Abstract
The epithelial-mesenchymal transition (EMT) and its reversal, MET, are fundamental processes involved in tumor cell invasion and metastasis. SEMA3F is a secreted semaphorin and tumor suppressor downregulated by TGFβ1 and ZEB1-induced EMT. Here we report that NRP2, the high-affinity receptor for SEMA3F and a co-receptor for certain growth factors, is upregulated during TGFβ1-driven EMT in lung cancer cells. Mechanistically, NRP2 upregulation was TβRI-dependent and SMAD-independent, occurring mainly at a post-transcriptional level involving increased association of mRNA with polyribosomes. ERK and AKT inhibition blocked NRP2 upregulation, while RNAi-mediated attenuation of ZEB1 reduced steady-state NRP2 levels. Additionally, NRP2 attenuation inhibited TGFβ1-driven morphologic transformation, migration/invasion, ERK activation, growth suppression and changes in gene expression. In a mouse xenograft model of lung cancer, NRP2 attenuation also inhibited locally invasive features of the tumor and reversed TGFβ1-mediated growth inhibition. In support of these results, in human lung cancer specimens with the highest NRP2 expression were predominantly E-cadherin negative. Furthermore, the presence of NRP2 staining strengthened the association of E-cadherin loss with high-grade tumors. Together, our results demonstrate that NRP2 contributes significantly to TGFβ1-induced EMT in lung cancer.
Keywords: Neuropilin-2, EMT, Lung cancer, TGF-beta, ZEB1
INTRODUCTION
Lung cancer accounts for nearly one-fifth of cancer deaths worldwide, with invasion, metastases and drug resistance representing major barriers to cure. The epithelial-mesenchymal transition (EMT), by which epithelial cells acquire a mesenchymal and invasive phenotype, contributes significantly to these barriers (1). TGFβ is a major inducer of the EMT (2), affecting lung cancer cells, acting either alone or in combination with cytokines or growth factors (3–5). In canonical TGFβ signaling, binding to the type I/II receptor (TβRI/TβRII) complex leads to phosphorylation of SMAD-2/3, their interaction with SMAD-4 and nuclear translocation (6). In contrast, non-canonical signaling is SMAD-independent involving AKT, ERK and other pathways (7, 8). In lung cancer, as with many malignant diseases, upregulation of TGFβ is a poor prognostic marker (9). Among other activities, TGFβ secretion by tumor cells and stromal components stimulates fibroblasts and extracellular matrix formation, while it inhibits anti-tumor immune responses (9). However, proliferation is also inhibited by TGFβ, suggesting why some tumors mutate or downregulate key TGFβ signaling components (10–12). The EMT process impairs growth of metastatic deposits, while its reversal, MET, is associated with increased proliferation and tumor growth (13). In human breast cancer, a partial EMT phenotype may be sufficient to facilitate invasion and vascular dissemination, as evidenced by circulating tumor cells and tumor cell clusters co-expressing epithelial and mesenchymal markers (14).
We previously demonstrated that SEMA3F, a secreted tumor suppressor isolated from a recurrent 3p deletion region in lung cancer (15), was downregulated by the EMT transcription factor, ZEB1 (16). The secreted class 3 semaphorins (SEMA3s) were discovered as inhibitors of nerve growth cone migration, and subsequently shown to function in various developmental and pathological processes, including cancer (17). Neuropilin-1 and 2 (NRP1/2) are high-affinity receptors for the SEMA3s, with SEMA3F binding predominantly to NRP2 (18, 19). In addition, both neuropilins were identified as co-receptors for VEGF and other selected growth factors (18, 20, 21). Importantly, neuropilins are overexpressed in several cancers, including lung, and their expression correlates with increased invasion and poor prognosis (22).
Because SEMA3F is downregulated during EMT in lung cancer cells, we asked what happens to NRP2. In contrast to SEMA3F, NRP2 is upregulated by TGFβ and contributes significantly to changes associated with induction of the EMT phenotype. Mechanistically, NRP2 upregulation was predominantly translational, involving increased mRNA binding of polyribosomes. Initial NRP2 upregulation was SMAD-independent and blocked by ERK and AKT inhibitors, while ZEB1 knockdown reduced steady-state NRP2 levels. In a xenograft model, we found that NRP2 contributes significantly to the invasive phenotype of lung cancer cells, as well as TGFβ1-mediated growth inhibition, which appears to be, at least in part, cell autonomous. In resected human lung cancers, high NRP2 staining together with low E-cadherin was associated with increased tumor grade. Moreover, tumors with the highest NRP2 scores were predominantly E-cadherin negative. Together, these results indicate that NRP2 upregulation plays an important role in TGFβ1–induced EMT.
Materials and Methods
Cell lines, reagents and expression constructs
NSCLC cell lines HCC-4006, NCI-H441, NCI-H358 and A549 were obtained from the Colorado Lung Cancer SPORE Cell Repository. Verification of NSCLC cell lines (by comparison to ATCC data) was carried out using microsatellite genotyping analysis performed by the University of Colorado DNA Sequencing and Analysis Core. Cells were grown in RPMI-1640 medium with 10% FCS and antibiotics (Invitrogen, Carlsbad, CA) at 37°C and 5% CO2.
Recombinant human TGFβ1 (5 or 10 ng/ml) was from R&D Systems (Minneapolis, MN). Inhibitors were from the following suppliers: SB431542 (StemGent, San Diego, CA), U0126 (Promega, Madison, WI), cycloheximide (Calbiochem, San Diego, CA), MKK-2206 (Selleckchem, Houston, TX), actinomycin-D (Sigma-Aldrich, St. Louis, MO), hygromycin, blasticidin, puromycin and doxycycline were from Invitrogen, Carlsbad, CA.
Stable integration of TGFβ1 under the control of a Tet/Dox inducible promoter was obtained with the Flp-In TRex system from Invitrogen. Empty vector or TGFβ1-transfected cells were selected and grown with 100 µg/ml hygromycin and 5 µg/ml blasticidin. Doxycycline was used at 100 ng/ml. SMAD7 in pRK5 was stably co-transfected with pcDNA3 and selected with 500 µg/ml G418. Stable shRNA transfections were generated from lentiviral particles (Sigma-Aldrich; Supplementary Table S1). Transfection was performed using 8 µg/ml Polybrene and cells were selected with 2 µg/ml puromycin.
RNA and protein analysis
Total RNA extraction protocol and gene expression analysis by quantitative real-time PCR analysis were described previously (4). Data were expressed as the percent of GAPDH. Primer sequences are provided in Supplementary Table S2.
Immunoblots were performed as described (4). Primary antibodies included: anti-Vimentin (3932S), anti-phospho SMAD2 (3101S), anti-phospho SMAD3/1 (9514S), anti-phospho SMAD1/5 (9516S), anti-phospho ERK1/2 (T202,Y204; 9101S), anti-AKT (4691S) and anti-phospho AKT (S473; 4060S) (1:1000) from Cell Signaling Technologies (Danvers, MA); anti-NRP1 (sc-7239) and anti-ZEB1 (sc-25388) antibodies (1:1000) from Santa Cruz Biotechnology; anti-NRP2 (AF2215; 1:1000) from R&D Systems; anti-E-cadherin (610181; 1:1000) from BD Biosciences (Franklin Lakes, NJ); anti-N-cadherin (ab12221; 1:1000) from Abcam (Cambridge, MA); anti-Myc tag (M4439) and anti-β-actin (A1978; 1:3000) from Sigma-Aldrich; anti-EpCAM (MS-144-PABX; 1:1000) from Neomarkers (Fremont, CA); anti-SMAD7 (42-0400; 1:1000) from Invitrogen and anti-ERK (V114A; 1:1000) from Promega. Secondary antibodies included: HRP-conjugated anti-rabbit (1858415) and anti-mouse (185843) from Thermo Fisher Scientific Inc (Rockford, IL); HRP-conjugated anti-goat (81–1620) was from Invitrogen. Detection was performed with Western Lightning Plus ECL reagent (PerkinElmer, Waltham, MA). E-cadherin and β-actin were detected using an Alexa488-conjugated anti-mouse antibody (A21200; Invitrogen) and signal was recorded using a Typhoon 9400 Image system (GE Healthcare). Band quantitation was performed on Typhoon images or by using infrared CW800 (926–32214) and CW680 (926–3221) IRDye-conjugated secondary antibodies and a LI-COR Odyssey fluorescence scanner (LI-COR Biosciences, Lincoln, NE). Autoradiographic bands were quantified using ImageJ freeware or LI-COR analysis software for IR fluorescence.
