Abstract
Haploinsufficiency of CHD7 (OMIM# 608892) is known to cause CHARGE syndrome (OMIM# 214800). Molecular testing supports a definitive diagnosis in approximately 65%–70% of cases. Most CHD7 mutations arise de novo, and no mutations affecting exon-7 have been reported to date. We report on an 8-year-old girl diagnosed with CHARGE syndrome that was referred to our laboratory for comprehensive CHD7 gene screening. Genomic DNA from the subject with a suspected diagnosis of CHARGE was isolated from peripheral blood lymphocytes and comprehensive Sanger sequencing, along with deletion/duplication analysis of the CHD7 gene using multiplex ligation-dependent probe amplification (MLPA), was performed. MLPA analysis identified a reduced single probe signal for exon-7 of the CHD7 gene consistent with potential heterozygous deletion. Long-range PCR breakpoint analysis identified a complex genomic rearrangement (CGR) leading to the deletion of exon-7 and breakpoints consistent with a replicative mechanism such as fork stalling and template switching (FoSTeS) or microhomology-mediated break-induced replication (MMBIR). Taken together this represents the first evidence for a CHD7 intragenic CGR in a patient with CHARGE syndrome leading to what appears to be also the first report of a mutation specifically disrupting exon-7. Although likely rare, CGR may represent an overlooked mechanism in subjects with CHARGE syndrome that can be missed by current sequencing and dosage assays.
Keywords: CHARGE syndrome, CHD7, FoSTeS/MMBIR, MLPA
INTRODUCTION
CHARGE association is the acronym for Coloboma of the eyes, Heart defect, Atresia choanae, Retarded growth and development, Genital abnormality, and Ear anomaly – anomalies found more frequently together than would be predicted if seen by chance occurrences as the probabilities of individual birth defects. The pattern of abnormalities differs among individuals with CHARGE syndrome due to clinical variability, and infants may often present with life-threatening medical conditions [Issekutz et al., 2005].
Clinical diagnosis of CHARGE syndrome is based on a combination of major and minor diagnostic criteria [Verloes, 2005]. However, molecular testing of the CHD7 gene is able to confirm the diagnosis in the majority of cases. CHD7 is a chromodomain helicase DNA binding protein 7 that plays a role in transcription activation and repression by chromatin remodeling. The Chromodomain Helicase DNA-binding (CHD) protein family consists of nine members that can be subdivided into three subfamilies, all containing two chromodomains, or domains modifying the chromatin organization, which are located proximally to the N-terminus of the protein, followed by a more distal sucrose non fermenting (SNF2)-like ATP-dependent catalytic helicase motif [Hall and Georgel, 2007; Marfella and Imbalzano, 2007]. CHD7 is highly conserved across species and orthologs have been identified in both vertebrates and invertebrates such as Xenopus, zebrafish, mouse, and chicken [Aramaki et al., 2007; Bajpai et al., 2010; Bosman et al., 2005]. The CHD7 gene is associated with the majority of CHARGE syndrome cases and consists of 38 exons, of which the first is noncoding, and spans 188kb of genomic DNA on chromosome 8q12.1–q12.2 [Vissers et al., 2004; Layman et al., 2010; Janssen et al., 2012].
Sequence and deletion/duplication analysis of the CHD7 coding region detects heterozygous mutations in 65%–80% of individuals with CHARGE syndrome. The majority are single nucleotide variant (SNV) alleles, including nonsense, frameshift deletions, or missense CHD7 mutations [Vissers et al., 2004; Layman et al., 2010; Janssen et al., 2012]. CHD7 mutations in human subjects are distributed along the entire coding sequence and do not appear to be correlated with specific aspects of the clinical phenotype [Vissers et al., 2004; Layman et al., 2010; Janssen et al., 2012]. However, no mutations have been found in exon-7 (56bp), which represents the boundary of the first chromodomain [Vissers et al., 2004; Layman et al., 2010; Janssen et al., 2012]. Most CHD7 mutations identified thus far have been shown to have arisen de novo although germline mosaicism has been reported in families with multiple affected siblings [Vissers et al., 2004; Layman et al., 2010; Janssen et al., 2012].
