% credibility intervals correspond to the full uncertainty surrounding the Ericameria rate (see Lancaster 2010 for details), a distribution also encompassing the mean rates and uncertainties of both Artemisia and Eupatorium (Fig. 4b). Both calibrations were set to shift the mean/median of each distribution away from, but still encompassing, the most extreme values in each respective selection of rates.
Table 1.
Substitution rates for the ITS region in seven Asteraceae representatives taken from the studies by Kay et al. (2006), Tremetsberger et al. (2012), and Lancaster (2010) with listed references
| Group | Calibration age | Rate (mean) | Reference |
| Artemisia | n/a | 1.69 × 10−9 | Lancaster 2010 |
| Ericameria | n/a | 2.17 × 10−9 | Lancaster 2010 |
| Eupatorium | 14.8 Ma | 2.51 × 10−9 | Schmidt and Shilling 2000 |
| Hawaiian Silverswords | 15.0 Ma | 3.0 × 10−9 | Baldwin and Sanderson 1998 |
| Dendroseris | 3.3 Ma | 5.0 × 10−9 | Sang et al. 1994 |
| Robinsonia | 4.0 Ma | 7.83 × 10−9 | Sang et al. 1995 |
| Cichorieae | 3.4–28.4 Ma | 1.08 × 10−8 | Tremetsberger et al. 2012 |
Substitution rates are listed as number of substitutions/site/myr.
Figure 4.
Prior distributions used for time-calibration of the Centipeda species-tree. (a) Gamma distribution of nuclear ITS rate. Gray lines indicate derived individual rates for five representatives of Asteraceae. (b) Normal distribution modeling the derived ITS rates for Eupatorium and Artemisia (solid lines) together with the rate and full uncertainty (dashed lines) of Ericameria (Lancaster 2010). (c) Prior for the mean rate of ETS and (d) the combined chloroplast regions. Black segments in distributions indicate upper and lower 2.5% and 97.5% prior credibility interval boundaries. Areas in solid gray show posterior distributions when sampling from the priors without data. All values are presented as substitutions/site/myr.
The two alternative calibration priors were integrated into the analysis file used by Nylinder et al. (2013), together with vague priors on the rates for plastid data (exponential decays with mean 1.0, Fig. 4c–d). The analyses were performed using Markov chain Monte Carlo (MCMC) in BEAST 1.7.5 (Drummond et al. 2012), first without data to evaluate cross prior influence (Fig. 4a–d), after which it was run three times using actual data for 100 million iterations each, logging parameters every 25,000 generations. Assessment of parameters reaching stationarity and adequate chain mixing (ESS values > 200) was done using Tracer 1.5 (Rambaut and Drummond 2004). The posterior distributions of trees were combined using LogCombiner (part of the BEAST 1.7.5-package) after removal of a proportion of each run as burn-in, summarized as a Maximum Clade Credibility (MCC) tree in TreeAnnotator (part of the BEAST 1.7.5-package), and visualized using FigTree 1.3.1 (Rambaut 2006).
Species-Tree Diffusion
Given that the implementation of the species-tree model in BEAST (*BEAST, Heled and Drummond 2010) does not extend to diffusion processes, a two-step approach was necessary to perform phylogeographic reconstructions of species ranges. First, we estimated a posterior distribution of species-trees using the *BEAST approach, and then following Pagel et al. (2004), we considered the extant species distributions as a fixed set of solutions to fit trait evolutionary models. Previous inference approaches for random walk models in continuous space have exploited data augmentation of the unobserved locations of ancestral nodes in the tree to compute the process likelihood (Lemey et al. 2010); such approaches that update internal node states need to be aware of how tree topologies change throughout the MCMC. As this information is lost for empirical tree sets, we adopted a recently developed analytical procedure to integrate out internal and root node states (Pybus et al. 2012), which has also proven to be more efficient for inference under high diffusion heterogeneity and for a large number of taxa. Similar to the data augmentation approach (Bouckaert et al. 2012), we here extended the integrated multivariate diffusion approach to accommodate arbitrarily shaped areas for tip locations, which we encoded as Keyhole Markup Language (KML) polygons. We complemented this development by implementing a novel transition kernel in BEAST to efficiently sample from disjoint areas, which may jointly represent a particular species range. To accomplish this, the transition kernel first proposes a new sample location drawn uniform randomly across the species range and then accepts or rejects this proposal location through a Metropolis-Hastings correction, allowing the previous location state and the current location state to lie in different disjoint areas.
The posterior distribution of calibrated species-trees was prepared for diffusion analysis by pruning the outgroup (Anisopappus Hook. & Arn.) in Mesquite 2.75 (Maddison and Maddison 2011). Details of sister group relationships for Centipeda are largely unknown (e.g., Bremer 1994; Nesom 1994; Walsh 2001; Panero 2005; Nylinder et al. 2013), and, as Anisopappus is present mainly in Africa with only the type species occurring elsewhere (China—A. chinensis Hook. & Arn.), we removed this accession to avoid introducing unnecessary bias into the diffusion process. Extant distributions of Centipeda species were shaped in Google Earth as polygons (Supplementary Figs. S1–S5, available at http://dx.doi.org/10.5061/dryad.69329) based on published information for each species (Atlas of Living Australia, http://www.ala.org.au; Walsh 2001), and converted into modified KML files (supplied as Supplementary Material) to be referenced by the analysis together with the posterior distribution of species-trees. The diffusion process was modeled by a relaxed random walk (RRW) process, which is an extension of a phylogenetic Brownian motion process that rescales the precision matrix by a branch-specific scalar that is drawn independently from an identical distribution, in our case a lognormal distribution centered on 1. We specified a prior exponential distribution on the standard deviation (SD) of the lognormal distribution with a mean of 2.712, (upper 97.5% credible interval of 10), to suggest a reasonable range for the diffusion rate heterogeneity (distance in km/time unit). Sensitivity to the constraint on SD was explored by increasing the power of the SD prior tenfold (mean 0.2712, upper credible interval 1.0). The analysis file was set up with both structured and random starting locations for each taxon (Supplementary Figs. S1–S5, available at http://dx.doi.org/10.5061/dryad.69329) to ensure independent convergences.
