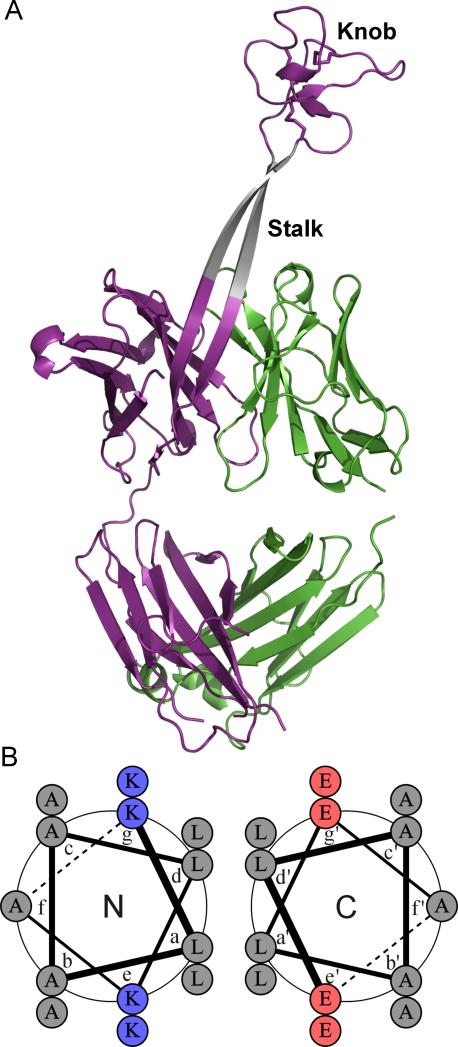
Antibody complementarity determining regions (CDRs) typically consist of hypervariable loops of 8-16 residues that are involved in antigen recognition. Unlike most mammalian antibodies, a subgroup of bovine antibodies contain an ultralong heavy chain CDR3 (CDR3H) region with 40-67 residues.[1] A recently solved X-ray crystal structure of one such bovine antibody, BLV1H12, revealed a novel structural motif in its CDR3H region that folds into a solvent exposed, antiparallel β-strand “stalk” (20 Å in length) that terminates in a folded “knob” domain stabilized by three disulfide bonds (Figure 1A).[2] This knob domain was shown to be involved in antigen binding by antibodies elicited to specific immunogens. Moreover, we showed that the solvent exposed, antiparallel β-strand “stalk” supports the substitution of the “knob” domain with functional polypeptides of various folds, which result in stably expressed antibody fusions with potent biologically activities.[3] We hypothesize that the rigid “stalk” region plays an important role in folding of this unique antibody family and engineered antibody fusions by separating the folded polypeptide within the ultralong CDR3H from the immunoglobulin framework, thereby preventing misfolding of either domain.
Figure 1.
Substitution of the β-strand “stalk” in bovine antibody BLV1H12 with a coiled-coil motif. (A) X-ray crystal structure of antibody BLV1H12 (PDB ID: 4K3D). (B) Helical wheel representation of the antiparallel heterodimeric coiled-coil.[8] The sequence of the ascending peptide with linkers at each end is: H2N-GGSGAKLAALKAKLAALKGGGGS-COOH; the sequence of the descending peptide with linkers at each end is: H2N-GGGGSELAALEAELAALEAGGSG-COOH.
The “stalk” region within the ultralong CDR3H of BLV1H12 represents an unusual protein motif, since long solvent exposed β-strands (seven residues on each strand) are rare. To date, partially solvent exposed, antiparallel β-strands (less than five residues on each strand) have been found only in certain bacterial aminoacyl-tRNA synthetases.[4] It is likely that this novel structure is templated by geometrical constraints imposed on the ends of the β-sheet by the “knob” domain and variable region interface. The β-sheet may be further stabilized by interstrand hydrogen bonds, side chain hydrophobic interactions and interactions with residues from adjacent CDR loops.
