Plants are the primary producers of the terrestrial ecosystems that dominate much of the natural environment. Occurring approximately 480 Ma (Sanderson 2003; Kenrick et al. 2012), the evolutionary transition of plants from an aquatic to a terrestrial environment was accompanied by several major developmental innovations. The freshwater charophyte ancestors of land plants have a haplobiontic life cycle with a single haploid multicellular stage, whereas land plants, which include the bryophytes (liverworts, hornworts, and mosses) and tracheophytes (also called vascular plants, namely, lycopods, ferns, and seed plants), exhibit a marked alternation of generations with a diplobiontic life cycle with both haploid and diploid multicellular stages and where the embryo remains attached to, and is nourished by, the gametophyte (Haig 2008). The interjection of a multicellular diploid phase into the land–plant life cycle was an important adaptation that enabled long-distance dispersal via mitotic spores where waterborne male gametes have restricted motility in dry terrestrial environments. Despite the similarity among land–plant life cycles, they differ in one significant aspect: in the three bryophyte groups, the haploid gametophytic stage is the dominant vegetative stage, whereas in vascular plants the diploid sporophyte dominates. A common assumption, and one implied by the tradition of referring to bryophytes as “lower plants”—in contrast to the “higher” tracheophytes—is that the bryophytes and their life cycle are primitive (Kato and Akiyama 2005). However, without a strong phylogenetic hypothesis of land–plant relationships, it is not clear which (if either) of the gametophyte or sporophyte was the dominant ancestral vegetative state present in the earliest land plants (Renzaglia et al. 2007; Qiu et al. 2012).
Early land plants have a relatively poor fossil record with few intermediate forms (Kenrick and Crane 1997; Wellman et al. 2003; Clarke et al. 2011), so most of the evidence for early land plant evolution has been based upon the patterns of morphological change that are implied by phylogenetic trees of relationships among extant land plant and algal groups. In this context, several recent studies based on large molecular data sets have converged upon a phylogenetic solution to land plant origins wherein tracheophytes are derived from bryophyte ancestors (Karol et al. 2001; Qiu et al. 2006; Gao et al. 2010; Karol et al. 2010; Chang and Graham 2011). In this hypothesis, the three bryophyte groups, namely liverworts, mosses, and hornworts, diverged sequentially and form a paraphyletic group with the hornworts sister to the tracheophytes. This phylogeny supports an intuitively elegant evolutionary trajectory whereby plants increased in morphological complexity from single-celled algae to seed plants via bryophyte intermediates (Karol et al. 2001; McCourt et al. 2004). Specifically, it implies that the gametophyte-dominant bryophyte life cycle was ancestral among land plants and that the complex modular growth form of the vascular plant sporophyte evolved from the simplistic bryophyte sporophyte that consists only of a single growth module (Kato and Akiyama 2005; Barthélémy and Caraglio 2007).
That the liverworts alone form the sister group to the remaining land plants has become the consensus opinion of land–plant relationships and such a tree is often invoked uncritically as the phylogeny of land plants (e.g., Bowman et al. 2007; Grewe et al. 2009; Wang et al. 2010; Clarke et al. 2011; Lenton et al. 2012; Bowman 2013), with studies typically citing the study of Qiu et al. (2006) as a major source of supporting evidence. The analyses presented by Qiu et al. (2006) are among the most data-rich (36 species for 67 chloroplast genes) and taxon-rich (193 taxa for 6 genes including 4 non-chloroplast ribosomal genes) and are claimed to strongly support the liverworts as the earliest-diverging land plant group. Indeed, as the authors stress, it is the congruence between the densely sampled data set and the data-rich genomic data set that is so compelling.
Interestingly, although support for liverworts as the earliest-diverging lineage among land plants is claimed to be strong in the Qiu et al. (2006) study, analyses of the amino acid translations of the chloroplast genome data presented in the same study actually contradict those of the gene sequence data, and instead strongly support a liverwort-plus-moss clade and a hornwort-plus-lycopod clade. The authors concluded that this conflict “likely reflect[ed] analytical artifacts […] dictated by functional selection at the second codon positions and amino acid sequences” (Qiu et al. 2006). However, the contradiction of this topology by chloroplast protein analyses is not an isolated case, as other studies of chloroplast proteins tend to support a monophyletic bryophytes (Nishiyama et al. 2004; Karol et al. 2010), or a moss-plus-liverwort clade when hornworts were not included (Nishiyama and Kato 1999). In addition, two studies of nuclear proteins also resolved a moss-plus-liverwort clade, although neither analysis included a hornwort representative (Finet et al. 2010; Wodniok et al. 2011). The problem underlying the conflict in many of these studies can be viewed as a question of where to place the charophyte root on a tree consisting of (in sequence) liverworts–mosses–hornworts–tracheophytes. If the root is placed between hornworts and tracheophytes then bryophytes will be monophyletic, if placed between mosses and hornworts it will make the mosses-plus-liverworts as the earliest-diverging lineages sister to the remaining land plants, whereas if it is placed on one of the branches leading to any of the three bryophyte groups then that group alone will form the earliest-diverging lineage.