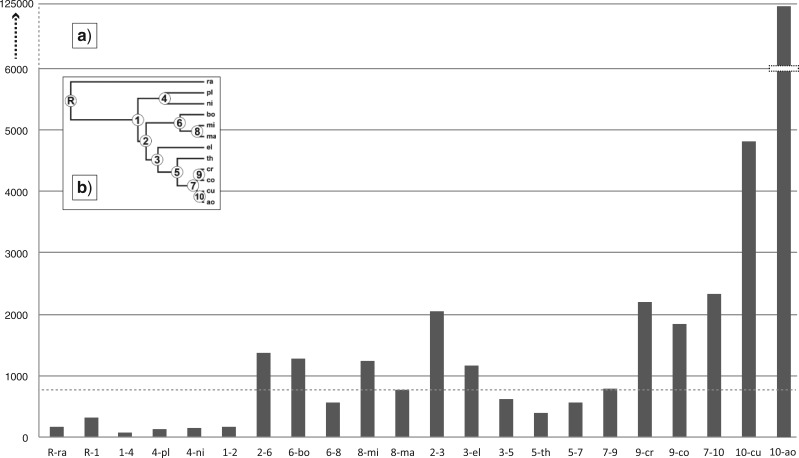
Five independent analyses of the diffusion inference were run for 50 million generations each, logging parameters every 50,000 generations. The diffused MCC tree with annotated diffusion estimates was visualized in SPREAD v.1.0.5 (Bielejec 2011), and ancestral areas projected as polygons on a geo-referenced map of Australia using Quantum GIS (http://www.qgis.org/). To visualize the spatial distribution of the genus at the time of the latest speciation event we summarized time slices of the species diffusion trees and obtained the 50%, 80%, and 95% highest posterior density (HPD) areas in SPREAD v.1.0.5 (Bielejec et al. 2011). The median diffusion rates between all adjacent nodes (or between nodes and terminals where applicable) in the MCC species-tree were extracted and summarized as rate per time unit (km/Ma) (Fig. 5a).
Figure 5.
(a) Summary of median diffusion rates between adjacent node pairs in the MCC species-tree, here presented on the y-axis as diffusion distance per time unit (km/Ma). Dotted line indicates median rate across all branches in the species-tree. (b) Outline of species-tree. Taxon abbreviations and node numbers follow those in Figure 6b.
Nucleotide Diversity
We also examined the nucleotide diversity between eastern and western parts of the ranges for those species with disjunct distributions separated by the NGGG desert belt. If vicariance caused by the increase in aridity of the NGGG belt had split these populations, then we expect to find similar amounts of nucleotide diversity on either side of this divide. On the contrary, if long distance dispersal (LDD) of a limited number of migrants had allowed recent colonization of one side of the NGGG belt from the other, we would instead expect nucleotide diversity to be lower on one side of this divide due to a founder effect (Nei et al. 1975). We used DnaSP v.5 (Librado and Rozas 2009) to estimate the average nucleotide diversity per site (π) after replacing ambiguities within each sequence with “N”.
Results
Node Ages of Species-Tree
The time-calibrated species-tree implementing the range of slowest evolving ITS rates (Fig. 6a) provide the oldest node estimates, which date the Centipeda stem age to the Late Miocene, ∼5.8 Ma (1.4–11.1 Ma, 95% HPD interval), with the first speciation (C. racemosa vs. the remainder of the genus) occurring at the transition into the Mid-Pleistocene at ∼1.9 Ma (770 ka–3.3 Ma). A subsequent speciation occurred at ∼960 ka (510 ka–1.6 Ma), an event from which the ancestor of the arid-centered species C. nidiformis and C. pleiocephala originated at ∼570 ka (205 ka–1.07 Ma). The stem age of a species pair consisting of C. borealis and C. minima is estimated to ∼840 ka (480 ka–1.4 Ma) with a crown age of ∼350 ka (151–650 ka), and a subspeciation of C. minima at ∼70 ka (17–165 ka). A stem age of the clade comprising the more temperate species (except for the arid representative C. thespidioides with a stem age of ∼390 ka (190–690 ka)) is estimated at ∼670 ka (330 ka–1.1 Ma) with the branching of C. elatinoides. The stem age of three species, C. crateriformis, C. cunninghamii, and C. aotearoana, is estimated to have occurred ∼98 ka (40–180 ka), with a split between the latter two at ∼24 ka (Present day–88 ka) and a subspeciation of C. crateriformis at ∼56 ka (Present day–120 ka).
Figure 6.
(a) MCC species-tree of Centipeda with node height estimates based on ITS rates for five representatives of Asteraceae (bold numbers, lower (red) bars for the 95% highest posterior distributions), and the derived ITS rate for Eupatorium (regular numbers, upper (blue) bars for 95% HPD). (b) Outline of species-tree with internal nodes labeled R + 1–10. Taxon names are abbreviated as: ra—C. racemosa, pl—C. pleiocephala, ni—C. nidiformis, bo—C. borealis, mi—C. minima ssp. minima, ma—C. minima ssp. macrocephala, el—C. elatinoides, th—C. thespidioides, cr—C. crateriformis ssp. crateriformis, co—C. crateriformis ssp. compacta, cu—C. cunninghamii, ao—C. aotearoana.
Ages inferred by implementing the distribution based on the more rapidly evolving ITS substitution rates are all estimated to be ∼40% younger than corresponding ages inferred when applying the distribution of the more slowly evolving Asteraceae ITS rates.
