Abstract
The advent of whole-exome next-generation sequencing (WES) has been pivotal for the molecular characterization of Mendelian disease; however, the clinical application of WES has remained relatively unexplored. We describe our experience with WES as a diagnostic tool in a three-year old female patient with a two-year history of episodic muscle weakness and paroxysmal dystonia who presented following a previous extensive but unrevealing diagnostic work-up. WES was performed on the proband and her two parents. Parental exome data was used to filter de novo genomic events in the proband and suspected mutations were confirmed using di-deoxy sequencing. WES revealed a de novo non-synonymous mutation in exon 21 of the calcium channel gene CACNA1S that has been previously reported in a single patient as a rare cause of atypical hypokalemic periodic paralysis. This was unexpected, as the proband’s original differential diagnosis had included hypokalemic periodic paralysis, but clinical and laboratory features were equivocal, and standard clinical molecular testing for hypokalemic periodic paralysis and related disorders was negative. This report highlights the potential diagnostic utility of WES in clinical practice, with implications for the approach to similar diagnostic dilemmas in the future.
Keywords: Hypokalemic periodic paralysis, CACNA1S, next generation sequencing, hypotonia
INTRODUCTION
Studies of Mendelian disease have been transformed by the advent of whole-exome next-generation sequencing (1), which has enabled the identification and molecular characterization of a wide spectrum of traits (2-5). More recently, however, the role of whole-exome sequencing in the armory of clinical genetics has come to the fore, with implications for the diagnostic and therapeutic aspects of disease (6-10).
Primary hypokalemic periodic paralysis (HypoKPP) is an autosomal dominant disorder typically characterized by acute, episodic, usually flaccid loss of skeletal muscle tone in the context of low serum potassium (between 1 and 3 mmol/L). The typical age of onset is between 5 and 20 years (11), with episodes that last hours to days and are often precipitated by carbohydrate-rich meals and rest after prolonged exercise. Myopathy may develop in some, but myotonia is not a prominent feature. HypoKPP is caused by mutations in the gene encoding a voltage gated calcium channel - CACNA1S in 50-75% of cases, or sodium channel - SCN4A in 8-10% of cases (11). Both genes are fairly large, and all reported disease-causing mutations involve substitution of highly conserved arginine residues found in voltage sensing segments. In CACNA1S, four mutations in exons 11 and 30 account for > 90% of pathogenicity; with a similar percentage of SCN4A cases accounted for by three mutations in exon twelve (12). This distribution of mutations has made targeted testing the mainstay of the current clinical testing paradigm (13). It is clear, however, that the full genetic spectrum of the disease is still unknown, as these two genes only account for about 80% of all cases (12, 14). A molecular diagnosis of HypoKPP is important for the future reproductive choices of the family, the testing of other family members at risk, and can influence appropriate pharmacologic therapy; for instance, although the diuretic acetazolamide is reported to be efficacious for the majority of patients with HypoKPP, it has been associated with less benefit or even worsening of symptoms in some patients with specific mutations in SCN4A(14). Further, individuals with HypoKPP are at an increased risk for post-operative complications related to anesthesia, including malignant hyperthermia. Therefore, a precise and secure molecular genetic diagnosis is important for clinical management, prognosis and genetic counseling.
We report a 3½-year-old female with early onset episodic muscle weakness associated with other neuromuscular abnormalities. After an extensive diagnostic evaluation, including equivocal therapeutic trials and negative clinical sequencing, WES was undertaken to evaluate the hypothesis that she had a rare, de novo mutation in a novel gene causing her symptoms. We advocated for this approach as a means of potentially reducing the time, effort, and cost associated with arriving at a genetic diagnosis.
MATERIALS and METHODS
Participants
Adult members of the study family, which consisted of the proband, mother, and father, provided written informed consent for themselves and their child for enrollment in the study. The study protocol was approved by the Baylor College of Medicine (BCM) Institutional Review Board.
