Abstract
Background & Aims
Increasing grade of pancreatic intraepithelial neoplasia (PanIN) has been associated with progression to pancreatic ductal adenocarcinoma (PDAC). However, the mechanisms that control progression from PanINs to PDAC are not well understood. We investigated the genetic alterations involved in this process.
Methods
Genomic DNA samples from laser-capture microdissected PDACs and adjacent PanIN2 and PanIN3 lesions from 10 patients with pancreatic cancer were analyzed by exome sequencing.
Results
Similar numbers of somatic mutations were identified in PanINs and tumors, but the mutational load varied greatly among cases. Ten of the 15 isolated PanINs shared more than 50% of somatic mutations with associated tumors. Mutations common to tumors and clonally related PanIN2 and PanIN3 lesions were identified as genes that could promote carcinogenesis. KRAS and TP53 were frequently altered in PanINs and tumors, but few other recurrently modified genes were detected. Mutations in DNA damage response genes were prevalent in all samples. Genes that encode proteins involved in gap junctions, the actin cytoskeleton, the mitogen-activated protein kinase signaling pathway, axon guidance, and cell cycle regulation were among the earliest targets of mutagenesis in PanINs that progressed to PDAC.
Conclusions
Early-stage PanIN2 lesions appear to contain many of the somatic gene alterations required for PDAC development.
Keywords: pancreas, tumorigenesis, LCM, whole genome amplification
INTRODUCTION
Pancreatic cancer is the fourth leading cause of cancer-associated mortality, accounting for over 35,000 deaths each year. Heterogeneous in form, 90% of pancreatic cancers are ductal pancreatic adenocarcinomas (PDAC) that present generally in the seventh decade of life1. Most cases have mild symptoms prior to diagnosis at late stage, locally advanced or metastatic disease2. Carcinoma of the exocrine pancreas is associated with a median survival of six-months and a five-year survival rate of 5%3. Approximately 15–20% of patients present with resectable disease and only 15–25% of surgically resected patients survive to five years4–5.
Despite the high incidence and poor survival associated with PDAC, few advances have been made in understanding the etiology and basic biology of pancreatic cancer and the mechanisms by which precursor lesions become early stage invasive tumors. Precursor lesions in the form of non-invasive pancreatic intraepithelial neoplasia (PanIN) are grouped into three grades according to increasing degree of cytological and architectural atypia6–7. PanIN1 lesions are further subdivided into flat (PanIN1A) and papillary types (PanIN1B). Additional loss of polarity, nuclear crowding, cell enlargement and hyperchromasia present in PanIN2s. Advanced PanIN3 lesions have severe nuclear atypia, luminal necrosis and manifest epithelial cell budding into the lumen of ducts7.
Evidence is strong for PanIN involvement in a cancer progression model. While PanIN1 lesions are frequently observed in normal pancreatic autopsy tissues, PanIN2 lesions are more common in tissue derived from neoplastic pancreata. PanIN3 lesions are rarely observed in pancreatic tissues in the absence of cancer. In addition, the full spectrum of PanINs has been observed prior to tumorigenesis in mouse models of pancreatic cancer8. Tumor recurrence at surgical margins containing unresected PanIN3 lesions further supports this model9. While it is thought that PDAC develops by stepwise progression through increasing grades of precursor lesions, the early genetic events that promote development of PanINs and progression to PDAC are not well defined. Identification of these early driver genes and pathways is expected to lead to improved selection of therapeutic targets and may result in improved early diagnosis of early stage PDAC or advanced precursor lesions. Here we report on exome sequencing of DNA isolated from pancreatic tumors and adjacent PanINs and on the identification of genes and pathways that contribute to PanIN development and progression to PDAC.
MATERIALS AND METHODS
Tissue selection and laser capture microdissection (LCM)
H&E stained sections of pancreatic cancer tissues were reviewed by pathologists and 10 cases containing high grade PanINs (P2 or P3) adjacent to tumor were selected. Ten 10 micron frozen pancreatic tissue sections were cut and stained with Cresyl Violet (LCM Staining Kit, Ambion; AM1935). PanIN, tumor and histologically normal regions were individually isolated by LCM using the Arcturus PixCell II microscope and CapSure Macro caps (Arcturus; LCM 0211).
Direct DNA extraction and amplification
Whole genome amplification was performed directly on LCM captured cells using a single-step procedure10. LCM cells were incubated for 10 min in 0.5X Repli-g D2 buffer (6.5µl) (Qiagen, CA,USA) and then in Repli-g Stop Solution (3.5µl). Cells were then mixed with Repli-g mini kit Master Mix (40ul) and incubated at 30°C for 16 hours. Four individual 50µl WGA reactions were pooled for each sample. DNA was quantified by Quant-iT PicoGreen analysis (Invitrogen, P7581) and qualitative multiplex PCR was performed (Sigma-Aldrich; P0982).
Exome Sequencing
DNA (3µg) was fragmented to ~200bp (Covaris E210) prior to assembly of adapter flanked Illumina indexed paired end libraries (NEB Next DNA Kit) using Illumina adapters (Illumina, CA, USA). Exome Capture was performed using Agilent SureSelect Human All Exon 50Mb kit (Agilent, CA, USA). Two indexed libraries were sequenced per lane on the Illumina HiSeq platform. 100 bp paired-end reads were aligned to hg19 using Novoalign (v2.07.13, Novocraft)11. Local sequence realignment was performed by GATK version 1.6–712 within the context of the TREAT framework13.
Somatic SNV and INDEL calling
Each PanIN and Tumor sample was compared to a corresponding normal sample using SomaticSniper14 for somatic SNVs or GATK’s Somatic INDEL Detector15. A minimum somatic score of 20 and >8x coverage was required in the reference normal sample. Genotypes were re-coded to take advantage of the multiple samples from the same individuals. For somatic variants with <30X read depth, ≥3 alternate reads supporting the variant call were required. For variants at ≥30X, alternate alleles exceeding 4% of all reads were required. Functional significance of mutations was predicted using SNPEffect (SnpEFF; http://snpeff.sourceforge.net/), and SIFT and PolyPhen, Genes were categorized into pathways using the model-based gene set analysis (mgsa)16. Analyses were performed using R-package version 1.2.0. with annotation from MSigDB.
RESULTS
Enrichment of genomic DNA from tumors and their adjacent PanINs
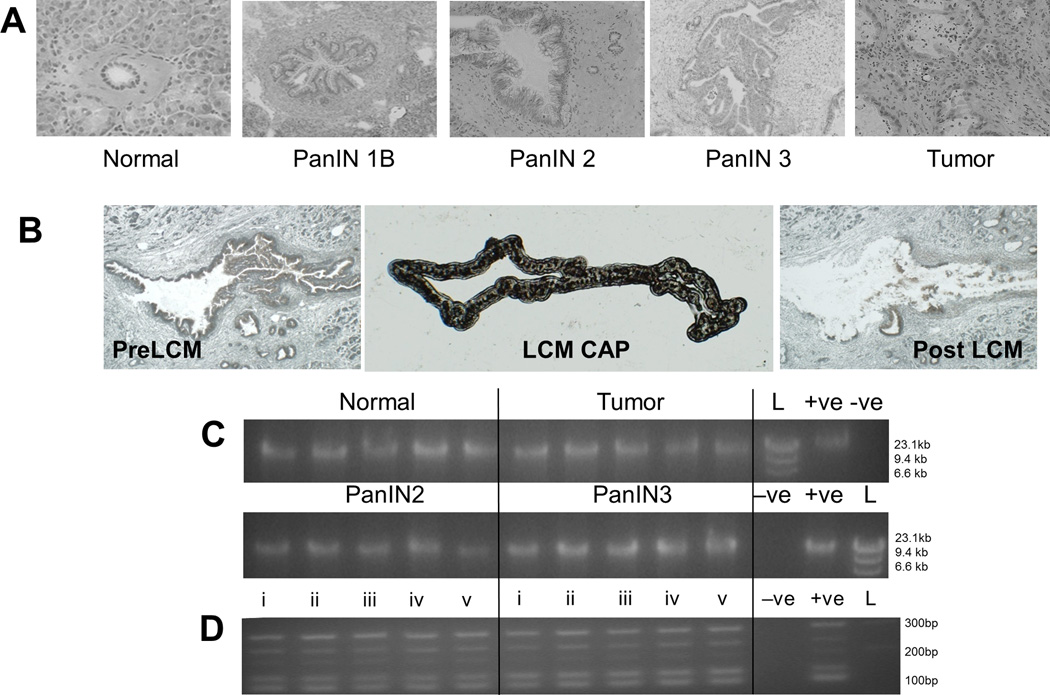
Evaluation of frozen pancreatic tumor tissue by a study pathologist identified specimens containing adjacent PanIN2 and/or PanIN3 lesions (Figure 1A). Epithelial cells from these lesions were purified by LCM (Figure 1B). The heterogeneity in shape and size of PanINs resulted in substantial variation in the number of cells collected. In general the number of cells constituting a PanIN lesion increased with grade and ranged from 20 to over 100 cells per 10µm section. To obtain sufficient genomic DNA for exome sequencing, a direct whole genome amplification (WGA) protocol was used, where laser captured cellular material was lysed on the cap membrane and DNA amplified directly with no intermediate DNA extraction10. This direct WGA method was applied to all tumor and PanIN tissues, yielding reproducible and consistent qualities of WGA DNA (Figure 1C). Minimal amplification bias was observed using a qualitative multiplex PCR assay (Figure 1D), indicating that the WGA DNA from small PanIN lesions was of sufficient quality for exome capture and sequencing. Germline DNA derived from peripheral blood mononuclear cells (PBMC) from the same pancreatic cancer cases was not amplified for this study.