Spatiotemporal Patterns of Speciation
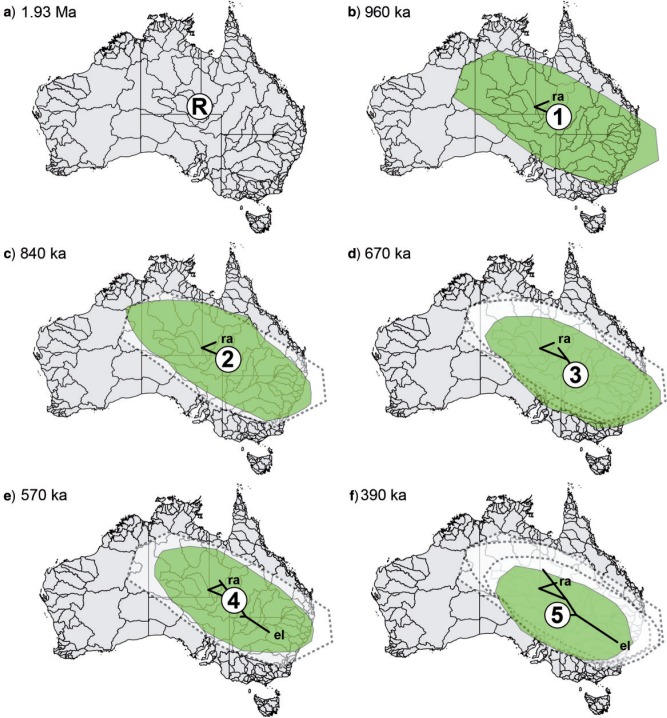
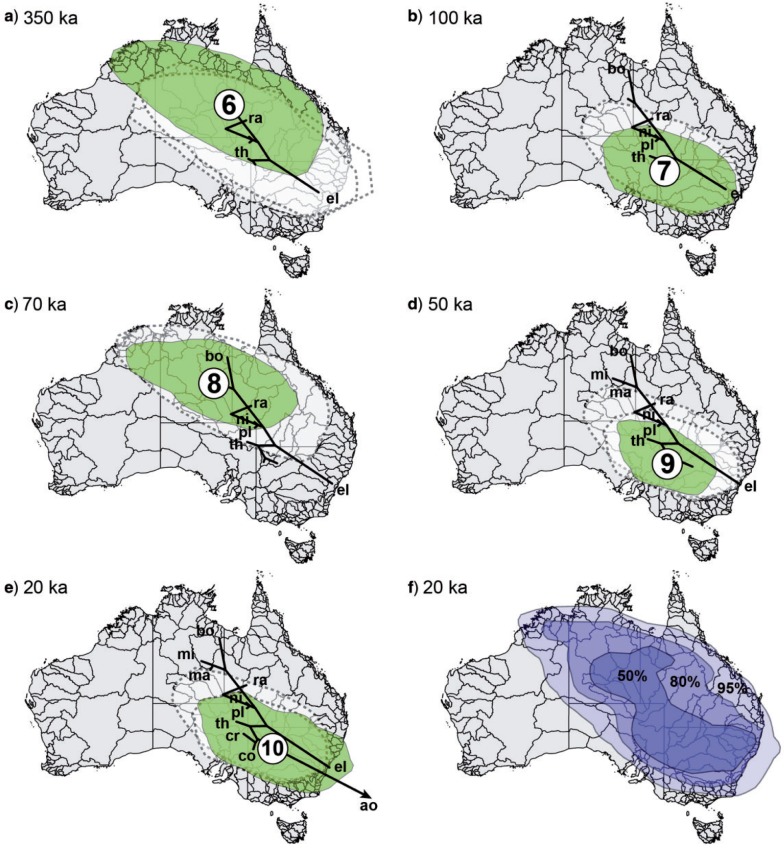
Geographic areas as locations for speciation events are presented as the 80% HPD values for each ancestral node (Figs. 7 and 8, nodes R + 1–10). The most likely region for the onset of diversification of Centipeda (node R, Fig. 7) was estimated to be the northwest (NW) limits of the Lake Eyre Basin. The point estimates for nodes 1–5 (Fig. 7) all occur in an area corresponding to the drainage channels of the Channel Country in SW Queensland and northeast (NE) South Australia, with the 80% HPD outer ranges reaching from the eastern edges of the Great Sandy Desert in the NW to the SE coast, and from the Lake Gairdner region in the SW to the edges of the arid zone in the NE. The ancestor of C. borealis and C. minima (Fig. 8a) is estimated to stem from the eastern edges of the Tanami Desert with an 80% HPD area from the NW coast along the Timor Sea to the SE limits of the arid zone, and from Lake Eyre in the SW to the Gulf of Carpentaria in the NE. Subspeciation of C. minima (Fig. 8c) is then estimated to have occurred in a more restricted part of the area inferred for the most recent common ancestor (MRCA) of C. minima and C. borealis but with a slight shift toward the north. The ancestral area of the MRCA of C. cunninghamii, C. crateriformis, and C. aotearoana, (Fig. 8b) is estimated to be the SE limits of the arid zone with a more or less circular 80% HPD area with a 500 km radius. The area of subspeciation of C. crateriformis (Fig. 8d) is estimated to have occurred at the NW border between New South Wales and South Australia, with a semi-circular 80% HPD area ∼350 km in radius, corresponding to the SE part of the arid zone. The MRCA of C. aotearoana and C. cunninghamii (Fig. 8e) is estimated to have originated close to the location of subspeciation in C. crateriformis, but shifted toward the central part of the SE dunefields and with a slightly larger 80% HPD area than the C. crateriformis subspecies ancestor. The 80% HPD area corresponds to that of the former, but also includes corresponding parts of the Great Dividing Range toward the coast.
Figure 7.
Snapshots of estimated ancestral node areas in the MCC tree at times of speciation as visualized using SPREAD. Node indicators (R + 1–5) and taxon abbreviations correspond to labeling in Figure 6b. All ages are presented as median age in millions (Ma) or thousands of years ago (ka). The 80% HPD area for nodes in each figure (a–f) is indicated as green polygons. Remnants of 80% HPD areas associated with closest connective nodes toward the root are indicated with dashed margins in figures (c–f). Branches relating to speciation events resulting in extant taxa are labeled with abbreviations. Map of Australia's river basins used for projections reproduced courtesy of Geoscience Australia (https://www.ga.gov.au/; last accessed December 19, 2013).