MATERIALS AND METHODS
Patient Report
The patient is an 8-year-old Caucasian female delivered at 35-weeks gestation via cesarean due to fetal inactivity and fluctuating heart rate. She presented at birth with multiple congenital anomalies including tetralogy of Fallot (TOF), for which she underwent a Blalock-Taussig shunt procedure at 12 hours of life followed by complete surgical repair at 16 months of age. In addition to TOF, other congenital heart malformations included a right aortic arch with a vascular ring and a cleft mitral valve. Immediately after birth the patient developed significant respiratory problems, which required intubation and constant monitoring in the neonatal intensive care unit (NICU) where she remained for more than 2 months. At two weeks of age, facial dysmorphic features and the overall clinical presentation appeared to be most consistent with typical CHARGE syndrome. In addition to the conotruncal defects, the patient presented with bilateral retinal colobomas apparently without major visual impairment. The patient also underwent Auditory Brainstem Response (ABR) testing that identified normal conduction of auditory stimuli in the right ear (wave I = 1.83msec; wave II = 4.39msec; wave V = 6.07msec; wave III–V latency difference = 1.69; wave I–V latency difference = 4.25) associated with moderate sensorineural hearing loss (30decibel at 500Hz; 55decibel at 4000Hz) in her right ear. The ABR test for the left ear failed to yield reliable measurements or response suggesting severe to profound primary sensorineural hearing loss in her left ear requiring hearing aids for both ears. Computed tomography (CT) scan analysis revealed complex congenital deformities of both middle and inner ears on each side, characterized by apparent anterior mallear fusion, vestibule hypoplasia with hypoplasia/absence of semicircular canals, cochlear dysplasia and an enlarged vestibular aqueduct on the left, and possible cochlear dysplasia on the right without a definitely enlarged vestibular aqueduct. There is also partial opacification of the middle ears and mastoids.
The patient was born with choanal atresia and an H-type tracheo-esophageal fistula, both of which were surgically repaired although the patient required enteral nutrition via gastrostomy tube (G-tube). The patient also presented with severe gastroesophageal reflux, which required a Nissen fundoplication. In addition, she presented with duodenal atresia in two sites, which was surgically repaired, followed by a duodenoplasty at 7 years of age. She developed osteoporosis and demonstrated motor, speech, and cognitive delays, requiring special education along with occupational, physical, and speech therapy. Prior to CHD7 analysis, the patient had a fluorescence in situ hybridization (FISH) study for 22q11.2 deletion, which was negative. The patient has also a healthy sibling, and the proband’s family history is otherwise unremarkable for CHARGE syndrome, heart defects, or other genetic syndromes.
GENETIC ANALYSIS
Genomic DNA from this individual was obtained after informed consent for clinical testing of the coding sequence and exon/intron boundaries of the CHD7 gene (RefSeq: NM_017780.3) and analyzed using polymerase chain reaction (PCR) and Sanger DNA sequencing as previously described, along with multiplex ligation-dependent probe amplification (MLPA) using a kit from MRC Holland (www.mlpa.com) in order to search for intragenic deletions or duplications as previously published [Wincent et al., 2008, 2009].
Long fragment PCR product (~7.3Kb) flanking the deleted region was obtained using the following primers: TGTTGCAGAAGGAATCTGGA (exon-7 intron/exon boundary forward) and TGCTTTGCAAACTCATTCCA (Intron 4 forward), CTGTCTGGGCCAGGTGTACT (Intron 7 reverse), ATGCCTGCTGGTCTGTTTCT (Intron 6 forward) and GCTTGAGCCACCCCTATTTA (Intron 6 reverse), TACCATTTCTCAAGTCAGGCCA (Intron 6/Intron 4 boundary reverse) linked to and M13 tail. Amplified junctional fragment PCR products were purified and sequenced as described above. Computational analysis was employed to examine the CHD7 genomic sequence encompassing the breakpoints observed in our patient sample.