Figure 1. LCM of PanIN and tumor yields high quality DNA after WGA.
(A) Representative examples of PanIN and tumor pathology. (B) LCM of ductal epithelial cells from PDAC. (C) Reproducibility of in situ WGA. Five replicate amplifications from normal, PanINs, and Tumor LCM samples yielded equivalent size and quantities of DNA, compared to non-LCM positive controls (+ve). −ve, PBS Negative control; L, size marker. (D) Multiplex PCR of five targets from five chromosomes. Five replicate WGA (i–v) from representative normal (left panel) and tumor (right panel) samples are shown.
Exome capture and sequence analysis
DNA from tumors, adjacent PanINs and normals from 10 PDAC cases were exome captured and sequenced. On average over 80% of baits yielded more than 20X sequence depth and more than 70% had 40X coverage. Variation in coverage did not correlate with the number of LCM purified cells and read duplication rate was only moderately higher for WGA material (~20% vs ~15%, data not shown). In order to evaluate dropout due to non-linear WGA the mean alternate allele frequency (AAF) for each sample, relative to corresponding non-amplified DNA from blood, was calculated using all heterozygous germline variants with >50 sequence reads (Supplementary Figure 1). WGA DNA from PanINs, tumors and non-WGA blood DNA all showed mean AAF of 46%, consistent with limited allele dropout. Based on these results, this direct in situ WGA methodology allows for comprehensive genomic interrogation of lesions with limiting cell numbers.
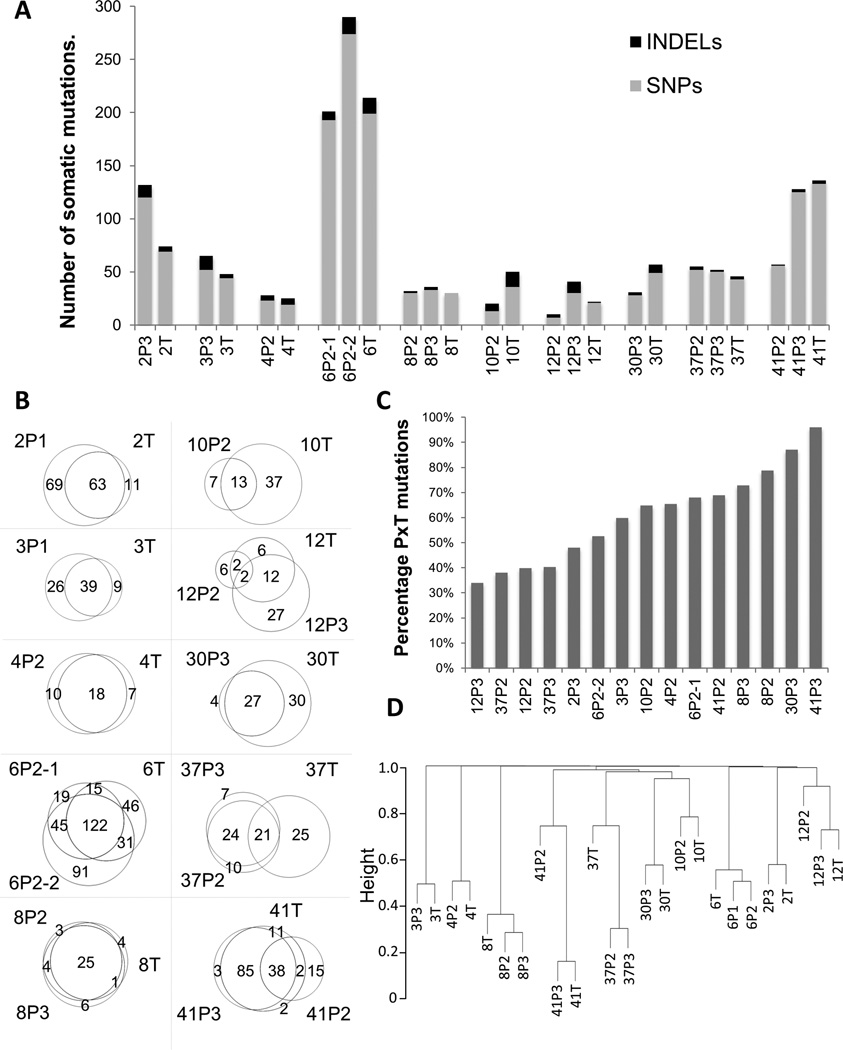
In total, 1053 somatic mutations altering protein-coding sequences were detected in 10 tumors and 15 adjacent PanIN specimens. These included 845 non-synonymous, 121 frame shifts, 51 nonsense, and 36 others. In total,937 genes contained at least one variant, many of which were reported previously mutated in cancer (COSMIC cancer database). Figure 2A shows the total numbers of single nucleotide variants (SNVs) and insertion or deletion (INDEL) variants for each case. Case 6 presented the most mutations in this study, with over 200 mutations present in each PanIN2 and the associated tumor. A total of 902 high confidence somatic SNVs were detected in the 25 samples. An average of 69.2 somatic SNVs was observed per sample, (48.3 omitting the outlier case 6) in line with previous studies in pancreatic cancer17. Excluding case 6 again, a trend towards fewer alterations in early stage disease was observed, with an average of 30.2 mutations per PanIN2 compared to 49.3 in tumors. Conversely, late grade PanIN3s presented increased numbers of mutations (62.6) compared to tumors. All somatic mutations from each case are shown in Supplementary Table 1. A total of 151 INDELs were observed within the 25 samples, for an average of six INDELs per specimen (Figure 2A, Supplementary Table 2).
Figure 2. Numbers of somatic mutations and relatedness of tumors and adjacent PanINs.
(A) Numbers of somatic mutations (gray) and indels (black) per sample (grouped by case). P2, PanIN2; P3, PanIN3; T, tumor. (B) Percentage commonality of PanINs with associated tumors. (C) Venn diagrams of somatic variants in each case. (D) Hierarchical clustering of tumors and PanINs using Euclidean distance measures for each possible comparison.
