Abstract
Surprisingly few protein kinases have been demonstrated in chloroplasts or mitochondria. Here we discuss the “activity of bc1 complex kinase” (ABC1K) protein family which we suggest locate in mitochondria and plastids, thus filling the kinase void. The ABC1Ks are atypical protein kinases and their ancestral function is the regulation of quinone synthesis. ABC1Ks have proliferated from 1–2 members in non-photosynthetic organisms to more than 16 members in algae and higher plants. In this review we reconstruct the evolutionary history of the ABC1K family, provide a functional domain analysis for angiosperms and a nomenclature for ABC1Ks in Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa) and maize (Zea mays). Finally, we hypothesize that targets of ABC1Ks include enzymes of prenyl-lipid metabolism as well as components of the organellar gene expression machineries.
The protein kinase-like superfamily
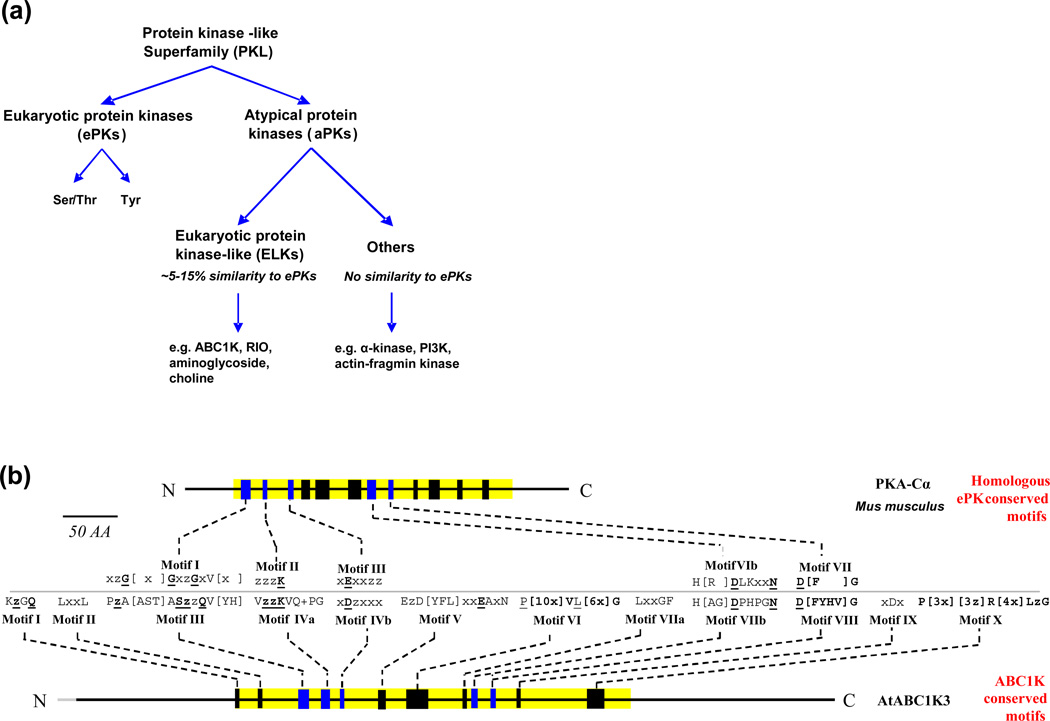
The protein kinase-like (PKL) superfamily encompasses all protein kinases and a subset of small molecule/metabolite kinases (eg phosphatidyl-inositol phosphate kinases) [1, 2]. The PKL superfamily displays an enormous variability in sequence and structure to match the wide array of target substrates. PKLs can be subdivided between the eukaryotic protein kinases (ePKs) prevalent in the eukaryotes, and the atypical protein kinases (aPKs), which predominate in the prokaryotes [2] (Figure 1A). Two-component kinases (ie histidine-aspartate kinases) form a separate family and are important in prokaryotes [3], but have been adapted by eukaryotes [4]. Sequence alignment analyses of numerous, diverse ePKs have established a ~250 amino acid ePK catalytic domain containing twelve subdomains [5, 6], while x-ray crystal structures of ePKs provide functional context for these subdomains (see e.g. [7–11]). The aPKs share little or no homology with ePKs, although crystal structures indicate that most maintain a similar overall protein kinase fold [12]. In silico sequence and structural studies of the entire PKL superfamily reveal only ~10 residues conserved across the ePKs and aPKs; and even these residues have been shown to be dispensable in certain PKLs [2, 12]. The ePK-like (ELK) group of aPKs, that share some sequence identity with ePKs (usually < 15%), have emerged as important regulatory kinases of bacteria [13–16] and include the activity of bc1 complex kinase” (ABC1K) family [17], RIO kinases [18, 19], aminoglycoside kinases [20], and others [2] (Figure 1A). In this article we will explore the evolutionary history of ABC1Ks, and provide support for their significance in plant mitochondria and plastids. We also propose a logical and complete numbering for all Arabidopsis, maize and rice ABC1K proteins based on phylogeny. We note that the ABC1K family has no relationship with the ATP-binding cassette (ABC) membrane transporter family.
Figure 1.
Categorical relationships and conserved motifs of the ABC1K protein domain, compared with the ePK family of kinases. (a) The diagram illustrates the categorization of the protein kinase-like superfamily. (b) The twelve conserved motifs found in the ABC1K domain from an alignment of 100 sequences of three eudicot species (Arabidopsis, M. truncatula, and P. trichocarpa) and four monocot species (Z. mays, O. sativa, B. distachyon, S. bicolor), are illustrated using the prototypical ePK of Mus musculus, protein kinase C alpha (PKA-Cα), and AtABC1K3. Five motifs are shared between ABC1Ks and ePKs and are colored in blue in the cartoon and are aligned to each other for comparison. Conserved motifs are described using the single letter code of amino acid residues and residues conserved in >75% of sequences are shown. Bold, underlined residues are conserved in 100% of the aligned sequences, z indicates a hydrophobic residue,×indicates any residue, + indicates a positive-charged residue. ABC1K motifs shared with the ePK motif: Motif III A part of the NTP-binding pocket, this motif is responsible for anchoring the α- and β-phosphate groups and positioning the γ-phosphate for catalysis. The sequence in ABC1Ks is divergent from the “GxGxxG” sequence seen in ePKs and other NTP-binding pockets. However, the glycine-rich loop of ePKs forms a turn that is easily substituted with the small side-chains of Ala and Ser, as seen in the ABC1K family [60]. Motif IVa Contains the invariant lysine helping to anchor the NTP by binding the α- and β-phosphates and also forms a salt bridge with the carboxyl group of the Asp in motif IVb. As in ePKs, this invariant lysine lies 14 residues downstream of the NTP-binding pocket (12–21 residues in ePKs [60]). Motif IVb The aspartic acid residue is homologous to the conserved glutamic acid residue of ePK motif III, necessary for stabilization of the interaction between Lys of motif III and the α- and β-phosphate of NTP. As in casein kinase 2 and a number of other ePKs, the conserved acidic residue lies exactly 13 residues downstream of the invariant lysine [6], however the pair of hydrophobic residues downstream of glutamic acid (+3 from Glu), required in ePKs, is not maintained in ABC1Ks, but they rather conserve a hydrophobic residue at the next position (+1 from Asp), suggesting a different local protein fold. Motif VIIb Referred to as the catalytic loop in ePKs, the conserved Asp serves as the catalytic base, activating the substrate hydroxyl group. Motif VIII Anchors the Mg2+ necessary for positioning the α- and β-phosphate of the NTP. The Asp chelates the divalent cation through the assistance of a hydrophobic bond with the conserved Gly.