Figure 8.
Snapshots of estimated ancestral node areas in the MCC tree at times of speciation as visualized in SPREAD. Node indicators (6–10) and taxon abbreviations correspond to labeling in Figure 6b. All ages are presented as median age in ka. The 80% HPD areas for nodes in each figure (a–e) are indicated as green polygons. Remnants of 80% HPD areas associated with closest connective nodes toward the root are indicated with dashed margins (a–e). Branches relating to speciation events resulting in extant taxa are labeled with abbreviations. Positions of taxon localities in (e) indicate most probable source areas for each taxon (C. aotearoana restricted to New Zealand omitted). (f) Summarized time slices of the species diffusion trees as 50%, 80%, and 95% HPD areas showing total area occupied by Centipeda at timing of latest speciation event (20 ka). Map of Australia's river basins used for projections reproduced courtesy of Geoscience Australia (https://www.ga.gov.au/; last accessed December 19, 2013).
The estimated areas of diversification for individual species are all positioned in a limited diagonal corridor of the arid zone, reaching from the eastern parts of the Northern Territory to the SE coast of New South Wales, with the exceptions of C. aotearoana, C. borealis, and C. elatinoides. The ancestor of Centipeda aotearoana is estimated to have radiated on the south island of New Zealand, most likely as a result of LDD from mainland Australia, the ancestor of C. borealis diverged in the tropical/monsoon zone south of the Gulf of Carpentaria, and C. elatinoides diverged on the temperate coastline near present day Sydney. Diffusion rates across the species-tree (Fig. 5) suggest low levels of movement for the ancestors to species C. racemosa, C. pleiocephala, and C. nidiformis, with increased rates inferred for all remaining species and ancestors.
Summarized time slices of the diffused posterior species-trees at the time of the latest speciation event (Fig. 8f) estimates the total area inhabited by Centipeda at that point in time (∼20 ka) to include most of Australia, except for the northernmost regions of Queensland and the Northern Territory, and the areas west of the NGGG desert belt.
Two taxa with more than one allele in each part of the range and with current disjunct distributions across the NGGG belt (C. minima ssp. macrocephala and C. thespesioides), show lower nucleotide diversity west of the NGGG belt than east of it. For ETS sequences in C. minima ssp. macrocephala, π = 0 ± 0 in the west (n = 10) versus π = 0.00148 ± 0.0007 (SD) in the east (n = 3). For cpDNA sequences in C. thespesioides, π = 0 ± 0 in the west (n = 5) versus π = 0.00191 ± 0.00052 (SD) in the east (n = 8). The mean in the west was lower than the mean in the east for three of four other available comparisons although these values lay within two SDs of the other mean.
Discussion
Spatiotemporal History of Centipeda
The family Asteraceae is believed to have originated some 42–36 Ma (for a more thorough discussion see Angiosperm Phylogeny Group 2009 and references listed therein), but a persistent lack of reliable fossils relating to the various subfamilies makes age estimation based on objective data difficult to achieve. Our use of secondary ITS substitution rates must therefore be considered sub-optimal and should best be treated with caution. Also, as the proper outgroup to Centipeda has yet to be established the true stem age may actually be younger than inferred in this study. By accommodating for as many derived ITS rates for Asteraceae as possible, and modeling them in separate slowly and rapidly evolving distributions, we avoid the recognized pitfalls when using absolute rates for age estimation (Graur and Martin 2004). It is therefore worthwhile to initiate a discussion on the evolution of the genus focused on the older ages as they suggest an upper limit for the estimated onset of speciation events, even for the stem node of Centipeda.
Patterns of speciation in Centipeda correlate strongly to the accumulated spatial increase of arid areas and the cyclic expansion of arid and semi-arid zones in Australia since the Mid-Pleistocene. The correlation persists even when extending the conclusions to encompass the entire 95% HPD uncertainty interval of the presumed age of diversification onset in Centipeda (Fig. 6), when inferred by slower ITS rates. The timing of the origin of Centipeda at the Miocene–Pliocene boundary is also broadly congruent with that of other major groups of Australian fauna and flora present in the arid zone (reviewed in Byrne et al. 2008). Inferred diffusion rates across the species-tree (Fig. 5) indicate the first order of diversification events in the genus to have occurred within a limited area. In comparison more recent events (since ∼840 ka) are all associated with elevated diffusion rates, particularly for taxa with a southeast origin (C. crateriformis, C. cunninghamii, and C. aotearoana), correlating to a Quaternary increase in climatic fluctuations and the development of aridity in southeast Australia (Hesse et al. 2004).
By the estimated time of origin of Centipeda palynofloral evidence suggests that open mosaic sclerophyll forests dominated the central parts of Australia with the presence of freshwater swamps (Martin 1990, 2006). By the early Pliocene, grasses also became more common in pollen deposits, indicating a shift toward more open landscapes in the central parts of Australia with the increased presence of grazing animals (Archer et al. 1994). Based on fossil evidence, it has been suggested that some taxa present in the arid zone to have originated during times of wetter climates, and co-existed in the same regions as vegetation with fewer adaptations to aridity (Martin 2006). Such a scenario makes it difficult to distinguish whether Centipeda underwent successive adaptive radiations, or stem from an ancestor living in mesic conditions that later went extinct. However, spatial patterns of speciation in Centipeda suggest the genus could have been pre-adapted to drought at the time of origin. Also, a change from the plesiomorphic character state of a pure perennial habit for the genus (Node R in Fig. 6b; Nylinder et al. 2013), to a majority of extant species being facultative perennials, correlates with a major change from wetter to drier conditions in the central parts of Australia before or at ∼1 Ma (Chen and Barton 1991; Fujioka et al. 2009).
