Abstract
We here report the first comparative proteomics of purified yeast post-Golgi vesicles (PGVs). Vesicle samples isolated from PGV-accumulating sec6-4 mutants were treated with isobaric tags (iTRAQ) for subsequent quantitative tandem mass spectrometric analysis of protein content. After background subtraction, a total of 66 vesicle-associated proteins were identified, including known or assumed vesicle residents as well as a fraction not previously known to be PGV associated. Vesicles isolated from cells lacking the polarity protein Sro7p contained essentially the same catalogue of proteins but showed a reduced content of a subset of cargo proteins, in agreement with a previously shown selective role for Sro7p in cargo sorting.
Keywords: exocytosis, Golgi, membrane trafficking, proteomics, vesicles, iTRAQ
Protein secretion in yeast is a polarized growth process by which newly synthesized proteins are transported through the secretory pathway to predestined sites at the plasma membrane. Following transport from the endoplasmic reticulum (ER) through the Golgi complex, secreted proteins are sorted into transport vesicles that move to the cell surface along polarized actin tracks, driven by myosin motor proteins (1). At the delivery site the vesicles fuse with the plasma membrane by the process of exocytosis. A large eight-protein complex, termed the exocyst, has a critical role in determining the attachment sites and tethers outbound vesicles to the plasma membrane (2,3). Subsequent fusion of the vesicles with the membrane is mediated by interaction with the plasma membrane target SNARE (t-SNARE) proteins Sso1/2p (4) and Sec9p (5), and the vesicle SNARE (v-SNARE) proteins Snc1/2p (6). SNARE proteins on opposing membranes form a complex which brings the membranes in close proximity, catalyzing fusion. Small GTPases are master regulators of the tethering and fusion processes by recruiting partner proteins (‘effectors’) to the membrane sites where they are bound. In yeast, the GTPase Sec4p has a key role in controlling the process of exocytosis (7,8). Many of the genes essential for secretion in yeast were identified in a screen for conditionally lethal mutations in which protein transport was arrested at various stages (9). Mutations in 10 of the isolated sec genes caused accumulation of post-Golgi trafficking vesicles when mutants were incubated at restrictive temperature. This phenotype has allowed the development of a procedure to isolate purified post-Golgi vesicles (PGVs) in sufficient amounts to perform detailed molecular analysis (10).
Sro7p is a yeast member of the family of Lethal giant larvae (Lgl) proteins, which are involved in cell polarity establishment in various organisms (11,12). In yeast, Sro7p plays a regulatory role in late exocytosis by physical interaction with myosins (13,14), the t-SNARE Sec9p (14,15), the exocyst subunit Exo84p (16) and Sec4p in its GTP loaded form (17). When exocytic vesicles arrive onto sites of plasma membrane growth, Sec4p-GTP is suggested to help initiating SNARE complex assembly via the transiently formed ternary complex (18). According to this model, the Sec4p-mediated signal will stimulate release of Sec9p from its inhibitory interaction with Sro7p, thereby allowing Sec9p to take part in SNARE complex formation and vesicle fusion with the plasma membrane. In Drosophila, the Lgl tumor suppressor protein is required to maintain epithelial cell polarity during embryogenesis and to establish asymmetric cell division and proper cell fate determination in the developing nervous system (11,19,20). An important aspect of Lgl function in these processes is its role in establishing polarized membrane domains by directing specific proteins to distinct regions of the plasma membrane. The asymmetric distribution of fate determinants in neural precursor cells involves intricate interactions with the Par polarity complex (21–23), but the detailed mechanisms for the role of Lgl in protein targeting are yet to be determined.
We have observed that yeast mutants lacking SRO7 become sensitive to NaCl stress (24) because of insufficient delivery of Ena1p, the sodium pumping ATPase, to the cell surface (25). Instead, Ena1p is directed to the vacuole for degradation, via the multivesicular body (MVB) pathway. This retargeting of Ena1p in sro7Δ mutants appears to occur at the late Golgi level. Consistent with this supposition, Ena1p-GFP is mainly detected in endosomes and vacuoles in sro7Δ mutants grown at high salinity, and we failed to detect Ena1p cargo in PGVs isolated from these mutants (25). In contrast, similarly treated sec mutants arrested in late exocytosis showed a clear accumulation of Ena1p in the isolated trafficking vesicles. Hence, missorting of Ena1p appeared not because of the well-established role for Sro7p in late exocytosis, but rather to a defect occurring in Golgi-mediated protein sorting (25). These findings indicated a second role for Sro7p in protein trafficking. Interestingly, this function occurs at Golgi – the main site for polarized protein sorting in the cell (26).
To shed further light on the role of Sro7p in protein secretion, we used quantitative proteomics to analyze the protein content in PGVs and as a means to specify what proteins are affected in their trafficking to the plasma membrane when SRO7 is lacking. Using this analysis we identified new residents of the PGVs and showed that Sro7p is required for correct secretion of a specific subset of cargo proteins in cells subjected to NaCl stress.
Results
Isolation and analysis of post-Golgi secretory vesicles
To isolate highly purified post-Golgi secretory vesicles we followed a modified version (5,27) of the well-established protocol of Walworth and Novick (10). Vesicles isolated by this procedure are morphologically homogenous and contain low levels of contaminating membranes. The protocol takes advantage of the accumulation of trafficking vesicles in the late acting secretory mutant, sec6-4, when shifted to restrictive temperature (37°C). As we also wanted to follow the secretion of the NaCl-induced sodium pump Ena1p (28,29), which is known to be missorted in sro7Δ cells under saline conditions (25), our protocol involved a shift of cells to high salinity medium to allow for identification of other cargo molecules that show dependence on Sro7p for correct delivery to the cell surface under these conditions. We first applied the protocol to a vesicle accumulating sec6-4 strain and a sec23-1 mutant that becomes depleted of vesicles at restrictive temperature because of an early block in secretion (30). This obstruction makes the sec23-1 mutant suitable for the preparation of ‘mock vesicle fractions’ that contain little genuine PGVs but contaminating proteins that co-purify with vesicle fractions in general (31). The vesicle-enriched P3 fraction obtained by differential centrifugation of lysed cells was layered onto a sorbitol gradient (5). Following velocity gradient centrifugation the gradients were fractionated and analyzed by immunoblotting for the presence of the SNARE proteins Sso1/2p and Snc1/2p (Figure 1A). Both markers yielded a coincident peak in the middle of the gradient for the sec6-4 mutant. A summary of the enrichment steps for a typical purification is given in Table 1. After gradient centrifugation the three pooled peak fractions showed a 41-fold enrichment of the PGV marker v-SNARE Snc1/2p over the total cell lysate. The pooled fractions were used for the subsequent proteomic analysis of the vesicles. Samples from sec23-1 and sec6-4 were solubilized, digested with trypsin and labeled for quantitative proteomic analysis using iTRAQ tags (isobaric tags for relative and absolute quantification) that generate amine-derivatized peptides (32). The derivatized peptides exhibit mass spectrometry (MS) signature ions that permit determination of their relative abundance in the original sample allowing for quantification of the parent proteins.
Figure 1. Distribution of the post-Golgi vesicle marker Snc1/2p and Sso1/2 in sorbitol vesicle gradient fractions from sec6-4 and sec23-1 strains.
A) Cells were cultured and lysates prepared and fractioned as described in Materials and Methods. Samples from each fraction of the gradient were heated in sample buffer at 95°C for 5 min, and resolved by SDS–PAGE on 12.5% Tris–HCl gels (Biorad). Separated proteins were transferred for western blot with antiserum against Sso1/2p and Snc1/2p. B) Distribution profile of Snc1/2p in the vesicle gradient fractions from sec6-4 and sec23-1 strains. The three peak fractions from the sec6-4 gradient were used for subsequent analysis. For the mock strain (sec23-1), fractions corresponding to the sec6-4 peak fractions were used. C) Relative strength of Snc1/2p signals from peak fractions 7, 8 and 9 of the sec6-4 and sec23-1 preparations.
Table 1.
Enrichment of the marker v-SNARE Snc1/2p during purification of sec6-4 PGVs
| Fraction | Protein (µg) | Snc1/2p intensity |
Specific intensity |
Fold purification |
|---|---|---|---|---|
| Lysate | 36.3 | 51.4 | 1.4 | |
| S2 | 7.5 | 18.1 | 2.4 | 1.7 |
| P2 | 4.2 | 27.1 | 6.4 | 4.6 |
| P3 | 2.3 | 47.7 | 17.4 | 12.2 |
| P4 | 0.31 | 18.1 | 58.4 | 41.1 |
Samples taken from each fraction during the purification process were subjected to SDS–PAGE and immunoblotting as described in Materials and Methods. The intensity of the vesicle marker Snc1/2p was analyzed using the odyssey quantification software and related to loaded amount of protein to determine ‘specific intensity’.
Abbreviations: S2 and P2 are supernatant and pellet, respectively, following a 13 000 × g spin of the S1 supernatant, which was recovered after a 700 × g spin of lysed sphaeroplasts. P3: pellet generated by a 100 000 × g spin of S2. P4: pellet remaining following a 100 000 × g spin of the pooled peak vesicle fractions from velocity gradient centrifugation of P3. For further details, see Materials and Methods.
Delimitation of vesicle proteins
Using this approach, the protein composition of purified vesicles was analyzed by LC-MS/MS (liquid chromatography-tandem mass spectrometry). From the 1382 peptides that were conclusively assigned, 242 different proteins were identified from sec6-4 control and sec23-1 mock fractions. A complete list of identified proteins is given in Table S1. Most proteins showed modest differences between control and mock values; about half of the set (51%) had normalized control/mock ratios between 2 and the observed lowest value, 0.4, and 15% had ratios ≤1. To define a set of proteins significantly enriched in the control vesicle fractions (depleted from the mock fraction), we applied a threshold ratio of >2.5, a conservative cutoff corresponding to the ratio for the vesicle t-SNARE, Sso2p. This protein catalogue, shown in Table 2, included 91 proteins with enrichment ratios between 2.5 and 50. The applied threshold selected a set of proteins significantly (Fisher’s exact test, Bonferroni correction, p < 0.1) enriched for specific cellular functions [comparison made to all proteins that had a chance of occurring in the set, i.e. excluding dubious open reading frames (ORFs)], many of them associated with plasma membrane-related activities (Figures 2 and S2). The observed enrichment for the v-SNARE and the vesiclemarker Snc2p (sixfold, Table 2) was in good agreement with the enrichment obtained for the marker by quantification of immunoblot signals (Figure 1B,C).