A protein kinase void in plant plastids and mitochondria
The coordination of complex processes in chloroplasts and mitochondria, such as photosynthesis and respiration, and their need for adaptations to changes in environmental conditions, likely involves an assortment of kinases. So far nearly 200 Arabidopsis chloroplast phosphoproteins have been identified in Arabidopsis leaves [21, 22]. The true number of kinase targets in chloroplast and non-photosynthetic plastids is likely much larger, considering the technical challenges to identify phosphopeptides, and considering that samples from only a small number of developmental states and (a)biotic conditions have been analyzed [23–25]. Phosphoproteome analysis of yeast mitochondria revealed a kinase network including 48 phosphoproteins involved in critical mitochondrial functions including carbohydrate metabolism, redox regulation, and apoptosis [26]. The phosphoproteome of plant mitochondria is poorly understood and fewer than 20 phosphoproteins have been detected in Arabidopsis [27].
So far, identified protein kinases are under-represented in the characterized proteomes of plant plastids and mitochondria [28, 29], likely because most attention has focused on ePKs. However, many kinases in plastids and mitochondria may be bacterial-derived kinases of the poorly annotated aPK family [2]. Indeed, because the ABC1Ks do not have many of the typical ePK features they often are not recognized as kinases (e.g. see [29]). Thus far in Arabidopsis, apart from ABC1Ks, 6–7 PKL kinases have been conclusively demonstrated to localize to the plastid or mitochondria, despite significant and systematic efforts [28, 29]. In addition, one chloroplast and one mitochondrial two-component sensor kinase have been identified, which are the chloroplast sensor kinase (CSK) [30] and pyruvate dehydrogenase kinase (PDK) [31], respectively. The experimentally identified ABC1Ks in Arabidopsis include seven kinases localized in plastids and one kinase in mitochondria (Table 1).
Table 1.
The nomenclature and subcellular localization of the ABC1K protein family in Arabidopsis, rice and maize
| Sub- familya |
Nameb | Locationc |
Arabidopsis homolog(s)d |
Tar- getPe |
Exp. loc.f |
Rice homolog(s)g |
Tar- getPe |
Exp. local.h |
Maize homolog(s)g |
Tar- getPe |
Exp. loc.j |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Photosynthetic-specific cladea | |||||||||||
| 1 | ABC1K1 | Plastid (PG) | AT4G31390k | P | PG | Os11g11000 | P | Plastid | GRMZM2G377115 | P | PG |
| 2 | ABC1K2 | Plastid | AT5G24970 | M | Leaf | Os07g27480 | M | N.d. | GRMZM5G817551n | M | N.d. |
| 3 | ABC1K3 | Plastid (PG) | AT1G79600k | P | PG | Os05g25840 | P | Plastid | GRMZM2G315125 | P | PG |
| GRMZM5G878070o | P | PG | |||||||||
| 4 | ABC1K4 | Plastid (Nuc) | AT2G39190 | P | Leaf | Os02g56200 | P | N.d | GRMZM5G855200p | C | Nuc |
| 5 | ABC1K5 | Plastid (PG) | AT1G71810k | P | PG | Os04g54790 | P | Plastid | GRMZM2G305007 | M | PG |
| 6 | ABC1K6 | Plastid (PG) | AT3G24190 | P | PG | Os02g57160 | P | Plastid | GRMZM2G157369 | P | PG |
| 7 | ABC1K7 | Plastid (PG) | AT3G07700 | C | PG | Os09g07660 | P | Plastid | GRMZM5G845129 | P | N.d. |
| 8 | ABC1K8 | Plastid (IE or Nuc) | AT5G64940l | P | IE, Nucj | Os02g36570 | P | Plastid | GRMZM2G040511 | P | Nuc |
| Ancestral cladea | |||||||||||
| 9 | ABC1K9 | Plastid (PG) | AT5G05200 | M | PG | Os07g12530 | P | Plastid | GRMZM2G031780 | M | PG |
| 10 | ABC1K10 | Mitoch. | AT1G11390 | M | Leaf | Os07g37180 | M | N.d. | GRMZM2G067520 | M | N.d. |
| AT1G61640 | M | N.d. | Os04g56510 | M | N.d | GRMZM2G087201 | M | N.d. | |||
| GRMZM2G368486 | S | N.d. | |||||||||
| Mitochondrial cladea | |||||||||||
| 11 | ABC1K11 | Mitoch. | AT5G24810 | M | Leaf | Os06g48770 | M | Leaf | GRMZM2G140917 | M | N.d. |
| 12 | ABC1K12 | Mitoch. | AT4G24810 | C | N.d | Os01g67720 | C | N.d. | GRMZM2G113264 | M | N.d. |
| AT5G50330 | C | N.d | GRMZM5G884972q | M | N.d. | ||||||
| 13 | ABC1K13 | Mitoch. (IM) | AT4G01660m | C | Pollen | Os01g21610 | S | N.d. | GRMZM2G003417 | C | N.d. |
| 14 | ABC1K14 | Mitoch. | AT1G65950 | M | Leaf | Os11g34750 | M | N.d. | GRMZM2G124553 | M | N.d. |
| Os11g34830 | M | N.d. | GRMZM2G040720 | M | N.d. | ||||||
| 15 | ABC1K15 | Mitoch. | AT2G40090 | M | Leaf | Os03g49140 | C | N.d. | GRMZM2G026180 | C | N.d. |
Abbreviations: C, cytosol; Exp. loc.; experimental localization; IE, inner envelope; IM, inner membrane; Mitoch., Mitochondria; N.d., not detected; Nuc, nucleoid; P, plastid; PG, plastoglobule; S, secreted.
Assigned name based on subfamilies.
Most likely subcellular location in Arabidopsis, rice or maize based on experimental data combined with predictions (see also PPDB: http://ppdb.tc.cornell.edu).
Gene/protein accession numbers are from Arabidopsis genome assembly TAIR10 (http://www.arabidopsis.org/).
The predicted subcellular location by TargetP based on most likely gene model for each locus.
Subcellular localization reported in the literature (PG [39], IE [40]). Leaf detected in Arabidopsis leaf samples. Pollen, this protein was reported in the pollen proteome [73].
Gene/protein accession numbers are from rice genome assembly v6 (http://rice.plantbiology.msu.edu/).
Experimental location based on mass spectrometry analysis of rice chloroplasts and leaves. (N.d. indicates that the protein was not detected in rice lea chloroplasts (Huang, Friso and van Wijk, unpublished)).
Gene/protein accession numbers are from maize genome assembly 5b.60 (http://www.maizesequence.org/).
Experimental location based on mass spectrometry analysis of maize subfractions and leaves by the van Wijk lab.
Null mutant results in conditional stress phenotype (Lundquist, Giacomelli and van Wijk, unpublished).