If we assume ages inferred from the more rapidly evolving distribution of ITS rates to more accurately represent the timing of speciation events in Centipeda, all diversification times appear to have occurred when the expansion of the central arid zone was already well under way (i.e., younger than ∼1 Ma; Chen and Barton 1991; Fujioka et al. 2009). This would shift speciation events to correlate more closely with the already disappearing palaeochannels, and supports the hypothesis of prolonged splits in populations tied to distinct ecological zones resulting in sympatric speciation (Kondrashov and Mina 1986; Rice 1987; Dieckmann and Doebeli 1999).
The current distribution of Centipeda in Australia includes western as well as central and eastern areas, with several species displaying disjunctions between the former two areas (C. cunninghamii, C. minima ssp. macrocephala, C. nidiformis, C. pleiocephala and C. thespidioides). Such disjunctions raise the question as to whether they could have arisen via vicariant fragmentation of more widespread ancestral distributions caused by the latest increase in aridification. However, our results suggest that this is unlikely because the inferred ancestral ranges (up until and including the most recent speciation events at ∼20 ka) do not include the western part of these species' current ranges (Fig. 8f). Such patterns could also result from local extinctions within more widespread distributions caused by the rise of the NGGG belt, but this is not compatible with the inferred ages of these species, which have evolved more recently than these deserts (Fujioka et al. 2009). Instead, the pattern of nucleotide diversity among alleles drawn from the eastern and western parts of two of these taxa with disjunct distributions support the idea of recent dispersal of a small number of individuals from east to west, which were consequently subject to founder effects in the western part of their ranges. It should be noted, however, that other explanations for the higher nucleotide diversity found in the east might also be possible, and we advocate for more rigorous tests if/when more data become available.
Comparison of Diversification to Other Organismal Groups
The majority of extant species in Centipeda are assumed to have originated since 500 ka, with the most recent taxa forming between 70 ka and 50 ka, a timespan that largely agrees with similar patterns for other organisms listed in a review of the Australian arid zone by Byrne et al. (2008). In the period from the early Pleistocene to ∼400 ka many Australian lineages are estimated to have diverged in east–west and/or north–south groups (e.g., Sminthopsis, Blacket et al. 2001); Eucalyptus, Byrne and Hines 2004; Nicolle 2008), and the north–(central)–south grouping of Centipeda species occurring at ∼840 ka corroborates this pattern. Of the lineages listed by Byrne et al. (2008) to have diversified after 400 ka, a majority show little or no genetic structure in relation to geography, which is also in agreement with our findings for all species divergences in east–west fractions by the NGGG belt (C. cunninghamii, C. crateriformis, C. minima, C. nidiformis, C. pleiocephala, C. racemosa, and C. thespidioides) (Nylinder et al. 2013), Figs. 3–5).
Diffusion Model Performance
The development of diffusion models for estimating biogeographic scenarios has proven a promising field of research. In this study we take a novel approach to estimating ancestral areas for the MRCAs of the extant species of Centipeda. On a continuous landmass such as Australia, and for a group such as Centipeda where species distributions and habitat preferences often and largely overlap, identification of areas for discrete analyses becomes extremely difficult. Continuous diffusion allows for averaging over a large number of range expansions/contractions while relaxation of the random walk is able to model the probability of a semi-restricted geographic relationship between a population and its offspring. In its current implementation the model assumes a uniform probability of occurrence of specimens within their respective species' distributions, which could potentially over-emphasize sampling of the fringes of the species distributions. We are aware that this may be an unrealistic representation of the actual variable densities of specimens within an area. However, the framework allows for potential but yet unimplemented solutions, such as including an accommodation for distribution heterogeneity by sampling from a set of point locations for each species, which would allow for a more direct way of using information stored in collection databases, or by defining a set of nested polygons representing different density intervals. Another potential extension includes optimization of multiple continuous and discrete parameters for each species, which together may constitute an individual “environmental envelope”, and would allow for the estimation of climate preferences of ancestral species similar to the approach of Yesson and Culham (2006), while integrating the ecological parameters directly into the likelihood framework.
Supplementary material
Data files and/or other supplementary information related to this article have been deposited at Dryad under http://dx.doi.org/10.5061/dryad.69329. The species-tree diffusion approach is currently available as part of the development branch of the BEAST source code repository (http://beast-mcmc.googlecode.com/svn/trunk), and will be distributed with the next public release.
Funding
This work was supported by the Swedish Research Council as a grant (to A.A.) for Angiosperm phylogeny; the European Union Seventh Framework Programme [FP7/2007-2013] under ERC Grant agreement no. 260864; the Swedish Research Council, the Royal Swedish Academy of Sciences, Lars Hiertas Minne fund, The Royal Physiographic Society in Lund, Helge Ax:son Johnsons fund, and the Lundgrenska fund (to B.E.P.); the National Science Foundation [DMS-1264153 to M.A.S. (in part)] and the National Institutes of Health [R01 HG006139 to M.A.S. (in part)]. The research leading to these results has received funding (to P.L. and M.A.S.) from the European Union Seventh Framework Programme [FP7/2007-2013] under ERC Grant agreement no. 260864.
Acknowledgments
The present investigation is part of an on-going research program on Asteraceae phylogeny. We are thankful to two anonymous reviewers for constructive comments and suggestions on the manuscript.