Commonality of tumors and adjacent PanINs
Of the somatic mutations detected, approximately 66% were common to tumors and adjacent PanINs (PxT) (Figure 2B). Mutations found only in tumor or only in PanIN totaled 10% and 24% of all variants, respectively. Commonality between PanINs and tumors was assessed for each PanIN by calculating the percentage of variants also present in the adjacent tumor (Figure 2C). While overlap between PanINs and associated tumor ranged from 34% to 96%, greater than 50% commonality with tumor was observed for ten of the 15 sequenced PanINs. The PanIN3s of cases 41 (41P3) and 30 (30P3) demonstrated the highest commonality with adjacent tumors, with 96% and 87% overlap, respectively. The PanIN2/3 pairs of cases 12 and 37 displayed the least commonality with just 34 to 40% homology with adjacent tumors. Conversely, PanIN 37P3 had much greater commonality with the adjacent 37P2 (77%), than either had with the associated tumor (Figure 2B). Separately, lineage was assessed using hierarchical clustering (Figure 2D). PanINs and tumors from the same cases were most closely related overall. However, distal branching of the 37T and 41P2 specimens from adjacent PanINs and tumors was observed fitting with the differential commonality shown in Figure 2B. Overall, these results suggest that all of the adjacent lesions arose from common ancestral lineages and that the majority of somatic mutations in tumors arose early in the progression at the PanIN2 stage or earlier.
KRAS and TP53 somatic mutations
As expected, KRAS and TP53 were the most commonly mutated genes, observed in nine and seven cases, respectively, with 60% of the cases containing mutations in both genes. Almost all PanINs (13 of 15) and tumors (9 of 10) presented with G12 KRAS mutations, which result in constitutively active forms of the GTPase18. Table 1 displays the algorithmic read depths and alternative allele frequencies at G12 for all samples studied. KRAS G12 mutations were initially detected in 16 of 25 samples (bold text) using our conservative algorithmic settings. However, visualization of all reads at the G12 position identified six additional low frequency KRAS mutations, which were all confirmed by Sanger sequencing using locked nucleic acids (LNA) to assure good analytical sensitivity (data not shown). Six cases had the relevant KRAS mutation in all samples, confirming KRAS mutations early in progression. G12V and G12D mutations were most abundant, observed in six and five cases respectively. Multiple different G12 mutations were observed within three cases (12, 30 and 37) and a KRAS Q61H mutation was also identified in the 6P2-2 of case 6. Three different G12 mutations were observed in case 37, consistent with the relatively low clonality between these lesions, and predicting early divergence of the tumor and adjacent PanINs prior to KRAS mutagenesis (Figure 2). In addition, the presence of two independent KRAS mutations in 37P2, 12P2 and 6P2-2 suggests heterogeneity exists within the premalignant lesions.
Table 1.
KRAS and TP53 Mutations.
| KRAS |
TP53 |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| chr12:25398284C>A | chr12:25398284C>T | chr12:25398285C>G | chr12:25398285C>A | chr12:25380275T>G | chr17:7574020TC>T | chr17:7577022G>A | chr17:7577094G>C | chr17:7577559G>A | chr17:7578406C>T | chr17:7579406G>T | ||
| G12 Read Depth | Alternative
Allele Reads |
|||||||||||
| G12V | G12D | G12R | G12C | Q61H | E204fs | R174* | R150P | S109F | R136H | S55* | ||
| 2P3 | 33 | 9 | 43,0 | 13,0 | 66,0 | 37,0 | 80,0 | 3,14 | 32,0 | |||
| 2T | 28 | 11 | 39,0 | 27,0 | 81,0 | 55,0 | 86,0 | 14,7 | 29,0 | |||
| 3P3 | 38 | 3 | 36,0 | 23,0 | 84,0 | 52,0 | 85,0 | 23,0 | 36,0 | |||
| 3T | 29 | 4 | 30,0 | 16,0 | 60,0 | 35,0 | 80,0 | 25,0 | 26,0 | |||
| 4P2 | 38 | 57,0 | 16,0 | 71,0 | 41,0 | 78,0 | 7,14 | 26,0 | ||||
| 4T | 11 | 29,0 | 19,0 | 76,0 | 46,0 | 88,0 | 9,7 | 35,0 | ||||
| 6P2-2 | 43 | 3 | 40,13 | 24,0 | 78,0 | 60,0 | 87,0 | 25,0 | 34,0 | |||
| 6P2-1 | 38 | 3 | 45,1 | 16,0 | 71,0 | 46,0 | 81,0 | 24,0 | 30,0 | |||
| 6T | 29 | 7 | 51,0 | 18,0 | 72,0 | 51,0 | 23,50 | 14,0 | 31,0 | |||
| 8P2 | 54 | 14 | 82,0 | 33,0 | 65,0 | 9,20 | 42,0 | 28,0 | 98,0 | |||
| 8P3 | 54 | 8 | 52,0 | 24,0 | 62,0 | 19,17 | 31,0 | 30,0 | 102,0 | |||
| 8T | 34 | 4 | 61,0 | 35,0 | 88,0 | 34,13 | 43,0 | 43,0 | 137,0 | |||
| 10P2 | 18 | 3 | 38,0 | 19,0 | 56,0 | 26,0 | 23,0 | 16,0 | 56,0 | |||
| 10T | 13 | 19,0 | 20,0 | 48,0 | 21,0 | 21,0 | 12,0 | 59,0 | ||||
| 12P2 | 40 | 4 | 5 | 42,0 | 21,0 | 56,0 | 28,0 | 3,0 | 25,0 | 79,1 | ||
| 12P3 | 55 | 19 | 54,0 | 10,0 | 26,0 | 12,0 | 5,0 | 8,0 | 47,0 | |||
| 12T | 52 | 8 | 57,0 | 17,0 | 64,0 | 49,0 | 30,0 | 31,0 | 77,17 | |||
| 30P3 | 18 | 2 | 22,0 | 13,0 | 38,0 | 24,0 | 20,0 | 20,0 | 30,0 | |||
| 30T | 17 | 3 | 29,0 | 5,6 | 22,0 | 7,0 | 24,0 | 16,0 | 30,0 | |||
| 37P2 | 40 | 12 | 7 | 42,0 | 19,0 | 80,0 | 36,0 | 34,0 | 27,0 | 80,0 | ||
| 37P3 | 39 | 15 | 40,0 | 10,0 | 62,0 | 24,0 | 28,0 | 23,0 | 88,0 | |||
| 37T | 8 | 5 | 20,0 | 4,0 | 22,32 | 30,0 | 11,0 | 8,0 | 23,0 | |||
| 41P2 | 27 | 9 | 1 | 46,0 | 27,0 | 88,0 | 52,0 | 39,1 | 42,1 | 120,1 | ||
| 41P3 | 27 | 10 | 56,0 | 24,0 | 60,0 | 35,0 | 31,0 | 13,0 | 81,0 | |||
| 41T | 28 | 3 | 48,0 | 46,0 | 122,0 | 50,0 | 55,0 | 62,0 | 136,0 | |||
Positions listed as Chrom:positionRef>Alt Comma separated numbers: Reference Allele, Alternative Allele Reads T: tumor; P2: PanIN2; P3: PanIN3; Indel: insertion or deletion
In the seven tumors with TP53 mutations, six displayed SNVs and one (40T) contained a 1bp deletion (Table 1). Sanger sequencing positively confirmed all seven mutations. TP53 mutations were observed only in the tumors from four of the seven cases (Table 2), but were present in both tumor and adjacent PanINs (two PanIN2s and two PanIN3s) from the three other cases. The presence of TP53 mutations in PanIN2 lesions, especially 4P2 and 8P2, which are highly clonal with the associated tumors, suggests that TP53 mutations may occur early in progression to tumor at relatively high frequency. Case 4 was the only case for which no KRAS mutation was detected, but a damaging missense R175H TP53 mutation was observed, which was also present in case 2. Two unique missense mutations (R282G and S241F) and two nonsense mutations (R306X and S945X) were also identified, each of which is associated with TP53 inactivation (www-p53.iarc.fr).
Table 2.