Mutant results in impaired cadmium tolerance [41]
Gene/protein number in genome assembly 4a53 is GRMZM2G045183
Gene/protein number in genome assembly 4a53 is GRMZM2G020627
Gene/protein number in genome assembly 4a53 is GRMZM2G008643
Gene/protein number in genome assembly 4a53 is GRMZM2G091267
Characterization of plastid protein kinases has emphasized the role of phosphorylation in plastid gene expression and regulation of the photosynthetic thylakoid electron transport. The state transition kinases, STN7 and STN8 localize to the thylakoid membrane system and phosphorylate subunits of the light-harvesting complex (LHC) and photosystem (PS), respectively, driving rapid alterations in light harvesting and electron transport in response to fluctuating environment [32–35]. Chloroplast stromal casein kinase IIα (cpCK2, also named plastid transcription kinase - PTK) is involved in regulation of plastid gene expression [29, 36], but based on phosphorylation motifs determined from chloroplast phosphoproteome analysis, also phosphorylates a subset of chloroplast metabolic proteins [23]. cpCK2 was shown to interact with CSK providing a link between redox sensing and plastid transcriptional control [37]. The identification of two-component sensor kinases in plastids and mitochondria emphasizes the bacterial ancestry of the organelles and justifies the expectation that many of the organellar kinases are bacterial-derived aPKs.
ABC1Ks in plastids and mitochondria
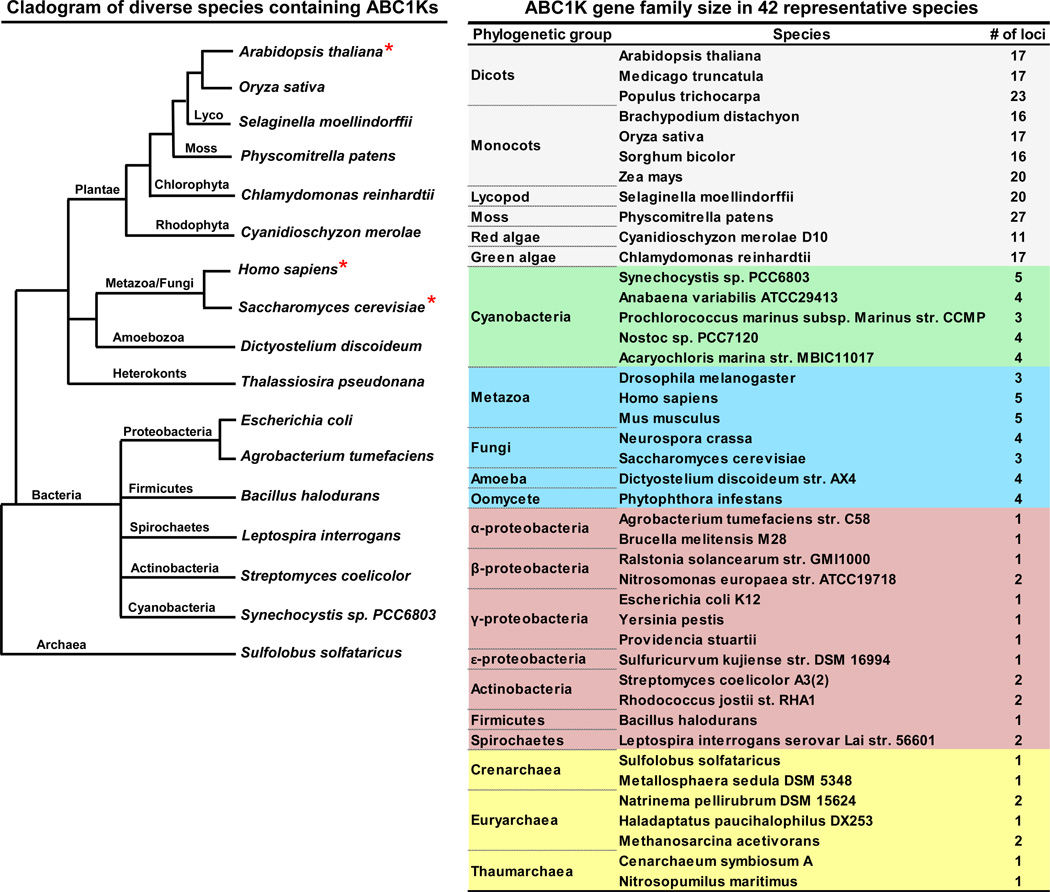
The ABC1Ks are an evolutionarily-ancient gene family, conserved throughout species of all three primary kingdoms (archaea, bacteria, and eukaryotes), but have greatly expanded in number in photosynthetic organisms. The gene family in Arabidopsis contains seventeen members (Table 1, Figure 2). Previously we assigned numbers to the few identified Arabidopsis and maize ABC1K proteins in the Plant Proteome Database (PPDB at http://ppdb.tc.cornell.edu/); here we propose a more logical and complete numbering for all Arabidopsis, maize and rice ABC1K proteins based on phylogeny (see below). Analyses of purified plastoglobules (PGs) from Arabidopsis chloroplasts identified six ABC1K proteins (AtABC1K1, 3–7) that localize predominantly to this plastid location [38–40]. A seventh protein (AtABC1K8/OSA1) was shown to localize to the inner plastid envelope [41]. Plastid localization of AtABC1K1,3,5 was confirmed by YFP fusions [29]. Furthermore, ABC1K13 is expected to be localized in mitochondria, based on the localization of its functional homolog in baker’s yeast (Saccharomyces cerevisiae) [17, 42]. Several other ABC1K proteins were observed in leaf or pollen samples (Table 1), but their subcellular localization was not determined. Proteome analysis of maize leaf fractions further supported plastid localization of eight ABC1K proteins; ZmABC1K1, ZmABC1K3 (2 homologues), ZmABC1K4, ZmABC1K5, ZmABC1K6, ZmABC1K8 and ZmABC1K9 were identified in maize proplastid and chloroplast fractions [43, 44], with ZmABC1K4 and ZmABC1K8 enriched in plastid nucleoids [45] (Table 1). Furthermore, seven rice ABC1K proteins were identified in chloroplasts (Table 1). Finally, TargetP, an in silico predictor of protein localization [46], predicts plastid or mitochondrial localization for most of the maize, rice and Arabidopsis ABC1K proteins (Table 1). Therefore, we suggest that most, if not all, ABC1K proteins in higher plants are located in plastids or mitochondria. Based on mass spectrometry analyses of leaf samples, as well as isolated chloroplast and mitochondrial fractions, ABC1Ks assigned to plastids are in general far more abundant than those assigned to mitochondria (see PPDB); this is in agreement with the notion that the plastids contribute much more protein biomass to the leaf than mitochondria.
Figure 2.
A phylogenetic tree and table illustrating the diversity of species containing ABC1K proteins and their evolutionary relationships. A proliferation of homologs in photosynthetic species, especially eukaryotic species, is seen in the table at right. The red asterisk indicates the three species with experimentally demonstrated functional homologs involved in UQ synthesis. Lengths in the phylogenetic tree are not meant to indicate evolutionary distance.