Table 2.
Proteins more than 2.5-fold enriched in contol (sec6-4) PGV fractions relative to the corresponding mock (sec23-1) fractions
| Protein | Molecular function | Cellular distribution | sec6-4/sec23-1 | Peptides found |
|---|---|---|---|---|
| CWP1 | Structural constituent of cell wall | Cell wall, membrane | 50.1 | 9 |
| PIR1 | Structural constituent of cell wall | Cell wall | 34 | 2 |
| YGP1 | Cell-wall-related secretory glycoprotein | Extracellular region | 29.5 | 9 |
| STL1 | Glycerol transporter | Plasma membrane, membrane | 15.7 | 3 |
| BGL2 | Glucan 1,3-beta-glucosidase activity | Cell wall | 15.6 | 8 |
| GAS5 | 1,3-beta-glucanosyltransferase activity | Cell wall, membrane fraction | 14.7 | 2 |
| CRH1 | Probable glycosidase | Cell wall | 13.5 | 5 |
| EHT1* | Medium-chain fatty acid ethyl ester synthase/esterase | Membrane, mitochondrial envelope, mitochondrion, cytoplasm | 13.2 | 13 |
| PST1 | Cell wall mannoprotein | Plasma membrane, cell wall, membrane | 12.3 | 5 |
| HXT7 | High-affinity hexose transporter | Plasma membrane, membrane, membrane fraction, cytoplasm, mitochondrion | 10.4 | 5 |
| YPT1 | GTPase activity | Membrane, endomembrane system, Golgi apparatus, endoplasmic reticulum, cytoplasm, mitochondrion | 9.4 | 13 |
| PHM7 | Phosphate metabolism protein | Cell periphery, cytoplasm, vacuole | 9.2 | 3 |
| PNS1 | Unknown | Plasma membrane, membrane fraction, membrane | 9 | 2 |
| CPR5 | peptidyl-prolyl cis–trans isomerase activity | Cytoplasm, endoplasmic reticulum | 8.9 | 3 |
| ERG6 * | Sterol 24-C-methyltransferase activity | Membrane, cytoplasm, lipid particle, endoplasmic reticulum, mitochondrial envelope, mitochondrion | 8.6 | 7 |
| HXT5 | Probable glucose transporter | Plasma membrane, membrane fraction, membrane | 8.2 | 6 |
| YRO2 | Unknown | Cellular bud, cytoplasm, mitochondrion | 8.1 | 5 |
| GPM1* | Phosphoglycerate mutase | Cytoplasm, mitochondrion | 8.0 | 14 |
| YPT31 | GTPase activity | Membrane, Golgi apparatus, cytoplasm, mitochondrion, mitochondrial envelope | 7.8 | 7 |
| KIC1 | Serine/threonine-protein kinase | Cellular bud, cytoplasm, site of polarized growth | 7.5 | 2 |
| ECM33 | Cell wall protein | Cell wall, plasma membrane, cytoplasm, membrane, membrane fraction, mitochondrion | 7.4 | 3 |
| SEC4 | Ras-related protein, GTPase activity | Membrane, membrane fraction, mitochondrial envelope, mitochondrion, cytoplasm, cytoplasmic membrane-bounded, cell cortex, cytoskeleton | 7.3 | 7 |
| AGP1 | General amino-acid permease | Plasma membrane | 7.2 | 2 |
| SLK19 * | Kinetochore protein | Cytoskeleton, nucleus, chromosome | 7.2 | 2 |
| VPS21 | Vacuolar protein sorting, GTPase activity | Membrane, endocytic intermediates, cytoplasm, mitochondrion, mitochondrial envelope | 6.8 | 2 |
| YPT7 | GTPase activity | Membrane, cytoplasm, mitochondrion, mitochondrial envelope, vacuole | 6.4 | 3 |
| PRC1 | Carboxypeptidase C activity | Cytoplasm, endoplasmic reticulum, vacuole | 6.3 | 2 |
| SNC2 | Synaptobrevin homolog 2 | Cytoplasm, cytoplasmic membrane-bounded, Golgi apparatus | 6.2 | 4 |
| ENA1 | Sodium transport ATPase | Plasma membrane, membrane, membrane fraction | 6.1 | 22 |
| PGK1 ** | Phosphoglycerate kinase | Cytoplasm, membrane fraction, mitochondrion | 5.8 | 22 |
| EMP47 * | Integral membrane protein | Cytoplasm, Golgi apparatus, cytoplasmic membrane-bounded | 5.8 | 2 |
| YJU3 | Serine hydrolase activity | Plasma membrane enriched fraction, cytoplasm, mitochondrion, mitochondrial envelope | 5.2 | 2 |
| TPI1** | Triosephosphate isomerase | Cytoplasm, membrane fraction, mitochondrion | 5.2 | 8 |
| KAP123 * | Importin subunit beta-4, protein carrier activity | Cytoplasm, membrane, endomembrane system, nucleus | 5.1 | 2 |
| AHP1 | Peroxiredoxin | Plasma membrane enriched fraction, cytoplasm | 5.0 | 2 |
| YJL171C | Uncharacterized protein | Cell wall, cytoplasm, membrane fraction, mitochondrion | 5 | 2 |
| GCN1 * | Translational activator | Cytoplasm, mitochondrion, ribosome | 4.9 | 2 |
| SOD1 * | Superoxide dismutase | Cytoplasm, mitochondrial envelope, mitochondrion, nucleus | 4.8 | 3 |
| CHS1 | Chitin synthase activity | Plasma membrane, membrane fraction, membrane | 4.8 | 2 |
| GPP1 * | Glycerol-3-phosphatase | Cytoplasm, nucleus | 4.8 | 7 |
| KAR2 | Glucose-regulated protein | Cytoplasm, endoplasmic reticulum | 4.7 | 9 |
| KRE2 | Glycolipid 2-α-mannosyltransferase | Cell wall, cytoplasm, Golgi apparatus | 4.6 | 3 |
| CDC42 | Cell division control protein, GTPase activity | Cellular bud, plasma membrane, membrane, site of polarized growth | 4.5 | 2 |
| ALD6** | Aldehyde dehydrogenase | Cytoplasm, mitochondrion | 4.4 | 5 |
| YLR413W | Cell membrane protein | Unknown | 4.4 | 3 |
| ERG4 * | delta24(24-1) sterol reductase activity | Cytoplasm, endoplasmic reticulum | 4.4 | 2 |
| GNP1 | High-affinity glutamine permease | Plasma membrane, cytoplasm, membrane, mitochondrion | 4.4 | 7 |
| HSP12 | 12 kDa heat shock protein | Plasma membrane, cytoplasm, plasma membrane enriched fraction, nucleus | 4.4 | 4 |
| GAS1 | 1,3-beta-glucanosyltransferase activity | Cell wall, plasma membrane, membrane, membrane fraction, mitochondrion, cytoplasm, nucleus | 4.3 | 6 |
| PDR15 | ATPase activity, coupled to transmembrane movement of substances | Membrane | 4.3 | 3 |
| PGI1** | Glucose-6-phosphate isomerase | Cytoplasm, membrane fraction, mitochondrion | 4.3 | 5 |
| RPL43A** | 60S Ribosomal protein L43 | Cytoplasm, ribosome | 4.2 | 2 |
| STE24 | CAAX prenyl protease, metalloendopeptidase activity | Membrane, endomembrane system, endoplasmic reticulum, cytosol, mitochondrion, mitochondrial envelope, nucleus | 4.2 | 2 |
| MRH1 | Unknown | Plasma membrane, membrane fraction, membrane, cytoplasm, mitochondrion | 4.1 | 2 |
| PDI1 | Protein disulfide isomerase | Cytoplasm, endoplasmic reticulum | 4 | 13 |
| ENO2 | Enolase | Plasma membrane, cytoplasm, membrane fraction, membrane, mitochondrion, vacuole | 4 | 17 |
| PDC1 ** | Pyruvate decarboxylase | Cytoplasm, nucleus | 3.9 | 14 |
| DRS2 | Probable phospholipid-transporting ATPase | Cytoplasm, trans-Golgi network | 3.7 | 2 |
| KTR3 | Probable mannosyltransferase | Membrane fraction | 3.7 | 5 |
| SPF1 | Probable cation-transporting ATPase, phosphorylative mechanism | Membrane, endomembrane system, Golgi apparatus, endoplasmic reticulum, mitochondrion, cytoplasm | 3.6 | 2 |
| SSP120 * | Unknown | Cytoplasm | 3.6 | 2 |
| YNL194C | Uncharacterized plasma membrane protein | Membrane, membrane fraction, cell cortex, endoplasmic reticulum, cytoplasm | 3.6 | 2 |
| GPP2 * | (DL)-glycerol-3-phosphatase 2 | Cytoplasm, nucleus | 3.6 | 2 |
| TDH3 | Glyceraldehyde-3-phosphate dehydrogenase | Cell wall, plasma membrane enriched fraction, cytoplasm, mitochondrion | 3.3 | 15 |
| RTN1 | Reticulon-like protein | Cell cortex, cytoplasm, membrane, endomembrane system, endoplasmic reticulum, Golgi apparatus, mitochondrion | 3.5 | 3 |
