Abstract
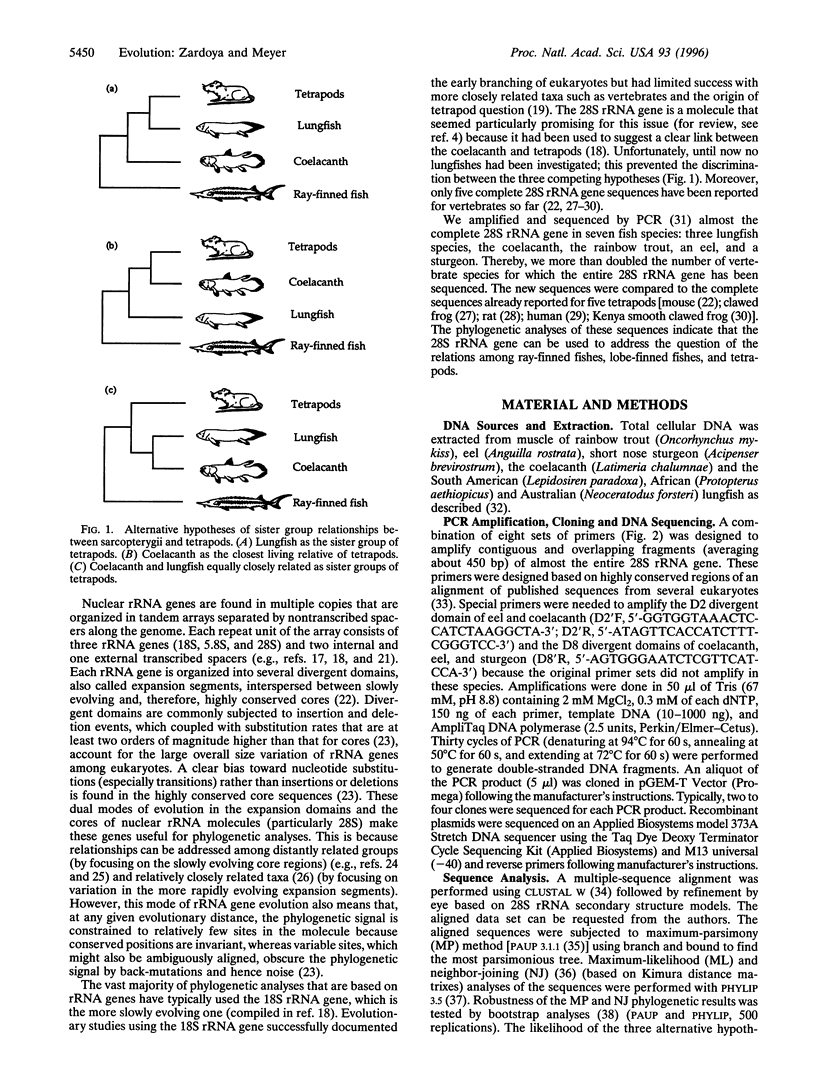
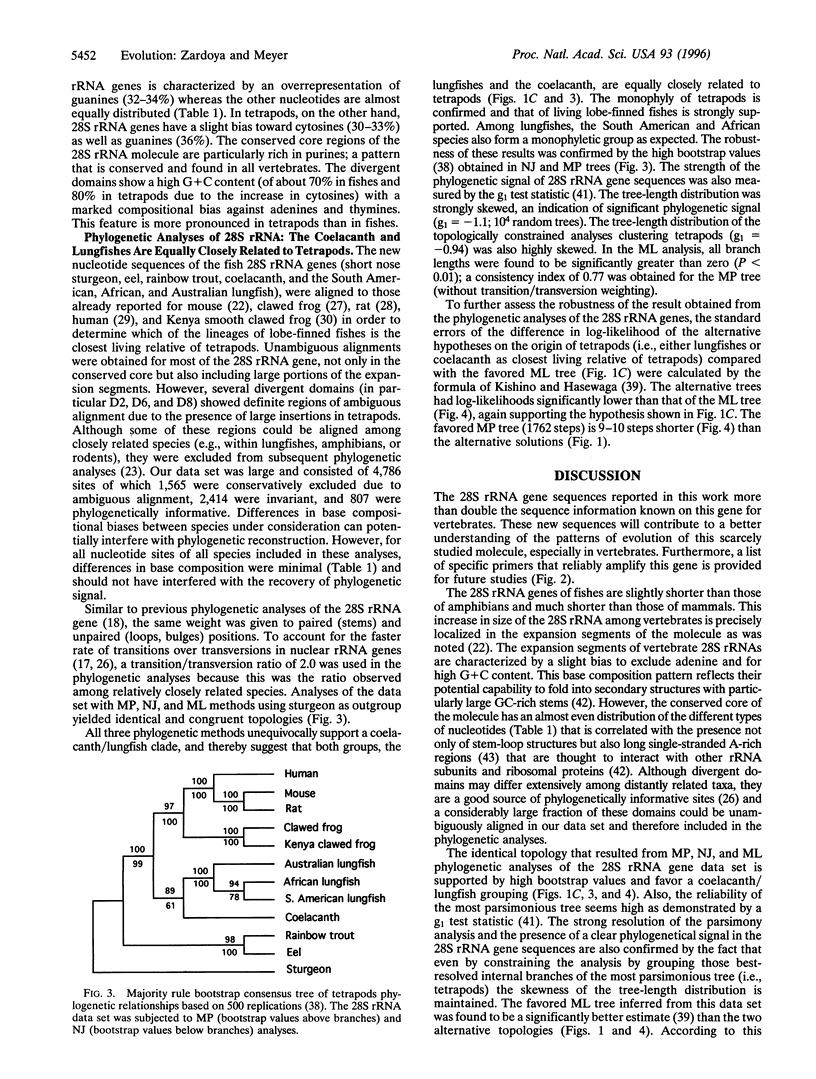
The origin of land vertebrates was one of the major transitions in the history of vertebrates. Yet, despite many studies that are based on either morphology or molecules, the phylogenetic relationships among tetrapods and the other two living groups of lobe-finned fishes, the coelacanth and the lungfishes, are still unresolved and debated. Knowledge of the relationships among these lineages, which originated back in the Devonian, has profound implications for the reconstruction of the evolutionary scenario of the conquest of land. We collected the largest molecular data set on this issue so far, about 3,500 base pairs from seven species of the large 28S nuclear ribosomal gene. All phylogenetic analyses (maximum parsimony, neighbor-joining, and maximum likelihood) point toward the hypothesis that lungfishes and coelacanths form a monophyletic group and are equally closely related to land vertebrates. This evolutionary hypothesis complicates the identification of morphological or physiological preadaptations that might have permitted the common ancestor of tetrapods to colonize land. This is because the reconstruction of its ancestral conditions would be hindered by the difficulty to separate uniquely derived characters from shared derived characters in the coelacanth/lungfish and tetrapod lineages. This molecular phylogeny aids in the reconstruction of morphological evolutionary steps by providing a framework; however, only paleontological evidence can determine the sequence of morphological acquisitions that allowed lobe-finned fishes to colonize land.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ajuh P. M., Heeney P. A., Maden B. E. Xenopus borealis and Xenopus laevis 28S ribosomal DNA and the complete 40S ribosomal precursor RNA coding units of both species. Proc Biol Sci. 1991 Jul 22;245(1312):65–71. doi: 10.1098/rspb.1991.0089. [DOI] [PubMed] [Google Scholar]