Commonly mutated genes.
| Tissue & | Gene | No. Cases | Impact $ | No. Exons | Amino acid + (Exon) | COSMIC # |
|---|---|---|---|---|---|---|
| T | SMAD4 | 2 | D/D | 12 | R361H(9) A452fs(11) | 417/9350 |
| PTPN5 | 2 | D/D | 15 | T125M(6) R438fs(13) | 27/4418 | |
| T, PxT | TP53 | 7 | all D | 12 | Table 1 | 22871/77279 |
| ATM | 2 | D/T | 63 | S1004N(20) C2801Y(57) | 384/8337 | |
| CTCF | 2 | D/D | 12 | R187C(4) E291G(9) | 71/4425 | |
| MAST4 | 2 | D/D | 29 | D626Y(19) E763V(22) | 9/4860 | |
| RNF43 | 2 | D/D | 10 | D140E(4) A169T(5) | 56/4240 | |
| C9orf174 | 2 | D/T | 51 | R1324W(42) S1520R(47) | 16/4148 | |
| NEB | 2 | D/T | 182 | E3400Q(74) H4431R(117) | 201/4283 | |
| RPGR | 2 | D/n | 19 | D668Y(15) Q1104K(16) | 24/4265 | |
| PxT | KRAS | 9 | all D | 6 | Table 1 | 24608/114312 |
| MUC16 | 4 | D/n/n/n | 84 | P6997R(3) P7895T(3) S11132G(5) Q13573K(56) | 392/4696 | |
| OTOF | 3 | D/T/T | 47 | P490fs(14) M607T(16) M607T(16) E1323K(32) | 99/4381 | |
| PABPC1 | 3 | D/D/T | 15 | D165G(3) K231E(5) E345*(8) | 32/4216 | |
| RBMX | 3 | D/D/T | 9 | A78T(4) P167A(5) G105fs(4) | 30/4265 | |
| SPTA1 | 3 | D/T/T | 52 | R468H(11) E846D(18) I2265T(49) | 232/4287 | |
| C15orf39 | 2 | D/D | 3 | G411fs(2) D575fs(2) | 16/4238 | |
| OSBPL9 | 2 | D/D | 23 | V51D(2) F233C(14) | 17/4216 | |
| VAV3 | 2 | D/D | 27 | E95K(2) G639fs(21) | 53/5465 | |
| GPX5 | 2 | D/T | 5 | A77V(2) T203M(5) | 15/4238 | |
| HLA-DRB1 | 2 | D/T | 6 | K118Q(3) S124A(3) | 9/4148 | |
| KIF4B | 2 | D/T | 1 | R742*(1) R762H(1) | 52/4248 | |
| KLHDC3 | 2 | D/T | 11 | F49C(2) N357T(11) | 7/4238 | |
| SH3RF1 | 2 | D/T | 12 | M518I(9) RP827S(11) | 30/4718 | |
| TCF7L2 | 2 | D/T | 13 | E344*(11) K385R(13) | 38/4734 | |
| SCLT1 | 2 | D/n | 21 | R417*(15) Splice Site(2) | 28/4671 | |
| SKA3 | 2 | D/n | 13 | K386R(8) Splice Site(7) | 19/4239 | |
| PxT, P | FCGBP | 4 | D/D/D/T | 36 | E390*(2) A1108T(6) S4284*(24) K3848E(28) | 139/4242 |
| ATP8B1 | 2 | D/D | 28 | D554E(16) D554E(16) | 50/4922 | |
| FN1 | 2 | D/D | 46 | E888V(18) R1207G(24) | 94/4730 | |
| LZTS1 | 2 | D/D | 3 | R36W(1) L113P(1) | 26/4238 | |
| OBSCN | 2 | D/D | 81 | C1188F(12) R4593C(52) | 184/4531 | |
| PAK2 | 2 | D/D | 15 | Q101H(4) K128R(4) | 12/5091 | |
| PGAP1 | 2 | D/D | 27 | F565C(18) I606N(20) | 37/4238 | |
| RBFOX1 | 2 | D/D | 14 | S27L(3) T118M(4) | 53/4238 | |
| APOB | 2 | D/T | 29 | N886H(18) V4227L(29) | 220/4306 | |
| GABRA5 | 2 | D/T | 11 | R221T(8) T412I(11) | 47/4238 | |
| KCNJ12 | 2 | D/T | 3 | (R261H(3) I262S(3) L211F(3)) E289Q(3) | 59/4239 | |
| LRP2 | 2 | D/T | 79 | L3587V(55) F4300I(70) | 237/4312 | |
| MST1 | 2 | D/T | 18 | P19S(1) T104S(3) | 33/4426 | |
| RYR2 | 2 | D/T | 105 | R389C(13) D3203E(68) | 352/4284 |
Tissue distribution of mutated genes; (T) tumors only, (PxT, T) PanINs and tumors of the same case and only tumors of additional cases, (PxT) PanINs and Tumors of the same case, (P, PxT) PanINs and tumors of the same case or only PanINs of additional cases, (P) only in PanINs. “Gene” refers to genes selected based on somatic mutation in two or more cases, predicted damaging (D) in one mutation, and listing in the COSMIC cancer database.
Impact: summary of SIFT/POLYPHEN analysis D = Predicted Damaging T = Predicted Tolerated.
Amino Acid alterations plus exon involved indicated in parenthesis:
= new stop site and fs = frame shift.
The frequencies of observed mutations in COSMIC for any cancer type (COSMIC all Cancers).
Commonly mutated genes
Sixty-seven genes were mutated in two or more cases (Supplementary Table 1). The large mucosal membrane protein genes; MUC16 and FCGBP were each recurrent in four cases, while OTOF, PABPC1, RBMX and SPTA1 were each mutated in three cases. PABPC1 and RBMX are involved in mRNA regulation, and SPTA1 directs cell shape and axon guidance. None have been previously linked with pancreatic cancer. An additional 58 genes were mutated in two cases. Table 2 lists 42 recurrently mutated genes previously implicated in cancer (COSMIC database) that had at least one predicted damaging mutation in the 25 samples. Only two of the commonly mutated genes (SMAD4 and PTPN5) were altered exclusively in tumors. While, SMAD4 has been heavily implicated late in pancreatic cancer development17, the protein tyrosine phosphatase, PTPN519, has not been linked previously to pancreatic cancer. The majority of the recurrently mutated genes carried PxT variants (common to tumor and PanIN of the same case). Seven recurrently altered genes were mutated in single tumors and also in adjacent tumor and PanIN combinations (T,PxT), but never in PanINs alone (P) (Table 2). ATM, which regulates the cellular response to DNA damage and has recently been implicated as a pancreatic cancer predisposition gene20, was mutated in two cases, both of which were validated by Sanger sequencing. Since the cases with ATM alterations did not contain any of the seven TP53 mutations, nine of the 10 tumors in this study contained aberrations in DNA damage response pathways. The CTCF transcriptional repressor/chromatin binding factor and the RNF43 E3-ubiquitin-protein ligase regulator of the WNT signaling pathway that has been linked to cell growth promotion and cancer21, were also in this group. PxT mutated genes included two Rho family regulators VAV3 and SH3RF1, the TCF7L2 WNT signaling factor, the oxidative damage protection gene GPX5 and three chromosomal structural regulating genes, KIF4B, KLHDC3 and SKA3. Additional PxT:P mutated genes included the LZTS1 tumor suppressor and the PAK2 Ser/Thr Kinase, as well as the structural regulatory proteins Fibronectin (FN1) and Obscurin (OBSCN), which have also been linked to cancer progression22,23.
Common driver genes in cancer
Mutations that were retained in progression from PanIN to tumor are likely enriched for alterations that drive tumor formation, whereas mutations specific to tumor are more likely drivers of progression. Table 3 lists the genes found to contain high confidence mutations in this study that are commonly mutated in cancer and listed in the COSMIC Gene Census database as cancer drivers. In addition to SMAD4, seven other known driver genes were mutated only in tumor samples including the transcription factors MYCL1, PAX8 and TRIP1124,25,26, the tyrosine receptor kinase NTRK3 and the mixed-lineage leukemia family member MLL2, previously linked with pancreatic cancer27. Nine PxT mutated genes were observed in addition to KRAS, including four transcription factors, DAXX, NCOA1, TCF7L2 and ZNF521. Of these genes TCF7L2 is associated with Wnt signaling and an increased risk of type 2 diabetes, and increased expression has been reported in pancreatic cancer28. In contrast, mutations in the established GNAS and CHEK2 cancer drivers29,30 were detected in PanIN lesions but not in adjacent tumors, highlighting independent progression of the PanINs and the tumors. Additional mutations involved the DNA replication repair gene BLM and the tumor suppressor CCDC6. Sanger sequencing was used to further validate these genes (Table 3). In addition to the multiple KRAS and TP53 mutations, alterations in MLL2, PAX8, TRIP11, LCP1, MALT1, GNAS and two each in ATM and SMAD4 faithfully validated. However, mutations in MLL3, CHEK2 and NSD1 failed to validate. The limited sensitivity of Sanger sequencing for low alternative allele frequencies resulting from heterogeneity in these lesions could explain the failure of these events to validate.