Whole-exome next-generation sequencing
Paired-end whole-exome-enriched libraries were prepared from genomic DNA isolated from the peripheral blood of the proband and parents using an in-house-developed capture reagent from Roche NimbleGen (Madison, WI, USA). Libraries were sequenced on the HiSeq 2000 platform (Illumina, San Diego, CA) and sequencing reads (with 20× or more coverage in 95% of targeted regions) were aligned to the March 2006 human reference assembly (NCBI36/hg18). Annotated high-quality variants were subsequently filtered to exclude common benign variants. Variants of interest were subsequently confirmed by di-deoxy sequencing. Details of the sequencing and filtering protocols are given as supplementary methods.
RESULTS
Clinical Report
The patient was a 3 ½-year-old female with a two-year history of episodic lower limb weakness, who had previously received an extensive but non-diagnostic evaluation. Her episodes manifested as difficulty with weight bearing, stumbling and clumsiness, and subjective descriptions of pain. They usually occurred in the morning and were often associated with prolonged sitting or compromised sleep. There was no association with meals or diet. Episodes occurred two to three times per week, with occasional daily attacks, lasting from 30 minutes to 4 hours. The frequency and intensity of attacks had been relatively stable over the previous two years.
The medical history included early-onset scoliosis, high arched feet, lower limb hypertonia, clumsy gait and frequent toe-walking. Her cognitive development was normal, but she had received developmental services between 18 months and two years of age for mild speech delay. She had a healthy two-year-old sister and there was no family history of similar symptoms, musculoskeletal or neurologic disease. On physical examination, she was non-dysmorphic with tight heel cords, pes cavus, increased plantar reflexes, lordosis, a positive Gower sign, and demonstrated stiff toe-walking and in-toeing of the right foot.
Her previous evaluations included basic blood chemistry and a complete blood count, serum copper, ceruloplasmin, thyroid stimulating hormone, and creatine kinase, all of which were normal. Her karyotype and chromosome microarray analysis (CMA)(15) were normal. Screens for inborn errors of metabolism, cerebrospinal fluid (CSF) studies and brain and spinal magnetic resonance imaging (MRI) were negative. An electromyogram (EMG) was normal and nerve conduction studies showed decreased amplitudes with normal latency and conduction velocities. Initial electroencephalogram (EEG) was normal and a 48-hour video EEG during an episode confirmed the episodes to be non-epileptogenic.
Previously considered diagnoses included paroxysmal dyskinesia (carbamazepine subjectively worsened the episodes); paroxysmal dystonia (a trial of L-dopa was unsuccessful) and hypokalemic periodic paralysis (an empiric trial of acetazolamide resulted in more frequent and intense episodes). Her initial serum electrolytes were normal, but a repeat serum potassium during an episode was 3.0 mmol/L. Subsequent potassium levels during episodes ranged from 2.2 to 3.3 mmol/L.
Clinical molecular testing
Clinical testing for hypokalemic periodic paralysis, consisting of evaluation of four CACNA1S mutations in exons 11 and 30, was negative. Sequencing revealed three benign variants in exon 11 only (see Supplementary Material). Similar DNA testing of exon 12 of SCN4A was also negative, as was sequencing of exons 13, 23, and 24, which contain variants for the allelic disorders Paramyotonia Congenita and Hyperkalemic periodic paralysis. Sequencing of all coding exons and adjacent intron sequences of CLCN1, which contains variants for Myotonia Congenita, revealed two variants of unknown significance (see Supplementary Material), both also found in one of the unaffected parents, who, after careful inquiry and focused examination, was confirmed to be completely asymptomatic. We therefore did not consider these variants sufficient to explain her clinical presentation.