RESULTS
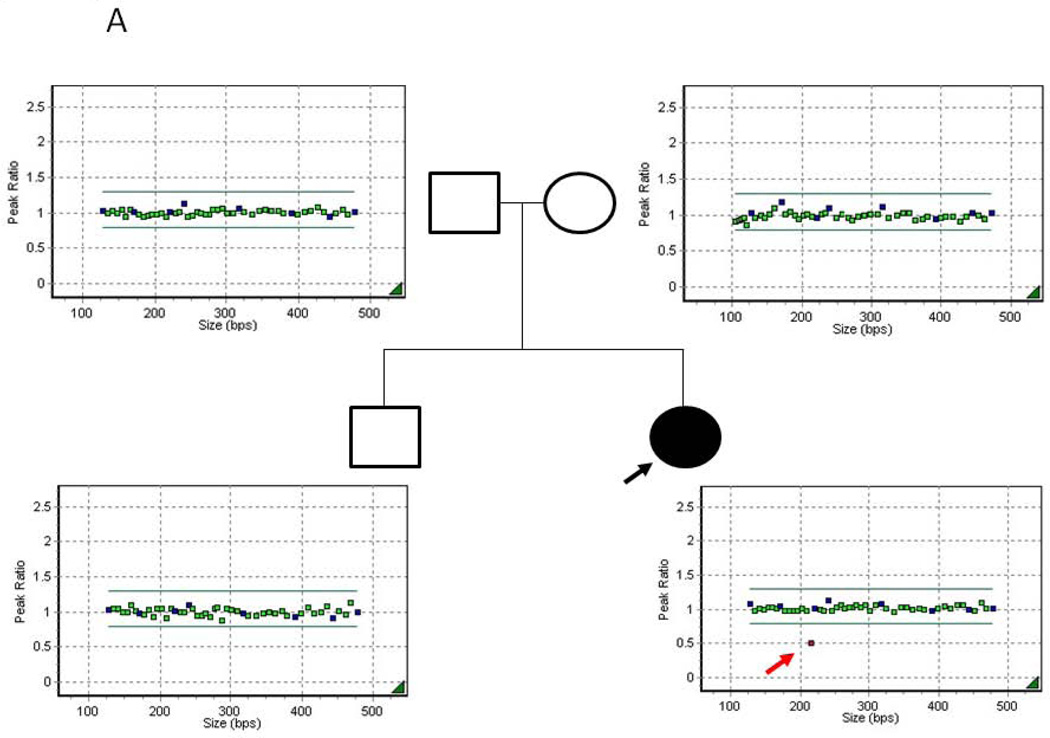
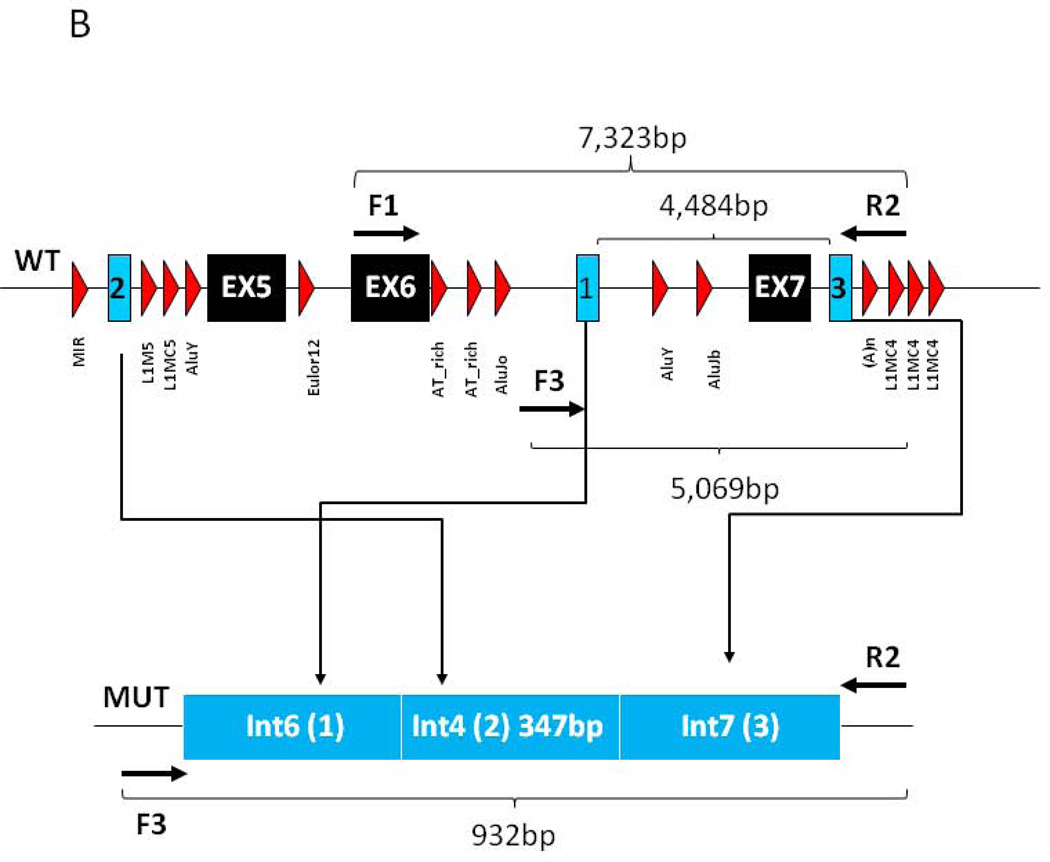
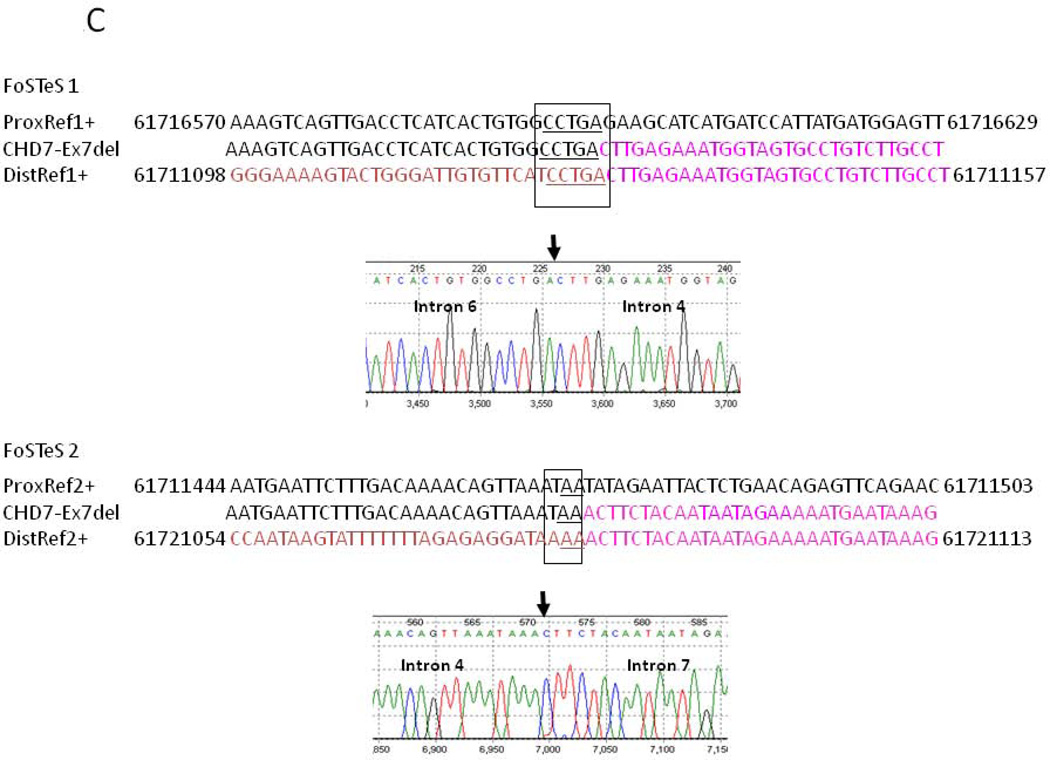
Sequence analysis did not identify any changes that could be classified as disease-causing while MLPA analysis, which was repeated on separate samples from the index case, showed consistently reduced signal for a single probe in exon-7, indicating a potential copy number variant (CNV) and possible heterozygous deletion of exon-7 of the CHD7 gene (Fig 1A). Neither pathogenic CNV nor single nucleotide variants (SNV) involving exon-7 has been previously reported in patients with CHARGE syndrome (www.chd7.org). Given that the possible deletion was suggested by a single probe and due to the proximity of the flanking exons, we employed a long range PCR approach as a secondary method, necessary for confirmation. Long fragment amplification