| PDR5 | Pleiotropic ABC efflux transporter of multiple drugs | Plasma membrane, cytoplasm, membrane, mitochondrion | 3.4 | 32 |
| VMA13 * | Vacuolar proton pump subunit H | Membrane, cytoplasm, vacuole | 3.4 | 2 |
| FKS1 | 1,3-β-glucan synthase activity | Plasma membrane, cell cortex, membrane, membrane fraction, cytoplasm, cytoskeleton, mitochondrion | 3.4 | 27 |
| DNF1 | Probable phospholipid-transporting ATPase | Plasma membrane, cytoplasm, mitochondrion | 3.3 | 5 |
| PTR2 | Peptide transporter activity | Plasma membrane, membrane fraction, membrane | 3.3 | 2 |
| KRE6 | Beta-glucan synthesis-associated protein | Cytoplasm, membrane, endomembrane system, endoplasmic reticulum, Golgi apparatus | 3.3 | 2 |
| APE3 * | Aminopeptidase Y | Cytoplasm, vacuole | 3.3 | 2 |
| PMA1 | Plasma membrane ATPase 1 | Plasma membrane, membrane fraction, membrane, cytoplasm, mitochondrion | 3.2 | 27 |
| FBA1 * | Fructose-bisphosphate aldolase | Cytoplasm, mitochondrion | 3.2 | 7 |
| MSC1 * | Meiotic sister chromatid recombination protein | Cytoplasm, membrane fraction, endoplasmic reticulum, mitochondrion | 3.2 | 4 |
| KTR1 | α-1,2-mannosyltransferase activity | Cytoplasm, Golgi apparatus | 3.2 | 5 |
| YOP1 | Unknown | Membrane | 3.1 | 3 |
| TSA1 * | Peroxiredoxin | Cytoplasm | 3 | 2 |
| FKS3 | 1,3-β-glucan synthase activity | Cytoplasm, mitochondrion | 3 | 2 |
| CHS3 | Chitin synthase activity | Cellular bud, plasma membrane, cytoplasm, cytoplasmic membrane-bounded, membrane fraction, membrane | 2.9 | 3 |
| SNQ2 | Xenobiotic-transporting ATPase activity | Plasma membrane, membrane, membrane fraction, cytoplasm, mitochondrion | 2.9 | 5 |
| ADH1 ** | Alcohol dehydrogenase | Cytoplasm, membrane fraction | 2.9 | 10 |
| FAA1 | Long-chain fatty-acid-CoA ligase activity | Membrane, membrane fraction, cytoplasm, mitochondrial envelope, mitochondrion | 2.9 | 10 |
| GRX2 * | Glutaredoxin-2, thiol-disulfide exchange intermediate activity | Cytoplasm, mitochondrion | 2.8 | 2 |
| TAT1 | Valine/tyrosine/tryptophan amino-acid permease | Plasma membrane, membrane | 2.8 | 3 |
| VPH1 | Subunit a of vacuolar-ATPase V0 domain | Membrane, cytoplasm, vacuole | 2.8 | 8 |
| SAC1 | Phosphoinositide phosphatase | Membrane, endomembrane system, endoplasmic reticulum, cytoplasm, Golgi apparatus, mitochondrion | 2.7 | 5 |
| SSS1 | Protein transport protein SSS1 | Membrane, endomembrane system, endoplasmic reticulum, cytosol | 2.6 | 2 |
| SSO2 | t-SNARE activity | Plasma membrane, membrane fraction, membrane, endoplasmic reticulum, cytoplasm | 2.6 | 2 |
| TSC13 | Enoyl reductase | Membrane, endomembrane system, endoplasmic reticulum, cytoplasm, mitochondrion | 2.6 | 2 |
| RHO1 | GTP-binding protein | Plasma membrane, cellular bud, site of polarized growth, membrane fraction, membrane, cytoplasm, mitochondrion, mitochondrial envelope, peroxisome | 2.5 | 4 |
Vesicles were prepared as described in Materials and Methods and analyzed by iTRAQ. Annotations were derived from the Gene Ontology classification system as represented in the SGD yeast GO slim component catalogue (http://www.yeastgenome.org/cgi-bin/GO/goSlimMapper.pl).
Likely contaminants have been indicated based on previously reported sub-cellular localization (*) or on their reported high abundance and cytosolic localization (**).
Figure 2. Sub-cellular enrichment of vesicle proteins prepared from sec6-4 (open bars) and sro7Δ (filled bars) strains.
Vesicle proteins were defined as proteins >2.5-fold more abundant in sec6-4 or sro7Δ fractions relative to the mock (sec23-1) strain. All sub-cellular compartment annotations were significantly enriched relative to the random expectation according to Fisher’s exact test (Bonferroni correction, p < 0.1) for either sec6-4 or sro7Δ. Localization annotations were from the Gene Ontology classification system as represented in the SGD yeast GO slim component catalogue (http://www.yeastgenome.org/cgi-bin/GO/goSlimMapper.pl).
To identify likely false positives in the assigned vesicle inventory, we first picked out high-abundance proteins previously shown to have a mainly cytosolic distribution. The selected seven proteins (marked with a double asterisk in Table 2) are all among the top 6% of the most abundant proteins in the cell (33). These proteins, primarily glycolytic enzymes, are very probable contaminants because they may co-purify with the enriched vesicle pool simply because of their extreme cellular abundance, exceeding the overall cellular abundance of most vesicle proteins by three orders of magnitude (33,34). We also identified 18 proteins, which, according to their presently known function/localization, are unlikely PGV cargo molecules (marked with a single asterisk). When eliminating the 25 assumed false positives (28%), a component list of 66 predicted high-confidence PGV residents remained.
Functional analysis of assigned vesicle proteins
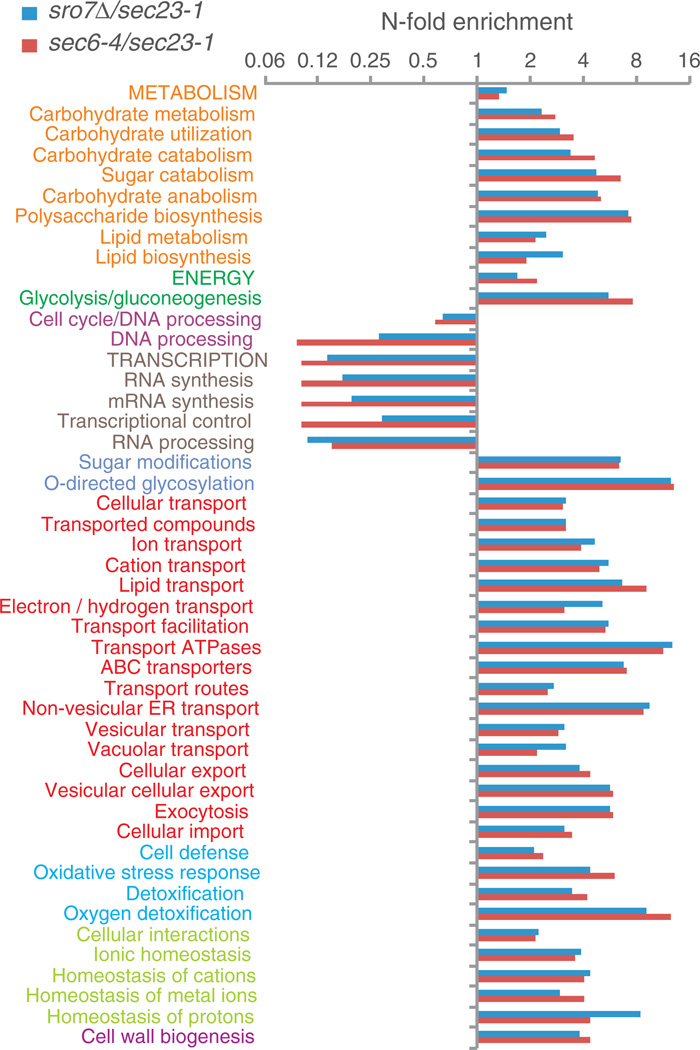
Analysis of the characteristics of the protein catalogue in Table 2 showed a defined set of functions, targeted for a distinct set of cellular compartments (Figures 2, 3 and S2). In particular, proteins found in, or associated to,membrane structures are significantly enriched (Fisher’s exact test, Bonferroni correction, p < 0.1) in the defined set as compared to the control group of proteins (determined as described above). The localization categories should be viewed with the awareness that most proteins are annotated to multiple categories (average of 3.6 in the current set). For example, among the 29 proteins annotated as mitochondrial residents (Figure S2, Table 2), 9 are also reported to be localized to the plasma membrane (Dnf1p, Ecm33p, Gnp1p, Mrh1p, Pdr5p, Pma1p, Snq1p, Yjl171cp, Yju8p), 2 are found in plasma membrane enriched fractions (Faa1p, Msc1p), 4 have roles in cell wall assembly (Fks1p, Fks3p, Gas1p, Sac1p), 3 are involved in cell polarity (Rho1p, Yro2p, Rtn1p) and 8 have regulatory or other functions in the secretory process (Ypt31, Spf1p, Ste24, Tsc13, Vps21, Ypt1, Ypt7 and Yro2). The remaining four (Eht1p, Erg6p, Gcn1p and Grx2p) have been indicated as putative contaminants in our preparations (see Table 2). Hence, most of the proteins in the mitochondrial category also have localizations compatible with PGV transport.
Figure 3. Functional classification of proteins in vesicles prepared from sec6-4 (red bars) and sro7Δ (blue bars) strains.
Annotations were derived from the functional ontology classification system hosted by the MIPS Comprehensive Yeast Genome Database (http://mips.helmholtz-muenchen.de/genre/proj/yeast/Search/Catalogs/catalog.jsp). Enrichment was determined as in Figure 2. Only functional processes with at least 15 annotated members were considered in the test.