References
- Angiosperm Phylogeny Group III. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot. J. Linn. Soc. 2009;161:105–121. doi:10.1111/j.1095-8339.2009.00996.x. [Google Scholar]
- Archer M., Hand S.J., Godhelp H. Patterns in the history or Australia's mammals and inferences about palaeohabitats. In: Hill R.S., editor. History of the Australian vegetation: cretaceous to recent. Cambridge: Cambridge University Press; 1994. pp. 80–103. [Google Scholar]
- Baldwin B.G., Sanderson M.J. Age and rate of diversification of the Hawaiian Silversword alliance (Compositae) Proc. Natl Acad. Sci. USA. 1998;95:9402–9406. doi: 10.1073/pnas.95.16.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielejec F., Rambaut A., Suchard M.A., Lemey P. SPREAD: spatial phylogenetic reconstruction of evolutionary dynamics bioinform. 2011 doi: 10.1093/bioinformatics/btr481. doi:10.1093 (available from: URL www.phylogeography.org/SPREAD) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacket M.J., Adams M., Cooper S.J.B., Krajewski C., Westerman M. Systematics and evolution of the dasyurid marsupial genus Sminthopsis: I. The Macroura species group. J. Mamm. Evol. 2001;8:149–170. [Google Scholar]
- Bouckaert R., Lemey P., Dunn M., Greenhill S.J., Alekseyenko A.V., Drummond A.J., Gray R.D., Suchard M.A., Atkinson Q.D. Mapping the origins and expansion of the Indo-European language family. Science. 2012;337:957–960. doi: 10.1126/science.1219669. doi:10.1126/science.1219669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer K. Asteraceae: cladistics and classification. Portland, Oregon: Timber Press; 1994. [Google Scholar]
- Brown A.H.D., Burdon J.J., Grace J.P. Genetic structure of Glycine canescens a perennial relative of soybean. Theor. Appl. Genet. 1990;79:729–736. doi: 10.1007/BF00224237. [DOI] [PubMed] [Google Scholar]
- Burridge C.P. Biogeographic history of geminate cirrhitoids (Perciformes: Cirrhitoidea) with east–west allopatric distributions across southern Australia, based on molecular data. Global Ecol. Biogeogr. 2000;9:517–525. doi:10.1046/j.1365-2699.2000.00204.x. [Google Scholar]
- Byrne M., Hines B. Phylogeographical analysis of cpDNA variation in Eucalyptus loxophleba (Myrtaceae) Aust. J. Bot. 2004;52:459–470. [Google Scholar]
- Byrne M., Yeates K., Joseph L., Kearney M., Bowler J., Williams A.J., Cooper S., Donnellan S.C., Keogh J.S., Leys R., Melville J., Murphy D.J., Porch N., Wyrwoll K.-H. Birth of a biome: insights into the assembly and maintenance of the Australian arid zone biota. Mol. Ecol. 2008;17:4398–4417. doi: 10.1111/j.1365-294X.2008.03899.x. doi:10.1111/j.1365-294X.2008.03899.x. [DOI] [PubMed] [Google Scholar]
- Chapman A.D. Numbers of living species in Australia and the world. Report to the Australian Biological Resources Survey. 2009 Available from: URL http://www.environment.gov.au/biodiversity/abrs/publications/other/species-numbers/2009/03-exec-summary.html#plants. [Google Scholar]
- Chen X.Y., Barton C.E. Onset of aridity and dune-building in central Australia: sedimentological and magnetostratigraphic evidence from Lake Amadeus. Palaeogeog. Palaeoclim. Palaeoecol. 1991;84:55–73. [Google Scholar]
- Clark P.U., Dyke A.S., Shakun J.D., Carlson A.E., Clark J., Wohlfarth B., Mitrovica J.X., Hostetler S.W., McCabe A.M. The last glacial maximum. Science. 2009;325:710–714. doi: 10.1126/science.1172873. [DOI] [PubMed] [Google Scholar]
- Close R.C., Moar N.T., Tomlinson A.I., Lowe A.D. Aerial dispersal of biological material from Australia to New Zealand. Int. J. Biometeor. 1978;22:1–19. [Google Scholar]
- Crisp M.D., Cook L.G. A congruent molecular signature of vicariance across multiple plant lineages. Mol. Phylogenet. Evol. 2007;43:1106–1117. doi: 10.1016/j.ympev.2007.02.030. [DOI] [PubMed] [Google Scholar]
- De Pauw E., Göbel W., Adam H. Agrometeorological aspects of agriculture and forestry in the arid zones. Agr. Forest. Meteorol. 2000;103:43–58. Available from: URL http://dx.doi.org/10.1016/S0168-1923(00)00118-0. [Google Scholar]
- Dieckmann U., Doebeli M. On the origin of species by sympatric speciation. Nature. 1999;400:354–357. doi: 10.1038/22521. [DOI] [PubMed] [Google Scholar]
- Drummond A.J., Suchard M.A., Xie D., Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. doi:10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulks L.K., Gilligan D.M., Beheregaray L.B. Islands of water in a sea of dry land: hydrological regime predicts genetic diversity and dispersal in a widespread fish from Australia's arid zone, the golden perch (Macquaria ambigua) Mol. Ecol. 2010;19:4723–4737. doi: 10.1111/j.1365-294X.2010.04848.x. doi: 10.1111/j.1365-294X.2010.04848.x. [DOI] [PubMed] [Google Scholar]
- Fensham R.J., Fairfax R.J. Water-remoteness for grazing relief in Australian arid-lands. Biol. Cons. 2008;141:1447–1460. [Google Scholar]
- Fried A.W., Smith N. Timescales and the role of inheritance in long-term landscape evolution, northern New England, Australia. Earth Surf. Proc. Land. 1992;17:375–385. [Google Scholar]
- Fujioka T., Chappell J., Fifield K., Rhodes E.J. Australian desert dune fields initiated with Pliocene–Pleistocene global climatic shift. Geology. 2009;37:51–54. [Google Scholar]