The question arises whether this unusual antiparallel β-strand motif is required for the unique “stalk-knob” structure of this ultralong bovine CDR3, or can it be substituted with other rigid motifs to afford a stable antibody with a similar architecture. One candidate for a “stalk” replacement is the coiled-coil, a highly versatile structural motif that plays an important structural and functional role in a variety of proteins.[5] The coiled-coil is a superhelix consisting of two or more α-helices with a repeated pattern, referred to as the heptad repeat, of buried hydrophobic residues sandwiched by exposed hydrophilic residues.[6] Substitution of the β-strand motif in the bovine antibody with a coiled-coil may also generate a rigid “stalk” that effectively separates the functional “knob” domain from the main framework of antibody. Moreover, in comparison to the solvent exposed β-strands, our detailed understanding of those factors that affect coiled-coil structure and stability may allow us to further engineer the “stalk” to modulate the chemical, physical, and biological properties of the antibody fusions Here we show that substitution of a heterodimeric coiled-coil for the β-strand “stalk” in BLV1H12 results in an antibody (Ab-coil) whose CDR3H folds into an antiparallel coiled-coil structure and that has thermodynamic stability comparable to that of BLV1H12. This coiled-coil “stalk” also allows generation of a functional antibody-bovine granulocyte colony-stimulating factor (bGCSF) fusion that stimulates GCSF-dependent cell proliferation with a potency similar to that of bGCSF.
Most naturally occurring coiled-coil motifs have parallel strands, but substitution of the antiparallel β-strands in the bovine antibody “stalk” region requires an antiparallel coiled-coil motif. Fujii and coworkers have designed a synthetic antiparallel coiled-coil by connecting positively (Base) and negatively (Acid) charged helical peptides with a glycine-based linker.[7] Both peptides are characterized by heptad repeats with leucine residues at the a and d sites and charged residues at the e and g sites, which promote and stabilize the coiled-coil structure (Figure 1B). These basic and acidic peptides were substituted for the ascending and descending β-strands of the “stalk” in BLV1H12, respectively, and are expected to adopt a heterodimeric coiled-coil structure when fused to the “knob” domain of BLV1H12. The substituted coiled-coil sequences contain 14 residues on each strand, which should give rise to a “stalk” approximately 21 Å in length, comparable to that of β-strand “stalk”. To optimize the folding and stability of the resulting antibody, flexible GGSG and GGGGS linkers were placed at each end of the coiled-coil sequences.
We expressed the Fab fragment of the bovine coiled-coil CDR3 variant in order to characterize its folding and stability. The Ab-coil Fab and BLV1H12 Fab (Ab-beta) were expressed in freestyle HEK293 cells by transient transfection. Proteins were purified by Ni-NTA chromatography and analyzed by SDS-PAGE and mass spectrometry (Figure S1-S3). Under non-reducing conditions, the Ab-beta Fab migrates as a single band of 53 kDa and Ab-coil Fab migrates at 55 kDa. In the presence of 50 mM dithiothreitol (DTT), the light chains of Ab-beta and Ab-coil Fabs migrate at 23 kDa; and the heavy chains of the Ab-beta and Ab-coil Fabs migrate at 30 kDa and 32 kDa, respectively, consistent with the “stalk-knob” sequences. The final yield of the Ab-coil Fab is ~17 mg/L, similar to that of the Ab-beta Fab. Both proteins are stable in PBS (pH 7.4), and can be concentrated to over 10 mg/mL without aggregation. We next measured the stabilities of the Ab- beta and Ab-coil Fabs using differential scanning fluorimetry (DSF) with SYPRO orange dye (Figure S4).[9] The experimental melting temperatures, Tm, are 74.6 ± 0.3°C for Ab-beta Fab and 74.1 ± 0.3°C for Ab-coil Fab. We previously found that DSF melting temperatures for Fabs track closely with those determined by differential scanning calorimetry.[10] These data, together with the expression and solubility data, suggest that substitution of the β-strand “stalk” with the antiparallel coiled-coil does not significantly affect the stability of antibody BLV1H12.