Among the nine proteins that were most highly (>10-fold) enriched in sec6-4 relative to mock preparations, seven were cellwall-associated proteins (Table 2). Bgl2p, the cell wall endo-β-1,3-glucanase (35) and a marker for secretory vesicles (10), belonged to this group. Notably however, three other vesicle markers, invertase (Suc2p), acid phosphatase, and exo-β-1,3-glucanase (Exg1p), were absent from our preparations, probably because of repressed production under our growth conditions (high concentration of glucose, phosphate and NaCl, respectively). A predominant fraction, 61% of the vesicle-associated proteins, was membrane proteins, many of which being established or presumed plasma membrane transporters (Dnf1p, Ena1p, Gnp1p, Hxt5p, Hxt7p, Agp1, Pdr5p, Pma1p, Ptr2p, Snq2p and Stl1p). Besides the major fraction of cargo proteins, the vesicles also held proteins with membrane identifying and regulatory roles. This group included the v-SNARE Snc2p and t-SNARE Sso2p, and a set of small GTPases that serve as spatial signals and regulators of membrane traffic. Snc1p and Sso1p were not confidently identified because of an insufficient number of identified unique peptides. Three of the identified GTPases, Sec4p, Cdc42p and Rho1p, are directly involved in controlling trafficking of PGVs and have an earlier established association with late-secretory vesicles (36–38). Rho1p is also a regulator of another vesicle resident, Fks1p, the catalytically active subunit of the 1,3-β-D-glucan synthase, involved in cell wall remodeling (39). Another regulatory subset, Ypt1p and Ypt31p, function as key controlling factors in trafficking to, within and from Golgi, whereas Vps21p and Ypt7p regulate aspects of endocytosis and Golgi–to-vacuole transport (40). Interestingly, previous immuno-electron microscopy has demonstrated that Ypt1p, one of the above GTPases that has no known role in exocytosis, is associated with Golgi-derived secretory vesicles (41). Our preparation also contained a lipid modifying phosphatidylinositol phosphatase (Sac1p) and two aminophospholipid translocases (Drs2p, Dnf1p) that may affect vesicle budding by regulating lipid composition of Golgi and plasma membrane (42,43). Another noteworthy group was constituted by mannosyltransferases involved in post-translational modification of proteins belonging to the KTR (Ktr1p, Ktr3p and Kre2p) family, having a role in cell wall mannoprotein synthesis and protein glycosylation at the Golgi (44). A vesicle-associated protein kinase was also uncovered, Kic1p that regulates 1,6-β-glucan levels in the cell wall (45). Notable is also the presence of a subunit, Sss1p, of the Sec61p translocon complex and two subunits of the vacuolar H+-ATPase (V-ATPase) (46); the V1 sector subunit Vma13p and the V0 transmembrane sector subunit Vph1p (see Discussion).
Comparative proteomics of vesicles isolated from sec6-4 and sro7Δ mutants
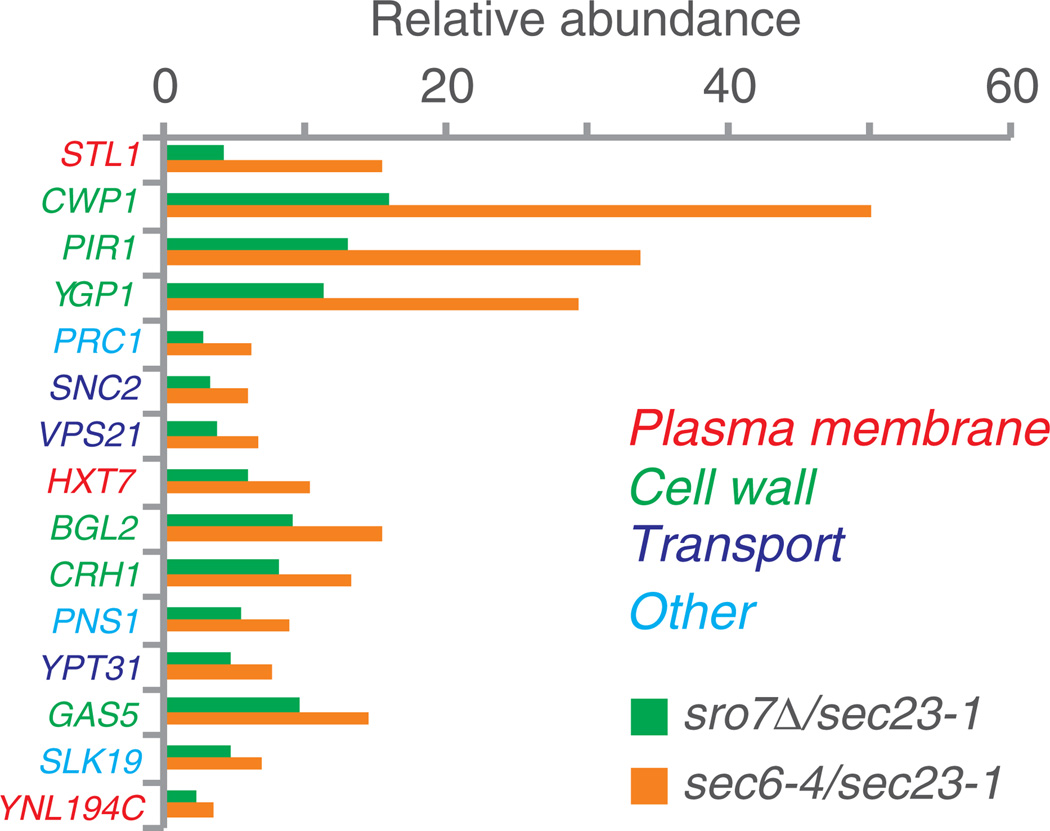
To identify proteins other than Ena1p that might display decreased trafficking to the plasma membrane in mutants lacking SRO7, we isolated PGVs from an sro7Δ strain subjected to the same salinity and temperature regime as previously described (Figure S1). The isolation of vesicles from sro7Δ mutants is facilitated by the fact that these mutants accumulate PGVs when exposed to high salinity (25) at which condition the missorting of Ena1p is also observed. All of the 91 assigned sec6-4 vesicle residents were present as high-confidence PGV proteins also in the corresponding sro7Δ fractions, demonstrating excellent reproducibility for the performed analysis (Table S2). Further analysis revealed no obvious cell function or component that was specifically affected by the deletion of SRO7 (Figures 2 and 3). However, a difference that stands out is the apparent depletion of a subset of proteins from the vesicles of the sro7Δ strain (Table S2). As Ena1p, the main plasma membrane sodium pump, was previously reported to be diverted from PGVs in sro7Δ mutants exposed to NaCl stress (25), we used the Ena1p distribution as an arbitrary threshold to define proteins that display depletion in sro7Δ vesicles. Somewhat surprisingly, Ena1p was only partly depleted in our present analysis of sro7Δ vesicles (sro7Δ/sec6-4 ratio of 0.7, Table 3), in contrast to the previous observations in which Ena1p failed to be detected insro7Δ PGVs, as determined by immunoblot of HA-tagged Ena1p (25). This discrepancy may at least partly stem from different experimental conditions; the present experiments were conducted at 37°C, whereas in the previous investigation the sro7Δ mutant was kept at 25°C. This modification is expected to affect the results, because the salt-sensitive phenotype of sro7Δ is weakened, although not abolished, at high temperatures (I. Wadskog, unpublished data). Using the Ena1p-based cutoff, 22 candidate proteins were identified as depleted in sro7Δ vesicles, showing sro7Δ/sec6-4 ratios from 0.3 to 0.7 (Table 3, Figure 4). Seven of these proteins (Cwp1p, Pir1p, Ygp1p, Bgl2p, Crh1p, Gas5p and Pst1p) are involved in cell wall functions. In agreement with these observations, secretion of the endo-β-1,3-glucanase, Bgl2p was previously shown to be defective in salt stressed sro7Δ mutants (25). It is also of interest that the vesicle fusion controlling Snc2p and Sec4p belong to the candidates for depletion in sro7Δ vesicles, as did three plasma membrane transporters: the glycerol proton symporter Stl1p, the main high-affinity glucose transporter Hxt7p and Ena1p. To validate the observed depletion of PGV residents in sro7Δ mutants, the distribution of selected cargo proteins was examined also by western blotting, using independently prepared vesicles from sro7Δ and sec6-4. The enrichment ratios obtained by immunoblotting (Figure 5A) were in agreement with those observed from iTRAQ data (Figure 5B), strongly corroborating the distribution shown in Table 3.
Table 3.
Proteins which are less abundant in sro7Δ vesicles than in sec6-4 vesicles
| Gene | Protein description | sro7Δ/sec23 | sec6-4/sec23-1 | sro7Δ/sec6-4 |
|---|---|---|---|---|
| STL1 | Sugar transporter | 4.5 | 15.7 | 0.3 |
| CWP1 | Cell wall protein CWP1 precursor | 16.1 | 50.1 | 0.3 |
| PIR1 | Protein PIR1 precursor | 13.2 | 34.0 | 0.4 |
| YGP1 | Protein YGP1 precursor | 11.5 | 29.5 | 0.4 |
| PRC1 | Carboxypeptidase Y precursor | 2.9 | 6.3 | 0.5 |
| SNC2 | Synaptobrevin homolog 2 | 3.5 | 6.2 | 0.6 |
| VPS21 | Vacuolar protein sorting-associated protein 21 | 4.0 | 6.8 | 0.6 |
| HXT7 | High-affinity hexose transporter | 6.2 | 10.4 | 0.6 |
| BGL2 | Glucan 1,3-beta-glucosidase precursor | 9.3 | 15.6 | 0.6 |
| CRH1 | Probable glycosidase CRH1 precursor | 8.4 | 13.5 | 0.6 |
| PNS1 | Protein PNS1 | 5.6 | 9.0 | 0.6 |
| YPT31 | GTP-binding protein | 4.9 | 7.8 | 0.6 |
| GAS5 | Glycolipid-anchored surface protein 5 precursor | 9.8 | 14.7 | 0.7 |
| SLK19* | Kinetochore protein | 4.8 | 7.2 | 0.7 |
| YNL194C | Uncharacterized plasma membrane protein YNL194C | 2.4 | 3.6 | 0.7 |
| EHT1* | Medium-chain fatty acid ethyl ester synthase/esterase 2 | 8.9 | 13.2 | 0.7 |
| PHM7 | Phosphate metabolism protein 7 | 6.3 | 9.2 | 0.7 |
| SEC4 | Ras-related protein | 5.1 | 7.3 | 0.7 |
| AGP1 | General amino-acid permease | 5.0 | 7.2 | 0.7 |
| YPT1 | GTP-binding protein | 6.7 | 9.4 | 0.7 |
| PST1 | Cell wall mannoprotein PST1 precursor | 8.8 | 12.3 | 0.7 |
| ENA1 | Sodium transport ATPase 1 | 4.4 | 6.1 | 0.7 |
The proteins were ranked according to their relative abundance in sro7Δ and sec6-4 vesicles, starting with the lowest sro7Δ/sec6-4 ratio. Annotations are from the SGD yeast GO slim component catalogue (http://www.yeastgenome.org/cgi-bin/GO/goSlimMapper.pl).