Identification of the ABC1K gene in yeast
The founding member of the ABC1K protein family, ABC1/COQ8 in yeast (hereafter called, ScCOQ8), is a nuclear-encoded protein required for ubiquinone (UQ) synthesis in the mitochondria. This gene was found to be necessary for redox activity of the mitochondrial bc1 complex involved in cellular respiration and was thus given the name abc1 (activity of bc1 complex) [47]. Loss of the ScCOQ8 gene causes a UQ deficiency and accumulation of the biosynthetic precursor 3-hexaprenyl-4-hydroxybenzoic acid, leading to instability of the bc1 complex and the lack of bc1 activity [48]. Missteps in the original analysis of ScCOQ8 gene function have caused confusion. The ScCOQ8 gene was initially believed to suppress a deleterious mutation in a cytochrome b translational activator (cbs2-223) leading to the incorrect conclusion that ScCOQ8p functions as a chaperone of cytochrome b [47, 49]. Not until a decade later was it found that ABC1 is the ScCOQ8 gene and that suppression of the translational activator mutant (cbs2-223) was due to a neighboring tRNATrp gene [48, 50]. It is thus currently accepted that ScCOQ8p is required specifically for regulation of UQ synthesis, and is not involved in chaperone activity.
Conservation of COQ8 function in UQ biosynthesis
Analysis of homologs of yeast ScCOQ8p from diverse species has revealed remarkable functional conservation in UQ biosynthesis. Loss of UbiB or aarF, ScCOQ8p homologs in Escherichia coli and Providencia stuartii, respectively, causes UQ deficiency and concomitant accumulation of 2-octaprenylphenol, indicating a block in the first monoxygenation step in their UQ biosynthetic pathway [51]. The enzyme catalyzing this monoxygenation step has not yet been identified in either E. coli or P. stuartii and is the postulated target of UbiB/AarF [51]. The functional homolog in Arabidopsis was found by complementation of the yeast ScCOQ8 deletion mutant with an Arabidopsis cDNA library; the only complementing cDNA was AtABC1K13 (At4g01660). This suggests that the Arabidopsis genome only encodes a single functional homolog of ScCOQ8, which is supported by the phylogenetic analysis of the angiosperm ABC1K homologs (see below). Deleterious mutations in the human ABC1K homolog, ADCK3 (for aarF-domain containing kinase 3), also displays a UQ deficiency [52, 53]. Functional homology of this gene was confirmed by successful complementation of the ScCOQ8 deletion mutant, when expressed along with a yeast mitochondrial transit peptide [17]. This heterologous expression of HsADCK3 in the ScCOQ8 mutant restored both UQ biosynthetic complex stability and phosphorylation of several enzymes of the pathway (COQ3, COQ4 and COQ7), suggesting that the kinase activity of this ABC1K protein can target multiple enzymes in the UQ pathway.
Phylogeny of the ABC1K proteins
Homologs of ScCOQ8p among the three branches of archaeal species (crenarchaea, euryarchaea, thaumarchaea) demonstrate strong BLAST hits (E-value < 3e−16). The presence of ABC1K homologs in all three branches of archaeal species, and throughout the bacterial kingdom, indicates their ancient origin prior to the archaea/bacterial split (Figure 3). Importantly, the archaea and many bacterial species do not synthesize UQ (a benzoquinone), but other types of prenylquinones (napthoquinones), in particular menaquinone [54], suggesting that the ancient function of ABC1Ks is regulation of quinones other than UQ. A striking proliferation of the ABC1K family is found in algae and land plants (11–27 homologs per species), whereas non-photosynthetic prokaryotes and non-photosynthetic eukaryotes consistently contain 1–2 and 3–5 homologs, respectively (Figure 2).
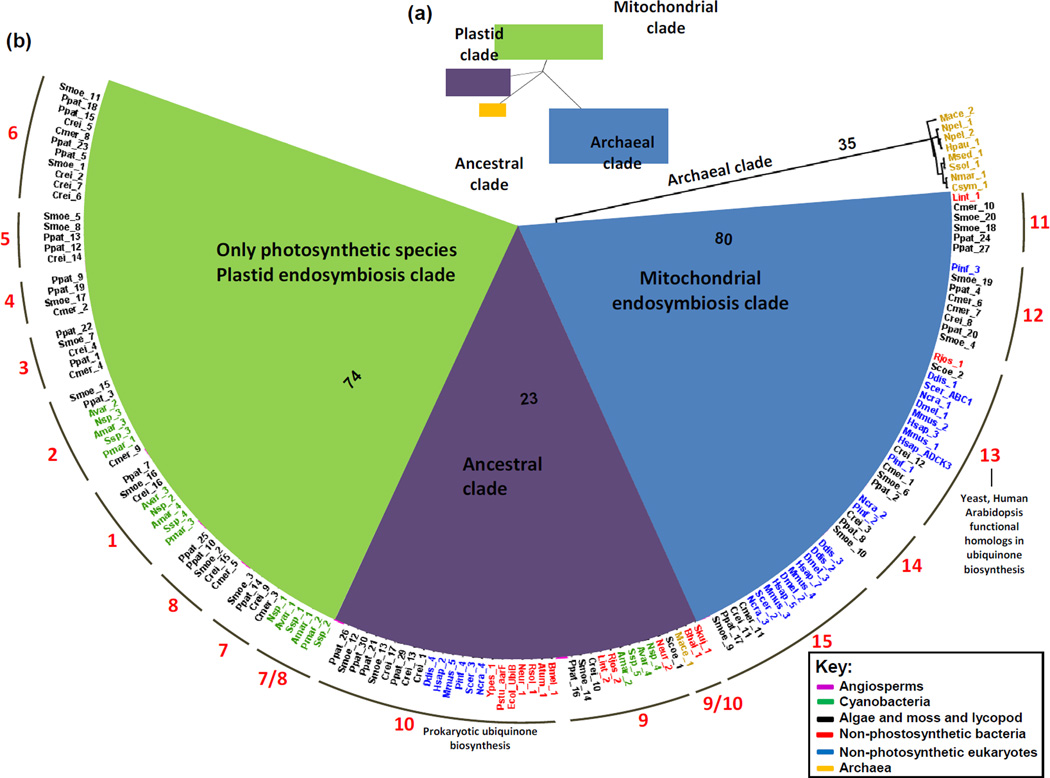
Figure 3.
Phylogenetic tree of the ABC1 kinase family among archaea, bacteria and eukaryotes. The full complement of 274 ABC1K proteins of 42 species, from diverse archaea, prokaryotes and eukaryotes were aligned using MUSCLE 3.5.1 (http://toolkit.tuebingen.mpg.de/muscle) and manually corrected in case of truncated protein sequences. The tree was constructed using the RAxML software tool at CIPRES (http://www.phylo.org/sub_sections/portal/) using the General Time Reversal model with 1000 bootstrap iterations and has been illustrated as an unrooted tree (a) and a proportional polar tree layout (b) using FigTree v1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/). (a), unrooted tree illustrating the three primary clades, in addition to an archaeal group forming a natural outgroup to the tree. The three clades can be categorized by their presumptive localizations and origins indicated by color; plastid endosymbiosis (green), mitochondrial endosymbiosis (blue), ancestral (purple), and outgroup (archaea specific clade) (orange). (b), Expansion of the tree shown in (a) illustrating the distribution of species. Bootstrap values are indicated for the four primary branches. Species names are colored to distinguish archaea, photosynthetic and non-photosynthetic bacteria and photosynthetic and non-photosynthetic eukaryotes. Angiosperm clades have been collapsed for legibility and are colored magenta. The 15 subfamilies are labeled around the perimeter of the wheel. Species are named as follows: Smoe, Selaginella moellindorffii; Ppat, Physcomitrella patens; Crei, Chlamydomonas reinhardtii; Cmer, Cyanidioschyzon merolae; Avar, Anabaena variabilis; Nsp, Nostoc sp.; Amar, Acarychloris marina; Ssp, Synechocystis sp.; Pmar, Prochlorococcus marinus; Ddis, Dictyostelium discoideum; Pinf, Phytophthora infestans; Hsap, Homo sapiens; Scer, Saccharomyces cerevisiae; Ncra, Neurospora crassa; Pstu, Providencia stuartii; Ypes, Yersinia pestis; Ecol, Escherichia coli; Neur, Nitrosomonas europaea; Rsol, Ralstonia solancearum; Bmel, Brucella melitensis; Atum, Agrobacterium tumefaciens; Lint, Leptospira interrogans; Scoe, Streptomyces coelicolor; Mace, Methanosarcina acetivorans; Bhal, Bacillus halodurans; Skuj, Sulfuricurvum kujiense; Mmus, Mus musculus; Dmel, Drosophila melanogaster; Rjos, Rhodococcus jostii; Nmar, Nitrosopumilus maritimus; Csym, Cenarchaeum symbiosum; Msed, Metallosphaera sedula; Ssol, Sulfolobus solfataricus; Hpau, Haladaptatus paucihalophilus; Npel, Natrinema pellirubrum.