Immunofluorescence and immunohistochemistry
For immunofluorescence, cells were processed as described (23). Slides were incubated with anti-ZEB1 or anti-NRP2 at 1:100 dilution. Alexa488-conjugated anti-goat (A11055) and Alexa594-conjugated anti-rabbit (A11072) secondary antibodies (1:200) were from Invitrogen. DAPI (1:50,000) was from Sigma-Adrich. Stained slides were mounted in Dako Fluorescent Mounting Medium (Dako). Images were captured with IPLab software on a BD CARVII spinning disc confocal microscope (BD Biosciences).
Immunohistochemistry procedure, analysis and scoring methods were performed as described (4). Primary antibodies were mouse anti-human E-cadherin (1:50), goat anti-human NRP2 (1:100), rabbit anti-human Ki67 (18-01912; 1:50) and rabbit anti-human pan-cytokeratin (18-0059; 1:50) from Invitrogen. Secondary antibodies (Vector Laboratories, Burlingame, CA) were biotinylated rabbit anti-goat (BA-5000), goat anti-rabbit (BA-1000) and rabbit anti-mouse (BA-9200) antibodies. For the TMA analysis, scores equal or below 10, for both NRP2 and E-cadherin staining, were considered negative. For the proliferation analysis in tumor xenografts, 3 fields/tumor were counted in four representative tumors per cohort.
Polyribosome separation on sucrose gradients was performed as described (24).
Migration/invasion assays
Transwell assays were performed using the BD Biocoat system following the manufacturer's instruction (BD Bioscience). Briefly, the lower chamber was filled with 5% FCS-containing RPMI-1640 as an attractant. Fifty-thousand H358 cells or 25,000 A549 cells were suspended in 0.5 mL serum free RPMI-1640 and added to the upper chamber followed by the addition of TGFβ or control solutions. H358 cells were incubated for 48 hours and A549 cells for 12 hours at 37°C, 5% CO2. Cells were then fixed with 4% para-formaldehyde, stained with crystal violet, rinsed and cells remaining on the upper side of the filter were removed. After allowing the filters to dry overnight, cells were counted under a SMZ1500 stereomicroscope (Nikon, Lewisville, TX). Each condition was performed in triplicate and 4 fields were counted per filter. ShControl transfectants were included in each experiment to assess reproducibility.
Proliferation assay
Control or NRP2 knockdown H358 Tr-TGFβ cells (5×104 cells/plate) were cultured in the presence or absence of doxycycline (100 ng/mL), trypsinized and counted at days 2 and 4. Doubling time was calculated using the following formula: T = t × ln(2)/(ln(N4)−ln(N2)) in which T is the doubling time, t is the time between two measurements, N4 is the final number of cells at day 4 and N2 is the number of cells measured at day 2. This experiment was repeated 3 times.
Tumor xenografts and human lung tumor microarrays
One million H358-shControl, H358-shNRP2-B, H358-Tr-TGFβ1-shControl or H358-Tr-TGFβ1-shNRP2-B cells suspended in 200 µl of 50% Matrigel were injected subcutaneously in the right flank of six week-old female athymic nude mice (10 mice per group; Hsd: Athymic Nude-Foxn1nu, Harlan, Dublin, VA). After sacrifice, tumors were removed and weighed. Half of each tumor was placed into OCT compound and the remainder fixed in 4% paraformaldehyde overnight, then incubated in 70% ethanol overnight prior to paraffin embedding. All animal experiments were approved by the MUSC Institutional Animal Care and Use Committee.
TMA slides containing 109 lung tumor samples and 10 normal lung tissues were obtained from US Biomax (Rockville, MD, ref: BC041115). These were processed for immunohistochemistry as described above.
Statistical analysis
Descriptive statistics were computed on TMA expression scores for all genes, as well as clinical characteristics. One-way ANOVA was used to test the association between EMT status (NRP2(+)/Ecadherin(−) or NRP2(−)/Ecadherin(+)) and clinical characteristics that are categorical variables namely sex, histology, and TNM stage. Correlations between the EMT status and age or tumor grade were analyzed using Fisher’s Exact Test. In addition to analyzing expression scores as continuous variables, each gene's expression level was also dichotomized: a score of 0–10 was considered negative and a score above 10 was considered positive; McNemar’s test was used to compare the dichotomous gene expression. Correlations between genes were analyzed using Spearman's statistics. The naïve p-values of these correlations were adjusted for multiple comparisons controlling for False Discovery Rate (FDR). Statistical analyses were performed using SAS/BASE and SAS/STAT software, Version 9.2 of the SAS System for Windows (SAS Institute Inc., Cary, NC, USA). To determine the relative effects of NRP2 shRNAs on migration/invasion, the log of the number of migrating cells per field was modeled as a function of shRNA (e.g. shNRP2-B vs. shNRP2-Y vs. shControl) and TGFβ (yes vs. no) using linear regression modeling. Model estimates were used to derive estimates of fold-changes comparing conditions, p-values for testing statistical significance of fold-change, and calculating 95% confidence intervals for fold-changes. Appropriateness of linear regression assumptions was assessed using residual plots. The significance of the differences between proliferation levels in xenografts and between E-cadherin scores in the TMA was assessed using the Student’s t-test.
RESULTS
NRP2 is upregulated by TGFβ1 in lung cancer cell lines
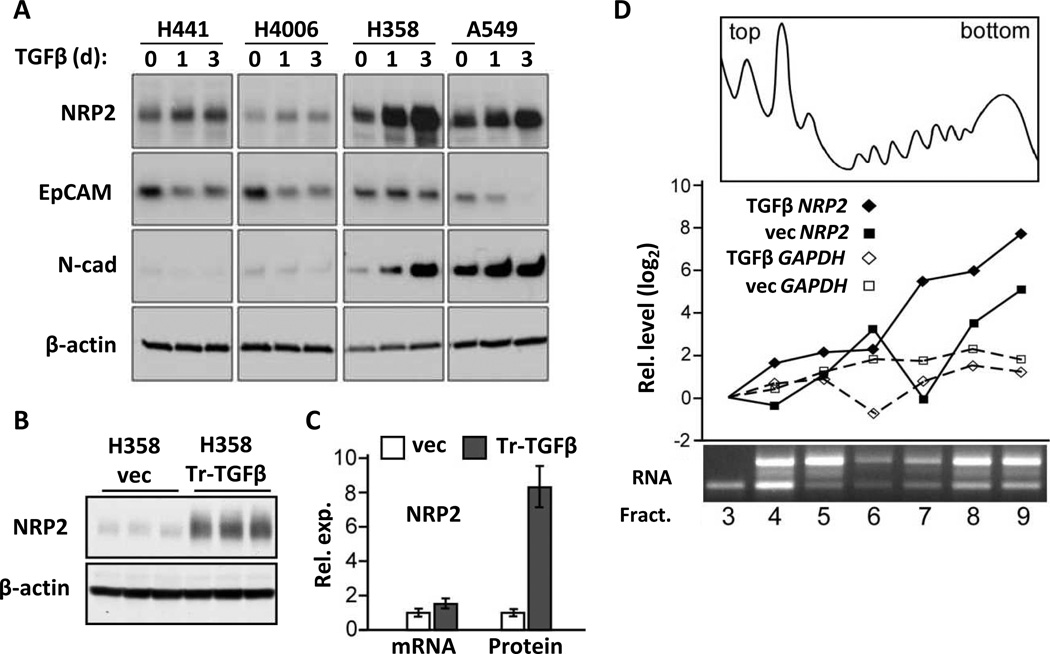
To determine if NRP1/2 levels were affected by EMT, we treated four NSCLC cell lines containing epithelial features with exogenous TGFβ1 for 1–3 days. Varying levels of increased NRP2 protein were evident along with expected changes in EMT markers (Fig. 1A). In contrast to NRP2, no consistent changes were observed in NRP1 and it was not further examined.
FIGURE 1.