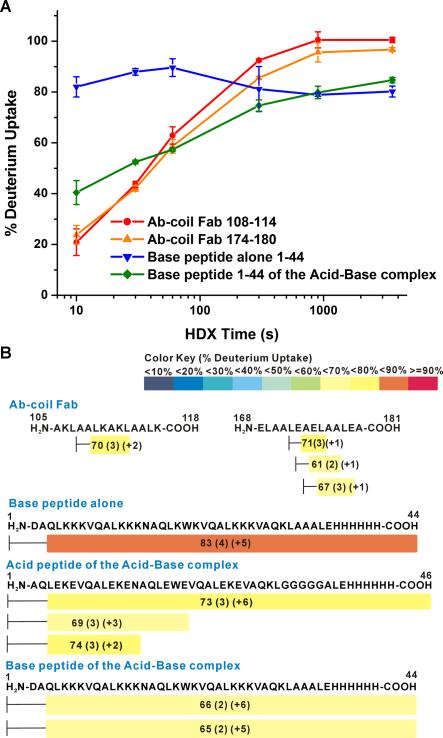
To further investigate whether the substituted peptides adopt a coiled-coil structure when incorporated into the “stalk” region of BLV1H12, we carried out a hydrogen deuterium exchange-mass spectrometry (HDX-MS) study of the Ab-coil Fab.[11] A previously characterized parallel heterodimeric coiled-coil consisting of an Acid and Base peptides was used as a control (Figure S5).[12] Consistent with the previous study, circular dichroism (CD) spectral analysis revealed that either the purified Acid or Base peptide alone forms an unfolded, disordered structure in solution, while their mixture in a 1:1 molar ratio results in a stable helical structure (Figure S6).[12] Deuterium incorporation measurements revealed that the backbone amides of the Base peptide alone exchange more than three times faster (0.174 ± 0.032 s-1) than those in the Acid-Base complex (0.052 ± 0.011 s-1) (Figure 2A). After 10 s in exchange buffer, the former has more than 80% deuterium incorporation, whereas the latter has only 40% deuterium uptake. This result is consistent with the CD analysis and indicates that in the presence of the Acid peptide, the Base peptide forms an α-helical structure. The HDX curves show that the deuterium exchange rates of the backbone amides within the coiled-coil regions of Ab-coil Fab (0.024 ± 0.009 s-1 for the ascending coil and 0.027 ±0.007 s-1 for the descending coil) are similar to those of the Acid-Base complex (Figure 2A) and also consistent with those of the α-helices in previous studies.[13] In addition, the average levels of deuterium incorporation into the coiled-coil regions are comparable to those of the Acid-Base complex, but are significantly lower than that of the Base peptide alone (Figure 2B). These results, together with the previous Tm and expression data, suggest that the substituted sequences fold into an antiparallel coiled-coil structure when substituted for the solvent exposed β-strands in the CDR3H of BLV1H12.
Figure 2.
Hydrogen deuterium exchange mass spectrometry (HDX-MS) analysis of Ab-coil Fab, Base peptide alone, and the Acid-Base complex. Antibody or peptide sample was diluted into D2O-containing exchange buffer (50 mM HEPES, pH 8.0, 150 mM NaCl) and incubated at 4°C for 10 to 3600 s. Measurements were repeated three times; all values were calculated based on experimental Dmax values. (A) Deuterium incorporation curves of the backbone amides within the coiled-coil regions of Ab-coil Fab and the Base peptide as measured by liquid chromatography-mass spectrometry (LC-MS) at multiple time points. (B) The average levels of deuterium incorporation for the coiled-coil regions of Ab-coil Fab, Base peptide alone, and the Acid-Base complex. The value within each peptide is the average % of deuterium incorporation over 6 time points (from 10 to 3600 s). The numbers in the parentheses are standard deviation and charge state of the analyzed peptide.
Next we explored whether the Ab-coil structure allows the correct folding of the fused polypeptide and generation of a functional antibody chimera in a similar fashion to BLV1H12. To test this notion, we first generated the full-length IgG forms of Ab-beta and Ab-coil. The resulting Ab-beta and Ab-coil IgGs were expressed, purified from mammalian cells by Protein A/G chromatography, and their structures confirmed by SDS-PAGE and mass spectrometry (Figure S7-S9). Both antibodies expressed in similar yields and had comparable solubilities. We then generated Ab-beta-bGCSF and Ab-coil-bGCSF fusion proteins by replacing the “knob” domain with bGCSF using GGGGS linkers at each end of the bGCSF as described previously.[3a] The resulting constructs were confirmed by SDS-PAGE and mass spectrometry (Figure S10-S15). The antibody-bGCSF fusion proteins expressed in mammalian cells afforded similar yields (~17 mg/L) and solubilities as BLV1H12, indicating that they are likely folded correctly.