The observation that chloroplast protein-coding gene sequence data (nucleotides) and their protein translations (amino acids) support conflicting phylogenies is profoundly important because both types of data are derived from the same observations and must have therefore evolved under the same evolutionary species tree. The conflict between nucleotides and amino acids must be due to erroneous assumptions in the models used to reconstruct the phylogenies. In this study, we characterize and diagnose the nature of this phylogenetic conflict within the context of the origin of land plants. We address this problem by reanalyzing the chloroplast protein-coding gene data of Karol et al. (2010) and the large taxon-dense, six gene, data set of Qiu et al. (2006)): the former because it contains an expanded sample of chloroplast data—additional lycopods and a second moss—not available at the time of the Qiu et al. (2006) study. Using conventional phylogenetic methods with both composition homogeneous (stationary) and heterogeneous (nonstationary) models, we show that the topological conflict between analyses of nucleotides and amino acids is statistically strong. That is, the nucleotide data of Karol et al. (2010) support a paraphyletic bryophytes whereas the amino acid translations of the same genes support a monophyletic bryophytes. To explore the source of the conflicting signal between the nucleotide data and the amino acid data, we implemented a nucleotide recoding scheme in which all possible synonymous substitutions among codon variants of each amino acid were eliminated by using nucleotide ambiguity codes (Criscuolo and Gribaldo 2010; Regier et al. 2010). This codon-degenerated data analysis allows us to demonstrate that the conflict between analyses of the original nucleotide data and the amino acid data can be attributed to the influence of a directional mutation pressure among synonymous substitutions that drives a composition bias in chloroplast protein-coding nucleotide sequences. Support for a paraphyletic bryophytes in the taxon-dense six gene study of Qiu et al. (2006) can be attributed to the same phenomenon in the two chloroplast genes of that data set. We conclude that support for the paraphyly of bryophytes in chloroplast protein-coding genes is likely to be a phylogenetic artifact, and that the bryophyte lineage is likely monophyletic as evidenced by the amino acid data.
Materials and Methods
Our initial strategy was to confirm the results of the analyses of Karol et al. (2010) and Qiu et al. (2006), before applying more complex models. Data used in the study of Karol et al. (2010) were obtained from the authors. The full data matrix consisted of 49 protein-coding genes from 43 taxa, with 35,382 aligned sites. After exclusion of the characters sets originally defined and excluded by Karol et al. (2010), the matrix was reduced in size to the 30,018 sites (Karol10-nuc) that were analyzed in the original study. The gene data were translated into 10,006 amino acids (Karol10-aa) using MacClade (version 4.08; Maddison and Maddision 2000). Original data matrices used in the Qiu et al. (2006) study were obtained from TreeBase study S1673: “multigene supermatrix” (M1693/M2935). The multigene supermatrix (Qiu06-MS) consisted of chloroplast (cp) SSU and LSU, mitochondrial LSU, nuclear SSU, and two chloroplast protein-coding genes, namely, rbcL and atpB, for 193 green algae and land plants.
Maximum-likelihood bootstrap analyses were conducted using MPI-compiled RAxML (version 7.0.4; Stamatakis 2006) and PAUP* (4.0b10; Swofford 2002). RAxML analyses consisted of 400 bootstrap replicates with default settings for parameter estimation accuracy, and using the amino acid model-specified composition frequencies (i.e., not + F). PAUP* bootstrap analyses were performed on the Karol10-nuc data as described in Karol et al. (2010), with the fixed parameter values of the GTR + I + Γ model estimated using MrModeltest (version 2.3; Nylander 2004) and NNI branch swapping. Bayesian MCMC analyses were performed using P4 (version 0.89; Foster 2004) and MrBayes (version 3.1.2; Ronquist and Huelsenbeck 2003). The node-discrete composition heterogeneity model (NDCH; Foster 2004) of P4, which allows composition to evolve across the tree, was used. Posterior probabilities less than 0.95 are considered low and to indicate only weak support of nodes, whereas those greater than 0.95 are considered strong indicators of monophyly. Marginal likelihoods were estimated according to Equation (16) of Newton and Raftery (1994) as implemented in P4. This estimator of the marginal likelihood was formulated to overcome some of the problems associated with the harmonic-mean estimator (Kass and Raftery 1995); problems with the harmonic-mean estimator have been noted elsewhere (Lartillot and Philippe 2006). Better-fitting models were those that had a higher marginal likelihood—log Bayes Factors (log BF) > 10 units calculated when comparing models are typically considered “very strong” support for the alternative model (2*(logeLalternative) − logeL(null)); Kass and Raftery 1995). The protein data set Karol10-aa was analyzed using the gcpREV model that was estimated from green plant chloroplast data (Cox and Foster 2013). Details of individual analyses are presented in the legends of Figures S1–S21 of the Supplementary Material which is available at http://dx.doi.org/10.5061/dryad.26qt6.
To test the influence of mutation-driven composition biases at each of the three codon positions of the protein-coding genes we performed codon-degenerated phylogenetic analyses. Each codon of the sequence data for each taxon in the Karol10-nuc, Qiu06-MS, and Qiu06-nuc matrices were manipulated through ambiguity recoding to eliminate all signal associated with synonymous substitutions, for example serine codons UCU and AGU both translate to WSN (Criscuolo and Gribaldo 2010; Regier et al. 2010; Supplementary Fig. S22). Triplets of sites defining codons that contain missing data that would translate to an ambiguous amino acid (“X”) were removed. Nucleotide composition and codon bias were analyzed with GCUA (McInerney 1998).
Results and Discussion
Conflict between Chloroplast Protein-Coding Genes and their Protein Translations
The best-fitting model to the protein-coding gene data (Karol10-nuc) was a codon-partitioned composition heterogeneous (NDCH) Bayesian analysis (Fig. 1a; Supplementary Fig. S1), which indicates that the mosses-plus-liverworts form the first-diverging clade, with the hornworts (Anthoceros) strongly supported as the sister group to the tracheophytes. Other nucleotide analyses also strongly support the hornwort-plus-tracheophyte clade (Supplementary Figs. S2–S7). Analyses of the protein translations (Karol10-aa) of the Karol10-nuc data consistently supported bryophytes as monophyletic (Fig. 1b; Supplementary Figs. S8–S12). However, the amino acid analyses showed no strongly supported resolution between the bryophyte, fern, lycopod, and seed plant clades, although each of these groups were themselves well supported. Further details of the phylogenetic results can be found in the Supplementary Material.