and junctional PCR product sequencing identified the breakpoint junctions in introns 6 and 7 (Figs 1B and 2C), consistent with a deletion of approximately 4484 nucleotides, which includes exon-7. Furthermore, sequence analysis of the PCR product encompassing the breakpoint junction revealed that approximately 347 nucleotides from intron 4 (human reference sequence GRCh37/hg19: chr8: 61711127–61711473) of the CHD7 gene were inserted between intron 6 and 7 breakpoints (Figs. 1B and 2C); all three distinct and usually separate genomic segments were juxtaposed in a direct orientation, and microhomology (5bp and 2bp) was observed at the breakpoint junctions (Figs. 1B and 2C). Samples from the unaffected parents and sib of the index cases were tested and showed normal signal for the MLPA exon-7 probe of the CHD7 gene (Fig 1A) and normal PCR fragment size (Figs. 2A–B). Taken together, these data support a de novo heterozygous deletion of exon-7 of the CHD7 gene in our patient, which is consistent with a clinical diagnosis of CHARGE syndrome.
Figure 1. MLPA analysis and genomic structure encompassing exon 7.
A) MLPA analysis of the proband and available first degree relatives. Filled symbol and black arrow indicates the proband. Unfilled symbols indicate unaffected relatives. The red arrow indicates the single probe with reduced signal in proband DNA. B) Genomic structure of CHD7 encompassing exon 7. Filled in black are the exons, in light blue the sequence composing the junctional fragment, while in red are the repetitive elements. Arrows indicate the position of the primers employed for the long range PCR analysis.
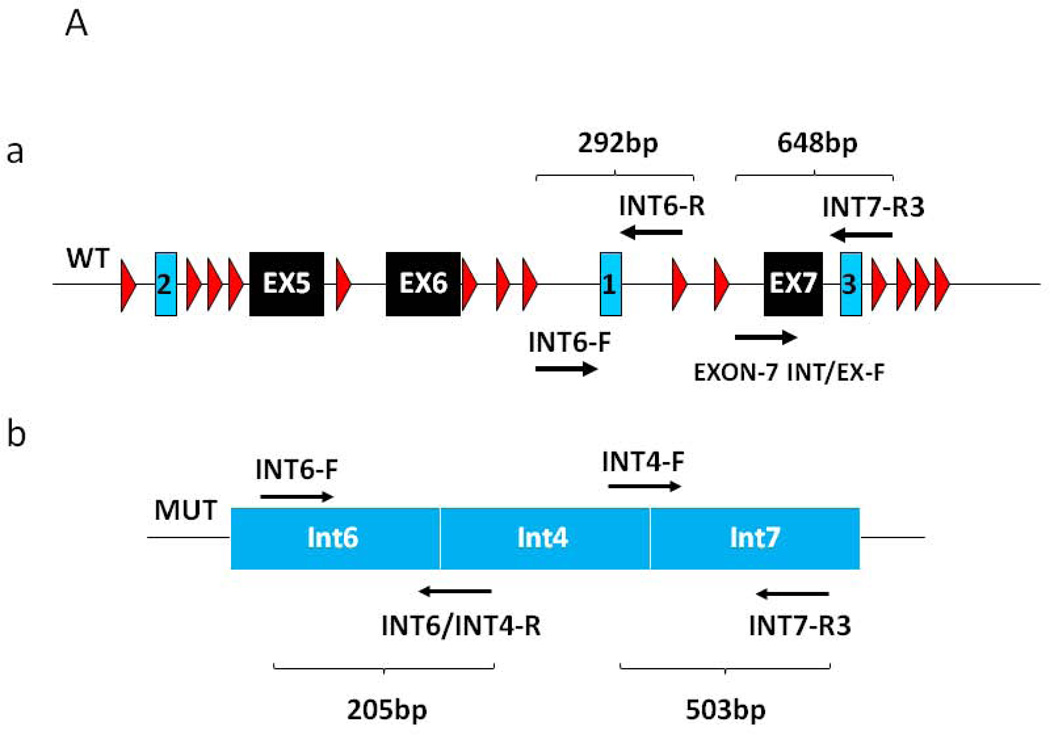
Figure 2. Multiplex PCR analysis of the wild type and mutant CHD7 alleles.