TGFβ1 upregulates NRP2 protein in NSCLC cell lines. (A) Western blot showing NRP2 levels in 4 NSCLC cell lines following 1 and 3 days of TGFβ1 (10 ng/ml) treatment. EpCAM and N-cadherin were used as epithelial and mesenchymal markers, respectively, and changes in their levels indicated an ongoing EMT response; β-actin: loading control. (B) Representative Western blot showing NRP2 levels (in triplicate) in H358 cells transfected with TGFβ1 (H358-Tr-TGFβ1) or control vector (vec) cultured for 72 h. (C) Bar graph indicating NRP2 mRNA and protein levels measured by qRT-PCR and densitometry, respectively, in three independent experiments. Error bars: s.d. (D) H358-vec and H358-Tr-TGFβ1 cells were treated with doxycycline, 100 ng/ml for 3days, lysed and fractionated on linear sucrose gradients. A representative A280 trace and electrophoretic analysis of RNA is shown in the top and bottom panels, respectively. The appearance of the fractions and mRNAs from control and H358-Tr-TGFβ1cells was essentially identical. NRP2 and GAPDH mRNA levels were measured in each fraction by qRT-PCR (middle). The Y-axis gives the relative expression on a log scale, normalized to the initial fraction, plotted against individual fractions from the gradient.
To explore NRP2 upregulation in more detail, we focused on two cell lines, H358 and A549. While both are epithelial in nature, A549 is partially shifted towards mesenchymal differentiation, as evidenced by lower E-cadherin, higher N-cadherin and higher ZEB1 levels (4). Moreover, the morphologic response and downregulation of E-cadherin to TGFβ occur more rapidly in A549 cells. As expected for a cell surface receptor, increased NRP2 protein was observed at the plasma membrane (Fig. S1A). Consistently, NRP2 upregulation was observed by 8 h (Fig. S1B), and with repeated exogenous TGFβ1 the effect was persistent at 10 days (Fig. S1C). Chronic TGFβ1 exposure was obtained by stable transfection in H358 cells (H358-Tr-TGFβ1), which while doxycycline-inducible, exhibited significant leaky expression. In the absence of dox, these cells produced 33% more activated TGFβ1 than vector-controls (Fig. S1D). This level was sufficient to upregulate NRP2 protein, as shown in Fig. 1B, while the addition of dox further increased both TGFβ1 and NRP2 (Fig. S1D and see Fig. 2A below).
FIGURE 2.
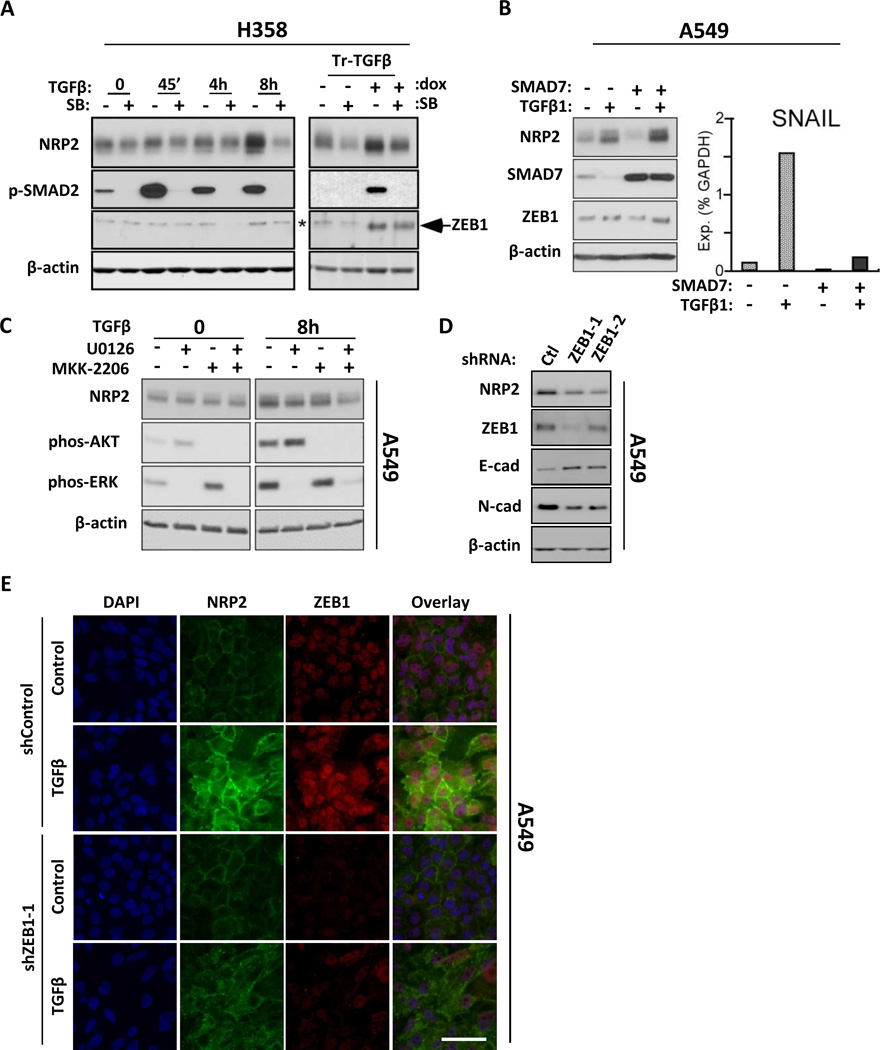
Induction and maintenance of NRP2 depends on non-canonical TGFβ signaling and involves ZEB1. (A) Left: H358 cells were treated with 10 µM SB431542 (SB) or vehicle for 60 min prior to TGFβ1 exposure for the indicated times. Right: H358-Tr-TGFβ cells were cultured with or without dox for 3 days, and for the final 24h exposed to SB431542, as indicated. Protein lysates were analyzed by Western blot. *background band in H358 samples. (B) Left: Control- or SMAD7-transfected A549 cells were exposed to TGFβ1 or vehicle for 24h and analyzed by Western blot. Right: SNAIL mRNA levels were measured by qRT-PCR in the same cultures. (C) Western blot analysis of lysates from A549 cells pre-treated for 30 min with vehicle, U0126 (10 µM), MKK-2206 (2 µM) or both, then exposed to TGFβ1 for indicated times. The full time course is shown in Fig. S2C. (D) A549 cells stably knocked down for ZEB1 with independent shRNAs (ZEB1-1 and ZEB1-2) analyzed for NRP2 and the indicated EMT markers. Ctl: control shRNA. (E) shZEB1-1 and shControl A549 cells were exposed to TGFβ1 for 48h and analyzed by confocal microscopy for NRP2 (green) and ZEB1 (red). Nuclei were stained with DAPI. Scale bar: 100 µm.
Compared to controls, steady-state NRP2 protein in H358-Tr-TGFβ1 cells was increased by ~8-fold, whereas mRNA was only 1.5-fold higher (Fig. 1C). Similar results were obtained with short-term exogenous TGFβ1, although the magnitude of the NRP2 protein upregulation was less (i.e., 2 to 4-fold). To gain further insight, we examined mRNA and protein stability following TGFβ1 stimulation and the addition of actinomycin-D or cycloheximide, respectively. However, neither was increased (Fig. S1E–F). We then asked whether TGFβ1 affected protein translation using sucrose gradient fractions from dox-treated control and H358-Tr-TGFβ1 cells (Fig. 1D). Increased translation should lead to an increase in the number of ribosomes associated with NRP2 mRNA. Indeed, TGFβ1 caused an approximate 4-fold increase of NRP2 mRNA in the heavy polysome fractions. In contrast, the distribution of GAPDH mRNA was unchanged. These results were reproducible and specific to TGFβ induction. Together, these data indicate that NRP2 protein upregulation predominantly involves increased mRNA-polyribosome association.
NRP2 upregulation, TGFβ signaling and ZEB1
Canonical TGFβ signaling leads to activation of TβRI and R-SMAD 2/3 phosphorylation, which can be blocked by SB431542. As shown in Fig. 2A, pre-treatment with SB431542 reduced baseline NRP2 expression due to endogenous TGFβ in control and uninduced H358-Tr-TGFβ1 cells. SB431542 also blocked NRP2 upregulation by exogenous TGFβ1 in both cell lines (Figs. 2A and S2A). When SB431542 was added to H358-Tr-TGFβ cells already exposed to doxycycline for 2 days, NRP2 levels dropped despite continued dox treatment (Fig. 2A). To determine if NRP2 upregulation was SMAD-dependent, the R-SMAD antagonist, SMAD7, was overexpressed in A549 cells prior to addition of TGFβ1 (Figs. 2B, S2B). After 24 h, SMAD7 blocked upregulation of SNAIL, a known SMAD target gene. However, SMAD7 did not inhibit NRP2 induction at either the protein or mRNA levels.