Figure 1.
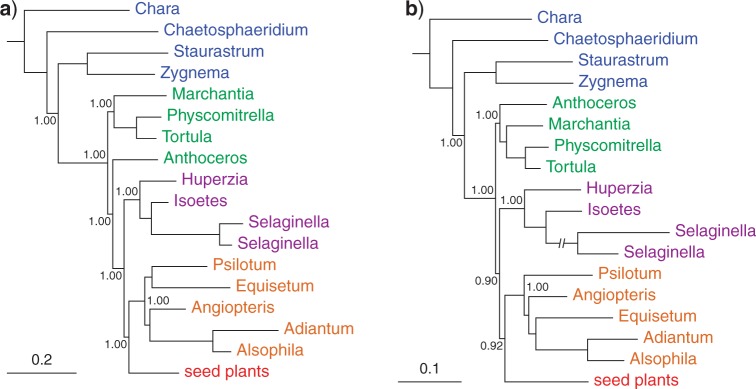
Summary trees of conflict between chloroplast protein-coding gene data (a) and their translated proteins (b) of the data set Karol10-nuc. Trees are depicted without the outgroup taxa (Chlorokybus and Mesosigma) and the seed plant clade is collapsed—full trees are presented in the Supplementary Material. Taxa are indicated as follows: algal charophytes (blue), bryophytes (Marchantia—liverwort, Physcomitrella and Tortula—mosses, Anthoceros—hornwort; green), lycopods (mauve), and ferns (orange). a) Bayesian MCMC (p4) with model 3*(GTR + I + Γ4 + CV2) + Rm + PP, marginal likelihood -Lh = 452713.8554 (Supplementary Fig. S1); b) Bayesian MCMC (p4) with model gcpREV + I + Γ4 + PP, marginal likelihood -Lh = 178265.1502 (Supplementary Fig. S9). The branch leading to Selaginalla species has been arbitrarily shortened (original length = 0.261519). All protein analyses maximally supported a monophyletic bryophytes (hornworts, mosses, liverworts) but only weak (<0.95 PP) support for relationships among tracheophyte groups (lycopods, ferns, seed plants) was observed.
Although the nucleotide data strongly support a tree with hornworts sister to the tracheophytes, analyses of the amino acid translations of the same data strongly support monophyletic bryophytes. As these two data types are different representations of the same species history, one (or perhaps both) set of analyses must have model assumptions that are wrong—here we attempt to diagnose this pathology.
Directional Mutation Bias, Nucleotide Composition, and Codon Bias
Directional mutational pressure, or biases in the mutation rate between the G/C and A/T nucleotide base pairs, causes base composition biases in the genome, the magnitude of which can vary among species (Sueoka 1988). Imbalances in the frequencies of synonymous codons (i.e., codon biases) are caused primarily by these composition biases at the nucleotide level rather than selective forces acting upon codons (Knight et al. 2001; Sharp et al. 2010). Consequently, patterns of nucleotide and protein change among species are driven by a directional mutation bias and shaped by purifying selection that varies among sites (Knight et al. 2001). Similar nucleotide composition biases among taxa can cause similarity (convergence) among nucleotide sequences, and even among protein sequences at sites that are selectively unconstrained. Among sites at the nucleotide level, the magnitude of the mutation-driven compositional bias is necessarily correlated with the substitution rate, such that sites that accommodate many selectively neutral substitutions (e.g., third codon positions of protein coding genes) can be expected to harbor greater bias than slowly evolving sites where substitution is selectively constrained (Wan et al. 2004). Moreover, because synonymous substitutions among redundant codons are free of selective constraint, nucleotide sequences evolve relatively rapidly when compared with protein sequences which are selectively constrained by function. Consequently, proteins can be expected to be less affected by sequence convergence caused by similar mutation-driven compositional biases.
From the perspective of phylogenetic practice, the observation that a directional mutation pressure that can vary among species drives patterns of neutral change in nucleotide sequences is important. All time- and site-homogeneous phylogenetic models assume that the base composition of sequences remains constant through time and among sites in a data partition. If this assumption is not met, analyses can result in incorrect solutions where taxa with a similar compositional biases are incorrectly joined in the tree (Lockhart et al. 1992; Foster 2004). But in addition, all current models assume the same target compositions among both nonsynonymous and synonymous substitutions in protein-coding nucleotide data.
Differential biases manifest at each of the three codon position sites in protein-coding sequences because these sites are under different substitutional constraints. Specifically, due to redundancy of the genetic code in protein-coding genes, substitutions at third positions of codons are mostly synonymous, whereas those at first codon positions are mostly nonsynonymous (except between synonymous codon variants of Leucine [TTR/CTN], Arginine [CGN/AGR], and Serine [TCN/AGY]), and almost all second codon position substitutions are nonsynonymous (except among synonymous codon variants of Serine [TCN/AGY]). These constraints are reflected in differences in substitution rate among the three codon positions of the Karol10-nuc data: in the codon site-specific analyses (Supplementary Fig. S1), the rate of substitution at third positions (2.2888) was 8.8x greater than at second positions (0.2614), and 5.1x greater than at first codon positions (0.4497). Because codon bias is causally linked with the amount of composition bias, it is possible to use codon bias as a qualitative indicator of the extent of variation in composition bias. One measure of codon bias is the relative synonymous codon usage (RSCU) value, which measures the frequency at which a codon is observed relative to the number of times that the codon would be observed in the absence of any codon usage bias (McInerney 1998). The first two principal components of a correspondence analysis of variation in RSCU values among the taxa of the Karol10-nuc are plotted in Figure 2. It is evident that the hornwort, Anthoceros, has variation in codon bias more similar to that of the seed plants and lycopods, than to the mosses and liverworts. This observation suggests that similar codon/composition biases may provide some of the explanation for why Anthoceros is resolved as more closely related to tracheophytes than the remaining bryophytes in the analyses of Karol10-nuc.