A) Relative position of the primers designed for the amplification of the wild type CHD7 allele fragments (a) and for mutant CHD7 allele fragments (b). B) Electrophoretic analysis of the multiplex PCR amplification along with the family pedigree. PCR fragment size is indicated. C) Computational analysis and electropherograms of the sequences composing the junctional fragment indicating the microhomology of the two breakpoints consistent with FoSTeS/MMBIR (FoSTeS 1 and 2). The position numbering of the nucleotide sequence refers to the genomic coordinates.
Computational analysis of the CHD7 genomic sequence encompassing the breakpoint identified regions of micro-homology (Fig 2C) consistent with this complex genomic rearrangement having been generated by replicative mechanism such as FoSTeS/MMBIR [Lee et al., 2007; Zhang et al., 2009; Hastings et al., 2009].
DISCUSSION
In this report, we provide the first evidence of a deletion of exon-7 of the CHD7 gene in an individual with a clinical diagnosis of CHARGE syndrome as a result of a complex genomic rearrangement consistent with a replicative mutational mechanism.13 In addition, this is the first report that involves specifically a deletion of exon-7, which represents the distal boundary of the first chromodomain of CHD7 [Vissers et al., 2004]. Interestingly, in the vicinity of breakpoint one and two, there are two AluY sequence elements, and although the rearrangements could not be explained by Alu-mediated recombinations, it has been recently hypothesized that AluY elements could facilitate template switching and annealing via their 31-bp microhomology between replication forks and restarting (priming) DNA replication in one of the multiple FoSTeS/MMBIR events that generated the complexity [Zhang et al., 2009; Hastings et al., 2009]. Previously, a de novo Alu retrotransposition-mediated deletion encompassing exons 8–12 of CHD7 was identified in a CHARGE syndrome subject [Udaka et al., 2007].
The FoSTeS/MMBIR mechanism represents an aberrant pattern of DNA replication during mitosis in which the active replication fork can stall and switch templates using complementary microhomologous DNA template to anneal and initiate DNA replication [Zhang et al., 2009]. Linearly, the replication forks could be parted by a significant genomic fragment although the DNA bending could favor their physical vicinity, allowing the utilization of the same replisome machinery. Therefore, the FoSTeS/MMBIR mechanism may represent a significant source of structural variations in the human genome and cause of human diseases, such as CHARGE.
ACKNOWLEDGMENTS
This work was supported by the Indiana University Health-Indiana University School of Medicine Strategic Research Initiative (MV and KGS) and in part by NINDS RO1NS058529 and NHGRI U54HG006542 (JRL).
Footnotes
DISCLOSURE
MV, ZN, PF, CME and ASW at the time of performing the project were at the Medical Genetics Laboratories, a Diagnostic Laboratory at Baylor College of Medicine. JRL is a paid consultant for Athena Diagnostics, has stock ownership in 23 and Me and Ion Torrent Systems, and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis (CMA) and clinical exome sequencing offered in the Medical Genetics Laboratory (MGL; http://www.bcm.edu/geneticlabs/). At the best of our knowledge there are no other conflicts of interest to disclose and all procedures have been compliant with ethical regulations.