Figure 4. Top 15 proteins depleted in vesicles from cells lacking SRO7.
Annotations and enrichment were determined as in Figure 2.
Figure 5. Abundance of selected vesicle proteins analyzed in Table 3.
A) An equal amount of protein from purified vesicles prepared from sro7Δ and sec6-4 mutants was subjected to immunoblotting as described in Materials and Methods. Protein bands of expected sizes were quantitated and results illustrated in the bar graphs (right). B) A bar graph of log(2) transformed ratio values comparing the sro7Δ/sec6-4 ratio from immunoblot with the corresponding iTRAQ values. Samples for iTRAQ analysis and immunoblot quantification were independently prepared.
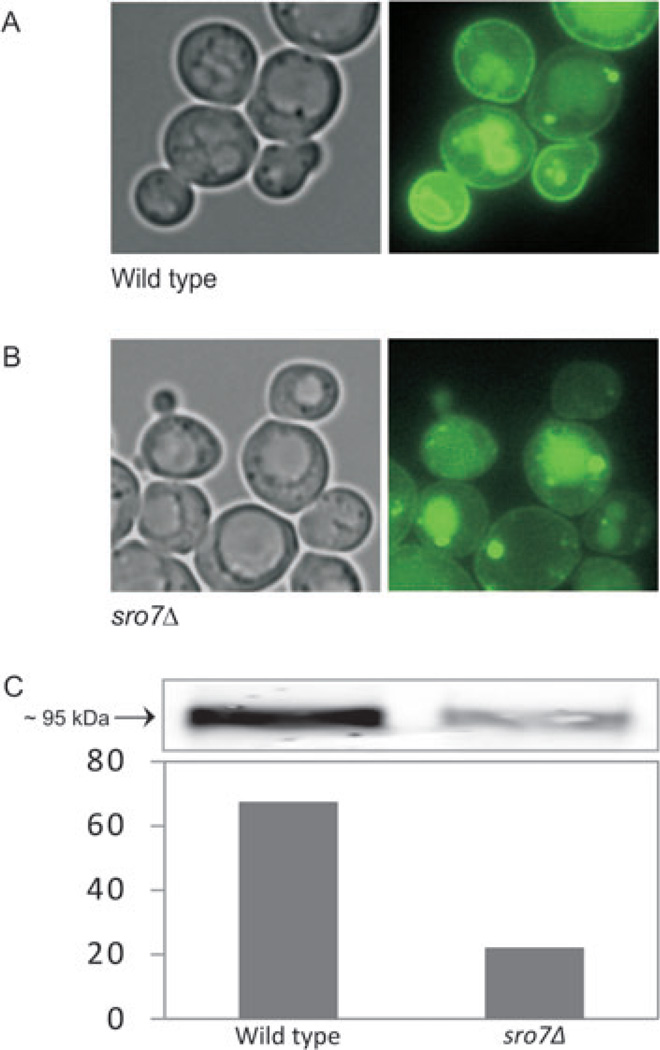
Stl1p was the protein most depleted in sro7Δ PGVs, showing a sro7Δ/sec6-4 ratio of 0.3. STL1 is a strongly salt-induced gene (47) that is subject to a temperature-alleviated glucose repression (48), two reasons on why Stl1p appears in our analysis. To further study the selective depletion of proteins in sro7Δ vesicles, we followed the fate of a GFP-tagged version of Stl1p in wild-type and sro7Δ cells, after consecutive shifts to high salinity and 37°C. Fluorescence micrographs, taken 2 h after induction of STL1-GFP production, showed a clear GFP signal from the plasma membrane of wild-type cells, whereas in sro7Δ cells there was a much weaker fluorescence from the membrane, most of the signal coming from the vacuole (Figure 6A). A selective exclusion of Stl1p-GFP from sro7Δ vesicles was also confirmed by western blotting of PGVs prepared from wild-type cells and sro7Δ mutants. This assay showed an sro7Δ/wild-type distribution of Stl1p-GFP of about 0.3 (Figure 6B), strongly supporting the depletion of Stl1p from sro7Δ PGVs shown by iTRAQ data (Table 3). We conclude that the described independent observations are all in agreement with a defective sorting of Stl1p in sro7Δ PGVs.
Figure 6. Localization of Stl1p-GFP in wild-type and sro7Δ cells.
Fluorescence images show the distribution of Stl1p-GFP in (A) wild type and (B) sro7Δ cells. Cells carrying a pUG35 STL1-GFP plasmid were grown to early exponential phase in medium with 500 µm methionine. NaCl was added to a final concentration of 0.6 m and incubation was continued for 2 h. Synthesis of Stl1p-GFP was induced by a shift to methionine-free medium (with 0.6 m NaCl) and incubation was continued for 2 h at 37°C. C) Distribution of Stl1p-GFP in purified PGVs from wild-type and sro7Δ strains. Cells transformed with the pUG35-STL1 plasmid were harvested after a 2 h shift to 0.6 m NaCl, followed by a 2 h shift to methionine free medium of the same salinity (to allow for induction of Stl1p-GFP) and 37°C. Purified PGVs were subjected to SDS–PAGE as described in Materials and Methods and Stl1p-GFP was detected by immunoblotting with anti-GFP antibody. Bands were quantitated (bar graph) and the Stl1p-GFP protein ratio in PGVs from sro7Δ relative to wild type estimated to 0.33.
As sro7Δ mutants apparently missort proteins encoded by two strongly salt-induced genes (ENA1 and STL1), we explored the possibility that Sro7p exerts a general control over the cellular response to NaCl stress. To this end, the NaCl tolerance of deletion mutants corresponding to each of the proteins showing depletion in sro7Δ PGVs was assayed. As demonstrated in Figure S3 the growth rate of these mutants was similar to that of wild-type cells giving no support for a generic role of Sro7p in the cellular NaCl response.
Discussion
Here we report on the yeast secretory proteome based on quantitative proteomics of purified PGVs. Using differential and velocity-gradient centrifugation, we purified secretory vesicles achieving a 41-fold enrichment over the crude cell homogenate. The purification compares favorably with previous reports (Ref. 27, #1190; Ref. 10, #1772) and the cargo enrichment is similar to that of a previous iTRAQ-based proteomic analysis of clathrin-coated vesicles from HeLa cells (31), in which the top 53 proteins considered to be bona fide vesicle proteins displayed a control/mock ratio of 2- to 12-fold. The performed analysis allowed us to study all of the PGVs that accumulate in late-secretory mutants, not just those associated with a particular marker protein. Using this approach we identified, after background subtraction, 66 proteins predicted to be specifically associated with exocytic vesicles (Table 2). As our report represents the first proteomic analysis of yeast PGVs, most of the identified proteins have not formerly been recognized as vesicle residents. However, a considerable share of the occupants is plasma-membrane proteins or cell wall-associated proteins and therefore expected regular traffickers of these vesicles. It should be stressed that the presented catalogue of proteins does not give a full account of PGV proteins. First, our experimental conditions – in particular the choice of carbon source (glucose) and the exposure of the cells to high salinity and temperature – will influence the selection of proteins that travels with the vesicles. Second, the preparation procedure, especially the fractioned centrifugation preceding the velocity-gradient step, may lead to loss of loosely associated proteins and explain the absence of proteins known to interact peripherally with the vesicle surface, such asMyo2p (49) and all the exocyst subunits except Sec3p (50). Finally, small proteins having close homologs may generate too few unique tryptic peptides to be reliably identified by MS analysis, leading to the failure of detecting the SNARE proteins Snc1p and Sso1. However, the absence of known coat-complex proteins was expected because the major routes from Golgi to the cell surface seem to involve mechanisms of vesicle formation that do not depend on clathrin and conventional adapter protein complexes (51,52). Coat proteins can, however, be involved when the secreted protein is recycled between Golgi and endosomes on its way to the cell surface. The trafficking of the chitin synthase, Chs3p, is awell-described example. This protein is kept in a reservoir by traveling between Golgi and early endosomes, and the recycling is mediated by clathrin and the AP-1 adaptor complex (53). Recruitment of Chs3p to the cell surface from this reservoir requires another and highly specific coat complex, composed of the two proteins Chs5p and Chs6p (54). Although coats are shed after vesicle budding, the complete absence of specific or classical coat proteins in our vesicle pools may reflect preparations, showing little contamination by vesicles that traffic endocytic and ER-to-Golgi pathways.
The presence of three α-1,2-mannosyltransferases in the vesicle cargo was a notable feature because these enzymes have a known Golgi-location, where they extend and modify O-linked and N-linked glycans that have been attached to membrane- and secreted proteins in the ER (44,55). Their residence in PGVs may simply reflect recycling between Golgi and the plasma membrane, or suggest that protein glycosylation is occurring during transport to and/or at the cell surface. Glycosylation may not only serve as a determinant specifying surface delivery of proteins (56), but has important roles for the stabilization and proper function of cell surface proteins, such as the glycoproteins of the outer cell wall layer (39). Another striking observation is the presence of two subunits of the vacuolar H+-ATPase in the vesicles. Although the V-ATPase has its main role in vacuolar acidification (46), it has been implicated in synaptic exocytosis, and V0 subunits are reported to interact with SNARE proteins on synaptic vesicles (57,58). Our results also strengthen the significance of the recently described genetic interaction between SEC9 and genes encoding V-ATPase subunits (59). Another seemingly paradoxical finding was the presence of a subunit of the translocon in the vesicle cargo. This channel translocates proteins across the ER membrane into the ER lumen and integrates proteins with transmembrane segments into the membrane (60). However, there is strong genetic and physical evidence that the translocon interacts with subunits of the exocyst (61,62), and it was suggested that this interaction may serve to co-ordinate translation and protein translocation into ER with the activity of the secretory machinery (63). These observations may serve as illustrating examples of the multiple, diverse sub-cellular localization for most of the identified proteins.