- Graur D., Martin W. Reading the entrails of chickens: molecular timescales of evolution and the illusion of precision. Trends Genet. 2004;20:80–86. doi: 10.1016/j.tig.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Heled J., Drummond A.J. Bayesian inference of species-trees from multilocus data. Mol. Biol. Evol. 2010;27:570–580. doi: 10.1093/molbev/msp274. doi: 10.1093/molbev/msp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse P.P., Magee J.W., van der Kaars S. Late Quaternary climates of the Australian arid zone: a review. Quatern. Int. 2004;118–119:87–102. [Google Scholar]
- Hughes J.M., Hillyer M.J. Mitochondrial DNA and allozymes reveal high dispersal abilities and historical movement across drainage boundaries in two species of freshwater fishes from inland rivers in Queensland, Australia. J. Fish Biol. 2006;68:270–291. [Google Scholar]
- Ingham J.A., Forster P.I., Crisp M.D., Cook L.G. Ancient relicts or recent dispersal: how long have cycads been in central Australia? Diversity Distrib. 2013;19:307–316. doi: 10.1111/j.1472-4642.2012.00936.x. [Google Scholar]
- Kay K.M., Whittall J.B., Hodges S.A. A survey of nuclear ribosomal internal transcribed spacer substitution rates across angiosperms: an approximate molecular clock with life history effects. BMC Evol. Biol. 2006;6:36. doi: 10.1186/1471-2148-6-36. doi:10.1186/1471-2148-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keast A. The Australian environment. In: Keast A., Crocker R.L., Christian C.S., editors. Biogeography and ecology in Australia. The Hague: Dr. W. Junk; 1959. pp. 9–35. [Google Scholar]
- Kliman R.M., Andolfatto P., Coyne J.A., Depaulis F., Kreitman M., Berry A.J., McCarter J., Wakeley J., Hey J. The population genetics of the origin and divergence of the Drosophila simulans complex species. Genetics. 2000;156:1913–1931. doi: 10.1093/genetics/156.4.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov A.S., Mina M.V. Sympatric speciation: when is it possible? Biol. J. Linn. Soc. 1986;27:201–223. [Google Scholar]
- Kuch U., Keogh J.S., Weigel J., Smith L.A., Mebs D. Phylogeography of Australia's king brown snake (Pseudechis australis) reveals Pliocene divergence and Pleistocene dispersal of a top predator. Naturwissenschaften. 2005;92:121–127. doi: 10.1007/s00114-004-0602-0. [DOI] [PubMed] [Google Scholar]
- Lancaster L.T. Molecular evolutionary rates predict both extinction and speciation in temperate angiosperm lineages. BMC Evol. Biol. 2010;10:162. doi: 10.1186/1471-2148-10-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier H.C., Edwards D.L., Knowles L.L. Phylogenetic structure of vertebrate communities across the Australian arid zone. J. Biogeogr. 2013;40:1059–1070. doi:10.1111/jbi.12077. [Google Scholar]
- Lemey P., Rambaut A., Drummond A.J., Suchard M.A. Bayesian phylogeography finds its roots. PLoS Comp. Biol. 2009;5(9):e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P., Rambaut A., Welch J.J., Suchard M.A. Phylogeography takes a relaxed random walk in continuous space and time. Mol. Biol. Evol. 2010;27:1877–1885. doi: 10.1093/molbev/msq067. doi:10.1093/molbev/msq067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon A.R., Lemmon E.R. A likelihood framework for estimating phylogeographic history on a continuous landscape. Syst. Biol. 2008;57:544–561. doi: 10.1080/10635150802304761. [DOI] [PubMed] [Google Scholar]
- Librado P., Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. Available from: URL http://www.ub.edu/dnasp/ [DOI] [PubMed] [Google Scholar]
- Maddison W.P., Maddison D.R. Mesquite: a modular system for evolutionary analysis. 2011 Version 2.75. Available from: URL http://mesquiteproject.org. [Google Scholar]
- Martin H.A. The palynology of the Namba Formation in the Wooltana-1 bore, Callabonna Basin (Lake Frome), South Australia, and its relevance to Miocene grasslands in central Australia. Alcheringa. 1990;14:247–255. [Google Scholar]
- Martin H.A. Cenozoic climatic change and the development of the arid vegetation in Australia. J. Arid Environ. 2006;66:533–563. [Google Scholar]
- Mast A.R., Givnish T.J. Historical biogeography and the origin of stomatal distributions in Banksia and Dryandra (Proteaceae) based on their cpDNA phylogeny. Am. J. Bot. 2002;89:1311–1323. doi: 10.3732/ajb.89.8.1311. [DOI] [PubMed] [Google Scholar]
- McGowran B., Holdgate G.R., Li Q., Gallagher S.J. Cenozoic stratigraphic succession in southeastern Australia. Aust. J. Earth Sci. 2004;51:459–496. [Google Scholar]
- Nanson G.C., Price D.M., Short S.A. Wetting and drying of Australia over the past 300 ka. Geology. 1992;20:791–794. [Google Scholar]
- Nei M., Maruyama T., Chakraborty R. The bottleneck effect and genetic variability in populations. Evolution. 1975;29:1–10. doi: 10.1111/j.1558-5646.1975.tb00807.x. [DOI] [PubMed] [Google Scholar]
- Nesom G.L. Subtribal classification of the Astereae (Asteraceae) Phytologia. 1994;76:193–274. [Google Scholar]
- Nicolle D. Systematic studies of the southern Australian Mallees ( Eucalyptus series Subulatae – Myrtaceae) 2008. PhD Thesis, Flinders University of South Australia. [Google Scholar]
- Nguyen T.T.T., Austin C.M., Meewan M.M., Schultz M.B., Jerry D.R. Phylogeography of the freshwater crayfish Cherax destructor Clark (Parastacidae) in inland Australia: historical fragmentation and recent range expansion. Biol. J. Linnean Soc. 2004;83:539–550. [Google Scholar]