Table 3.
Common driver genes in cancer.
| Gene | No. Cases | Case | Tissue | Chr. | Position | Mutation # | Sangervalidation $ | Amino acid | Impact | Ref Allele & | Allele 1 & | Allele 2 & | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | GOLGA5 | 1 | 2 | T | 14 | 93276655 | T - C | / | F350S | D | T 22 | T 22 | C 8 |
| MLL2 | 1 | 37 | T | 12 | 49433874 | TG - T | YES | H2560fs | D | TG 7 | TG 2 | T 6 | |
| MYCL1 | 1 | 8 | T | 1 | 40363341 | C - A | / | Q266H | T | C 120 | C 88 | A 31 | |
| NTRK3 | 1 | 6 | T | 15 | 88476311 | G - T | / | C599* | D | G 80 | G 47 | T 16 | |
| PAX8 | 1 | 10 | T | 2 | 114002179 | G - A | YES | R72W | D | G 197 | G 32 | A 11 | |
| SLC45A3 | 1 | 4 | T | 1 | 205628369 | GAG - G | / | Y551fs | D | GAG 16 | GAG 2 | G 4 | |
| SMAD4 | 2 | 37 | T | 18 | 48591919 | G - A | YES | R361H | D | G 31 | G 16 | A 12 | |
| 10 | T | 18 | 48603054 | C - CT | YES | A452fs | D | C 14 | C 8 | CT 6 | |||
| TRIP11 | 1 | 30 | T | 14 | 92470155 | C - T | YES | E1389K | D | C 110 | C 45 | T 15 | |
| PxT, T | ATM | 2 | 10 | T | 11 | 108214082 | G - A | YES | C2801Y | D | G 49 | G 33 | A 14 |
| 41 | PxT | 12 | 108142067 | G - A | YES | S1004N | T | G 146 | G 90 | A 44 | |||
| TP53 | 7 | Table 1 | 17 | YES | |||||||||
| PxT | DAXX | 1 | 6 | PxT | 6 | 33288845 | A - T | / | F161Y | D | A 11 | A 10 | T 8 |
| KRAS | 9 | Table 1 | 12 | YES | D | ||||||||
| LCP1 | 1 | 2 | PxT | 13 | 46730643 | G - A | YES | R141W | D | G 72 | G 60 | A 26 | |
| MALT1 | 1 | 3 | PxT | 18 | 56376751 | A - G | YES | E264G | T | A 77 | A19 | G 20 | |
| MLL3 | 1 | 2 | PxT | 7 | 151860002 | T - TA | ND | F3553fs | D | T 53 | T 8 | TA 15 | |
| NCOA1 | 1 | 6 | PxT | 2 | 24930303 | A - C | / | K655T | D | A 32 | A 23 | C 6 | |
| NUP98 | 1 | 6 | PxT | 11 | 3727776 | A - T | / | L942M | T | A 51 | A 35 | T 14 | |
| PDE4DIP | 1 | 4 | PxT | 1 | 144952220 | C - T | / | A135T | T | C 18 | C 9 | T 15 | |
| TCF7L2 | 2 | 6 | PxT | 10 | 114925320 | A - G | / | K385R | T | A 37 | A 11 | G 10 | |
| 2 | PxT | 10 | 114912131 | G - T | / | E344* | D | G 31 | G 60 | T 18 | |||
| ZNF521 | 1 | 6 | PxT | 18 | 22804449 | C - T | / | V1145I | D | C 89 | C 64 | T 26 | |
| P | BLM | 1 | 6 | P | 15 | 91358439 | A - T | / | K1395I | D | A 32 | A 18 | T 11 |
| CCDC6 | 1 | 37 | P | 10 | 61572516 | G - A | / | P242S | D | G 37 | G 53 | A 37 | |
| CHEK2 | 1 | 2 | P | 22 | 29121277 | G - T | ND | T133K | T | G 41 | G 53 | A 24 | |
| CLTCL1 | 1 | 2 | P | 22 | 19209058 | C - A | / | A880S | D | C 43 | C 69 | A 27 | |
| GNAS | 1 | 41 | P | 20 | 57484421 | G - A | YES | R186H | D | G 32 | G 57 | A 23 | |
| KDM6A | 2 | 37 | P | X | 44922970 | C - T | / | Q611* | D | C 9 | C 6 | T 7 | |
| 6 | P | X | 44910988 | AA - A | / | Q230fs | D | AA | AA 3 | A 6 | |||
| NACA | 1 | 12 | P | 12 | 57114403 | GA - G | / | S304fs | D | GA 46 | GA 10 | G 19 | |
| NSD1 | 1 | 2 | P | 5 | 176719127 | C - T | ND | A1875V | D | C 47 | C 49 | T 14 | |
Columns: Tissue, No. Cases (number of cases with mutations in this gene), Case (the pancreatic cancer case in This Study), Amino Acid, Chr. (Chromosome), Position (nucleotide), Impact (SIFT/Polyphen). “Gene” refers to genes selected based on listing in the COSMIC census database of common driver genes.
Mutation: reference – mutated nucleotide;
Sanger Validation: YES (mutations confirmed by Sanger sequencing), ND (not detected), / (not evaluated).
Allelic nucleotide and read depth for the Reference normal (Ref Allele) and the alternative alleles for the PanIN and Tumor tissues (Allele 1, Allele 2)
Commonly mutated genes in pancreatic cancer
A comparison of the 937 genes with mutations in this study with genes frequently mutated in pancreatic cancer from two other pancreatic exome sequencing studies17,31 identified 40 genes in common (Table 4). Mutations in the key pancreatic cancer driver genes KRAS, TP53, SMAD4 and ATM were observed in the three studies. However, apart from KRAS and TP53, these mutations were not identical. The majority of the recurrently mutated genes contained PxT mutations, suggesting that many of these commonly mutated genes may be early targets of mutation during tumor development. Of these, the potential tumor suppressor gene CSMD1, linked with aggressive carcinomas32, had not been previously linked to pancreatic cancer. Two unrelated motor protein genes, DNAH8 and MYO1E, were mutated only in individual tumors in this study and also observed in two cases each in the previous studies. While no link to pancreatic cancer is reported, MYO1E has been linked to TP53-associated DNA damage response33. A number of common mutated genes, including the Mucin (MUC) and RYR family’s members, are more likely passenger events due to the large size of the associated genes, rather than common driver events in pancreatic cancer. A further six genes in Table 4 were mutated solely in PanINs. This group may be enriched for passenger genes that do not contribute to tumor development or progression. However, no absolute conclusions can be drawn on the basis of a single case. For instance, NAV3 is a putative tumor suppressor previously linked to pancreatic and other cancers34.
Table 4.