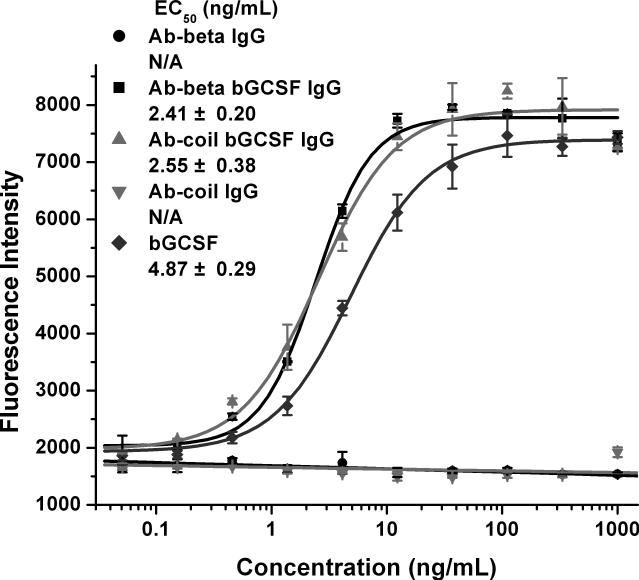
We next examined the biological activity of the Ab-coil bGCSF fusion protein using mouse NFS-60 cells that are growth-dependent on GCSF.[14] Both Ab-beta bGCSF and Ab-coil bGCSF stimulate NFS-60 cell proliferation in a dose-dependent manner (Figure 3), whereas Ab-beta and Ab-coil by themselves have no proliferative activities, indicating that the observed activities of the antibody-bGCSF fusion proteins result from the fused bGCSF. The potencies of the antibody-bGCSF fusion proteins (EC50: 2.41 ± 0.20 ng/mL for Ab-beta bGCSF, and 2.55 ± 0.38 ng/mL for Ab-coil bGCSF) are comparable to that of bGCSF (EC50: 4.87 ± 0.29 ng/mL). In addition, Ab-beta bGCSF Fab and Ab-coil bGCSF Fab stimulate NFS-60 cell proliferation in a dose-dependent fashion (Figure S16). The EC50 is 1.59 ± 0.12 ng/mL for Ab-beta bGCSF Fab and 1.28 ± 0.07 ng/mL for Ab-coil bGCSF Fab. Thus, grafting of bGCSF onto the substituted coiled-coil “stalk” in the CDR3H of the Ab-coil does not appear to affect the activity of this cytokine, indicating that like the β-strand “stalk”, the antiparallel heterodimeric coiled-coil allows the creation of a functional antibody chimera by supporting the folding of the biologically active fused polypeptide.
Figure 3.
Ab-coil bGCSF fusion protein stimulates proliferation of mouse NFS-60 cells in a dose-dependent manner. Cells cultured in RPMI-1640 medium with 10% FBS and 0.05 mM 2-mercapoethanol were treated with various concentrations of bGCSF, Ab-beta IgG, Ab-coil IgG, Ab-beta bGCSF IgG and Ab-coil bGCSF IgG. Cell viability was quantified using an Alamar Blue (Invitrogen) assay; measurements were repeated three times.
In conclusion, we have shown that by substituting coiled-coil sequences for the solvent exposed, antiparallel β-strands in the “stalk” region of bovine antibody BLV1H12, one can generate a novel extended CDR3H consisting of an antiparallel heterodimeric coiled-coil terminating in a folded domain. It is likely that this unique structure can be inserted into the CDRs of other antibodies as well, including human antibodies. Importantly, relative to the bovine-derived β-strand “stalk”, the coiled-coil motif provides a versatile structural module for further engineering the properties of functional antibody fusions. The β-strand and coiled-coil motifs may also allow generation of novel antibody chimeras containing two or more polypeptides by fusing these motifs into other CDR loops, opening the door to engineered antibodies with dual activities. Future studies will include structural characterization of the coiled-coil antibody, fusion of other cytokines, growth factors and peptide hormones into the ultralong CDR3H region, and generation of antibodies containing two or more CDR protein fusions.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) Grant R01 GM062159 (to P.G.S.). This manuscript is number 25001 of The Scripps Research Institute.
Footnotes
Supporting information for this article is available on the WWW under http://www.angewandte.org
Contributor Information
Yong Zhang, Department of Chemistry, The Scripps Research Institute 10550 N. Torrey Pines Road, La Jolla, CA 92037 (USA).