Non-canonical TGFβ signaling includes ERK and AKT pathways (7, 8). In A549 cells, inhibition of ERK or AKT with U0126 and MKK-2206, respectively, impaired NRP2 upregulation by TGFβ1, while combining the inhibitors was more effective (Figs. 2C, and S2C for full time-course). Similar results were obtained with the individual inhibitors in H358 cells, although the combination did not result in a greater effect.
We previously reported that ZEB1 inhibits expression of the tumor suppressor, SEMA3F, which uses NRP2 as its high-affinity receptor (16). ZEB1 is also up-regulated by TGFβ1 in NSCLC cell lines (e.g., as shown in Fig. 2A) and is the EMT transcription factor best correlated with steady-state mesenchymal features (4, 25). In A549 cells, shRNA targeting of ZEB1 led to reduced steady-state levels of NRP2 (Fig. 2D). ZEB1 knockdown also reduced NRP2 levels after treatment with exogenous TGFβ in A549 cells (Figs. 2E, S2D), and after 5 days of dox-induction in H358-Tr-TGFβ1 cells (Fig. S2E). However, in short-term experiments up to 8 h, NRP2 upregulation occurred without a detectable change in ZEB1 (Fig. S2C). Moreover, ZEB1 levels were unaffected by the MEK and AKT inhibitors that blocked NRP2 upregulation. These results suggest that while ZEB1 may not be involved in the initial phase of NRP2 upregulation, it contributes to NRP2 maintenance. Although SNAIL reportedly contributes to ZEB1 upregulation (26), blocking SNAIL with SMAD7 had no effect on ZEB1 levels after 24 h of TGFβ (Fig. 2B).
NRP2 knockdown impairs downstream TGFβ1 responses
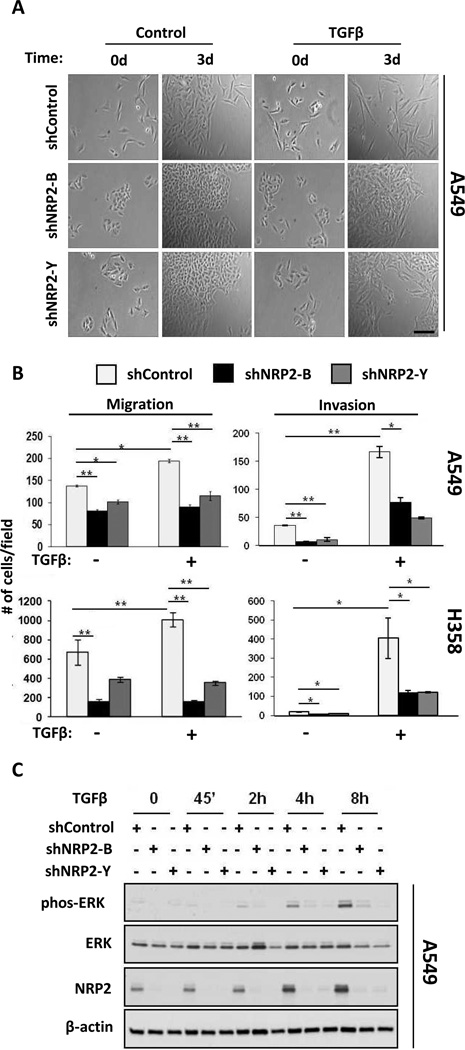
To assess the effects of NRP2 on TGFβ1 activities, we stably transfected control and NRP2-targeting shRNAs into H358 and A549 cells (Figs. S3A, B). Morphologically, NRP2 knockdown inhibited the mesenchymal transformation of A549 cells (Fig. 3A); similar results were obtained in H358 cells treated with exogenous TGFβ1 and H358-Tr-TGFβ induced with doxycycline (Fig. S4). Using transwell assays, NRP2 knockdown inhibited both baseline and TGFβ1-stimulated migration (Fig. 3B). Likewise, invasion through Matrigel-coated membranes was impaired.
FIGURE 3.
TGFβ–mediated changes in EMT features are attenuated by NRP2 knockdown. (A) Phase contrast images of A549 cells stably knocked-down for NRP2 with independent shRNAs following zero or three days of culture in the presence or absence of TGFβ1. Scale bar: 100 µm. (B) Migration (left) and invasion (right) assays in A549 (top) and H358 cells (bottom) expressing control and shNRP2 lentiviruses and treated or not with TGFβ1. Significant differences are indicated by: (*) p ≤ 0.05, (**) p ≤ 0.001. Error bars: s.e.m. (C) A549 shControl and NRP2-knockdown cells were exposed to exogenous TGFβ1 for the indicated times. Lysates were analyzed for ERK1/2 phosphorylation and NRP2 induction.
The effect of NRP2 knockdown on TGFβ1-induced changes in gene expression and signaling was also examined. NRP2 knockdown blunted the suppression of epithelial genes, as well as upregulation of mesenchymal genes (Fig. S3C). As indicated above, TGFβ1 signaling led to ERK and AKT phosphorylation. While the kinetics differed, NRP2 knockdown consistently inhibited ERK phosphorylation (Figs. 3C, S3D). NRP2 knockdown also inhibited AKT phosphorylation, but only in H358 cells. In hepatic stellate cells, NRP1 knockdown was shown to shift SMAD phosphorylation from SMAD2/3 to SMAD1/5, and NRP2 was stated to have a similar effect (27). However, in multiple experiments under a variety of conditions, NRP2 knockdown had no effect on SMAD2/3 or SMAD1/5 phosphorylation in these lung cancer cells (Fig. S3E). In contrast, NRP1 knockdown did enhance SMAD1/5 phosphorylation without affecting phospho-SMAD2/3, although long exposure times were required to detect this (Fig. S3G). Together, we conclude that NRP2 knockdown inhibits morphology, cell migration/invasion, gene expression and non-canonical signaling changes induced by TGFβ1, whereas SMAD phosphorylation was unaffected, at least under the conditions studied.