Yeast has at least two different routes for transport from Golgi to the cell surface (27,64,65). The situation in polarized epithelial cells is more complex. These cells have apical and basolateral cell surfaces separated by junctions and the two membrane regions have distinct physiological functions and protein composition. To establish and maintain this polarity, the majority of apical and basolateral proteins are sorted in trans-Golgi and directed to the appropriate surfaces via multiple pathways (26). Drosophila lgl mutants fail to establish correct epithelial polarity during embryogenesis; proteins that are determinants of the apical membrane become mislocalized to the basolateral domain (11). The specific role of Lgl in restricting proteins to the apical domain of the cell membrane has not yet been fully elucidated. We have previously reported that mutants lacking SRO7, the yeast Lgl ortholog, become salt sensitive because of mistargeting of the main sodium pump, Ena1p, at high salinity (24,25). Rather than being routed to the cell membrane, Ena1p is predominantly sent to the vacuole for degradation in these mutants. To identify other possible candidates for mistargeting, we here performed comparative proteomics of vesicles isolated from sec6-4 and sro7Δ strains at restrictive conditions. This analysis demonstrated that Ena1p is only partly depleted from sro7Δ vesicles (Table 3), which contrasts with a previously reported failure of detecting Ena1p in sro7Δ PGVs (25). The discrepancy may partly reflect different experimental setup, as previously discussed. However, some vesicle-mediated transport of Ena1p to the plasma membrane has to occur in sro7Δ mutants, because thesemutants are lessNaCl sensitive than ena1Δ mutants (24), which are completely devoid of the sodium pump. Data from the iTRAQ-based analysis (Table 3), supported by independently performed immunoblots (Figure 5), showed that a subset of proteins was more strongly depleted than Ena1p in sro7Δ vesicles. A substantial fraction of these 22 proteins comprised cell-wall-associated proteins, involved in remodeling of the carbohydrate chains (Gas5p, Bgl2 and Crh1p), or constituting cell wall proteins (Cwp1, Pir1p, Ygp1p and Pst1p). Another noteworthy observation was the depletion of the v-SNARE Snc2p and the vesicle-associated Rab GTPase, Sec4p, in sro7Δvesicles. As mutants defective in Snc1/2p (6) and Sec4p (27) accumulate PGVs, the reduced presence of these fusion controlling proteins may be a contributing factor for the vesicle accumulation seen in the sro7Δ strain at high salinity (25). This depletion might add to effects from the established role of Sro7p as a regulator of SNARE function through its interaction with Sec9p (8,66). The protein that was most under-represented in the sro7Δ vesicles was the glycerol/H+ symporter, Stl1p. Similarly to ENA1, STL1 has a role in the osmotic stress response and is strongly and transiently induced by salt stress (47). The mobilization of Stl1p serves to prevent leakage of glycerol, an osmoregulator that accumulates in cells subjected to hyper-osmotic stress. Based on the fate of Stl1p in sro7Δ mutants (Figure 6), it is tempting to suggest that Stl1p may travel together with Ena1p and like the sodium pump be subject to diversion from the exocytic pathway in sro7Δ mutants. However, none of the other proteins that were under represented in sro7Δ PGVs appear to be important for NaCl tolerance (Figure S3), showing that Sro7p has a more general role than sorting of proteins involved in sodium control.
The transit of proteins from Golgi to the cell surface has been much less studied than trafficking of the early steps of the secretory pathway. We believe that the proteomic analysis presented here may provide a useful basis for further elucidation of exocytic processes. The use of this technique to examine the effect of the polarity protein Sro7p on sorting into the vesicle compartment illustrates that the analysis can be applied to study effects of particular factors on the composition of the proteome.
Materials and Methods
Strains and media
The Saccharomyces cerevisiae strains used in these experiments were BY2: Mat a; ura3-52, BY37: Mat a; ura3-52; sec6-4, BY47: Mat a; ura3-52; sec23-1, and Mat a; ura3-52; sro7Δ: LEU2 (BY2 background). Cultures were grown in YPD (1%Bacto Yeast Extract and 2%Bacto Peptone; Difco laboratories) or SD medium (0.17% YNB, 0.5% ammonium sulfate) with 2% glucose and with the addition of 5 m NaCl to a final concentration of 0.6 m where stated.
Purification of secretory vesicles
Cells were cultured at 25°C to early exponential phase (optical density at 610 nm (OD610) of 0.6–0.7) in 750 mL cultures of YPD medium. Following a shift to 0.6 m NaCl, incubation was continued at 25°C for 2h and cultures were then shifted to 37°C for another 2 h. The cells were harvested after addition of NaN3 and NaF (20 mm final concentration). To account for the different degree of vesicle accumulation in the strains, 600 OD610 units were harvested from the sec6-4 mutant, while 1800 OD610 units were taken from the wild type, sec23-1 or the sro7Δ strains for the subsequent purification. Cells were washed in ice-cold Tris buffer (10 mm Tris–HCl pH 7.5, 10 mm NaN3, 10mm NaF) and then treated by previously described procedures with a few modifications (5,10,27). In short, cells were resuspended in sphaeroplast buffer (0.1 m Tris–HCl pH 7.5, 1.4 m sorbitol, 10 mm NaN3, 0.001% beta-mercaptoethanol, 0.1 mg/mL zymolyase 100T) (Seikagaku) and incubated at 37°C for 30 min. The sphaeroplasts were collected at 2000 × g and resuspended in lysis buffer containing 0.8 m sorbitol, 10 mm triethanolamine with 1 mm ethylenediaminetetraacetic acid (EDTA) [adjusted to pH 7.2 with acetic acid (TEAE)], and protease inhibitor cocktail (Roche Cat. No. 1873580). The sphaeroplasts were lysed by gentle resuspension and pipetting in lysis buffer, and spun at 700 × g for 10 min generating the S1 supernatant. The S1 fraction was centrifuged at 13 000 × g for 20 min yielding the S2 supernatant and P2 pellet. This supernatant was centrifuged at 100 000 × g for 1 h at 4°C in a 70.1 Ti rotor (Beckman) generating the P3 membrane pellet and the S3 supernatant. The P3 pellet was kept in lysis buffer on ice to dissolve for 2 h. The resuspended, vesicle enriched P3 fraction was further purified by velocity gradient centrifugation. An 11mL linear sorbitol gradient was preparedwith 1.2 mL steps of 40%, 37.5%, 35%, 32.5% 30%, 27.5% 25%, 22.5% and 20% sorbitol (w/v) dissolved in 10 mm triethanolamine acetate with 1 mm EDTA (pH 7.2). The resuspended P3 fraction was layered on top of the gradient and spun for 1 h at 71 000 × g at 4°C in an SW41 rotor (Beckman) and then fractionated from top to bottom in 16 fractions. The SNARE proteins Sso1/2p and Snc1/2p were detected in the gradient fractions by immunoblotting.
Sample preparation and iTRAQ analysis
iTRAQ analysis was carried out essentially as described by Borner et al. (31). Selected peak vesicle fractions from each gradient were concentrated by centrifugation for 1 h at 100 000 × g and 4°C in a 70.1 Ti rotor (Beckman). The resulting P4 vesicle pellets were resuspended in 2.5% SDS, 50 mm Tris, pH 8.0, heated to 65°C for 2min and centrifuged at 16 000 × g for 2 min. Protein in the supernatant was precipitated with 5 volumes of ice-cold acetone at −20°C overnight. Precipitated proteins were solubilized in 100-µL 25-mm triethylammonium bicarbonate (TEAB), pH 8.5, 8 m urea, 2% Triton-X-100 and 0.1% SDS (labeling buffer). Protein concentration was determined using the BCA Protein Assay Kit (Pierce). Samples were adjusted to equal protein concentration with 0.5 m TEAB, 75 µg per sample condition was used for the labeling procedure. Each sample was reduced and alkylated as previously described (32) before digestion with mass spectrometry grade trypsin (Promega). Digestion was performed overnight and stopped by adding 0.1% formic acid and the total of one iTRAQ reagent vial (Applied Biosystems) resolved in 70 µL ethanol to each peptide mixture. Labeled peptides were pooled, lyophilized and resuspended in 250 µL 25mm ammonium formate, 20% acetonitrile, pH 2.8. Strong cation exchange chromatography (SCX) on the peptide fractionation was performed with an Ä ktaPurifier (Amershan Pharmacia biotech) using a Polysulfoethyl A column 100 mm × 2.1 mm × 5 µm 300 Å(PolyLC). Peptideswere eluted by a 0–100% gradient toward 500 mm ammonium formate, 20% acetonitril, pH 2.8. Collected fractions were lyophilized and resolved in 0.1% formic acid.
Mass spectrometry analysis
Two-microliter sample injections were made with a HTC-PAL autosampler (CTC Analytics AG) equipped with a Cheminert valve (0.25 mm bore, C2V-1006D-CTC, Valco Instruments Co), connected to an Agilent 1200 binary pump (Agilent Technologies). The peptides were trapped on a precolumn (45 × 0.075 mm i.d.) packed with 3 m C18-bonded particles and separated on a reversed phase column, 200 × 0.050 mm, both columns were packed in-house with 3 µm Reprosil-Pur C18-AQ particles (Dr. Maisch GmbH, Ammerbuch-Entringen). A 40-min gradient 10–50% acetonitrile in 0.2% formic acid was used for separation of the peptides. Nanoflow LC-MS/MS was performed on a hybrid linear LTQ-Orbitrap mass spectrometer (LTQ Orbitrap XL, Thermo Scientific) equipped with a nanospray sourcemodified in-house. The spectrometer was operated in a data-dependentmode, both MS and MS/MS scans were obtained in Orbitrap. For each MS, the three most intense doubly and triply charged ions were fragmented by higher energy C-trap dissociation (HCD). MS/MS were collected in profiled mode with the first mass fixed m/z at 100 Da to include the iTRAQ reporter ions in the every scan.