Figure 2.
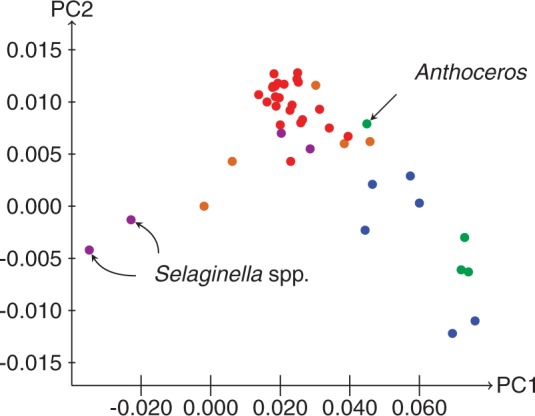
Correspondence analysis of the first and second principal components of the variation of RSCU among the taxa in the Karol10-nuc chloroplast gene data set. Algae: blue; bryophytes: green; ferns: orange; lycopods: mauve; seed plants: red.
Degenerate Codon Analyses
To further investigate the effect of composition biases induced by synonymous substitutions, we replaced the codons of all the gene sequences with degenerate codons that use nucleotide ambiguity codes to eliminate the distinction between synonymous codons (Criscuolo and Gribaldo 2010; Regier et al. 2010). By removing the effect of the synonymous substitutions at each codon, we can see the effect of such biases. Comparisons of exchange rates and compositions between nonsynonymous substitutions of the codon-degenerated data and all substitutions indicate that nonsynonymous and synonymous substitutions are evolving under quite different processes (Fig. 3; Supplementary Material). ML bootstrap analyses of the Karol10-nuc codon-degenerate data under a time-homogeneous model completely eliminates (0% bootstrap proportion) support for the relationship of the hornwort Anthoceros to the tracheophytes (Fig. 4; Supplementary Fig. S13), in stark contrast to the ML bootstrap analyses of the nondegenerated data (Supplementary Fig. S3) that supports the same relationship at 100% bootstrap proportion. Likewise, MCMC analyses of the Karol10-nuc codon-degenerate data reduces the posterior probability of the hornwort-plus-tracheophyte clade to low values ( < 0.50–0.59 PP Supplementary Figs. S14–S17) compared with their being maximally supported in similar analyses of the nondegenerated data (1.00 PP in each of Supplementary Figs. S4–S7). These analyses indicate that the conflict between Karol10-nuc data and their translated proteins, Karol10-aa, with regard to the placement of the hornwort Anthoceros, is due to the influence of synonymous substitutions and, as indicated by Figure 2, is most likely in part the result of differences in composition manifest in these substitutional changes.
Figure 3.
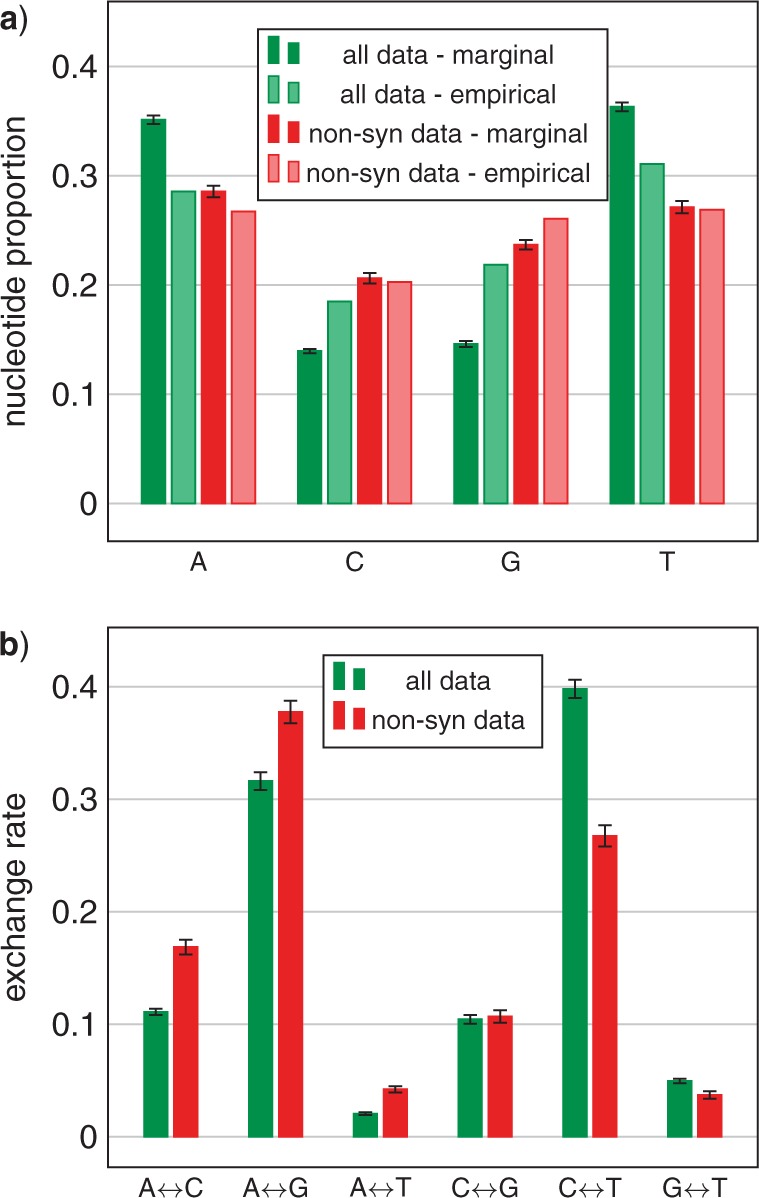
Comparison of mean model parameter values compared between data of all substitutions (all) and the codon-degenerated data consisting of only nonsynonymous substitutions (non-syn) of the Karol10-nuc data: a) composition frequencies, both empirical values and marginal estimates, and b) marginal estimates of substitution exchange rates. Marginal estimates are mean posterior values from the homogeneous MCMC analyses shown in Supplementary Figures S6 and S15 for “all” and “non-syn” data respectively. The error bars represent the 95% confidence interval (1.96 standard deviations of the mean of the posterior sample).