TGFβ1 suppression of tumor growth is reversed by NRP2 knockdown
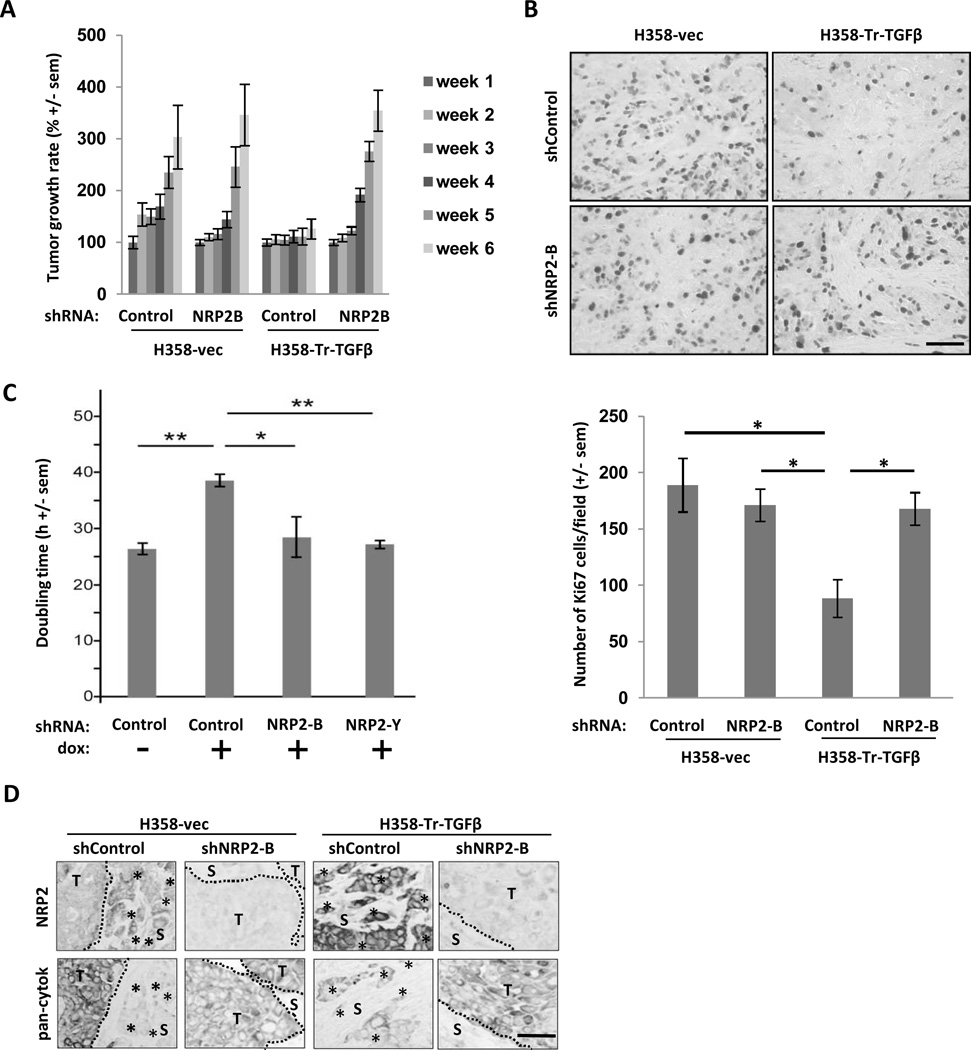
To explore the consequences of NRP2 knockdown, we used subcutaneous xenografts of control and uninduced H358-Tr-TGFβ1 cells stably transfected with non-targeting or NRP2 shRNAs. In the presence of wild-type NRP2 levels, the growth of H358-Tr-TGFβ1 xenografts was substantially slowed (Fig. 4A). This is consistent with the ability of TGFβ1 to inhibit epithelial cell proliferation while driving the EMT process (6, 28). Ki67 staining demonstrated that proliferation was significantly inhibited by TGFβ (Fig. 4B). NRP2 knockdown, which was confirmed by immunohistochemistry (Fig. S5), completely restored growth of H358-Tr-TGFβ1 xenografts, while it had no effect on the growth of control tumors. To determine if growth inhibition was autocrine in nature, at least in part, we examined the consequences of TGFβ exposure and NRP2 knockdown in vitro. These results paralleled the in vivo findings demonstrating that NRP2 knockdown relieved the growth inhibition resulting from TGFβ1 (Fig. 4C).
FIGURE 4.
NRP2 knockdown restores growth to TGFβ1–inhibited xenografts and reduces invasive morphology. (A) Control and H358-Tr-TGFβ cells, infected with shNRP2-B or control shRNA lentiviruses, were injected subcutaneously into nu/nu mice (n=10 animals/cohort). Tumor volumes were determined over 6 weeks and the relative tumor volume (compared to tumor volume at week 1) is given for each cohort. Error bars: s.e.m. (B) Top: Formalin-fixed tumor sections from (A) were immunostained for Ki67. Bottom: Mean number of Ki67 positive nuclei/field from 12 fields/cohort. *: Significant differences (p ≤ 0.05). Error bars: s.e.m. (C) H358-Tr-TGFβ1 cells transfected with control or NRP2 shRNAs were cultured during 2 to 4 days −/+ doxycycline, trypsinized and counted to estimate the doubling time. *: Significant differences (p ≤ 0.05). Error bars: s.e.m. (D) Representative images of NRP2 and pan-cytokeratin immunostaining in each cohort described in (A). T = Tumor; S = Stroma; dotted lines indicate tumor-stromal interface; *: clusters of tumor cells separated from primary tumor mass. Scale bars: 100 µm
In control and H358-Tr-TGFβ1 tumors expressing non-targeting shRNA, NRP2 staining was intensified in isolated tumor cells and tumor cell clusters in the stroma (Figs. 4D, S5). These cells showed reduced staining for the epithelial marker, pan-cytokeratin, suggesting they had undergone a partial mesenchymal transition and were invasive. As anticipated from our morphologic observations in vitro (Fig. 3A, S4), NRP2 knockdown resulted in more regularly-shaped tumor aggregates and fewer cells that appeared invasive. Thus, we conclude that NRP2 is important for TGFβ1-mediated effects on proliferation and migration. However, while the changes induced by TGFβ1 are autocrine, at least in part, we cannot exclude additional paracrine effects from the tumor micro-environment.
Increased NRP2 expression correlates with tumor grade and E-cadherin in patient samples
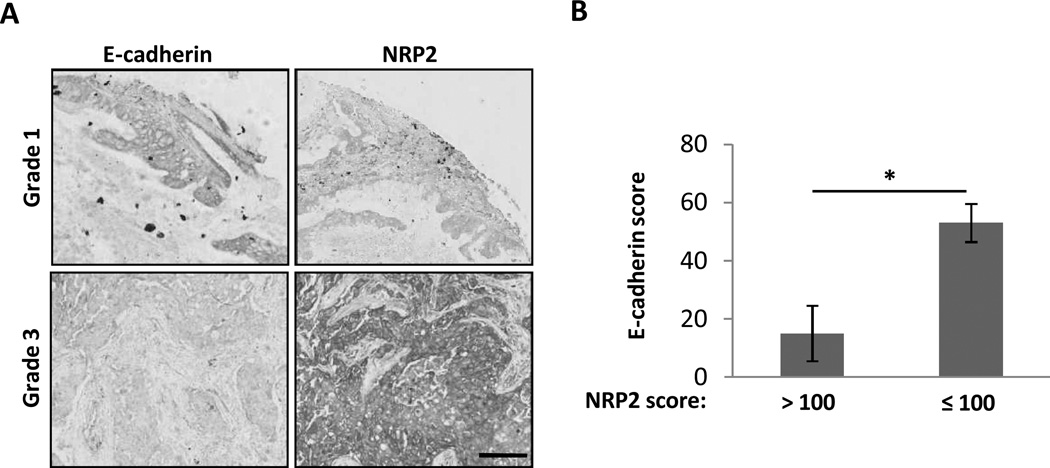
In previous studies, we showed that E-cadherin, an epithelial marker frequently lost during the EMT process, is absent in 10 to 30% of lung tumors (4, 29)). However, this likely underestimates the number of tumors transiently progressing through the EMT. In other studies, we reported that NRP2 levels significantly increase in early lung lesions (30). Based on our current findings and previous results, we hypothesized that tumors with high NRP2 and low E-cadherin would be more aggressive. This was examined using a microarray containing 109 resected human lung cancers. Overall, NRP2 and E-cadherin were expressed in 83 (76%) and 69 (63%) tumors, respectively. Of 22 NRP2(+)/Ecad(−) tumors, 9 were grade 3, while the rest were grade 2, and the association with tumor grade was significant (Fisher’s exact test, p-value = 0.0049) (Table 1; Figs. 5A, S6A). Conversely, all NRP2(−) /Ecad(+) tumors were grade 1 or 2. In both scenarios, the use of NRP2 as a marker strengthened the association of E-cadherin with tumor grade. We also hypothesized that high NRP2 scores would correlate with E-cadherin loss. In fact, tumors with the highest NRP2 scores (>100) were predominantly E-cadherin negative (7/9, p = 0.0252), while tumors with NRP2 scores less than 100 had higher mean E-cadherin staining (53 +/− 7 vs. 15 +/− 10, p=0.0043) (Table 2, Figs. 5B, S6B). Thus, the upregulation of NRP2 during TGFβ-induced EMT in lung cancer cell lines, and its association with E-cadherin loss and more aggressive tumors in the lung cancer tissue microarrays may reflect the same underlying biology.
TABLE 1.
Association of tumor grade with NRP2 and E-cadherin status
| Grade | NRP2 (−) E-cad (+) n (%) | NRP2 (+) E-cad (−) n (%) |
|---|---|---|
| Grade 1 | 2 (17) | 0 (0) |
| Grade 2 | 10 (83) | 13 (59) |
| Grade 3 | 0 (0) | 9 (40) |
| Total | 12 (100) | 22 (100) |
FIGURE 5.