REFERENCES
- Aramaki M, Kimura T, Udaka T, Kosaki R, Mitsuhashi T, Okada Y, Takahashi T, Kosaki K. Embryonic expression profile of chicken CHD7, the ortholog of the causative gene for CHARGE syndrome. Birth Defects Res A Clin Mol Teratol. 2007;79:50–57. doi: 10.1002/bdra.20330. [DOI] [PubMed] [Google Scholar]
- Bajpai R, Chen DA, Rada-Iglesias A, Zhang J, Xiong Y, Helms J, Chang CP, Zhao Y, Swigut T, Wysocka J. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature. 2010;463:958–962. doi: 10.1038/nature08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosman EA, Penn AC, Ambrose JC, Kettleborough R, Stemple DL, Steel KP. Multiple mutations in mouse Chd7 provide models for CHARGE syndrome. Hum Mol Genet. 2005;14:3463–3476. doi: 10.1093/hmg/ddi375. [DOI] [PubMed] [Google Scholar]
- Hall JA, Georgel PT. CHD proteins: a diverse family with strong ties. Biochem Cell Biol. 2007;85:463–476. doi: 10.1139/O07-063. [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issekutz KA, Graham JM, Prasad C, Smith IM, Blake KD. An epidemiological analysis of CHARGE syndrome: preliminary results from a Canadian study. Am J Med Genet A. 2005;133A:309–317. doi: 10.1002/ajmg.a.30560. [DOI] [PubMed] [Google Scholar]
- Janssen N, Bergman JE, Swertz MA, Tranebjaerg L, Lodahl M, Schoots J, Hofstra RM, van Ravenswaaij-Arts CM, Hoefsloot LH. Mutation update on the CHD7 gene involved in CHARGE syndrome. Hum Mutat. 2012;33:1149–1160. doi: 10.1002/humu.22086. [DOI] [PubMed] [Google Scholar]
- Layman WS, Hurd EA, Martin DM. Chromodomain proteins in development: lessons from CHARGE syndrome. Clin Genet. 2010;78(1):11–20. doi: 10.1111/j.1399-0004.2010.01446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Marfella CG, Imbalzano AN. The Chd family of chromatin remodelers. Mutat Res. 2007;618(1–2):30–40. doi: 10.1016/j.mrfmmm.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udaka T, Okamoto N, Aramaki M, Torii C, Kosaki R, Hosokai N, Hayakawa T, Takahata N, Takahashi T, Kosaki K. An Alu retrotransposition-mediated deletion of CHD7 in a patient with CHARGE syndrome. Am J Med Genet A. 2007;143:721–726. doi: 10.1002/ajmg.a.31441. [DOI] [PubMed] [Google Scholar]
- Verloes A. Updated diagnostic criteria for CHARGE syndrome: a proposal. Am J Med Genet A. 2005;133A:306–308. doi: 10.1002/ajmg.a.30559. [DOI] [PubMed] [Google Scholar]
- Vissers LE, van Ravenswaaij CM, Admiraal R, Hurst JA, de Vries BB, Janssen IM, van der Vliet WA, Huys EH, de Jong PJ, Hamel BC, Schoenmakers EF, Brunner HG, Veltman JA, van Kessel AG. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nature Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- Wincent J, Holmberg E, Strömland K, Soller M, Mirzaei L, Djureinovic T, Robinson K, Anderlid B, Schoumans J. CHD7 mutation spectrum in 28 Swedish patients diagnosed with CHARGE syndrome. Clin Genet. 2008;74:31–38. doi: 10.1111/j.1399-0004.2008.01014.x. [DOI] [PubMed] [Google Scholar]
- Wincent J, Schulze A, Schoumans J. Detection of CHD7 deletions by MLPA in CHARGE syndrome patients with a less typical phenotype. Eur J Med Genet. 2009;52(4):271–272. doi: 10.1016/j.ejmg.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Zhang F, Khajavi M, Connolly AM, Towne CF, Batish SD, Lupski JR. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet. 2009;41:849–853. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]