Figure 4.
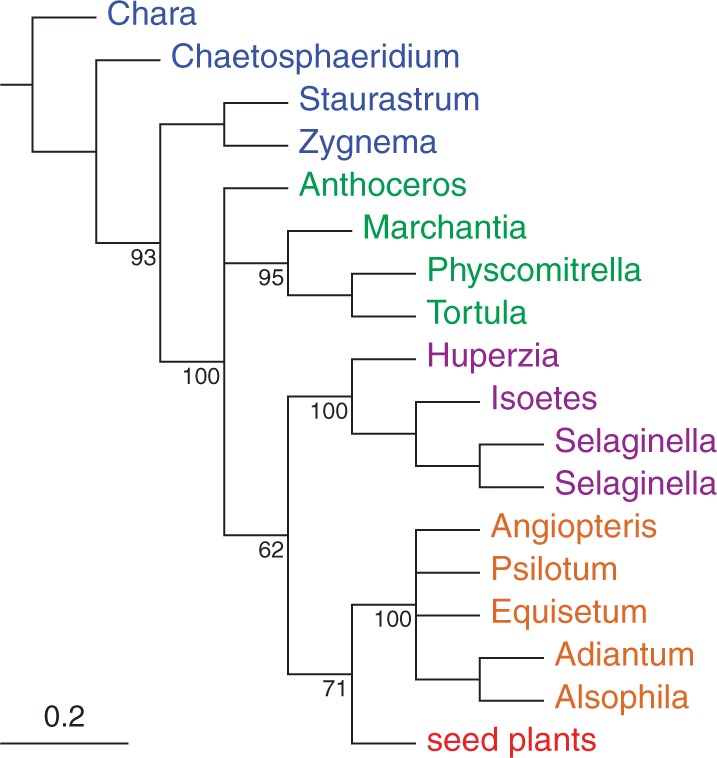
Summary tree of the maximum-likelihood bootstrap analysis of the codon-degenerated Karol10-nuc data. Tree structure summation and taxon colors follow those detailed in the legend of Figure 1.
Synonymous Substitutions are Unreliable Indicators of Phylogeny among Major Land Plant Groups
For analyses of relatively deep, ancient, phylogenetic relationships, it is common to eliminate all third codon positions prior to analyses (e.g., Goremykin et al. 2003; Gibson et al. 2005; Saitoh et al. 2006; Dávalos et al. 2012): a decision sometimes justified by assuming the sites are substitutionally “saturated” that is, so much change has occurred at these sites that no historically accurate information remains. Even if third codon positions retain reliable information for shallow (more recent) divergences, they have been shown to retain less inferential power for more ancient relationships (Townsend 2007). However, it is the bias that is often associated with long branches and saturation, rather than saturation per se, that is potentially phylogenetically misleading, and faster changing sites are likely to harbor greater biases. Accordingly, here we note that it is synonymous substitutions in total among all three codon positions, and not just those at third codon positions, that provide evidence for the sister group relationship of Anthoceros to the tracheophytes. When third codon positions only are codon-degenerated, Anthoceros is again resolved as the sister group to tracheophytes but without strong posterior probability support (0.89 PP Supplementary Fig. S18) or ML bootstrap support (51% Supplementary Fig. S19). This result indicates that the synonymous substitutions at first and second codon positions among Leucine, Arginine, and Serine synonymous codon families contain signal supporting the relationship: exactly what would be expected if the causative factor were similar composition biases driven by mutation pressure acting at a genomic level among all synonymous substitutions. Inagaki et al. (2004)) observed a similar conflict between the phylogenies of first and second codon positions and their protein translations due to substitutions between synonymous variants of Leucine, Arginine, and Serine codons in two chloroplast photosystem genes. Moreover, as similar observations have been made in animal nuclear gene data (Regier et al. 2010; Regier and Zwick 2011; Rota-Stabelli et al. 2012), it is likely a feature of protein-coding genes in general.
Despite having identified composition bias induced by synonymous substitutions as the cause of the conflict between protein-coding nucleotide sequences and their protein translations in the Karol et al. (2010) data, the question remains whether this signal is a reliable indicator of phylogeny. That is, whether the similar composition and codon biases resulting from synonymous substitutions between the hornwort and tracheophytes (Fig. 2) evolved in an immediate ancestor of both groups to the exclusion of other bryophytes. If so, then the bryophyte monophyly supported by the amino acid data must be an artifact. One possible cause of such an artifact, as suggested by Qiu et al. (2006), is convergent protein functional constraint, in this case between hornworts and other bryophytes. However, this seems unlikely to have occurred as the functional constraint would need to act in concert at the level of the whole chloroplast genome (i.e., among many proteins), and chloroplasts are highly specialized to a few metabolic pathways (Allen 2002), such as light harvesting, which are not known to be specifically correlated with the bryophyte life style. On the other hand, if the relationship between hornworts and tracheophytes is itself an artifact, then we have a known genome-wide acting causative agent in mutation pressure-driven compositional bias that is demonstrably present in the taxa in question, and a mechanism through which the bias can be seen to act (i.e., through synonymous substitutions) and is known to cause phylogenetic error. Hence, we consider it highly likely that the synonymous substitutions are unreliable and hence that the bryophytes are better interpreted as monophyletic as supported by the amino acid data.