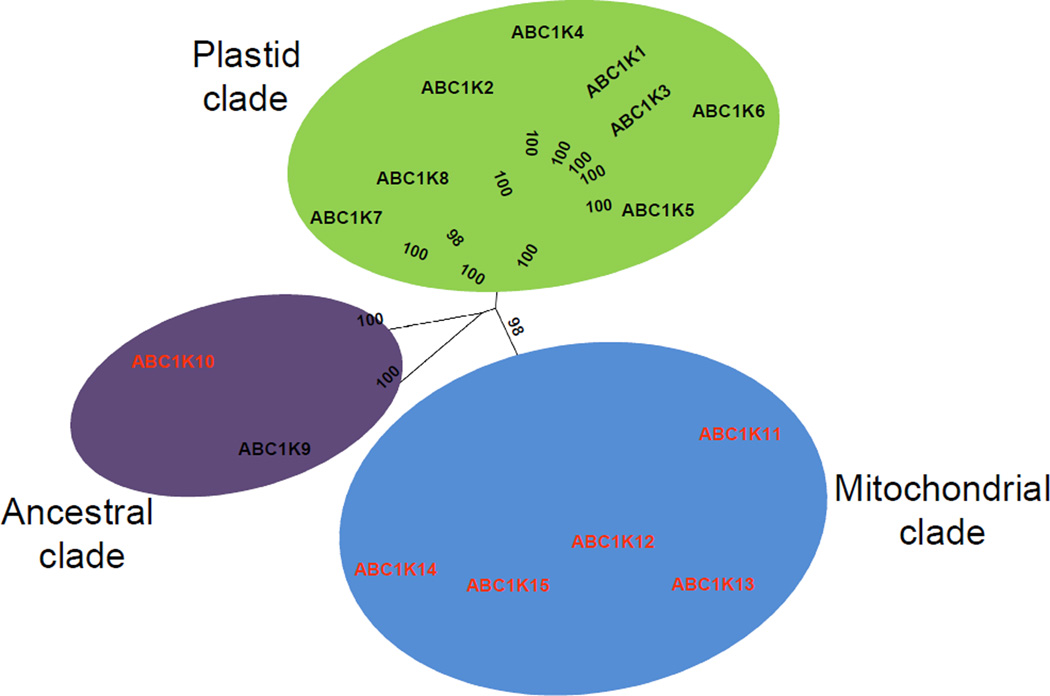
Construction of a phylogenetic tree from the ABC1K proteins of 42 diverse species of archaea, bacteria, and eukaryotes reveals a division into 15 subfamilies, which we name ABC1K1 to ABC1K15 (Figure 3). Immediately apparent is the fact that the archaeal homologs of multiple species all group in a clade evolutionarily distant from the homologs of bacteria and eukaryotes (except for one of two homologs of archaeal Methanosarcina acetivorans). This creates a natural outgroup incorporated into the phylogenetic tree, here assigned the archaeal clade (Figure 3). Within each subfamily, the seven angiosperm species (four monocots and three eudicots) all collapse into their own subclade (Figure 3). Immediately sister to each angiosperm clade are homologs from lycopod (Selaginella moellendorffii) and moss (Physcomitrella patens) with sequences from green and red algae also closely related, indicating that each of the 15 ABC1K families arose with the emergence of photosynthetic eukaryotes. Strikingly, the phylogenetic tree divides into three clear primary clades characterized by evolutionary origins and sub-cellular localization (Figure 3).
The first clade comprises eight subfamilies (1–8) and is specific for photosynthetic organisms. Cyanobacteria harbor three of the eight photosynthetic-specific subfamilies (1, 2, and 7) and it is likely that plastid endosymbiosis resulted in the introduction of these three proteins to a photosynthetic ancestor (see [55]) which subsequently expanded into the current eight members. The presence of algae in six of the eight subfamilies indicates that expansion of the plastid clade occurred very early in the development of the photosynthetic eukaryotic lineage. It is interesting that a majority of the plastid ABC1Ks (ABC1K2 to ABC1K6) of algae and plants appear to be derived from the ancestral ABC1K2 of cyanobacteria, suggesting that they may have closely related or overlapping targets (Figures 3 and 4). Most of the higher plant proteins in this clade were identified in plastid fractions.
Figure 4.
Phylogenetic tree of the angiosperm ABC1K proteins. The unrooted tree was based on the amino acid sequence alignment of the 126 ABC1K proteins from three eudicot species (Arabidopsis, M. truncatula, and P. trichocarpa) and four monocot species (Z. mays, O. sativa, B. distachyon, S. bicolor) which was manually corrected in the case of the truncated protein products. The tree was generated as outlined in figure 3. The three clades identified and illustrated in figure 3 are indicated here using the same color code and each of the 15 subfamilies are labeled. Subcellular localization of each subfamily (black – plastids, red – mitochondria) has been determined by experimental evidence or, in the absence of experimental evidence, by TargetP prediction. Bootstrap values are indicated.
The second clade consists of the subfamilies 11–15, which are all likely targeted to the mitochondria based on localization predictions, and because they were observed by proteomics in leaves or pollen (high in mitochondrial content) but not in isolated chloroplasts(Table 1). Furthermore, in the case of subfamily 13 (which corresponds with ScCOQ8p and its functional homologs in Arabidopsis and humans) experimental evidence supports the mitochondrial location assignment, since ScCOQ8p of subfamily 13 localizes to the inner mitochondrial envelope [17] and the UQ pathway localizes within the mitochondria [56]. Except for Arabidopsis mitochondrial ABC1K13, experimental localization data is lacking for plant proteins in this clade. However, in silico analysis of proteins by TargetP predicts mitochondrial targeting for most of the Arabidopsis, maize and rice members, and none of the plant proteins have been observed in plastids (Table 1). Furthermore, homologs of this clade are prevalent in both photosynthetic and non-photosynthetic plant species, as would be expected from genes of a mitochondrial origin derived from endosymbiosis prior to the divergence of the plants [57]. It is not surprising, that homologs of actinobacteria (Rhodococcus jostii and Streptomyces coelicolor), and spirochaetes (Leptospira interrogans) are present in this clade rather than the α-proteobacteria, the presumed mitochondrial ancestors, considering the influence of a fluid evolutionary model of prokaryotic genomes. This model posits that the ancestral mitochondrial donor genome did not contain the current set of α-proteobacterial genes because of mutations, gene loss and horizontal gene transfer over > 1.5 billion years of genome evolution [58].