Analysis of NRP2 and E-cadherin expression from a TMA with 109 human lung cancers. (A) Examples of Grade 1 and Grade 3 tumors stained for E-cadherin and NRP2. Scale bar: 200 µm. (B) Mean score for E-cadherin staining in tumors divided according to high (> 100) or low (≤ 100) NRP2 scores. *: Significant difference (p ≤ 0.05). Error bars: s.e.m.
TABLE 2.
NRP2 score vs. E-cadherin status
| NRP2 score |
E-cad (−) n (%) | E-cad (+) n (%) |
|---|---|---|
| > 100 | 7 (78) | 2 (22) |
| ≤ 100 | 34 (34) | 66 (66) |
DISCUSSION
Our results demonstrate that NRP2 is specifically upregulated in TGFβ-responsive lung cancer cells and that it contributes significantly to development of the EMT phenotype. NRP2 induction was rapid, stable with continued TGFβ and present at the plasma membrane. Changes in NRP2 were evident primarily at the protein level with modest increases in mRNA. Experiments with cycloheximide or actinomycin-D treatment indicated that neither NRP2 protein nor mRNA were stabilized by TGFβ. Instead, we observed a reproducible increase in the association of NRP2 mRNA with fractions containing the heaviest polyribosomes, consistent with increased translation. In comparison, the polysomal association of GAPDH mRNA was not altered by TGFβ.
Mechanistically, how increased NRP2 protein translation occurs is unknown. The translation of many proteins is regulated by micro-RNAs (31). Using the miRiam target prediction tool (32), we identified 17 miRs or miR families and 39 high-probability binding sites in the 3’UTR of the major NRP2 isoform (Fig. S7A). However, using overlapping luciferase reporter constructs for the NRP2 3’UTR, we failed to detect consistent changes after exposure to exogenous TGFβ. Nevertheless, predicted binding sites for the TGFβ-regulated miR-15b and miR-16 were present, and although both were expressed at quite different levels, they were inhibited by TGFβ. Further studies will be required to determine their significance. In addition, TGFβ-regulated protein translation during EMT has been shown to occur through heterogeneous nuclear ribonucleoprotein E1 (hnRNP E1), which binds a structural element in the 3' untranslated region of Dab2 and ILEI (33). Stable knockdown of hnRNP E1 in NMuMG cells was associated with NRP2 upregulation (Fig. S7B). More work will be required to determine if hnRNP E1 or another hnRNP has a direct role in the observed NRP2 upregulation.
In the present study, we found that early NRP2 upregulation was SMAD-independent. This is based on the inability of SMAD7 to block NRP2 increases, whereas a known SMAD target gene, SNAIL, was inhibited. Among non-canonical signaling pathways activated by TGFβ, we found that ERK and AKT inhibitors impaired NRP2 upregulation and this effect was greater when the inhibitors were combined. Mechanistically, both TβRI and TβRII can be phosphorylated on tyrosine residues that serve as Shc recruitment sites leading to activation of ERK (34). ERK activation is an essential component in TGFβ-mediated EMT (35), leading in part to increased FRA-1/c-JUN heterodimers and increased AP-1 activity (36–38). In some cases, ERK activation is dependent only on TβRII (39). However, our results demonstrating that NRP2 upregulation is blocked by SB431542 indicates that TβRI is required. TβRI also interacts with PI3K-p85, and TGFβ exposure activates AKT with multiple downstream consequences (40, 41). There is also cross-talk between the AKT and ERK / AP-1 pathways, since AKT inhibition of GSK3 leads to reduced c-JUN degradation (42). Furthermore, AKT2 phosphorylates and inactivates hnRNP E1 (33), which potentially contributes to NRP2 upregulation. In contrast to ERK and AKT inhibitors, p38 and JNK blockade had no effect on NRP2 levels.
Changes in gene expression, a hallmark of EMT, result in part from the upregulation of transcriptional repressors, including ZEB1, that bind E-box elements in genomic DNA (43). Previously, we found that ZEB1 was the repressor best correlated with EMT features and EGFR inhibitor resistance in NSCLC cell lines (4, 44). In addition, we reported that ZEB1 bound E-box sites in the SEMA3F promoter and suppressed its expression (16). Recently, we found that ZEB1 preferentially inhibits acetylation of histone H3 lysine 27 in the promoter region of ZEB1 target genes (45). In the current study, ZEB1 knockdown inhibited NRP2 expression. However, ZEB1 levels were unchanged during early NRP2 induction, and ERK and AKT inhibitors, which blocked NRP2 upregulation, had no effect on ZEB1. Together, these results suggest that ZEB1 acts as a maintenance factor for NRP2. A similar requirement for ZEB1 in maintenance of a stable mesenchymal phenotype was observed in breast cancer (46). ZEB2 was identified as a Smad-interacting protein (47) and subsequent studies demonstrated that both ZEB proteins bind activated R-Smads (48, 49). Our studies do not exclude a role for SMAD-ZEB1 interactions in the maintenance of NRP2 or the possibility that maintenance and induction involve different TGFβ-dependent pathways.
The EMT process is characterized by changes in morphology, loss of junctional complexes, increased migration/invasion, decreased proliferation and alterations in gene expression (1). NRP2 knockdown inhibited these TGFβ-induced changes. In hepatic stellate cells exposed to TGFβ1, NRP1 knockdown was shown to reduce SMAD2/3 and increase SMAD1/5 phosphorylation (27). These authors stated that NRP2 knockdown elicited a similar response. However, we found that in lung cancer cells, only NRP1 knockdown affected SMAD phosphorylation. Thus, the two NRPs are not equivalent in this context. The absence of a SMAD effect prompted us to examine non-canonical pathways. Importantly, NRP2 deficiency consistently inhibited TGFβ-mediated ERK activation, while there was no effect on JNK or p38. The kinetics of ERK activation by TGFβ are known to be context dependent (8) and this was apparent in the NSCLC cell lines. NRP2 also influences ERK signaling in response to class 3 semaphorins or VEGF (23, 50, 51), but to our knowledge a role in non-canonical TGFβ signaling has not been reported.
In vivo, we found that TGFβ impaired the growth of xenograft tumors by reducing proliferation, as previously reported (25, 52). This was confirmed by in vitro growth assays. Our results indicate that the anti-proliferative effect of TGFβ has an autocrine component, at least in part, although we cannot exclude an additional paracrine effect. Reduced levels of TβRII have been described in about 40% of primary NSCLCs, often associated with 5’CpG methylation (53). However, a recent study reported that drug-resistant NSCLCs are linked to high TβRII expression and EMT features, concomitant with reduced proliferation, which may facilitate resistance to cytotoxic therapy (54). Moreover, elevated expression of TGFβ has also been correlated with poor prognosis (Sterlacci (9)et al., Hum Pathol 2011). Thus, the consequences of TGFβ signaling in lung cancer are context-dependent.
In xenograft tumors, increased NRP2 staining was present in invasive-appearing isolated tumor cells and small tumor cell clusters in the stroma displaying decreased pan-cytokeratin. Of note, NRP2 knockdown resulted in more regularly-shaped tumor aggregates and fewer cells that appeared invasive. Recent studies have confirmed the importance of the EMT process in tumor cell invasion and dissemination, while demonstrating that EMT reversal is critical for the subsequent proliferation of these cells in metastatic sites (13, 14, 55). Furthermore, a partial EMT phenotype appears sufficient to generate circulating tumor cells and tumor cell clusters (14), reminiscent of what we observed in the stroma. In tumor microarrays from resected human lung cancer specimens, we speculated that high NRP2 expression would be associated with reduced E-cadherin and less differentiation. Indeed, tumors with the highest NRP2 scores were predominantly E-cadherin negative, while reduced NRP2 was associated with higher E-cadherin staining. Moreover, the presence of NRP2 staining strengthened the association of E-cadherin loss with high-grade tumors.