Analyses of the Taxon-Rich Multigene Data Set of Qiu et al. (2006)
Because both the taxon-rich (193 taxa) multigene (Qiu06-nuc) and multi-genome data set (Qiu06-MS) of Qiu et al. (2006) strongly supported a nonmonophyletic bryophytes with liverworts the earliest branching land plant lineage and hornworts the sister group to the tracheophytes, these analyses were seen as convincing evidence for the paraphyly of bryophytes and the derivation of tracheophytes from a bryophyte-like ancestor. In addition, these analyses were seemingly persuasive as they included ribosomal RNA genes from all three genomic compartments (nuclear SSU, mitochondrial LSU, and chloroplast SSU and LSU). However, when the signal associated with synonymous substitutions of the two chloroplast protein-coding genes is removed by codon-degenerate ambiguity recoding, the ML bootstrap support for a paraphyly of the bryophytes is < 50% (Supplementary Fig. S20). We also note that the high taxon-richness per se is not necessary to derive a paraphyletic bryophytes tree under the analytical assumptions of the original authors. In fact, reducing the numbers of land plant taxa to only 14—two in each major land–plant group spanning the most ancestral node—is still sufficient sampling to result in good ML bootstrap support for the same topology among major land–plant groups as when all 193 taxa were included (Supplementary Fig. S21). Consequently, we suggest (for the same reasons argued above) that the support for the paraphyly of bryophytes, specifically the support for the hornworts as the sister group to the tracheophytes, in these data is an artifact caused by convergent base composition induced by synonymous substitutions.
Conclusions
Our analyses show that the support for bryophyte paraphyly in the protein-coding gene analysis of Karol et al. (2010)) and the taxon-dense six-gene analysis of Qiu et al. (2006)) can be attributed to compositional biases induced by synonymous substitutions. Consequently, we consider the monophyly of the bryophytes obtained in our new analyses of the amino acid data of Karol et al. (2010)) to be the best supported hypothesis. To explain the apparent conflict with the previously published trees, we suggest that previous results are caused by a failure to correct for a mutation pressuredriven compositional bias, present mainly in synonymous substitutions of protein-coding genes, that is convergent between hornworts and tracheophytes. Mutation-driven compositional biases associated with synonymous substitutions may be common in protein-coding gene sequences and hence potentially a general source of phylogenetic artifacts.
Whereas the data we analyzed here are mainly restricted to the chloroplast, we note that a monophyletic bryophyte group does not conflict with current land plant estimates of phylogeny based on nuclear data (Wodniok et al. 2011)—although crucially those data lack a representative of the hornworts. If, as anticipated, the chloroplast phylogeny is congruent with species phylogeny, then the observation that bryophytes form a monophyletic group is of the highest importance for our understanding of the evolution of plants on land. In particular, although it is now clear that the land–plant sporophyte evolved by intercalation of a multicellular diploid phase in the life cycle of a charophytic ancestor (i.e., favoring the antithetic hypothesis over the homologous theory; Haig 2008), if the bryophytes form a monophyletic group, there is no longer a clear implication that the complex sporophyte of the tracheophytes evolved from the simplistic, single module, sporophyte present in bryophytes.
Supplementary Material
Data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.26qt6.
Funding
Fundação para a Ciência e a Technologia (FCT), Portugal (PTDC/BIA-BCM/099565/2008, PTDC/BIA-EVF/113129/2009 to C.J.C.). European Regional Development Fund (ERDF) through the COMPETE—Operational Programme Competitiveness and national funds through FCT, under the project Pest-C/MAR/LA0015/2011(in part). European Research Council Advanced Investigator Programme (to T.M.E.).
Acknowledgments
We would like to thank Ron DeBry, Harald Schneider, and an anonymous reviewer for their helpful criticism during the preparation of this article.