The third, central clade with subfamilies 9 and 10 contains the majority of the non-photosynthetic bacterial ABC1Ks, and their homologs in cyanobacteria, metazoa, and plants; this clade represents the ancestral group of ABC1Ks, derived not from organelle endosymbiosis, but from the common ancestor of bacteria, archaea, and the nuclear genome of eukaryotes. However, subfamily 9 contains the photosynthetic prokaryotes (cyanobacteria) which are missing in subfamily 10. Conversely, subfamily 10 contains non-photosynthetic eukaryotes, which are missing in subfamily 9.
Cyanobacteria lack UQ, instead using plastoquinone 9 (PQ-9) for both photosynthetic and respiratory electron transport [59]. The ABC1Ks with demonstrated roles in UQ biosynthesis divide between subfamilies 10 and 13, consistent with the absence of cyanobacteria in these subfamilies. Furthermore, these ABC1Ks in non-photosynthetic prokaryotes (aarF in P. stuartii and UbiB in E. coli) fall into subfamily 10 (ancestral clade), whereas these ABC1Ks in eukaryotes (HsADCK3, ScCOQ8, and AtABC1K13) fall into subfamily 13 (mitochondrial clade). This may be a reflection of differences in biosynthetic pathways and requirements for regulation.
Remarkably, PQ-9 biosynthesis appears to have arisen from the UQ biosynthesis in ancestral proteobacteria [60]; both PQ and UQ are evolutionary younger benzoquinones with higher redox potentials than the more ancient naphtaquinones [59]. This is supported by: (i) the Synechocystis genome encodes for homologs of the E. coli UQ biosynthetic pathway (Ubi X, D, H, E), (ii) the two pathways both derive their head group from 4-hydroxybenzoate, rather than homogentisate as in eukaryotic PQ-9 synthesis, and (iii) the PQ-9 prenyl-transferase of Synechocystis functionally complements the ubiA deletion mutant in E. coli [60]. It can be expected that the regulatory ABC1K of UQ synthesis was similarly co-opted for PQ-9 synthesis in cyanobacteria. Thus it appears that plants, through endosymbiosis of plastid and mitochondrial ancestors, have inherited multiple pathways for quinone metabolism along with the corresponding regulatory ABC1Ks, resulting in the proliferation of the ABC1K family in photosynthetic eukaryotes.
Defining the ABC1K protein domain
Based on the alignment of 100 full length ABC1K protein sequences from the seven angiosperm species used in the phylogenetic tree (Figure 3), a common ABC1K domain is observed, spanning ~350 residues and containing twelve conserved motifs (Figure 1B). Furthermore, eight of the ten key residues of the PKL superfamily identified in [2] are present in the ABC1K family and correspond to motifs III, IVa, IVb, VIIb, and VIII, involved in ATP binding and orientation (III, IVa and IVb), catalysis (VIIb), and Mg2+ chelation (VIII) [6, 8, 11, 61]. The other seven motifs of the ABC1K domain (I, II, V, VI, VIIa, IX, X) do not have homologous sequences in ePKs, but can be found in a number of proteins outside of the PKL superfamily with diverse enzyme activities. The significance of these ABC1K motifs is unknown and will likely require crystallization studies and mutational analysis.
Motifs conserved with ePKs
The nucleotide-binding pocket (motif III) in the ABC1K family is unusual in that the first two characteristic Gly residues of nucleotide-binding pockets are both replaced with Ala (see also [29]). Between these two alanines is a single residue, either another Ala, or a serine or threonine. An invariant serine and glutamine, flanking two hydrophobic residues also appears in the ABC1K nucleotide-binding pocket. It cannot be concluded from the primary sequence alone which nucleotide (NTP) the ABC1Ks prefer as co-factor. The invariant lysine of PKLs (in motif II) lies in ABC1K motif IVa, fourteen residues downstream of motif III and is immediately downstream of three hydrophobic residues, as in the ePKs. The lysine helps to anchor the NTP by binding its α- and β-phosphates and positions the γ-phosphate for catalysis [6]. Motif IVb in ABC1Ks is characterized by an invariant (acidic) Asp residue, which we suggest is homologous to the invariant (acidic) Glu of ePKs. An acidic residue is necessary for stabilization of the Lys-NTP interaction [6]. The catalytic motif (VIIb) contains a consensus HADPHPGN sequence in the ABC1K family. The Asp is 100% conserved among tested angiosperms and is likely homologous to the conserved Asp of ePK catalytic motifs, responsible for activating the substrate hydroxyl group via nucleophilic attack [6]. Mutation of this residue in ScCOQ8 resulted in the abc1 mutant phenotype [17]. The histidines in the consensus sequence are conserved in all Arabidopsis ABC1Ks, except for ABC1K14 and ABC1K12 where the His positions are replaced with either Asn or Gln, indicating an absolute requirement for an amine-containing side chain at that position. Motif VIII comprises the D[FYHV]G motif which anchors the Mg2+ necessary for positioning NTP, using the invariant Asp to chelate this divalent cation [6]. The C-terminal motif of the ABC1K domain (motif X) deserves special mention because the motif in family 13 (PPEExxSLHRKxxG) is homologous to a motif in a number of proteins from diverse species including the RsbU phosphatase 2C of Bacillus subtilis [17]. X-ray crystallography studies of the RsbU protein have indicated that the sequence is critical for homodimerization by stabilizing the protein through helix-helix interaction [62]. Point mutations in motif X of the HsABC1K13, (G549S, E551K) and ScCOQ8 (G475D) cause UQ deficiency, indicating a critical function for this motif in ABC1K function [17, 52, 53]. The homodimerization of the RsbU phosphatase facilitates binding with the RsbT serine kinase, creating a complex that mediates stress responses to environmental and nutritional signals in B. subtilis [63]. The other ABC1K families show divergent variants of this motif. Thus motif X in the ABC1Ks may similarly mediate protein-protein interactions necessary for mediating stress responses integral to ABC1K function (see below).
Domain architecture
Land plant and red algae ABC1Ks in subfamily 11 (green algae have no observed homolog in this subfamily) contain a C-terminal β-lactamase domain, which in bacteria catalyzes hydrolysis of the β-lactam ring of penicillin. Identification of the intact catalytic motifs suggests that the lactamase domain is active [64]. Several β-lactamase domain proteins have been found in plants with other enzymatic activities such as glyoxylase proteins in rice and Arabidopsis [65] and an Arabidopsis DNA ligase [66]. The function of β-lactamase domains in plants and other eukaryotic species is unknown, but it has been suggested that the fusion of the β-lactamase domain to ABC1K confers a unique mechanism of autoregulation of kinase activity [64].