- Nylander J.A.A., Olsson U., Alström P., Sanmartín I. Accounting for phylogenetic uncertainty in biogeography: a Bayesian approach to dispersal-vicariance analysis of the thrushes (Aves: Turdus) Syst. Biol. 2008;57:257–268. doi: 10.1080/10635150802044003. [DOI] [PubMed] [Google Scholar]
- Nylinder S., Cronholm B., de Lange P.J., Walsh N., Anderberg A.A. Species-tree phylogeny and character evolution in the genus Centipeda (Asteraceae): evidence from DNA sequences from coding and non-coding loci from the plastid and nuclear genomes. Mol. Phylogenet. Evol. 2013 doi: 10.1016/j.ympev.2013.03.020. (in press). Available from: URL http://dx.doi.org/10.1016/j.ympev.2013.03.020. [DOI] [PubMed] [Google Scholar]
- Orr M.R., Smith T.B. Ecology and speciation. Trends Ecol. Evol. 1998;13:502–506. doi: 10.1016/s0169-5347(98)01511-0. [DOI] [PubMed] [Google Scholar]
- Pagel M. The maximum likelihood approach to reconstructing ancestral character states of discrete characters on phylogenies. Syst. Biol. 1999;48:612–622. [Google Scholar]
- Pagel M., Meade A., Barker D. Bayesian estimation of ancestral character states on phylogenies. Syst. Biol. 2004;53:673–684. doi: 10.1080/10635150490522232. doi:10.1080/10635150490522232. [DOI] [PubMed] [Google Scholar]
- Panero J.L. New combinations and infrafamilial taxa in the Asteraceae. Phytologia. 2005;87:1–14. [Google Scholar]
- Pianka E.R. Zoogeography and speciation of Australian desert lizards: an ecological perspective. Copeia. 1972;1972:127–145. [Google Scholar]
- Powney G.D., Grenyer R., Orme C.D.L., Owens I.P.F., Meiri S. Hot, dry and different: Australian lizard richness is unlike that of mammals, amphibians and birds. Global Ecol. Biogeogr. 2010;19:386–396. doi:10.1111/j.1466-8238.2009.00521.x. [Google Scholar]
- Pybus O.G., Suchard M.A., Lemey P., Bernardin F.J., Rambaut A., Crawford F.W., Gray R.R., Arinaminpathy N., Stramer S.L., Busch M.P., Delwart E.L. Unifying the spatial epidemiology and molecular evolution of emerging epidemics. Proc. Natl Acad. Sci. USA. 2012;109:15066–15071. doi: 10.1073/pnas.1206598109. doi:10.1073/pnas.1206598109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. 2006. FigTree. v1.3.1 (available from: URL http://tree.bio.ed.ac.uk/software/figtree).
- Rambaut A., Drummond A.J. 2004. Tracer. v1.4 (available from: URL http://beast.bio.ed.ac.uk/Tracer).
- Ree R.H., Moore B.R., Webb C.O., Donoghue M.J. A likelihood framework for inferring the evolution of geographic range on phylogenetic trees. Evolution. 2005;59:2299–2311. [PubMed] [Google Scholar]
- Rice W.R. Speciation via habitat specialization: the evolution of reproductive isolation as a correlated character. Evol. Ecol. 1987;1:301–314. doi:10.1007/BF02071555. [Google Scholar]
- Ronquist F. Dispersal–vicariance analysis: a new approach to the quantification of historical biogeography. Syst. Biol. 1997;46:195–203. [Google Scholar]
- Ronquist F., Sanmartín I. Phylogenetic methods in biogeography. Annu. Rev. Ecol. Evol. Syst. 2011;42:441–464. doi:10.1146/annurev-ecolsys-102209-144710. [Google Scholar]
- Sang T., Crawford D.J., Kim S.-C., Stuessy T.F. Radiation of the endemic genus Dendroseris (Asteraceae) on the Juan Fernandez Islands: evidence from sequences of the ITS regions of nuclear ribosomal DNA. Am. J. Bot. 1994;81:1494–1501. [Google Scholar]
- Sang T., Crawford D.J., Stuessy T.F., Silva-O M. ITS sequences and phylogeny of the genus Robinsonia (Asteraceae) Syst. Bot. 1995;20:55–64. [Google Scholar]
- Sanmartín I., Ronquist F. New solutions to old problems: widespread taxa, redundant distributions and missing areas in event-based biogeography. Anim. Biodivers. Conserv. 2002;25:75–93. [Google Scholar]
- Schluter D. Evidence for ecological speciation and its alternative. Science. 2009;323:737–741. doi: 10.1126/science.1160006. [DOI] [PubMed] [Google Scholar]
- Schmidt G.J., Schilling E.E. Phylogeny and biogeography of Eupatorium (Asteraceae: Eupatorieae) based on nuclear ITS sequence data. Am. J. Bot. 2000;87:716–726. [PubMed] [Google Scholar]
- Toon A., Mather P.B., Baker A.M., Durrant K.L., Hughes J.M. Pleistocene refugia in an arid landscape: analysis of a widely distributed Australian passerine. Mol. Ecol. 2007;16:2525–2541. doi: 10.1111/j.1365-294X.2007.03289.x. [DOI] [PubMed] [Google Scholar]
- Tremetsberger K., Gemeinholzer B., Zetzsche H., Blackmore S., Kilian N., Talavera S. Divergence time estimation in Cichorieae (Asteraceae) using a fossil-calibrated relaxed molecular clock. Org. Divers. Evol. 2012;13:1–13. doi:10.1007/s13127-012-0094-2. [Google Scholar]
- Walsh N.G. A revision of Centipeda (Asteraceae) Muelleria. 2001;15:33–64. [Google Scholar]
- Woolfit M., Bromham L. Increased rates of sequence evolution in endosymbiotic bacteria and fungi with small effective population sizes. Mol. Biol. Evol. 2003;20:1545–1555. doi: 10.1093/molbev/msg167. doi:10.1093/molbev/msg167. [DOI] [PubMed] [Google Scholar]
- Woolfit M., Bromham L. Population size and molecular evolution on islands. Proc. R. Soc. B. 2005;272:2277–2282. doi: 10.1098/rspb.2005.3217. doi:10.1098/rspb.2005.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesson C., Culham A. A phyloclimatic study of Cyclamen. BMC Evol. Biol. 2006;6:72. doi: 10.1186/1471-2148-6-72. doi:10.1186/1471-2148-6-72. [DOI] [PMC free article] [PubMed] [Google Scholar]