Whole-exome next generation sequencing
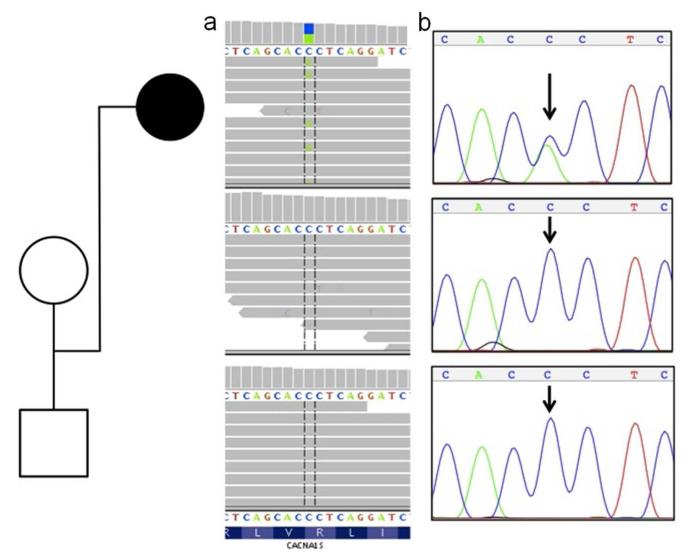
We then considered whether the proband’s clinical presentation may not have been caused by the typical molecular causes for HypokPP. Rather than ad hoc sequencing of potential candidate genes, we carried out WES (see materials and methods) on the patient and her parents with the goal of providing a molecular diagnosis. In our analysis, we first utilized a trio-based model of a fully-penetrant dominant disorder, in which inherited variants were excluded from the list of rare deleterious variants identified in the patient (Table 1). This subtractive process revealed a total of five de novo variants in the patient. We were surprised to find among these a single variant in the CACNA1S gene (Figure 1a) that resulted in an arginine to serine amino-acid change in exon 21 (c.2691G>T; p.R897S). Sanger di-deoxy sequencing confirmed the mutation as de novo in the proband (Figure 1b). This variant was predicted to be deleterious by PolyPhen2 (16) and has been reported as a pathogenic mutation in a single patient with severe, early-onset, hypokalemic periodic paralysis (17). None of the other identified gene variants (see Supplementary Material) were known to be disease-causing or relevant to the patient’s phenotype.
Table 1.
Identification of de novo variants in affected proband.
| Variants | Proband | Mother | Father |
|---|---|---|---|
| Total | 26986 | 26437 | 27146 |
| NSa/SSb/Indelc | 13279 | 12755 | 13317 |
| Noveld | 1301 | 834 | 1241 |
| De novo | 5 | - | - |
NS - nonsynonymous;
SS - splice-site;
indel - frameshift insertion/deletion.
Novel variants were defined as those not found in 1000 Genomes Pilot 1 or an internal exome database.
Figure 1.
De novo CACNA1S mutation in the affected trio. Whole-exome sequence reads (horizontal grey bars) (1a) show a C>A (blue/green block) change in approximately one-half of overlapping reads (vertical hashed lines) at the first position of an arginine codon (R) in the affected proband (top panel), and homozygous wild-type (C) reads for both unaffected parents at the same site (lower two panels). Sanger sequencing (1b) independently confirmed this as a de novo mutation in the proband.
The previous report of CACNA1S R897S (17), involved a 2-year old male who also had an atypical presentation of HypoKPP with early onset and predominant lower extremity involvement.
Our patient had several noteworthy differences (Table 2), which made it difficult to associate her presentation with the previous case report. The reason for these differences remains unclear, however, the clinical similarities between the two patients are sufficient to confirm the R897S variant as pathogenic and suggest that a genotype-phenotype correlation may exist with early age of onset, predominant lower limb involvement, and a spectrum of atypical symptoms.
Table 2.