Commonly mutated genes in pancreatic cancer.
| Gene | This Study $ | ICGC (99) $ | Jones (24) $ | Total | Common Site |
Case | Tissue | Chr. | Position | Amino acid | Impact | Allele 1 | Allele 2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | SMAD4 | 2 | 16 | 8 | 26 | n | 37 | T | 18 | 48591919 | R361H | D | G 16 | A 12 |
| n | 10 | T | 18 | 48603054 | A452fs | D | C 6 | CT 8 | ||||||
| DNAH8 | 1 | 1 | 1 | 3 | n | 10 | T | 6 | 38723768 | I476M | D | A 67 | G 26 | |
| MYO1E | 1 | 2 | 0 | 3 | n | 37 | T | 15 | 59564510 | I48V | D | T 60 | C 19 | |
| NTRK3 | 1 | 1 | 1 | 3 | n | 6 | T | 15 | 88476311 | C599* | D | G 47 | T 16 | |
| T, PxT | TP53 | 7 | 33 | 18 | 58 | y | Table 1 | |||||||
| ATM | 2 | 5 | 0 | 7 | n | 10 | T | 11 | 108214082 | C2801Y | D | G33 | A 14 | |
| n | 41 | PxT | 11 | 108142067 | S1004N | T | G 90 | A 44 | ||||||
| NEB | 2 | 5 | 1 | 8 | n | 41 | PxT | 2 | 152474902 | E3400Q | D | C 143 | G 107 | |
| n | 37 | T | 2 | 152421598 | H4431R | T | T 103 | C 32 | ||||||
| PxT | SLITRK3 | 1 | 1 | 1 | 3 | n | 6 | PxT | 3 | 164906758 | G621* | D | C 20 | A 10 |
| ATP10A | 1 | 1 | 2 | 4 | n | 2 | PxT | 15 | 25959351 | T605M | D | G 18 | A 16 | |
| CDH4 | 1 | 2 | 0 | 3 | n | 6 | PxT | 20 | 60499411 | P476S | D | C 24 | T 8 | |
| CSMD1 | 1 | 5 | 1 | 7 | n | 2 | PxT | 8 | 4494898 | D90N | D | C 46 | T 26 | |
| DPP6 | 1 | 1 | 3 | 5 | n | 8 | PxT | 7 | 154593134 | R393* | D | C 31 | T 32 | |
| FNIP1 | 1 | 1 | 1 | 3 | n | 6 | PxT | 5 | 131008104 | L650S | D | A 39 | G 13 | |
| HMCN1 | 1 | 4 | 0 | 5 | n | 41 | PxT | 1 | 186063449 | C3413F | D | G26 | T 56 | |
| KRAS | 9 | 94 | 24 | 118 | y | Table 1 | ||||||||
| LRFN5 | 1 | 0 | 2 | 3 | n | 2 | PxT | 14 | 42355997 | R57W | D | C 26 | T 18 | |
| MLL3 | 1 | 6 | 4 | 11 | n | 2 | PxT | 7 | 151860002 | F3553fs | D | T 11 | TA 18 | |
| MUC16 | 4 | 5 | 2 | 11 | n | 12 | PxT | 19 | 8993006 | Splice Site | N | C 21 | A 28 | |
| n | 12 | P | 19 | 9066456 | P6997R | N | G 110 | C 110 | ||||||
| n | 6 | PxT | 19 | 9063763 | P7895T | N | G 34 | T 13 | ||||||
| n | 2 | PxT | 19 | 8999458 | Q13573K | D | G 62 | T 10 | ||||||
| n | 41 | PxT | 19 | 9048237 | S11132G | N | T 164 | C 86 | ||||||
| ODZ4 | 1 | 1 | 1 | 3 | n | 41 | PxT | 11 | 78381167 | G2075R | D | C 158 | G 70 | |
| PKHD1L1 | 1 | 2 | 0 | 3 | n | 2 | PxT | 8 | 110454274 | N1415H | D | A 37 | C 8 | |
| RYR1 | 1 | 1 | 1 | 3 | n | 41 | PxT | 19 | 38991484 | M2490V | T | A 18 | G 4 | |
| RYR2 | 2 | 2 | 2 | 6 | n | 6 | PxT | 1 | 237870277 | D3203E | T | T 6 | A 9 | |
| n | 37 | P | 1 | 237604778 | R389C | D | C 161 | T45 | ||||||
| RYR3 | 1 | 4 | 0 | 5 | n | 10 | T | 15 | 34145258 | N4542K | T | T 98 | A 34 | |
| SCN2A | 1 | 1 | 1 | 3 | n | 41 | PxT | 2 | 166187962 | D758H | D | G 92 | C 54 | |
| SPATA17 | 1 | 1 | 1 | 3 | n | 41 | PxT | 1 | 217975170 | P328L | D | C 35 | T 33 | |
| SYCE1 | 1 | 3 | 1 | 5 | n | 2 | PxT | 10 | 135368531 | P341S | D | G48 | A 24 | |
| USP34 | 1 | 2 | 0 | 3 | n | 6 | PxT | 2 | 61622085 | K219T | D | T 12 | G 9 | |
| PxT, P | APOB | 2 | 1 | 1 | 4 | n | 2 | P | 2 | 21225615 | V4227L | T | C 24 | A 14 |
| n | 6 | PxT | 2 | 21245863 | N886H | D | T 22 | G 19 | ||||||
| FN1 | 2 | 1 | 1 | 4 | n | 4 | PxT | 2 | 216259428 | R1207G | D | T 36 | C 19 | |
| n | 2 | P | 2 | 216271900 | E888V | D | T70 | A 25 | ||||||
| OBSCN | 2 | 2 | 1 | 5 | n | 37 | P | 1 | 228433195 | C1188F | D | G 104 | T 20 | |
| n | 4 | PxT | 1 | 228505380 | R4593C | D | C 42 | T 42 | ||||||
| P | CDH23 | 1 | 2 | 0 | 3 | n | 37 | P | 10 | 73553299 | P2205L | D | C 77 | T 32 |
| GPR133 | 1 | 1 | 2 | 4 | n | 8 | P | 12 | 131456051 | K79M | D | A 50 | T 35 | |
| LAMA1 | 1 | 1 | 1 | 3 | n | 12 | P | 18 | 7042183 | K407fs | D | CAG 14 | C 7 | |
| NAV3 | 1 | 1 | 1 | 3 | n | 41 | P | 12 | 78594300 | V2233L | D | G 97 | T 35 | |
| NBAS | 1 | 3 | 0 | 4 | n | 6 | P | 2 | 15613348 | L575M | D | A 21 | T 10 | |
| PCDHGA1 | 1 | 1 | 1 | 3 | n | 2 | P | 5 | 140711164 | E305* | D | G 15 | T 32 | |
Columns: Tissue, Amino Acid, Chr. (Chromosome), Position (nucleotide), Impact (SIFT/Polyphen), Allele 1 and Allele 2 (nucleotide and read depth) as presented as in Table 3; “Genes” refers to genes commonly mutated in This Study, (ICGC) Biankin et al. (24) and Jones et al. (11).
The number of cases mutated in each study is shown;. “Common site” refers to identical mutations in the studies; “Case” refers to the specific case in This Study with a mutation in the specific gene.
Pathway Analysis
To identify commonly mutated pathways involved in development of pancreatic cancer, genes with mutations occurring in PxT or tumor only were annotated by pathway (Supplementary Table 3). The most significant pathways are summarized in Supplementary Table 4. NCAM, Insulin and PDGF signaling pathways were implicated in the early development of pancreatic cancer in part because of the involvement of KRAS mutations. Gap junction signaling, chemokine signaling and regulation of the actin cytoskeleton were altered predominantly by PxT mutations in each of the ten cases. Similarly, MAPK signaling and cell cycle regulation pathways were modified in all cases, but equal numbers of PxT and T mutations were detected. The recently highlighted Axon guidance pathway in the ICGC study31 was also observed early in two separate pathway sets (Kegg and Reactome). PxT mutations were observed in 17 genes in the neuroactive ligand receptor-interaction pathway in 8 cases and in 12 independent genes in olfactory signaling in 7 cases. Mechanistically, genes in these pathways are major components of the superfamily of rhodopsin-like G protein-coupled receptors (GPCRs), which can induce cascades of responses related to carcinogenesis35. Due to the limited number of genes mutated only in tumors, few pathways were associated only with tumors. However, mutations unique to tumors were enriched in genes involved in Wnt and TGF-beta signaling, consistent with the loss of response to these ligands late in tumor progression36.
DISCUSSION
A detailed understanding of the genes and signaling pathways that influence pancreatic cancer onset and progression is required to improve early diagnosis, prevention and therapy. However, identification of the important driver events has proven difficult due to the heterogeneous nature of somatic mutations and accumulation of many passenger mutations. Recent whole exome studies have verified the prominent involvement of KRAS mutations, the late presentation of TP53 mutations and the frequent involvement of SMAD4 in pancreatic tumor progression17,31. While important, these studies were limited by the use of bulk extracted or macrodissected tumor tissues for genomic interrogation. Given the low cellularity of these tumors, this can lead to inclusion of adjacent non-tumor tissues and subsequent incorporation of large numbers of passenger mutations in studies. In addition, because these studies focused on advanced stage tumors, little information about genetic alterations involved in progression from premalignant lesions to invasive tumors was obtained.