The role of NRP2 in tumor growth is almost certainly complex and context-dependent. In contrast to our results in lung cancer cells, NRP2 expression was associated with increased proliferation in a colorectal cancer model treated with TGFβ (56). Presumably, these cells had escaped TGFβ-mediated growth inhibition. Other studies have shown that NRP2 knockdown inhibits survival, but not proliferation (57–59). In addition, use of an anti-NRP2 function blocking antibody reduced tumor lymphatic vessels and the number of lung metastases without affecting primary tumor growth (60). Altogether, our results suggest that in lung cancer cells, NRP2 is upregulated and contributes significantly to TGFβ-mediated EMT. Although NRP2 knockdown inhibited the invasive phenotype, it also rescued growth suppressed by TGFβ. While this would be a clinically undesired effect, these cells may be more sensitive to cytotoxic chemotherapy or other agents. Lastly, it would appear that the suppression of SEMA3F and upregulation of NRP2 by TGFβ represents a coordinated program contributing to the acquisition of a motile, invasive phenotype.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the University of Colorado Lung Cancer SPORE grant, NCI-CA58187, and by the Medical University of South Carolina Hollings Cancer Center Support Grant, Biostatistics Core NCI-CA138313. Imaging facilities for this research were supported, in part, by Cancer Center Support Grant P30 CA138313 to the Hollings Cancer Center, Medical University of South Carolina. Cell line genotyping was performed by the University of Colorado Cancer Center DNA Sequencing & Analysis Core, supported in part by grant P30 CA046934.
Footnotes
The authors disclose no potential conflicts of interest
References
- 1.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 2.Heldin CH, Vanlandewijck M, Moustakas A. Regulation of EMT by TGFbeta in cancer. FEBS Lett. 2012;586:1959–1970. doi: 10.1016/j.febslet.2012.02.037. [DOI] [PubMed] [Google Scholar]
- 3.Thomson S, Petti F, Sujka-Kwok I, Epstein D, Haley JD. Kinase switching in mesenchymal-like non-small cell lung cancer lines contributes to EGFR inhibitor resistance through pathway redundancy. Clin Exp Metastasis. 2008;25:843–854. doi: 10.1007/s10585-008-9200-4. [DOI] [PubMed] [Google Scholar]
- 4.Gemmill RM, Roche J, Potiron VA, Nasarre P, Mitas M, Coldren CD, et al. ZEB1-responsive genes in non-small cell lung cancer. Cancer letters. 2011;300:66–78. doi: 10.1016/j.canlet.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao Z, Fenoglio S, Gao DC, Camiolo M, Stiles B, Lindsted T, et al. TGF-beta IL-6 axis mediates selective and adaptive mechanisms of resistance to molecular targeted therapy in lung cancer. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15535–15540. doi: 10.1073/pnas.1009472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siegel PM, Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 7.Mu Y, Gudey SK, Landstrom M. Non-Smad signaling pathways. Cell Tissue Res. 2012;347:11–20. doi: 10.1007/s00441-011-1201-y. [DOI] [PubMed] [Google Scholar]
- 8.Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sterlacci W, Wolf D, Savic S, Hilbe W, Schmid T, Jamnig H, et al. High transforming growth factor beta expression represents an important prognostic parameter for surgically resected non-small cell lung cancer. Hum Pathol. 2012;43:339–349. doi: 10.1016/j.humpath.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 10.Anumanthan G, Halder SK, Osada H, Takahashi T, Massion PP, Carbone DP, et al. Restoration of TGF-beta signalling reduces tumorigenicity in human lung cancer cells. British journal of Cancer. 2005;93:1157–1167. doi: 10.1038/sj.bjc.6602831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halder SK, Cho YJ, Datta A, Anumanthan G, Ham AJ, Carbone DP, et al. Elucidating the mechanism of regulation of transforming growth factor beta Type II receptor expression in human lung cancer cell lines. Neoplasia (New York, NY. 2011;13:912–922. doi: 10.1593/neo.11576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malkoski SP, Haeger SM, Cleaver TG, Rodriguez KJ, Li H, Lu SL, et al. Loss of transforming growth factor beta type II receptor increases aggressive tumor behavior and reduces survival in lung adenocarcinoma and squamous cell carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:2173–2183. doi: 10.1158/1078-0432.CCR-11-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer cell. 2012;22:725–736. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roche J, Boldog F, Robinson M, Robinson L, Varella-Garcia M, Swanton M, et al. Distinct 3p21.3 deletions in lung cancer and identification of a new human semaphorin. Oncogene. 1996;12:1289–1297. [PubMed] [Google Scholar]
- 16.Clarhaut J, Gemmill RM, Potiron VA, Ait-Si-Ali S, Imbert J, Drabkin HA, et al. ZEB-1, a repressor of the semaphorin 3F tumor suppressor gene in lung cancer cells. Neoplasia (New York, NY. 2009;11:157–166. doi: 10.1593/neo.81074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rehman M, Tamagnone L. Semaphorins in cancer: biological mechanisms and therapeutic approaches. Semin Cell Dev Biol. 2013;24:179–189. doi: 10.1016/j.semcdb.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Bagri A, Tessier-Lavigne M, Watts RJ. Neuropilins in tumor biology. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:1860–1864. doi: 10.1158/1078-0432.CCR-08-0563. [DOI] [PubMed] [Google Scholar]
- 19.Chen H, Chedotal A, He Z, Goodman CS, Tessier-Lavigne M. Neuropilin-2, a novel member of the neuropilin family, is a high affinity receptor for the semaphorins Sema E and Sema IV but not Sema III. Neuron. 1997;19:547–559. doi: 10.1016/s0896-6273(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 20.Soker S, Takashima S, Miao HQ, Neufeld G, Klagsbrun M. Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. Cell. 1998;92:735–745. doi: 10.1016/s0092-8674(00)81402-6. [DOI] [PubMed] [Google Scholar]
- 21.Giger RJ, Urquhart ER, Gillespie SK, Levengood DV, Ginty DD, Kolodkin AL. Neuropilin-2 is a receptor for semaphorin IV: insight into the structural basis of receptor function and specificity. Neuron. 1998;21:1079–1092. doi: 10.1016/s0896-6273(00)80625-x. [DOI] [PubMed] [Google Scholar]
- 22.Potiron VA, Roche J, Drabkin HA. Semaphorins and their receptors in lung cancer. Cancer letters. 2009;273:1–14. doi: 10.1016/j.canlet.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potiron VA, Sharma G, Nasarre P, Clarhaut JA, Augustin HG, Gemmill RM, et al. Semaphorin SEMA3F affects multiple signaling pathways in lung cancer cells. Cancer research. 2007;67:8708–8715. doi: 10.1158/0008-5472.CAN-06-3612. [DOI] [PubMed] [Google Scholar]
- 24.Johannes G, Sarnow P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. Rna. 1998;4:1500–1513. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Argast GM, Krueger JS, Thomson S, Sujka-Kwok I, Carey K, Silva S, et al. Inducible expression of TGFbeta, snail and Zeb1 recapitulates EMT in vitro and in vivo in a NSCLC model. Clin Exp Metastasis. 2011;28:593–614. doi: 10.1007/s10585-011-9394-8. [DOI] [PubMed] [Google Scholar]
- 26.Dave N, Guaita-Esteruelas S, Gutarra S, Frias A, Beltran M, Peiro S, et al. Functional cooperation between Snail1 and twist in the regulation of ZEB1 expression during epithelial to mesenchymal transition. The Journal of biological chemistry. 2011;286:12024–12032. doi: 10.1074/jbc.M110.168625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao Y, Szabolcs A, Dutta SK, Yaqoob U, Jagavelu K, Wang L, et al. Neuropilin-1 mediates divergent R-Smad signaling and the myofibroblast phenotype. The Journal of biological chemistry. 2010;285:31840–31848. doi: 10.1074/jbc.M110.151696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Massague J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bremnes RM, Veve R, Gabrielson E, Hirsch FR, Baron A, Bemis L, et al. High-throughput tissue microarray analysis used to evaluate biology and prognostic significance of the E-cadherin pathway in non-small-cell lung cancer. J Clin Oncol. 2002;20:2417–2428. doi: 10.1200/JCO.2002.08.159. [DOI] [PubMed] [Google Scholar]