Devrishi Goswami, Department of Molecular Therapeutics, The Scripps Research Institute, Jupiter, FL, 33458 (USA).
Danling Wang, California Institute for Biomedical Research (Calibr), 11119 N. Torrey Pines Road, La Jolla, CA 92307 (USA).
Tsung-Shing Andrew Wang, Department of Chemistry, The Scripps Research Institute 10550 N. Torrey Pines Road, La Jolla, CA 92037 (USA).
Shiladitya Sen, Department of Chemistry and Biochemistry, The Ohio State University, 100 West 18th Avenue, Columbus, Ohio 43210 (USA).
Thomas J. Magliery, Department of Chemistry and Biochemistry, The Ohio State University, 100 West 18th Avenue, Columbus, Ohio 43210 (USA)
Patrick R. Griffin, Department of Molecular Therapeutics, The Scripps Research Institute, Jupiter, FL, 33458 (USA)
Feng Wang, Department of Chemistry, The Scripps Research Institute 10550 N. Torrey Pines Road, La Jolla, CA 92037 (USA).
Peter G. Schultz, Department of Chemistry, The Scripps Research Institute 10550 N. Torrey Pines Road, La Jolla, CA 92037 (USA).
References
- 1.a Berens SJ, Wylie DE, Lopez OJ. Int. Immunol. 1997;9:189–199. doi: 10.1093/intimm/9.1.189. [DOI] [PubMed] [Google Scholar]; b Lopez O, Perez C, Wylie D. Immunol. Rev. 1998;162:55–66. doi: 10.1111/j.1600-065x.1998.tb01429.x. [DOI] [PubMed] [Google Scholar]; c Saini SS, Allore B, Jacobs RM, Kaushik A. Eur. J. Immunol. 1999;29:2420–2426. doi: 10.1002/(SICI)1521-4141(199908)29:08<2420::AID-IMMU2420>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]; d Saini SS, Farrugia W, Ramsland PA, Kaushik AK. Int. Immunol. 2003;15:845–853. doi: 10.1093/intimm/dxg083. [DOI] [PubMed] [Google Scholar]; e Zhao Y, Jackson SM, Aitken R. Dev. Comp. Immunol. 2006;30:175–186. doi: 10.1016/j.dci.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 2.Wang F, Ekiert DC, Ahmad I, Li W, Zhang Y, Bazirgan O, Torkamani A, Raudsepp T, Mwangi W, Criscitiello MF, Wilson IA, Schultz PG, Smider VV. Cell. 2013;153:1379–1393. doi: 10.1016/j.cell.2013.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a Zhang Y, Wang D, de Lichtervelde L, Sun SB, Smider VV, Schultz PG, Wang F. Angew. Chem. Int. Ed. 2013;52:8295–8298. doi: 10.1002/anie.201303656. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Zhang Y, Wang D, Welzel G, Wang Y, Schultz PG, Wang F. ACS Chem. Biol. 2013;8:2117–2121. doi: 10.1021/cb4004749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a Fukai S, Nureki O, Sekine S, Shimada A, Tao J, Vassylyev DG, Yokoyama S. Cell. 2000;103:793–803. doi: 10.1016/s0092-8674(00)00182-3. [DOI] [PubMed] [Google Scholar]; b Nureki O, Vassylyev DG, Tateno M, Shimada A, Nakama T, Fukai S, Konno M, Hendrickson TL, Schimmel P, Yokoyama S. Science. 1998;280:578–582. doi: 10.1126/science.280.5363.578. [DOI] [PubMed] [Google Scholar]; c Cusack S, Yaremchuk A, Tukalo M. EMBO J. 2000;19:2351–2361. doi: 10.1093/emboj/19.10.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a Apostolovic B, Danial M, Klok HA. Chem. Soc. Rev. 2010;39:3541–3575. doi: 10.1039/b914339b. [DOI] [PubMed] [Google Scholar]; b Burkhard P, Stetefeld J, Strelkov SV. Trends Cell Biol. 2001;11:82–88. doi: 10.1016/s0962-8924(00)01898-5. [DOI] [PubMed] [Google Scholar]; c Gazi AD, Charova SN, Panopoulos NJ, Kokkinidis M. Cell Microbiol. 