MS data acquisition and protein quantification
RAW MS files were converted into a centroid and profiled mxXML file using ReadAw (http://sourceforge.net/projects/sashimi/files/). Both files were further processed into two peaklists using mzXML2Search (Part of the Trans-Proteomics-pipeline, http://sashimi.sourceforge.net/software_tpp.html). Database searches were performed on the centroid peaklist using mascot (v2.2.01, Matrixscience) against all Saccharomyces cerevisiae proteins in the swissprot database (release 55.3) containing 6833 entries. Search parameters were peptide mass tolerance 5 p.p.m. and a 0.5 Da mass window for the MS/MS. One trypsin miss cleavage was allowed, fixed modification methylthio (C), iTRAQ4plex (K and N-terminal), variable modification oxidation of methionine. Protein identifications were excepted when based on >1 unique peptides with >95% confidence. iTRAQ quantification was performed using the perl script version of i-tracker (67), extracting the area under the different iTRAQ reporter ions from the profiled peak list. Peptide identification and quantification were combined using an in-house script, all data were exported into Microsoft Excel for statistical analysis. Combined iTRAQ ratios were set to one for each peptide identification, ratios were calculated for sro7Δ/sec23-1, sec6-4/sec23-1 and sro7Δ/sec6-4. Average of the log2 for all assigned peptide ratios per proteins was used for protein quantification, and fold regulation was defined as ±1 SD from the median of all quantified proteins in a sample-to-sample comparison (32). For determination of relative abundance of proteins in control and mock PGVs (control/mock ratios), normalization of protein abundance was performed after excluding ribosomal proteins, because ribosomes are in the same size range as exocytic vesicles and therefore expected co-purificants (68).
Western blot analysis
The prepared P4 pellets were resuspended in lysis buffer and samples were heated in sample buffer at 95°C for 5 min (or 15 min at 65°C for STL1-GFP) and resolved by SDS–PAGE on 12.5 or 10% Tris–HCl gels (Biorad). Separated proteins were transferred to nitrocellulose membrane for western blot with rabbit antiserum against Sso1p/2p, Snc1p/2p, Bgl2p, Cwp1p, Gas1p ormouse anti-GFP antibody (Roche Cat. No. 11814460001). Secondary antibody was either Alexa Fluor 680 goat anti-rabbit or Alexa Fluor 680 goat anti-mouse (Invitrogen). The blots were analyzed with the Odyssey Infrared Imaging System (LI-COR) and quantitated using the odyssey quantification software (version 2.1).
Fluorescence microscopy
For fluorescence microscopy, mid-exponential cultures of wild-type and sro7Δ cells carrying Stl1p-GFP expressed from the MET25 promoter of a pUG35 plasmid (kindly provided by Dr. Anders Brandt) were shifted to fresh methionine-free medium with 0.6 m NaCl and were incubated for 2 h at 37°C prior to microscopy. Cells were viewed with a LEICA DMRXA microscope (Deerfield) using a Fluotar lens and the Leica 513852 filter for the GFP channel. Photographs were captured using a Hamamatsu ORCA cooled CCD camera (Bridgewater).
Supplementary Material
Acknowledgments
This work was supported by grants from the Swedish Research Council, Carl Trygger Foundation, Helge Ax:son Johnson Foundation, and The Royal Society of Arts and Sciences in Göteborg. We are indebted to Dr. Sjoerd van der Post at the Proteomics Core Facility, Göteborg, for generous expert help with iTRAQ analysis and data, and to Dr. Joakim Norbeck for helpful discussions. The generous gifts of the pUG35-STL1-GFP plasmid from Dr. Anders Brandt (Carlsberg laboratory), the Gas1p antibody from Dr. Laura Popolo (University of Milan) and the Cwp1p antibody from Dr. Frans Klis (University of Amsterdam) are greatly appreciated.
Footnotes
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1: Western blot showing the distribution of Snc1/2p and Sso1/2p in sorbitol vesicle gradient fractions from the sro7Δ strain Samples were prepared and treated as in Figure 1 and Materials and Methods. Proteins were resolved by SDS–PAGE and analyzed by Snc1/2p and Sso1/2p immunoblots.
Figure S2: Breakdown by known localization of the 91 proteins identified in Table 2 Annotations are from the SGD yeast GO slim component catalogue (http://www.yeastgenome.org/cgi-bin/GO/goSlimMapper.pl). Sub-cellular compartments are organized on the basis of number of proteins with the indicated annotation, in clockwise falling order.
Figure S3: Relative generation time (h) of deletion mutants The relative generation time was calculated as (LN [WT/mutant]). Cells were cultured in defined medium in microtiter plates at 30°C and OD was recorded automatically as previously described (69). Medium contained no added NaCl or 0.85 m NaCl. The relative generation time of the salt-sensitive sro7Δ is given as a reference.
Table S1: All proteins identified by iTRAQ analysis
Table S2: Protein abundance in PGV fractions from sec6-4 cells as compared to sro7Δ cells
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Pruyne D, Legesse-Miller A, Gao L, Dong Y, Bretscher A. Mechanisms of polarized growth and organelle segregation in yeast. Annu Rev Cell Dev Biol. 2004;20:559–591. doi: 10.1146/annurev.cellbio.20.010403.103108. [DOI] [PubMed] [Google Scholar]
- 2.Hsu SC, TerBush D, Abraham M, Guo W. The exocyst complex in polarized exocytosis. Int Rev Cytol. 2004;233:243–265. doi: 10.1016/S0074-7696(04)33006-8. [DOI] [PubMed] [Google Scholar]
- 3.TerBush DR, Maurice T, Roth D, Novick P. The Exocyst is a multi-protein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- 4.Aalto MK, Ronne H, Keranen S. Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. Embo J. 1993;12:4095–4104. doi: 10.1002/j.1460-2075.1993.tb06093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brennwald P, Kearns B, Champion K, Keranen S, Bankaitis V, Novick P. Sec9 is a SNAP-25-like component of a yeast SNARE complex that may be the effector of Sec4 function in exocytosis. Cell. 1994;79:245–258. doi: 10.1016/0092-8674(94)90194-5. [DOI] [PubMed] [Google Scholar]
- 6.Protopopov V, Govindan B, Novick P, Gerst JE. Homologs of the synaptobrevin/VAMP family of synaptic vesicle proteins function on the late secretory pathway in S. cerevisiae. Cell. 1993;74:855–861. doi: 10.1016/0092-8674(93)90465-3. [DOI] [PubMed] [Google Scholar]
- 7.Salminen A, Novick PJ. The Sec15 protein responds to the function of the GTP binding protein, Sec4, to control vesicular traffic in yeast. J Cell Biol. 1989;109:1023–1036. doi: 10.1083/jcb.109.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu H, Rossi G, Brennwald P. The ghost in the machine: small GTPases as spatial regulators of exocytosis. Trends Cell Biol. 2008;18:397–404. doi: 10.1016/j.tcb.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- 10.Walworth NC, Novick P. Purification and characterization of constitutive secretory vesicles from yeast. J Cell Biol. 1987;105:163–174. doi: 10.1083/jcb.105.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wirtz-Peitz F, Knoblich JA. Lethal giant larvae take on a life of their own. Trends Cell Biol. 2006;16:234–241. doi: 10.1016/j.tcb.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Brennwald P, Rossi G. Spatial regulation of exocytosis and cell polarity: yeast as amodel for animal cells. FEBS Lett. 2007;581:2119–2124. doi: 10.1016/j.febslet.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kagami M, Toh-e A, Matsui Y. Sro7p, a Saccharomyces cerevisiae counterpart of the tumor suppressor l(2)gl protein, is related to myosins in function. Genetics. 1998;149:1717–1727. doi: 10.1093/genetics/149.4.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gangar A, Rossi G, Andreeva A, Hales R, Brennwald P. Structurally conserved interaction of Lgl family with SNAREs is critical to their cellular function. Curr Biol. 2005;15:1136–1142. doi: 10.1016/j.cub.2005.05.046. [DOI] [PubMed] [Google Scholar]
- 15.Lehman K, Rossi G, Adamo JE, Brennwald P. Yeast homologues of tomosyn and lethal giant larvae function in exocytosis and are associated with the plasma membrane SNARE, Sec9. J Cell Biol. 1999;146:125–140. doi: 10.1083/jcb.146.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Wang P, Gangar A, Zhang J, Brennwald P, Terbush D, Guo W. Lethal giant larvae proteins interact with the exocyst complex and are involved in polarized exocytosis. J Cell Biol. 2005;170:273–283. doi: 10.1083/jcb.200502055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grosshans BL, Andreeva A, Gangar A, Niessen S, Yates JR, Brennwald P, Novick P. The yeast lgl family member Sro7p is an effector of the secretory Rab GTPase Sec4p. J Cell Biol. 2006;172:55–66. doi: 10.1083/jcb.200510016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hattendorf DA, Andreeva A, Gangar A, Brennwald PJ, Weis WI. Structure of the yeast polarity protein Sro7 reveals a SNARE regulatory mechanism. Nature. 2007;446:567–571. doi: 10.1038/nature05635. [DOI] [PubMed] [Google Scholar]
- 19.Vasioukhin V. Lethal giant puzzle of Lgl. Dev Neurosci. 2006;28:13–24. doi: 10.1159/000090749. [DOI] [PubMed] [Google Scholar]
- 20.Bilder D. Epithelial polarity and proliferation control: links from the Drosophila neoplastic tumor suppressors. Genes Dev. 2004;18:1909–1925. doi: 10.1101/gad.1211604. [DOI] [PubMed] [Google Scholar]
- 21.Betschinger J, Mechtler K, Knoblich JA. The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature. 2003;422:326–330. doi: 10.1038/nature01486. [DOI] [PubMed] [Google Scholar]
- 22.Plant PJ, Fawcett JP, Lin DC, Holdorf AD, Binns K, Kulkarni S, Pawson T. A polarity complex of mPar-6 and atypical PKC binds, phosphorylates and regulates mammalian Lgl. Nat Cell Biol. 2003;5:301–308. doi: 10.1038/ncb948. [DOI] [PubMed] [Google Scholar]
- 23.Wirtz-Peitz F, Nishimura T, Knoblich JA. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 2008;135:161–173. doi: 10.1016/j.cell.2008.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsson K, Böhl F, Söström I, Akhtar N, Strand D, Mechler B, Grabowski R, Adler L. The Saccharomyces cerevisiae SOP1 and SOP2 genes, which act in cation homeostasis, can be functionally substituted by the Drosophila lethal(2)giant larvae tumor suppressor gene. J Biol Chem. 1998;273:33610–33618. doi: 10.1074/jbc.273.50.33610. [DOI] [PubMed] [Google Scholar]