In this study we have addressed these issues by isolating pure populations of cells using LCM and by focusing on PanINs and adjacent tumors. Robust methodology was used to reproducibly amplify, exome capture and sequence DNA from small numbers of purified cells. While WGA can introduce mutations and sequence bias resulting in loss of certain genomic regions, we show here that the in situ WGA technique can result in representative somatic mutation profiles. Specifically, 80% of the exome was consistently captured to sufficient sequence depth from as few as 100 cells. Allelic drop out following WGA was also shown to be limited based on comparable mean AAF of germline SNPs in amplified DNA and related unamplified DNA from blood (Supplementary Figure 1). LCM guided by pathology review was expected to reduce normal cell contamination, increasing cellularity. In an attempt to calculate tumor cellularity for each sample we evaluated the mean AAF for all heterogeneous somatic variants with >50X read-depth. However, the level of cellularity predicted using this model varied substantially across samples, ranging from 25-to-72% (data not shown), and was generally much lower than expected for defined PanIN lesions (Fig. 1A). Further analysis showed the mean AAF was highly correlated with commonality, suggesting that this measure more likely represents heterogeneity within each sample, consistent with the existence of multiple KRAS mutations in certain samples (Table 1). Thus, the inability to establish co-existence of somatic variants in individual cellular populations and the associated under-representation of the genome due to a limited and varied number of somatic events present at 50X read-depth, make estimation of tumor cellularity for this study challenging.
The simultaneous analysis of tumors and adjacent precursor lesions from a series of pancreatic cases distinguishes this study from other pancreatic tumor exome sequencing studies. The identification of identical somatic mutations present in tumor and adjacent PanINs (PxT; 66% of SNVs) internally validates these events and provides strong evidence that these aberrations arose in common clonal ancestors. This also suggests that PxTs, especially those in highly clonal tumors and PanINs, are enriched for drivers of tumorigenesis. The high frequency of the commonly mutated genes (KRAS, TP53) in this study and the presentation of the mutations at different stages of PanIN progression (KRAS in early PanINs, TP53/SMAD3/4 in Tumors) are consistent with results from exome sequencing of non-amplified samples. The additional identification of TP53 mutations in two PanIN2s also showed that driver mutations in TP53 can occur early during the development of pancreatic tumors, emphasizing the benefit of evaluating PanIN/tumor combinations. Mutations presenting solely in PanINs (24%) could not be implicated in cancer development, while mutations found only in tumors (10%) were either involved in driving cancer progression or represent bystander events. This limited population of unique mutations in individual samples suggests that WGA did not introduce large numbers of non-specific mutations.
While every PanIN had some somatic mutations in common with associated tumor samples (ranging from 34–96%), the commonality varied considerably. In particular, the PanINs of cases 41, 30 and 8 shared over two thirds of their rare somatic mutations with adjacent tumor. This may reflect very recent divergence of the tumor and PanIN clones from a common ancestor. Alternatively, it is possible that the PanIN lesions are actually components of these tumors that have spread by cancerization of ducts, although there was no correlation between clonality and the proximity of tumor and adjacent PanIN. Conversely, the PanINs of cases 12 and 37 presented with least commonality with their adjacent tumors (34–40%), predicting more distal divergence of the tumor or independent histological progression from a mutated background. Importantly, highly clonal lesions were very informative, not only in identifying PxT mutations as candidate drivers, but also in further defining the roles of P only and T only mutations in tumor progression.
Development of pancreatic cancer involves a compendium of genetic mutations. Whether a stepwise accumulation of mutations is required for the transition from pre-malignant to malignant lesions is an area of great debate. In this study we show that PanIN2 lesions often contain as many mutations as PanIN3 and invasive tumor samples, even when accounting for commonality/clonality. This raises the possibility that PanIN2 lesions may transition directly to tumor without forming PanIN3 lesions. Alternatively, since mutational load is similar, it is possible that premalignant lesions may require epigenetic modifications, aneuploidy, or expression-based alterations to progress to invasive tumor. Further studies of highly clonal lesions will provide further insight into these important issues.
Despite the high frequency of KRAS and TP53 mutations in pancreatic cancer, mutations in these genes may not be necessary for development of pancreatic tumors. While case 4 presented no detectable KRAS mutations, an early TP53 mutation was detected in the PanIN2 (4P2). Only 10P2 had no detectable KRAS or TP53 mutations. The coverage across these genes was sufficient to identify mutations present in a high proportion of cells in each sample, however, mutations present only in subclones within these lesions could have been missed. Additionally, exome sequencing may have overlooked potential large deletions of TP53. One limitation of this study is that seven of 24 G12 KRAS mutations were overlooked in the initial algorithmic calling, because the variants did not pass quality filters due to lower sequence depth of the alternative alleles. However, the seven overlooked mutations were detected upon re-analysis of sequences at the G12 position and were validated by LNA Sanger sequencing. While this suggests that mutations in other genes may have been overlooked in this study, due to low sequence coverage, the more conservative filtering applied in this study was preferred because the number of false positive variants was controlled. Mutations in other genes in the TP53 and KRAS signaling pathways may have accounted for the loss of cell cycle control and the enhanced proliferation needed for PanIN and tumor growth. One potentially significant observation from this study is that additional key modulators of cellular response to DNA damage are mutated (ATM (cases 10 and 41) and MDC1 (case 3)) in the three cases where no TP53 mutations were observed. Additional mutations in TOP2A, CHEK2, FANCB, FAN1, POLH, RECQL4, APLF, DCLRE1B, BLM, HELQ, TOPBP1 and MRE11a which also influence DNA repair or the cell cycle response to DNA damage, suggest that defective DNA damage response signaling is an early event in PanIN and tumor development. Pathways involving gap junction, actin cytoskeleton, chemokine, MAPK, cell cycle and axon guidance signaling also impacted all cases in this study with a predominant early presentation.
Despite resection to clear surgical margins during tumor excision, loco-regional recurrence is common in pancreatic cancer37. The clinical significance of clonality of PanINs with adjacent tumors may have implications for surgical resection of pancreatic cancers where there is persistence of PanIN2 lesions at the surgical margins. Current clinical guidelines do not necessitate the expanded resection of PanIN2 areas. However, the results of this study emphasize the high probability of clonality of PanIN2s with adjacent tumors, the accumulation of multiple predicted somatic driver mutations of pancreatic cancer in PanINs, and the potential for PanIN2s to progress directly to tumor. Further studies, on the basis of these findings, are needed to determine whether PanIN2 and PanIN3 precursors at margins should be routinely resected.
Our results demonstrate extreme heterogeneity of mutational burden across different patients. Excluding known factors such as TP53 and KRAS mutations, very few genes were mutated in multiple tumors or individuals. While this study may not have been large enough to identify other commonly mutated genes, alignment of our somatic mutation lists with those of previous data sets from global cancer (COSMIC) and specific pancreatic studies17,31, allowed identification of a large number of genes that may contribute to pancreatic tumor formation. Additional studies are needed to identify specific genes and or pathways that influence PanIN development and progression to tumor.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge the contributions of Yean (Kit) Lee of the Mayo Medical Genome Facility for work on the exome capture and library assembly protocols and Donna Muzny, Jennifer Drummond and David Wheeler of the Human Genome Sequencing Center at the Baylor College of Medicine for their help in initial exome sequencing.
Grant Support: This work was supported by a grant from The Lustgarten Foundation for Pancreatic Research, an NIH Specialized Program of Research Excellence (SPORE) in Pancreatic Cancer (CA102701), the Mayo Clinic Center for Cell Signaling in Gastroenterology (NIDDK P30DK084567), the Minnesota Partnership for Biotechnology and Medical Genomics, and the Mayo Clinic Center for Individualized Medicine.
Abbreviations
- PanIN
Pancreatic Intraepithelial Neoplasia
- PDAC
Pancreatic Ductal Adenocarcinoma
- WGA
Whole Genome Amplification
- LCM
Laser Capture Microdissection
- LNA
Locked Nucleic Acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: All authors disclose no potential conflicts of interest.