Kinase activity among the ABC1Ks
A direct demonstration of kinase activity in the ABC1K protein family has proven difficult. However, indirect results from point mutants in predicted kinase residues of the yeast and human ABC1K genes [17, 52, 53, 67] and an in-gelo study of the Arabidopsis AtABC1K8/OSA1 gene have reinforced the hypothesized protein kinase activity [41]. ScCOQ8p-dependent isoelectric point shifts of several subunits of the yeast UQ biosynthetic complex (COQ3, COQ5 and COQ7) have been detected in 2D IEF-SDS-PAGE gels, suggesting that ScCOQ8p either directly or in-directly phosphorylates several enzymes of the UQ biosynthesis complex in yeast [17, 67]. Biochemical phenotypes of point mutations in conserved kinase subdomains have further supported the participation of ScCOQ8p in UQ biosynthetic complex phosphorylation. Five of eight point mutants demonstrating the ABC1K deletion phenotype were mutated in residues of shared ePK-ABC1K motifs. In particular, mutation of the invariant lysine-216 (motif IVa) to an alanine caused dramatically reduced steady-state levels of the protein. Similarly, several deleterious mutations in the human ADCK3, causing UQ deficiency, are point mutants in conserved kinase subdomains [52, 53].
Experimental studies of plant ABC1Ks and mRNA co-expression analysis
In non-photosynthetic organisms, the ABC1K family has only been studied in relation to UQ biosynthesis. Yet, proliferation of the ABC1K family in photosynthetic eukaryotes implies an expansion of functions and targets.
Leaf proteome analysis showed that the six most abundant ABC1K proteins in Arabidopsis were located in thylakoid-associated lipoprotein particles, called plastoglobules (PGs) (Table 1). Based on genome-wide co-expression analysis of these six ABC1Ks [40], we suggested that they have regulatory functions concerning formation, maintenance and optimization of photosynthetic performance through regulation of specific sets of enzymes involved in carotenoid biosynthesis, photoacclimation, senescence, and plastid gene expression. CrABC1K6 (EYE3) in the green algae Chlamydomonas reinhardtii, was located in carotenoid-rich lipophilic plastid particles (the pigment granule) in the eyespot of Chlamydomonas. Null mutants in CrABC1K6 failed to develop pigment granules or eyespots. The Arabidopsis homolog of CrABC1K6 (AtABC1K6) is located in chloroplast PGs [40] (Table 1), consistent with the belief that PGs serve as the precursors of the eyespot pigment granule [68]. Additionally, AtABC1K6 tightly co-expressed with zeaxanthin epoxidase (ZEP), suggesting a regulatory role in the xanthophyll cycle [40]. Based on various data (Table 1), we believe that higher plant ABC1K4 is located in the nucleoid, where it may help regulate plastid gene expression. The functions of mitochondrial ABC1K proteins include regulation of UQ biosynthesis and are otherwise unclear, but we speculate that it includes regulation of mitochondrial gene expression.
Other experimental studies of plant ABC1K genes have emphasized a role in various types of abiotic stress tolerance, including the heavy metal cadmium. AtABC1K8 (OSA1), encoding for a chloroplast envelope protein, was transcriptionally upregulated in response to cadmium, and loss of AtABC1K8 expression rendered plants more susceptible to cadmium toxicity, high-light, and H2O2 [41]. Even under optimal growth conditions, these AtABC1K8 mutants displayed elevated biochemical markers of oxidative stress (e.g. SOD activity). Likewise, a homolog of the PG-localized AtABC1K7 from the heavy-metal over-accumulator species Brassica juncea was also up-regulated in response to 24 hours of treatment with cadmium [69]. The maize homolog ZmABC1K8 expressed primarily in green tissue, with highest expression levels in fully mature leaves, and its expression was enhanced in response to cadmium [70]. Conversely, ZmABC1K8 mRNA accumulation was down-regulated by treatment with ABA, H2O2and darkness, and did not respond to cold-treatment. It was suggested that heterologous expression of wheat ABC1K13 in Arabidopsis conferred enhanced tolerance against a wide variety of stresses, but no evidence was presented for accumulation of the transgenic protein [71]. mRNA analysis of rice ABC1K genes suggested highest expression in leaf tissue and varying responses to abiotic stresses [72].
Conclusions and future directions
Regulation of quinone synthesis is the ancient (archaeal) function of the ABC1K family. Plants, through endosymbiosis of plastid and mitochondrial ancestors, have inherited pathways for quinone metabolism along with the corresponding regulatory ABC1Ks. The requirement for additional quinolic and other prenyl-lipids likely further drove the expansion of the ABC1K family in algae and higher plants. These ABC1Ks localize in plastids and mitochondria in which they represent the majority of known kinases. The direct targets of ABC1Ks are not known, but likely include enzymes of prenyl-lipid metabolism (eg carotenoids) and components of the organellar gene expression machineries. Systematic analysis of ABC1K targets will be critical in defining the functional significance of the ABC1K family in photosynthetic organisms.
Acknowledgements
PKL was in part funded by an NIH Chemistry-Biology Interface (CBI) training grant (# 5T32GM008500). Research in the van Wijk lab is supported primarily by the National Science Foundation (MCB-1021963, IOS-0701736 and IOS-0922560).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cheek S, et al. Sequence and Structure Classification of Kinases. Journal of Molecular Biology. 2002;320:855–881. doi: 10.1016/s0022-2836(02)00538-7. [DOI] [PubMed] [Google Scholar]
- 2.Kannan N, et al. Structural and Functional Diversity of the Microbial Kinome. PLoS Biol. 2007;5:e17. doi: 10.1371/journal.pbio.0050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casino P, et al. The mechanism of signal transduction by two-component systems. Curr Opin Struct Biol. 2010;20:763–771. doi: 10.1016/j.sbi.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Schaller GE, et al. Two-component systems and their co-option for eukaryotic signal transduction. Curr Biol. 2011;21:R320–R330. doi: 10.1016/j.cub.2011.02.045. [DOI] [PubMed] [Google Scholar]
- 5.Hanks SK, et al. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 6.Hanks S, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. The FASEB Journal. 1995;9:576–596. [PubMed] [Google Scholar]
- 7.Dirk B. Protein kinases - structure and function. FEBS Letters. 1995;369:57–61. doi: 10.1016/0014-5793(95)00580-3. [DOI] [PubMed] [Google Scholar]
- 8.Taylor SS, Radzio-Andzelm E. Three protein kinase structures define a common motif. Structure (London, England: 1993) 1994;2:345–355. doi: 10.1016/s0969-2126(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 9.Bowyer JR, et al. Carboxyl-terminal processing of the D1 protein and photoactivation of water-splitting in photosystem II. Partial purification and characterization of the processing enzyme from Scenedesmus obliquus and Pisum sativum. J Biol Chem. 1992;267:5424–5433. [PubMed] [Google Scholar]
- 10.Taylor SS, et al. PKA: a portrait of protein kinase dynamics. Biochimica et Biophysica Acta (BBA) - Proteins & Proteomics IPK'2003. Inhibitors of protein kinases and Workshop: Phosphoryl-transfer mechanisms. 2004;1697:259–269. doi: 10.1016/j.bbapap.2003.11.029. [DOI] [PubMed] [Google Scholar]