Comparison of clinical features reported with the CACNA1S R897S mutation.
| This patient | Chabrier et al (17) | |
|---|---|---|
| Age at Onset | 1 year | 1 year |
| Age at Diagnosis | 4 years | 2 years |
| Perinatal period | IUGR* | Neonatal hypotonia |
| Muscle presentation | Predominantly lower limb Persistent hypertonia |
Global hypotonia (lower limbs > than upper) |
| Maximum frequency of attacks | Daily | Daily |
| Temporal relation of attacks | Morning | Evening |
| Initial hypokalemia (with attack) |
3.0 mmol/L | 3.1 mmol/L |
| Additional clinical features | Scoliosis | |
| Pes Cavus | ||
| Toe-walking |
Intrauterine Growth Restriction
DISCUSSION
Molecular confirmation of the diagnosis provided evidence for the necessity of long-term potassium supplementation. Gradual titration of her potassium dose resulted in a decrease in the frequency and intensity of her episodes. The molecular diagnosis also allowed us to mitigate concerns that the family’s youngest child might also be affected and provided reassurance to the parents that their recurrence risk in future pregnancies was low; this can now be determined through pre-natal testing.
This report illustrates the type of case for which WES is well suited - potentially avoiding invasive, expensive, and time-consuming diagnostic evaluations in patients with atypical presentations and equivocal diagnoses. A very important consideration is whether making an actionable diagnosis justifies the cost of WES. We found that, if applied judiciously, WES may actually result in cost savings. Conservatively, the cost for the exome analyses, including the patient and both parents, reagents, equipment and expertise to carry out, interpret and confirm the sequencing results was two to three thousand (US) dollars less than the cost of the previous diagnostic work-up (Supplementary Table), and would have been even more cost effective had we included the costs for hospitalizations, emergency room visits, medications, and physician services. Moreover, the cost of the WES technology is steadily decreasing, and for those patients whose clinical evaluation is likely to involve extensive, expensive, or invasive diagnostic testing to evaluate a broad differential diagnosis, it has been reasonably argued that WES should be considered early in the diagnostic approach (18).
We identified a rare, de novo cause for a genetic disease that was not detected by clinical molecular testing strategies. For CACNA1S-associated HypoKPP, a tiered approach of targeted mutation testing is used, followed by sequencing of the exons 11 and 30. This is sufficient to identify the vast majority of positive cases. Full sequencing of CACNA1S or SCN4A for hypokalemic periodic paralysis is not routinely done on a clinical basis, and the R897S CACNA1S mutation, having only been described once before in the medical literature, is not routinely tested. As a result, our patient’s mutation was not identified by available clinical sequencing. Were our suspicion for hypokalemic periodic paralysis higher, we might have considered the extensive task of sequencing the 44 and 26 exons of the CACNA1S and SCN4A genes (respectively) in their entirety; however, we viewed this as low-yield from a research standpoint given the clinical presentation, and on a clinical basis, this would have been challenging to accomplish. Full sequencing of individual genes in diseases with a restricted and established disease-allele spectrum can be cost prohibitive for clinical laboratories given the lower likelihood of finding rare pathogenic mutations, and accordingly, this is usually reserved for clinically unambiguous cases, (of which our patient was not). Thus, for unusual clinical presentations, WES is likely to be a viable alternative to costly stepwise clinical sequencing of suspected disease genes because WES allows one to simultaneously query all known genes and pathogenic variants, as well as to identify mutations within novel candidate genes.
In conclusion, by identifying a de novo mutation as the cause of HypokPP in this atypical case, we have demonstrated the potential utility of WES in the armory of clinical diagnostics. Viewed in the context of previous reports (6-8), there is now a growing body of evidence to suggest that WES should be considered as a diagnostic option in disorders for which clinical and molecular diagnoses are challenging, especially those for which clinical management may be affected (18).
Supplementary Material
Acknowledgments
The authors would like to thank the family for their participation in the study. This study was funded by US National Human Genome Research Institute (NHGRI) grant 2-U54HG003273-09 to R.A.G.
Footnotes
Conflict of interest statement: J.R.L. is a consultant for Athena Diagnostics, has stock ownership in 23andMe and Ion Torrent Systems, and is a co-inventor on multiple United States and European patents for DNA diagnostics. R.A.G. is a founding shareholder in Seq-Wright, Inc. Some of the authors are based in the Department of Molecular and Human Genetics at Baylor College of Medicine, which derives revenue from genetic laboratory testing, including whole-exome sequencing. The remaining authors have no conflicts of interest to declare.