2009;11:719–729. doi: 10.1111/j.1462-5822.2009.01297.x. [DOI] [PubMed] [Google Scholar]; d Marsden HR, Kros A. Angew Chem Int Edit. 2010;49:2988–3005. doi: 10.1002/anie.200904943. [DOI] [PubMed] [Google Scholar]; e Peckham M. Biochem. Soc. Trans. 2011;39:1142–1148. doi: 10.1042/BST0391142. [DOI] [PubMed] [Google Scholar]; f Eckert DM, Kim PS. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 6.a Hadley EB, Gellman SH. J. Am. Chem. Soc. 2006;128:16444–16445. doi: 10.1021/ja067178r. [DOI] [PubMed] [Google Scholar]; b Harbury PB, Zhang T, Kim PS, Alber T. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]; c Hill RB, Raleigh DP, Lombardi A, Degrado WF. Acc. Chem. Res. 2000;33:745–754. doi: 10.1021/ar970004h. [DOI] [PMC free article] [PubMed] [Google Scholar]; d McClain DL, Woods HL, Oakley MG. J. Am. Chem. Soc. 2001;123:3151–3152. doi: 10.1021/ja004099l. [DOI] [PubMed] [Google Scholar]; e Oakley MG, Hollenbeck JJ. Curr. Opin. Struct. Biol. 2001;11:450–457. doi: 10.1016/s0959-440x(00)00232-3. [DOI] [PubMed] [Google Scholar]; f Woolfson DN. Fibrous Proteins: Coiled-Coils, Collagen and Elastomers. 2005;70:79–112. [Google Scholar]
- 7.Suzuki N, Fujii N. Tetrahedron Lett. 1999;40:6013–6017. [Google Scholar]
- 8.Grigoryan G, Keating AE. Curr. Opin. Struct. Biol. 2008;18:477–483. doi: 10.1016/j.sbi.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a Lavinder JJ, Hari SB, Sullivan BJ, Magliery TJ. J. Am. Chem. Soc. 2009;131:3794–3795. doi: 10.1021/ja8049063. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Wang F, Sen S, Zhang Y, Ahmad I, Zhu X, Wilson IA, Smider VV, Magliery TJ, Schultz PG. Proc. Natl. Acad. Sci. U.S.A. 2013;110:4261–4266. doi: 10.1073/pnas.1301810110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Sun SB, Sen S, Kim NJ, Magliery TJ, Schultz PG, Wang F. J. Am. Chem. Soc. 2013;135:9980–9983. doi: 10.1021/ja402927u. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.a Chalmers MJ, Busby SA, Pascal BD, He Y, Hendrickson CL, Marshall AG, Griffin PR. Anal. Chem. 2006;78:1005–1014. doi: 10.1021/ac051294f. [DOI] [PubMed] [Google Scholar]; b Chalmers MJ, Busby SA, Pascal BD, West GM, Griffin PR. Expert Rev. Proteomics. 2011;8:43–59. doi: 10.1586/epr.10.109. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Pascal BD, Willis S, Lauer JL, Landgraf RR, West GM, Marciano D, Novick S, Goswami D, Chalmers MJ, Griffin PR. J. Am. Soc. Mass Spectrom. 2012;23:1512–1521. doi: 10.1007/s13361-012-0419-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Shea EK, Lumb KJ, Kim PS. Curr. Biol. 1993;3:658–667. doi: 10.1016/0960-9822(93)90063-t. [DOI] [PubMed] [Google Scholar]
- 13.a Liu YQ, Smith DL. J. Am. Soc. Mass Spectrom. 1994;5:19–28. doi: 10.1016/1044-0305(94)85080-1. [DOI] [PubMed] [Google Scholar]; b Wintrode PL, Friedrich KL, Vierling E, Smith JB, Smith DL. Biochemistry. 2003;42:10667–10673. doi: 10.1021/bi034117m. [DOI] [PubMed] [Google Scholar]; c Wagner DS, Melton LG, Yan YB, Erickson BW, Anderegg RJ. Protein Sci. 1994;3:1305–1314. doi: 10.1002/pro.5560030817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a Bai Y, Ann DK, Shen WC. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7292–7296. doi: 10.1073/pnas.0500062102. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Shirafuji N, Asano S, Matsuda S, Watari K, Takaku F, Nagata S. Exp. Hematol. 1989;17:116–119. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.