- 25.Wadskog I, Forsmark A, Rossi G, Konopka C, öyen M, Goksör M, Ronne H, Brennwald P, Adler L. The yeast tumor suppressor homologue Sro7p is required for correct targeting of the sodium transporting ATPase to the plasma membrane. Mol Biol Cell. 2006;17:4988–5003. doi: 10.1091/mbc.E05-08-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez-Boulan E, Kreitzer G, Musch A. Organization of vesicular trafficking in epithelia. Nat Rev Mol Cell Biol. 2005;6:233–247. doi: 10.1038/nrm1593. [DOI] [PubMed] [Google Scholar]
- 27.Harsay E, Bretscher B. Parallel pathways to the cell surface in yeast. J Cell Biol. 1995;131:297–310. doi: 10.1083/jcb.131.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haro R, Garciadeblas B, Rodriguéz-Navarro A. A novel P-type ATPase from yeast involved in sodium transport. FEBS Lett. 1991;291:189–191. doi: 10.1016/0014-5793(91)81280-l. [DOI] [PubMed] [Google Scholar]
- 29.Wieland J, Nietsche AM, Strayle J, Steiner H, Rudolph HK. The PMR2 gene cluster encodes functionally distinct isoforms of a putative Na+ pump in the yeast plasma membrane. EMBO J. 1995;14:3870–3882. doi: 10.1002/j.1460-2075.1995.tb00059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaiser CA, Schekman R. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 1990;61:723–733. doi: 10.1016/0092-8674(90)90483-u. [DOI] [PubMed] [Google Scholar]
- 31.Borner GH, Harbour M, Hester S, Lilley KS, Robinson MS. Comparative proteomics of clathrin-coated vesicles. J Cell Biol. 2006;175:571–578. doi: 10.1083/jcb.200607164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 34.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 35.Mrsa V, Klebl F, Tanner W. Purification and characterization of the Saccharomyces cerevisiae BGL2 gene product, a cell wall endo-beta-1,3-glucanase. J Bacteriol. 1993;175:2102–2106. doi: 10.1128/jb.175.7.2102-2106.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abe M, Qadota H, Hirata A, Ohya Y. Lack of GTP-bound Rho1p in secretory vesicles of Saccharomyces cerevisiae. J Cell Biol. 2003;162:85–97. doi: 10.1083/jcb.200301022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goud B, Salminen A, Walworth NC, Novick PJ. A GTP-binding protein required for secretion rapidly associates with secretory vesicles and the plasma membrane in yeast. Cell. 1988;53:753–768. doi: 10.1016/0092-8674(88)90093-1. [DOI] [PubMed] [Google Scholar]
- 38.Wedlich-Soldner R, Altschuler S, Wu L, Li R. Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science. 2003;299:1231–1235. doi: 10.1126/science.1080944. [DOI] [PubMed] [Google Scholar]
- 39.Lesage G, Bussey H. Cell wall assembly in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2006;70:317–343. doi: 10.1128/MMBR.00038-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park HO, Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol Mol Biol Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mulholland J, Wesp A, Riezman H, Botstein D. Yeast actin cytoskeleton mutants accumulate a new class of Golgi-derived secretary vesicle. Mol Biol Cell. 1997;8:1481–1499. doi: 10.1091/mbc.8.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Natarajan P, Wang J, Hua Z, Graham TR. Drs2p-coupled aminophospholipid translocase activity in yeast Golgimembranes and relationship to in vivo function. Proc Natl Acad Sci U S A. 2004;101:10614–10619. doi: 10.1073/pnas.0404146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roth MG. Phosphoinositides in constitutivemembrane traffic. Physiol Rev. 2004;84:699–730. doi: 10.1152/physrev.00033.2003. [DOI] [PubMed] [Google Scholar]
- 44.Lussier M, Sdicu AM, Bussey H. The KTR and MNN1 mannosyltransferase families of Saccharomyces cerevisiae. Biochim Biophys Acta. 1999;1426:323–334. doi: 10.1016/s0304-4165(98)00133-0. [DOI] [PubMed] [Google Scholar]
- 45.Sullivan DS, Biggins S, Rose MD. The yeast centrin, cdc31p, and the interacting protein kinase, Kic1p, are required for cell integrity. J Cell Biol. 1998;143:751–765. doi: 10.1083/jcb.143.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kane PM. The long physiological reach of the yeast vacuolar H+-ATPase. J Bioenerg Biomembr. 2007;39:415–421. doi: 10.1007/s10863-007-9112-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferreira C, van Voorst F, Martins A, Neves L, Oliveira R, Kielland-Brandt MC, Lucas C, Brandt A. A member of the sugar transporter family, Stl1p is the glycerol/H+ symporter in Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:2068–2076. doi: 10.1091/mbc.E04-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferreira C, Lucas C. Glucose repression over Saccharomyces cerevisiae glycerol/H+ symporter gene STL1 is overcome by high temperature. FEBS Lett. 2007;581:1923–1927. doi: 10.1016/j.febslet.2007.03.086. [DOI] [PubMed] [Google Scholar]
- 49.Schott DH, Collins RN, Bretscher A. Secretory vesicle transport velocity in living cells depends on the myosin-V lever arm length. J Cell Biol. 2002;156:35–39. doi: 10.1083/jcb.200110086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boyd C, Hughes T, Pypaert M, Novick P. Vesicles carry most exocyst subunits to exocytic sites marked by the remaining two subunits, Sec3p and Exo70p. J Cell Biol. 2004;167:889–901. doi: 10.1083/jcb.200408124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Black MW, Pelham HR. A selective transport route from Golgi to late endosomes that requires the yeast GGA proteins. J Cell Biol. 2000;151:587–600. doi: 10.1083/jcb.151.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Costaguta G, Stefan CJ, Bensen ES, Emr SD, Payne GS. Yeast Gga coat proteins function with clathrin in Golgi to endosome transport. Mol Biol Cell. 2001;12:1885–1896. doi: 10.1091/mbc.12.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valdivia RH, Baggott D, Chuang JS, Schekman RW. The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev Cell. 2002;2:283–294. doi: 10.1016/s1534-5807(02)00127-2. [DOI] [PubMed] [Google Scholar]
- 54.Wang CW, Hamamoto S, Orci L, Schekman R. Exomer: a coat complex for transport of select membrane proteins from the trans- Golgi network to the plasma membrane in yeast. J Cell Biol. 2006;174:973–983. doi: 10.1083/jcb.200605106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goto M. Protein O-glycosylation in fungi: diverse structures and multiple functions. Biosci Biotechnol Biochem. 2007;71:1415–1427. doi: 10.1271/bbb.70080. [DOI] [PubMed] [Google Scholar]
- 56.Proszynski TJ, Simons K, Bagnat M. O-glycosylation as a sorting determinant for cell surface delivery in yeast. Mol Biol Cell. 2004;15:1533–1543. doi: 10.1091/mbc.E03-07-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Galli T, McPherson PS, De Camilli P. The V0 sector of the V-ATPase, synaptobrevin, and synaptophysin are associated on synaptic vesicles in a Triton X-100-resistant, freeze-thawing sensitive, complex. J Biol Chem. 1996;271:2193–2198. doi: 10.1074/jbc.271.4.2193. [DOI] [PubMed] [Google Scholar]
- 58.Hiesinger PR, Fayyazuddin A, Mehta SQ, Rosenmund T, Schulze KL, Zhai RG, Verstreken P, Cao Y, Zhou Y, Kunz J, Bellen HJ. The v-ATPase V0 subunit a1 is required for a late step in synaptic vesicle exocytosis in Drosophila. Cell. 2005;121:607–620. doi: 10.1016/j.cell.2005.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Williams DC, Novick PJ. Analysis of SEC9 suppression reveals a relationship of SNARE function to cell physiology. PLoS One. 2009;4:e5449. doi: 10.1371/journal.pone.0005449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Osborne AR, Rapoport TA, van den Berg B. Protein translocation by the Sec61/SecY channel. Annu Rev Cell Dev Biol. 2005;21:529–550. doi: 10.1146/annurev.cellbio.21.012704.133214. [DOI] [PubMed] [Google Scholar]
- 61.Lipschutz JH, Lingappa VR, Mostov KE. The exocyst affects protein synthesis by acting on the translocationmachinery of the endoplasmic reticulum. J Biol Chem. 2003;278:20954–20960. doi: 10.1074/jbc.M213210200. [DOI] [PubMed] [Google Scholar]
- 62.Toikkanen JH, Miller KJ, Soderlund H, Jantti J, Keranen S. The beta subunit of the Sec61p endoplasmic reticulum translocon interacts with the exocyst complex in Saccharomyces cerevisiae. J Biol Chem. 2003;278:20946–20953. doi: 10.1074/jbc.M213111200. [DOI] [PubMed] [Google Scholar]
- 63.Guo W, Novick P. The exocyst meets the translocon: a regulatory circuit for secretion and protein synthesis? Trends Cell Biol. 2004;14:61–63. doi: 10.1016/j.tcb.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 64.David D, Sundarababu S, Gerst JE. Involvement of long chain fatty acid elongation in the trafficking of secretory vesicles in yeast. J Cell Biol. 1998;143:1167–1182. doi: 10.1083/jcb.143.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harsay E, Schekman R. A subset of yeast vacuolar protein sorting mutants is blocked in one branch of the exocytic pathway. J Cell Biol. 2002;156:271–285. doi: 10.1083/jcb.200109077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Novick P, Medkova M, Dong G, Hutagalung A, Reinisch K, Grosshans B. Interactions between Rabs, tethers, SNAREs and their regulators in exocytosis. Biochem Soc Trans. 2006;34:683–686. doi: 10.1042/BST0340683. [DOI] [PubMed] [Google Scholar]
- 67.Shadforth IP, Dunkley TP, Lilley KS, Bessant C. i-Tracker: for quantitative proteomics using iTRAQ. BMC Genomics. 2005;6:145. doi: 10.1186/1471-2164-6-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takamori S, Holt M, Stenius K, Lemke EA, Gronborg M, Riedel D, Urlaub H, Schenck S, Brugger B, Ringler P, Muller SA, Rammner B, Grater F, Hub JS, De Groot BL, et al. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 69.Warringer J, Blomberg A. Automated screening in environmental arrays allows analysis of quantitative phenotypic profiles in Saccharomyces cerevisiae. Yeast. 2003;20:53–67. doi: 10.1002/yea.931. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.