REFERENCES
- 1.Teer JK, Mullikin JC. Exome sequencing: the sweet spot before whole genomes. Hum Mol Genet. 2010;19:R145–151. doi: 10.1093/hmg/ddq333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng SB, Bigham AW, Buckingham KJ, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ng SB, Buckingham KJ, Lee C, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Sullivan J, Bitu CC, Daly SB, et al. Whole-Exome sequencing identifies FAM20A mutations as a cause of amelogenesis imperfecta and gingival hyperplasia syndrome. Am J Hum Genet. 2011;88:616–620. doi: 10.1016/j.ajhg.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lupski JR, Reid JG, Gonzaga-Jauregui C, et al. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med. 2010;362:1181–1191. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goh V, Helbling D, Biank V, et al. Next-generation Sequencing Facilitates the Diagnosis in a Child With Twinkle Mutations Causing Cholestatic Liver Failure. J Pediatr Gastroenterol Nutr. 2011;54:291–294. doi: 10.1097/MPG.0b013e318227e53c. [DOI] [PubMed] [Google Scholar]
- 7.Murdock DR, Clark GD, Bainbridge MN, et al. Whole-exome sequencing identifies compound heterozygous mutations in WDR62 in siblings with recurrent polymicrogyria. Am J Med Genet A. 2011;155:2071–2077. doi: 10.1002/ajmg.a.34165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Worthey EA, Mayer AN, Syverson GD, et al. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2010;13:255–262. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- 9.Bainbridge MN, Wiszniewski W, Murdock DR, et al. Whole-genome sequencing for optimized patient management. Sci Transl Med. 2011;3:87re83. doi: 10.1126/scitranslmed.3002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzaga-Jauregui C, Lupski JR, Gibbs RA. Human genome sequencing in health and disease. Annu Rev Med. 2012;63:35–61. doi: 10.1146/annurev-med-051010-162644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sternberg D, Tabti N, Hainque B, et al. GeneReviews. Seattle (WA): University of Washington; Seattle: 2002. Hypokalemic Periodic Paralysis. [Google Scholar]
- 12.Sternberg D, Maisonobe T, Jurkat-Rott K, et al. Hypokalaemic periodic paralysis type 2 caused by mutations at codon 672 in the muscle sodium channel gene SCN4A. Brain. 2001;124:1091–1099. doi: 10.1093/brain/124.6.1091. [DOI] [PubMed] [Google Scholar]
- 13.Raja Rayan DL, Hanna MG. Skeletal muscle channelopathies: nondystrophic myotonias and periodic paralysis. Curr Opin Neurol. 2010;23:466–476. doi: 10.1097/WCO.0b013e32833cc97e. [DOI] [PubMed] [Google Scholar]
- 14.Miller TM, Dias da Silva MR, Miller HA, et al. Correlating phenotype and genotype in the periodic paralyses. Neurology. 2004;63:1647–1655. doi: 10.1212/01.wnl.0000143383.91137.00. [DOI] [PubMed] [Google Scholar]
- 15.Cheung SW, Shaw CA, Yu W, et al. Development and validation of a CGH microarray for clinical cytogenetic diagnosis. Genet Med. 2005;7:422–432. doi: 10.1097/01.gim.0000170992.63691.32. [DOI] [PubMed] [Google Scholar]
- 16.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chabrier S, Monnier N, Lunardi J. Early onset of hypokalaemic periodic paralysis caused by a novel mutation of the CACNA1S gene. J Med Genet. 2008;45:686–688. doi: 10.1136/jmg.2008.059766. [DOI] [PubMed] [Google Scholar]
- 18.Biesecker L. Editorial comment on “Whole Exome Sequencing Identifies Compound Heterozygous Mutations in WDR62 in Siblings With Recurrent Polymicrogyria#x201D; Am J Med Genet Part A. 2011;155:2069–2070. doi: 10.1002/ajmg.a.34183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.