References
- Allen J.F. The function of genomes in bioenergetic organelles. Phil. Trans. R. Soc. Lond. B. 2002;358:19–38. doi: 10.1098/rstb.2002.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthélémy D., Caraglio Y. Plant architecture: a dynamic, multilevel and comprehensive approach to plant form, structure and ontogeny. Ann. Botany. 2007;99:375–407. doi: 10.1093/aob/mcl260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J.L. Walkabout on the long branches of plant evolution. Curr. Opin. Plant Biol. 2013;16:70–77. doi: 10.1016/j.pbi.2012.10.001. [DOI] [PubMed] [Google Scholar]
- Bowman J.L., Floyd S.K., Sakakibara K. Green genes - comparative genomics of the green branch of life. Cell. 2007;129:229–234. doi: 10.1016/j.cell.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Chang Y., Graham S.W. Inferring the higher-order phylogeny of mosses (Bryophyta) and relatives using a large, multigene plastid data set. Am. J. Botany. 2011;98:839–849. doi: 10.3732/ajb.0900384. [DOI] [PubMed] [Google Scholar]
- Clarke J.T., Warnock R.C.M., Donoghue P.C.J. Establishing a time-scale for plant evolution. New Phytol. 2011;192:266–301. doi: 10.1111/j.1469-8137.2011.03794.x. [DOI] [PubMed] [Google Scholar]
- Cox C.J., Foster P.G. A 20-state empirical amino-acid substitution model for green plant chloroplasts. Mol. Phylogenet. Evol. 2013;68:218–210. doi: 10.1016/j.ympev.2013.03.030. [DOI] [PubMed] [Google Scholar]
- Criscuolo A., Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dávalos L.M., Cirranello A.L., Geisler J.H., Simmons N.B. Understanding phylogenetic incongruence: lessons from phyllostomid bats. Biol. Rev. 2012;87:991–1024. doi: 10.1111/j.1469-185X.2012.00240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finet C., Timme R.E., Delwiche C.F., Marlétaz F. Multigene phylogeny of the green lineage reveals the origin and diversification of land plants. Curr. Biol. 2010;20:2217–2222. doi: 10.1016/j.cub.2010.11.035. [DOI] [PubMed] [Google Scholar]
- Foster P.G. Modeling compositional heterogeneity. Syst. Biol. 2004;53:485–495. doi: 10.1080/10635150490445779. [DOI] [PubMed] [Google Scholar]
- Gao L., Su. Y.-J., Wang T. Plastid genome sequencing, comparative genomics, and phylogenomics: current status and prospects. J. Syst. Evol. 2010;48:77–93. [Google Scholar]
- Gibson A., Gowri-Shankar V., Higgs P.G., Rattray M. A comprehensive analysis of mammalian mitochondrial genome base composition and improved phylogenetic methods. Mol. Biol. Evol. 2005;22:251–264. doi: 10.1093/molbev/msi012. [DOI] [PubMed] [Google Scholar]
- Goremykin V.V., Hirsch-Ernst K.I., Wölfl S., Hellwig F.H. Analysis of the Amborella trichopoda chloroplast genome sequence suggests that Amborella is not a basal angiosperm. Mol. Biol. Evol. 2003;20:1499–1505. doi: 10.1093/molbev/msg159. [DOI] [PubMed] [Google Scholar]
- Grewe F., Viehoever P., Weisshaar B., Knoop V. A trans-splicing group I intron and tRNA-hyperediting in the mitochondrial genome of the lycophyte Isoetes engelmannii. Nucleic Acids Res. 2009;37:5093–5104. doi: 10.1093/nar/gkp532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D. Homologous versus antithetic alternation of generations and the origin of sporophytes. Bot. Rev. 2008;74:395–418. [Google Scholar]
- Inagaki Y.I., Simpson A.G.B., Dacks J.B., Roger A.J. Phylogenetic artifacts can be caused by leucine, serine, and arginine codon usage heterogeneity: dinoflagellate plastid origins as a case study. Syst. Biol. 2004;53:582–593. doi: 10.1080/10635150490468756. [DOI] [PubMed] [Google Scholar]
- Karol K.G., Arumuganathan K., Boore J.L., Duffy A.M., Everett K.D.E., Hall J.D., Hansen S.K., Kuehl J.V., Mandoli D.F., Mishler B.D., Olmstead R.G., Renzaglia K.S., Wolf P.G. Complete plastome sequences of Equisetum arvense and Isoetes flaccida : implications for phylogeny and plastid genome evolution of early land plant lineages. BMC Evol. Biol. 2010;10:321. doi: 10.1186/1471-2148-10-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karol K.G., McCourt R.M., Cimino M.T., Delwiche C.F. The closest living relatives of land plants. Science. 2001;294:2351–2353. doi: 10.1126/science.1065156. [DOI] [PubMed] [Google Scholar]
- Kass R.E., Raftery A.E. Bayes factors. J. Am. Statist. Assoc. 1995;90:773–795. [Google Scholar]
- Kato M., Akiyama H. Interpolation hypothesis for origin of the vegetative sporophyte of land plants. Taxon. 2005;54:443–450. [Google Scholar]
- Kenrick P., Crane P. The origin and early diversification of land plants. Washington, DC: Smithsonian Institutional Press; 1997. pp. 1–437. [Google Scholar]
- Kenrick P., Wellman C.H., Schneider H., Edgecombe G.D. A timeline for terrestrialization: consequences for the carbon cycle in the Paleozoic. Phil. Trans. R. Soc. B. 2012;367:519–536. doi: 10.1098/rstb.2011.0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight R.D., Freeland S.J., Landweber L.F. A simple model based on mutation and selection explains trends in codon and amino-acid usage and GC composition within and across genomes. Genome Biol. 2001;2:1–12. doi: 10.1186/gb-2001-2-4-research0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N., Philippe H. Computing Bayes factors using thermodynamic integration. Syst. Biol. 2006;55:195–207. doi: 10.1080/10635150500433722. [DOI] [PubMed] [Google Scholar]
- Lenton T.M., Crouch M., Johnson M., Pires N., Dolan L. First plants cooled the Ordovician. Nature Geosci. 2012;5:86–89. [Google Scholar]