- 30.Lantuejoul S, Constantin B, Drabkin H, Brambilla C, Roche J, Brambilla E. Expression of VEGF, semaphorin SEMA3F, and their common receptors neuropilins NP1 and NP2 in preinvasive bronchial lesions, lung tumours, and cell lines. J Pathol. 2003;200:336–347. doi: 10.1002/path.1367. [DOI] [PubMed] [Google Scholar]
- 31.Maroney PA, Yu Y, Nilsen TW. MicroRNAs, mRNAs, and translation. Cold Spring Harb Symp Quant Biol. 2006;71:531–535. doi: 10.1101/sqb.2006.71.043. [DOI] [PubMed] [Google Scholar]
- 32.Lagana A, Forte S, Russo F, Giugno R, Pulvirenti A, Ferro A. Prediction of human targets for viral-encoded microRNAs by thermodynamics and empirical constraints. J RNAi Gene Silencing. 2010;6:379–385. [PMC free article] [PubMed] [Google Scholar]
- 33.Chaudhury A, Hussey GS, Ray PS, Jin G, Fox PL, Howe PH. TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat Cell Biol. 2010;12:286–293. doi: 10.1038/ncb2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee MK, Pardoux C, Hall MC, Lee PS, Warburton D, Qing J, et al. TGF-beta activates Erk MAP kinase signalling through direct phosphorylation of ShcA. Embo J. 2007;26:3957–3967. doi: 10.1038/sj.emboj.7601818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davies M, Robinson M, Smith E, Huntley S, Prime S, Paterson I. Induction of an epithelial to mesenchymal transition in human immortal and malignant keratinocytes by TGF-beta1 involves MAPK, Smad and AP-1 signalling pathways. J Cell Biochem. 2005;95:918–931. doi: 10.1002/jcb.20458. [DOI] [PubMed] [Google Scholar]
- 36.Talotta F, Mega T, Bossis G, Casalino L, Basbous J, Jariel-Encontre I, et al. Heterodimerization with Fra-1 cooperates with the ERK pathway to stabilize c-Jun in response to the RAS oncoprotein. Oncogene. 2010;29:4732–4740. doi: 10.1038/onc.2010.211. [DOI] [PubMed] [Google Scholar]
- 37.Kustikova O, Kramerov D, Grigorian M, Berezin V, Bock E, Lukanidin E, et al. Fra-1 induces morphological transformation and increases in vitro invasiveness and motility of epithelioid adenocarcinoma cells. Mol Cell Biol. 1998;18:7095–7105. doi: 10.1128/mcb.18.12.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin S, Dimitri CA, Yoon SO, Dowdle W, Blenis J. ERK2 but not ERK1 induces epithelial-to-mesenchymal transformation via DEF motif-dependent signaling events. Mol Cell. 2010;38:114–127. doi: 10.1016/j.molcel.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bandyopadhyay B, Han A, Dai J, Fan J, Li Y, Chen M, et al. TbetaRI/Alk5-independent TbetaRII signaling to ERK1/2 in human skin cells according to distinct levels of TbetaRII expression. J Cell Sci. 2011;124:19–24. doi: 10.1242/jcs.076505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yi JY, Shin I, Arteaga CL. Type I transforming growth factor beta receptor binds to and activates phosphatidylinositol 3-kinase. The Journal of biological chemistry. 2005;280:10870–10876. doi: 10.1074/jbc.M413223200. [DOI] [PubMed] [Google Scholar]
- 41.Lamouille S, Derynck R. Emergence of the phosphoinositide 3-kinase-Akt-mammalian target of rapamycin axis in transforming growth factor-beta-induced epithelial-mesenchymal transition. Cells Tissues Organs. 2011;193:8–22. doi: 10.1159/000320172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG., Jr The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer cell. 2005;8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 43.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 44.Witta SE, Gemmill RM, Hirsch FR, Coldren CD, Hedman K, Ravdel L, et al. Restoring E-cadherin expression increases sensitivity to epidermal growth factor receptor inhibitors in lung cancer cell lines. Cancer research. 2006;66:944–950. doi: 10.1158/0008-5472.CAN-05-1988. [DOI] [PubMed] [Google Scholar]
- 45.Roche J, Nasarre P, Gemmill R, Baldys A, Pontis J, Korch C, Guilhot J, Ait-Si-ali S, Drabkin H. Global decrease of histone H3K27 acetylation in ZEB1-induced epithelial to mesenchymal transition in lung cancer cells. Cancers. 2013;5:334–356. doi: 10.3390/cancers5020334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gregory PA, Bracken CP, Smith E, Bert AG, Wright JA, Roslan S, et al. An autocrine TGF-beta/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol Biol Cell. 2011;22:1686–1698. doi: 10.1091/mbc.E11-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verschueren K, Remacle JE, Collart C, Kraft H, Baker BS, Tylzanowski P, et al. SIP1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5'-CACCT sequences in candidate target genes. The Journal of biological chemistry. 1999;274:20489–20498. doi: 10.1074/jbc.274.29.20489. [DOI] [PubMed] [Google Scholar]
- 48.Remacle JE, Kraft H, Lerchner W, Wuytens G, Collart C, Verschueren K, et al. New mode of DNA binding of multi-zinc finger transcription factors: deltaEF1 family members bind with two hands to two target sites. Embo J. 1999;18:5073–5084. doi: 10.1093/emboj/18.18.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Postigo AA, Depp JL, Taylor JJ, Kroll KL. Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. Embo J. 2003;22:2453–2462. doi: 10.1093/emboj/cdg226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kessler O, Shraga-Heled N, Lange T, Gutmann-Raviv N, Sabo E, Baruch L, et al. Semaphorin-3F is an inhibitor of tumor angiogenesis. Cancer research. 2004;64:1008–1015. doi: 10.1158/0008-5472.can-03-3090. [DOI] [PubMed] [Google Scholar]
- 51.Bielenberg DR, Hida Y, Shimizu A, Kaipainen A, Kreuter M, Kim CC, et al. Semaphorin 3F, a chemorepulsant for endothelial cells, induces a poorly vascularized, encapsulated, nonmetastatic tumor phenotype. The Journal of clinical investigation. 2004;114:1260–1271. doi: 10.1172/JCI21378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wendt MK, Tian M, Schiemann WP. Deconstructing the mechanisms and consequences of TGF-beta-induced EMT during cancer progression. Cell Tissue Res. 2012;347:85–101. doi: 10.1007/s00441-011-1199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang HT, Chen XF, Wang MH, Wang JC, Qi QY, Zhang RM, et al. Defective expression of transforming growth factor beta receptor type II is associated with CpG methylated promoter in primary non-small cell lung cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004;10:2359–2367. doi: 10.1158/1078-0432.ccr-0959-3. [DOI] [PubMed] [Google Scholar]
- 54.Huang S, Holzel M, Knijnenburg T, Schlicker A, Roepman P, McDermott U, et al. MED12 controls the response to multiple cancer drugs through regulation of TGF-beta receptor signaling. Cell. 2012;151:937–950. doi: 10.1016/j.cell.2012.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ocana OH, Corcoles R, Fabra A, Moreno-Bueno G, Acloque H, Vega S, et al. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer cell. 2012;22:709–724. doi: 10.1016/j.ccr.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 56.Grandclement C, Pallandre JR, Valmary Degano S, Viel E, Bouard A, Balland J, et al. Neuropilin-2 expression promotes TGF-beta1-mediated epithelial to mesenchymal transition in colorectal cancer cells. PLoS One. 2011;6:e20444. doi: 10.1371/journal.pone.0020444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gray MJ, Van Buren G, Dallas NA, Xia L, Wang X, Yang AD, et al. Therapeutic targeting of neuropilin-2 on colorectal carcinoma cells implanted in the murine liver. J Natl Cancer Inst. 2008;100:109–120. doi: 10.1093/jnci/djm279. [DOI] [PubMed] [Google Scholar]
- 58.Dallas NA, Gray MJ, Xia L, Fan F, van Buren G, 2nd, Gaur P, et al. Neuropilin-2-mediated tumor growth and angiogenesis in pancreatic adenocarcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:8052–8060. doi: 10.1158/1078-0432.CCR-08-1520. [DOI] [PubMed] [Google Scholar]
- 59.Samuel S, Gaur P, Fan F, Xia L, Gray MJ, Dallas NA, et al. Neuropilin-2 mediated beta-catenin signaling and survival in human gastro-intestinal cancer cell lines. PLoS One. 2011;6:e23208. doi: 10.1371/journal.pone.0023208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Caunt M, Mak J, Liang WC, Stawicki S, Pan Q, Tong RK, et al. Blocking neuropilin-2 function inhibits tumor cell metastasis. Cancer cell. 2008;13:331–342. doi: 10.1016/j.ccr.2008.01.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.