- Gonzalez I. L., Gorski J. L., Campen T. J., Dorney D. J., Erickson J. M., Sylvester J. E., Schmickel R. D. Variation among human 28S ribosomal RNA genes. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7666–7670. doi: 10.1073/pnas.82.22.7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M., Li W. H. Molecular phylogeny of the kingdoms Animalia, Plantae, and Fungi. Mol Biol Evol. 1989 Mar;6(2):109–122. doi: 10.1093/oxfordjournals.molbev.a040536. [DOI] [PubMed] [Google Scholar]
- Gutell R. R., Weiser B., Woese C. R., Noller H. F. Comparative anatomy of 16-S-like ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- Hadjiolov A. A., Georgiev O. I., Nosikov V. V., Yavachev L. P. Primary and secondary structure of rat 28 S ribosomal RNA. Nucleic Acids Res. 1984 Apr 25;12(8):3677–3693. doi: 10.1093/nar/12.8.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M., Iida Y., Yano T., Takaiwa F., Iwabuchi M. Phylogenetic relationships among eukaryotic kingdoms inferred from ribosomal RNA sequences. J Mol Evol. 1985;22(1):32–38. doi: 10.1007/BF02105802. [DOI] [PubMed] [Google Scholar]
- Hassouna N., Michot B., Bachellerie J. P. The complete nucleotide sequence of mouse 28S rRNA gene. Implications for the process of size increase of the large subunit rRNA in higher eukaryotes. Nucleic Acids Res. 1984 Apr 25;12(8):3563–3583. doi: 10.1093/nar/12.8.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges S. B., Hass C. A., Maxson L. R. Relations of fish and tetrapods. Nature. 1993 Jun 10;363(6429):501–502. doi: 10.1038/363501b0. [DOI] [PubMed] [Google Scholar]
- Hillis D. M., Dixon M. T. Ribosomal DNA: molecular evolution and phylogenetic inference. Q Rev Biol. 1991 Dec;66(4):411–453. doi: 10.1086/417338. [DOI] [PubMed] [Google Scholar]
- Hillis D. M., Huelsenbeck J. P. Signal, noise, and reliability in molecular phylogenetic analyses. J Hered. 1992 May-Jun;83(3):189–195. doi: 10.1093/oxfordjournals.jhered.a111190. [DOI] [PubMed] [Google Scholar]
- Kishino H., Hasegawa M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J Mol Evol. 1989 Aug;29(2):170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- Kocher T. D., Thomas W. K., Meyer A., Edwards S. V., Päbo S., Villablanca F. X., Wilson A. C. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6196–6200. doi: 10.1073/pnas.86.16.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson A., Wilson A. C. Patterns of ribosomal RNA evolution in salamanders. Mol Biol Evol. 1989 Mar;6(2):131–154. doi: 10.1093/oxfordjournals.molbev.a040539. [DOI] [PubMed] [Google Scholar]