- Lockhart P.J., Howe C.J., Bryant D.A., Beanland T.J., Larkum A.W.D. Substitutional bias confounds inference of cyanelle origins from sequence data. J. Mol. Evol. 1992;34:15–162. doi: 10.1007/BF00182392. [DOI] [PubMed] [Google Scholar]
- Maddison D.R., Maddision W.P. MacClade 4. Sunderland, MA: Sinauer Associates; 2000. [Google Scholar]
- McCourt R.M., Delwiche C.F., Karol K.G. Charophyte algae and land plant origins. T.R.E.E. 2004;19:661–666. doi: 10.1016/j.tree.2004.09.013. [DOI] [PubMed] [Google Scholar]
- McInerney J.O. GCUA (General Codon Usage Analysis) Bioinformatics. 1998;14:372–373. doi: 10.1093/bioinformatics/14.4.372. [DOI] [PubMed] [Google Scholar]
- Newton M.A., Raftery A.E. Approximate Bayesian inference with the weighted likelihood bootstrap. J. R. Stat. Soc. Ser. B. 1994;56:3–48. [Google Scholar]
- Nishiyama T., Kato M. Molecular phylogenetic analysis among bryophytes and tracheophytes based on combined data of plastid coded genes and the 18S rRNA gene. Mol. Biol. Evol. 1999;16:1027–1036. doi: 10.1093/oxfordjournals.molbev.a026192. [DOI] [PubMed] [Google Scholar]
- Nishiyama T., Wolf P.G., Kugita M., Sinclair R.B., Sugita M., Sugiura C., Wakasugi T., Yamada K., Yoshinaga K., Yamaguchi K., Ueda K., Hasebe M. Chloroplast phylogeny indicates that bryophytes are monophyletic. Mol. Biol. Evol. 2004;21:1813–1819. doi: 10.1093/molbev/msh203. [DOI] [PubMed] [Google Scholar]
- Nylander J.A.A. MrModeltest 2.3. Uppsala, Sweden: Evolutionary Biology Centre, Uppsala University; 2004. Program distributed by the author. [Google Scholar]
- Qiu Y.-L., Li L., Wang B., Chen Z., Knoop V., Groth-Malonek M., Dombrovska O., Lee J., Kent L., Rest J., Estabrook G.F., Hendry T.A., Taylor D.W., Testa C.M., Ambros M., Crandall-Stotler B., Duff R.J., Stech M., Frey W., Quandt D., Davis C.C. The deepest divergences in land plants inferred from phylogenomic evidence. Proc. Natl. Acad. Sci. U.S.A. 2006;103:15511–15516. doi: 10.1073/pnas.0603335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y.-L., Taylor A.B., McManus H.A. Evolution of the life cycle in land plants. J. Syst. Evol. 2012;50:171–194. [Google Scholar]
- Regier J.C., Shultz J.W., Zwick A., Hussey A., Ball B., Wetzer R., Martin J.W., Cunningham C.W. Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature. 2010;463:1079–1083. doi: 10.1038/nature08742. [DOI] [PubMed] [Google Scholar]
- Regier J.C., Zwick A. Sources of signal in 62 protein-coding nuclear genes for higher-level phylogenetics of arthropods. PLoS One. 2011;6:1–21. doi: 10.1371/journal.pone.0023408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzaglia K.S., Schuette S., Duff R.J., Ligrone R., Shaw A.J., Mishler B.D., Duckett J.D. Bryophyte phylogeny: advancing the molecular and morphological frontiers. Bryologist. 2007;110:179–213. [Google Scholar]
- Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rota-Stabelli O., Lartillot N., Philippe H., Pisani D. Serine codon usage bias in deep phylogenomics: pancrustacean relationships as a case study. Syst. Biol. 2012 doi: 10.1093/sysbio/sys077. Advance Access published 6 September 2012. [DOI] [PubMed] [Google Scholar]
- Saitoh K., Sado T., Mayden R.L., Hanzawa N., Nakamura K., Nishida M., Miya M. Mitogenomic evolution and interrelationships of the cypriniformes (Actinopterygii: Ostariophysi): the first evidence toward resolution of higher-level relationships of the world's largest freshwater fish clade based on 59 whole mitogenome sequences. J. Mol. Evol. 2006;63:826–841. doi: 10.1007/s00239-005-0293-y. [DOI] [PubMed] [Google Scholar]
- Sanderson M.J. Molecular data from 27 proteins do not support a precambrian origin of land plants. Am. J. Bot. 2003;90:954–956. doi: 10.3732/ajb.90.6.954. [DOI] [PubMed] [Google Scholar]
- Sharp P.M., Emery L.R., Zeng K. Forces that influence the evolution of codon bias. Phil. Trans. R. Soc. B. 2010;365:1203–1212. doi: 10.1098/rstb.2009.0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Sueoka N. Directional mutation pressure and neutral evolution. Proc. Natl. Acad. Sci. U.S.A. 1988;85:2653–2657. doi: 10.1073/pnas.85.8.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D.L. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer Associates; 2002. Version 4. [Google Scholar]
- Townsend J.P. Profiling phylogenetic informativeness. Syst. Biol. 2007;56:222–231. doi: 10.1080/10635150701311362. [DOI] [PubMed] [Google Scholar]
- Wan X.-F., Xu D., Kleinhofs A., Zhou J. Quantitative relationship between synonymous codon usage bias and GC composition across unicellular genomes. BMC Evol. Biol. 2004;4:19. doi: 10.1186/1471-2148-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Yeun L.H., Xue J.-Y., Yang L., Ané J.-M., Qiu Y.-L. Presence of three mycorrhizal genes in the common ancestor of land plants suggests a key role of mycorrhizas in the colonization of land by plants. New Phytol. 2010;186:514–525. doi: 10.1111/j.1469-8137.2009.03137.x. [DOI] [PubMed] [Google Scholar]
- Wellman C.H., Osterloff P.L., Mohiuddin U. Fragments of the earliest land plants. Nature. 2003;425:282–285. doi: 10.1038/nature01884. [DOI] [PubMed] [Google Scholar]
- Wodniok S., Brinkmann H., Glöckner G., Heidel A.J., Philippe H., Melkonian M., Becker B. Origin of land plants: do conjugating green algae hold the key? BMC Evol. Biol. 2011;11:104. doi: 10.1186/1471-2148-11-104. [DOI] [PMC free article] [PubMed] [Google Scholar]