Author Contributions: S.J.M., C.S., L.Z., R.R.W., G.V. and F.J.C. planned the project, S.J.M., J.F.L., J.L.W., B.R.K. and B.W.E. carried out the experiments. S.N.H., S.J.M., M.K., S.E.E., R.A.G., B.R.K., G.V. and F.J.C. analyzed the data and S.J.M., S.N.H., G.V. and F.J.C. wrote the manuscript. All authors edited and commented on the manuscript.
REFERENCES
- 1.Fitzgerald TL, Hickner ZJ, Schmitz M, Kort EJ. Changing incidence of pancreatic neoplasms: a 16-year review of statewide tumor registry. Pancreas. 37:134–138. doi: 10.1097/MPA.0b013e318163a329. [DOI] [PubMed] [Google Scholar]
- 2.Cubilla AL, Fitzgerald PJ. Pancreas cancer (non-endocrine) Clin Bull. 2008;8:91–99. [PubMed] [Google Scholar]
- 3.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 4.Smeenk HG, Tran TC, Erdmann J, et al. Survival after surgical management of pancreatic adenocarcinoma: does curative and radical surgery truly exist? Langenbecks Arch Surg. 2005;390:94–103. doi: 10.1007/s00423-004-0476-9. [DOI] [PubMed] [Google Scholar]
- 5.Sohn TA, Yeo CJ, Cameron JL, et al. Resected adenocarcinoma of the pancreas-616 patients: results, outcomes, and prognostic indicators. J Gastrointest Surg. 2000;4:567–579. doi: 10.1016/s1091-255x(00)80105-5. [DOI] [PubMed] [Google Scholar]
- 6.Kloppel G, Hruban RH, Longnecker DS, et al. Ductal Adenocarcinoma of the Pancreas, WHO Classification of Tumors: Pathology and Genetics of the Digestive System. Lyon: IARC Press; 2000. [Google Scholar]
- 7.Hruban RH, Takaori K, Klimstra DS, et al. An illustrated consensus on the classification of PanIN and intraductal papillary mucinous neoplasms. Am J Surg Pathol. 2004;28:977–987. doi: 10.1097/01.pas.0000126675.59108.80. [DOI] [PubMed] [Google Scholar]
- 8.Hingorani SR, Petricoin EF, Maitra A, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 9.Kim J, Reber HA, Dry SM, et al. Unfavourable prognosis associated with K-ras gene mutation in pancreatic cancer surgical margins. Gut. 2006;55:1598–1605. doi: 10.1136/gut.2005.083063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy SJ, Cheville JC, Zarei S, et al. Mate-Pair sequencing of WGA DNA following LCM of prostate cancer. DNA Res. 2012;19:395–406. doi: 10.1093/dnares/dss021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asmann YW, Middha S, Hossain A, et al. TREAT: a bioinformatics tool for variant annotations and visualizations in targeted and exome sequencing data. Bioinformatics. 2012;28:277–278. doi: 10.1093/bioinformatics/btr612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larson DE, Harris CC, Chen K, et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics. 2012;28:311–317. doi: 10.1093/bioinformatics/btr665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng PC, Henikoff S. Predicting the effects of amino-acid substitutions on protein function. Annu Rev Genomics Hum Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- 16.Bauer S, Going Gagneuer J. Bayesian: model-based gene set analysis of genome-scale data. Nucleic Acids Res. 2010;38:3523–3535. doi: 10.1093/nar/gkq045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janakiraman M, Vakiani E, Zeng Z, et al. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res. 2010;70:5901–5911. doi: 10.1158/0008-5472.CAN-10-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukherjee S, Poddar R, Deb I, Paul S. Dephosphorylation of specific sites in the kinase-specificity sequence domain leads to ubiquitin-mediated degradation of the tyrosine phosphatase STEP. Biochem J. 2011;440:115–125. doi: 10.1042/BJ20110240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts NJ, Jiao Y, Yu J, et al. ATM Mutations in Patients with Hereditary Pancreatic Cancer Cancer Discovery. 2012;2:41–46. doi: 10.1158/2159-8290.CD-11-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugiura T, Yamaguchi A, Miyamoto K. A cancer-associated RING finger protein, RNF43, is a ubiquitin ligase that interacts with a nuclear protein, HAP95. Exp Cell Res. 2008;314:1519–1528. doi: 10.1016/j.yexcr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 22.Perry NA, Shriver M, Mameza MG, et al. Loss of giant obscurins promotes breast epithelial cell survival through apoptotic resistance. FASEB J. 2012;26:2764–2775. doi: 10.1096/fj.12-205419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waalkes S, Atschekzei F, Kramer MW, et al. Fibronectin 1 mRNA expression correlates with advanced disease in renal cancer. BMC Cancer. 2010;10:503–508. doi: 10.1186/1471-2407-10-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morton CC, Nussenzweig MC, Sousa R, et al. Mapping and characterization of an X-linked processed gene related to MYCL1. Genomics. 1989;4:367–375. doi: 10.1016/0888-7543(89)90344-3. [DOI] [PubMed] [Google Scholar]
- 25.Nikiforov YE, Nikiforova MN. Molecular genetics and diagnosis of thyroid cancer. Nat Rev Endocrinol. 2011;7:569–580. doi: 10.1038/nrendo.2011.142. [DOI] [PubMed] [Google Scholar]
- 26.Chang KH, Chen Y, Chen TT, et al. A thyroid hormone receptor coactivator negatively regulated by the retinoblastoma protein. Proc Natl Acad Sci USA. 1997;94:9040–9045. doi: 10.1073/pnas.94.17.9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ansari KI and Mandal SS. Mixed lineage leukemia: roles in gene expression, hormone signaling and mRNA processing. FEBS Journal. 2010;277:1790–1804. doi: 10.1111/j.1742-4658.2010.07606.x. [DOI] [PubMed] [Google Scholar]
- 28.Frietz S, Wang R, Yao L et al. Cell type-specific binding patterns reveal that TCF7L2 can be tethered to the genome by association with GATA3. Genome Biol. 2012;26; 13(9):R52. doi: 10.1186/gb-2012-13-9-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu J, Matthaei H, Maitra A, et al. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci Transl Med. 2011;3 doi: 10.1126/scitranslmed.3002543. 92ra66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antoni L, Sodha N, Collins I, Garrett MD. CHK2 kinase: cancer susceptibility and cancer therapy- two sides of the same coin? Nat Rev Cancer. 2007;7:925–936. doi: 10.1038/nrc2251. [DOI] [PubMed] [Google Scholar]
- 31.Biankin AV, Waddell N, Kassahn KS et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamal M, Shaaban AM, Zhang L et al. Loss of CSMD1 expression is associated with high tumour grade and poor survival in invasive ductal breast carcinoma. Breast Cancer Res Treat. 2010;121:555–563. doi: 10.1007/s10549-009-0500-4. [DOI] [PubMed] [Google Scholar]
- 33.Adamsen BL, Kravik KL, Clausen OP, De Angelis PM. Apoptosis, cell cycle progression and gene expression in TP53-depleted HCT116 colon cancer cells in response to short-term 5-fluorouracil treatment. Int J Oncol. 2007;31:1491–1500. [PubMed] [Google Scholar]
- 34.Bleeker FE, Lamba S, Rodolfo M, et al. Mutational profiling of cancer candidate genes in glioblastoma, melanoma and pancreatic carcinoma reveals a snapshot of their genomic landscapes.Hum. Mutat. 2009;30:E451–E459. doi: 10.1002/humu.20927. [DOI] [PubMed] [Google Scholar]
- 35.Wei P, Tang H, Li D. Insights into Pancreatic Cancer Etiology from Pathway Analysis of Genome-Wide Association Study Data. PLOS One. 7:e46887. doi: 10.1371/journal.pone.0046887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simeone DM, Pham T, Logsdon CD. Disruption of TGFbeta signaling pathways in human pancreatic cancer cells. Ann Surg. 2000;232:73–80. doi: 10.1097/00000658-200007000-00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim J, Reber HA, Dry SM, et al. Unfavourable prognosis associated with K-ras gene mutation in pancreatic cancer surgical margins. Gut. 2006;55:1598–1605. doi: 10.1136/gut.2005.083063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.