- Le H. L., Lecointre G., Perasso R. A 28S rRNA-based phylogeny of the gnathostomes: first steps in the analysis of conflict and congruence with morphologically based cladograms. Mol Phylogenet Evol. 1993 Mar;2(1):31–51. doi: 10.1006/mpev.1993.1005. [DOI] [PubMed] [Google Scholar]
- Lenaers G., Maroteaux L., Michot B., Herzog M. Dinoflagellates in evolution. A molecular phylogenetic analysis of large subunit ribosomal RNA. J Mol Evol. 1989 Jul;29(1):40–51. doi: 10.1007/BF02106180. [DOI] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- Meyer A., Dolven S. I. Molecules, fossils, and the origin of tetrapods. J Mol Evol. 1992 Aug;35(2):102–113. doi: 10.1007/BF00183221. [DOI] [PubMed] [Google Scholar]
- Meyer A., Wilson A. C. Origin of tetrapods inferred from their mitochondrial DNA affiliation to lungfish. J Mol Evol. 1990 Nov;31(5):359–364. doi: 10.1007/BF02106050. [DOI] [PubMed] [Google Scholar]
- Olsen G. J., Woese C. R. Ribosomal RNA: a key to phylogeny. FASEB J. 1993 Jan;7(1):113–123. doi: 10.1096/fasebj.7.1.8422957. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994 Nov 11;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vawter L., Brown W. M. Rates and patterns of base change in the small subunit ribosomal RNA gene. Genetics. 1993 Jun;134(2):597–608. doi: 10.1093/genetics/134.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware V. C., Tague B. W., Clark C. G., Gourse R. L., Brand R. C., Gerbi S. A. Sequence analysis of 28S ribosomal DNA from the amphibian Xenopus laevis. Nucleic Acids Res. 1983 Nov 25;11(22):7795–7817. doi: 10.1093/nar/11.22.7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobori S., Hasegawa M., Ueda T., Okada N., Nishikawa K., Watanabe K. Relationship among coelacanths, lungfishes, and tetrapods: a phylogenetic analysis based on mitochondrial cytochrome oxidase I gene sequences. J Mol Evol. 1994 Jun;38(6):602–609. doi: 10.1007/BF00175880. [DOI] [PubMed] [Google Scholar]