- 11.Dissmeyer N, Schnittger A. The age of protein kinases. Methods in molecular biology (Clifton, NJ) 2011;779 doi: 10.1007/978-1-61779-264-9_2. [DOI] [PubMed] [Google Scholar]
- 12.Scheeff ED, Bourne PE. Structural Evolution of the Protein Kinase-Like Superfamily. PLoS Comput Biol. 2005;1:e49. doi: 10.1371/journal.pcbi.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krupa A, Srinivasan N. Diversity in domain architectures of Ser/Thr kinases and their homologues in prokaryotes. BMC Genomics. 2005;6:129. doi: 10.1186/1471-2164-6-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang C-C. Bacterial signalling involving eukaryotic-type protein kinases. Molecular Microbiology. 1996;20:9–15. doi: 10.1111/j.1365-2958.1996.tb02483.x. [DOI] [PubMed] [Google Scholar]
- 15.Shi L, et al. The serine, threonine, and/or tyrosine-specific protein kinases and protein phosphatases of prokaryotic organisms: a family portrait. FEMS Microbiology Reviews. 1998;22:229–253. doi: 10.1111/j.1574-6976.1998.tb00369.x. [DOI] [PubMed] [Google Scholar]
- 16.Kennelly PJ. Protein kinases and protein phosphatases in prokaryotes: a genomic perspective. FEMS Microbiology Letters. 2002;206:1–8. doi: 10.1111/j.1574-6968.2002.tb10978.x. [DOI] [PubMed] [Google Scholar]
- 17.Xie LX, et al. Expression of the human atypical kinase ADCK3 rescues coenzyme Q biosynthesis and phosphorylation of Coq polypeptides in yeast coq8 mutants. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2011;1811:348–360. doi: 10.1016/j.bbalip.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LaRonde-LeBlanc N, Wlodawer A. A Family Portrait of the RIO Kinases. Journal of Biological Chemistry. 2005;280:37297–37300. doi: 10.1074/jbc.R500013200. [DOI] [PubMed] [Google Scholar]
- 19.LaRonde-LeBlanc N, Wlodawer A. The RIO kinases: An atypical protein kinase family required for ribosome biogenesis and cell cycle progression. Biochimica et Biophysica Acta (BBA) - Proteins & Proteomics Inhibitors of Protein Kinases (4th International Conference, Inhibitors of Protein Kinases) and Associated Workshop: Modelling of Specific Molecular Recognition Processes (Warsaw, Poland, June 25–29, 2005) 2005;1754:14–24. doi: 10.1016/j.bbapap.2005.07.037. [DOI] [PubMed] [Google Scholar]
- 20.Toth M, et al. Source of Phosphate in the Enzymic Reaction as a Point of Distinction among Aminoglycoside 2″-Phosphotransferases. Journal of Biological Chemistry. 2009;284:6690–6696. doi: 10.1074/jbc.M808148200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rokka A, et al. Thylakoid phosphoproteins: identification of phosphorylation sites. Methods Mol Biol. 2011;684:171–186. doi: 10.1007/978-1-60761-925-3_15. [DOI] [PubMed] [Google Scholar]
- 22.Baginsky S, Gruissem W. The Chloroplast Kinase Network: New Insights from Large-Scale Phosphoproteome Profiling. Molecular Plant. 2009;2:1141–1153. doi: 10.1093/mp/ssp058. [DOI] [PubMed] [Google Scholar]
- 23.Reiland S, et al. Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol. 2009;150:889–903. doi: 10.1104/pp.109.138677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lohrig K, et al. Phosphorylation site mapping of soluble proteins: bioinformatical filtering reveals potential plastidic phosphoproteins in Arabidopsis thaliana. Planta. 2009;229:1123–1134. doi: 10.1007/s00425-009-0901-y. [DOI] [PubMed] [Google Scholar]
- 25.Sugiyama N, et al. Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol Syst Biol. 2008;4:193. doi: 10.1038/msb.2008.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reinders Jr, et al. Profiling Phosphoproteins of Yeast Mitochondria Reveals a Role of Phosphorylation in Assembly of the ATP Synthase. Molecular & Cellular Proteomics. 2007;6:1896–1906. doi: 10.1074/mcp.M700098-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Ito J, et al. A survey of the Arabidopsis thaliana mitochondrial phosphoproteome. Proteomics. 2009 doi: 10.1002/pmic.200900064. [DOI] [PubMed] [Google Scholar]
- 28.Schliebner I, et al. A Survey of Chloroplast Protein Kinases and Phosphatases in Arabidopsis thaliana. Curr Genomics. 2008;9:184–190. doi: 10.2174/138920208784340740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bayer RG, et al. Chloroplast-localized protein kinases: a step forward towards a complete inventory. Journal of Experimental Botany. 2012;63:1713–1723. doi: 10.1093/jxb/err377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puthiyaveetil S, et al. The ancestral symbiont sensor kinase CSK links photosynthesis with gene expression in chloroplasts. Proc Natl Acad Sci U S A. 2008;105:10061–10066. doi: 10.1073/pnas.0803928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thelen JJ, et al. Pyruvate dehydrogenase kinase from Arabidopsis thaliana: a protein histidine kinase that phosphorylates serine residues. Biochemical Journal. 2000;349:195–201. doi: 10.1042/0264-6021:3490195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pesaresi P, et al. Dynamics of reversible protein phosphorylation in thylakoids of flowering plants: The roles of STN7, STN8 and TAP38. Biochim Biophys Acta. 2010 doi: 10.1016/j.bbabio.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 33.Tikkanen M, Aro EM. Thylakoid protein phosphorylation in dynamic regulation of photosystem II in higher plants. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbabio.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 34.Turkina MV, Vener AV. Identification of phosphorylated proteins. Methods Mol Biol. 2007;355:305–316. doi: 10.1385/1-59745-227-0:305. [DOI] [PubMed] [Google Scholar]
- 35.Rochaix JD. Regulation of photosynthetic electron transport. Biochim Biophys Acta. 1807:375–383. doi: 10.1016/j.bbabio.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 36.Turkeri H, et al. Phylogenetic and functional features of the plastid transcription kinase cpCK2 from Arabidopsis signify a role of cysteinyl SH-groups in regulatory phosphorylation of plastid sigma factors. Febs J. 2011;279:395–409. doi: 10.1111/j.1742-4658.2011.08433.x. [DOI] [PubMed] [Google Scholar]
- 37.Puthiyaveetil S, et al. Transcriptional control of photosynthesis genes: the evolutionarily conserved regulatory mechanism in plastid genome function. Genome Biol Evol. 2010;2:888–896. doi: 10.1093/gbe/evq073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vidi PA, et al. Tocopherol cyclase (VTE1) localization and vitamin E accumulation in chloroplast plastoglobule lipoprotein particles. J Biol Chem. 2006;281:11225–11234. doi: 10.1074/jbc.M511939200. [DOI] [PubMed] [Google Scholar]
- 39.Ytterberg AJ, et al. Protein profiling of plastoglobules in chloroplasts and chromoplasts; a surprising site for differential accumulation of metabolic enzymes. Plant Physiol. 2006;140:984–997. doi: 10.1104/pp.105.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lundquist P, et al. The functional network of the Arabidopsis thaliana plastoglobule proteome based on quantitative proteomics and genome-wide co-expression analysis. Plant Physiol. 2012 doi: 10.1104/pp.111.193144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jasinski M, et al. AtOSA1, a member of the Abc1-like family, as a new factor in cadmium and oxidative stress response. Plant Physiol. 2008;147:719–731. doi: 10.1104/pp.107.110247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cardazzo B, et al. Isolation of an Arabidopsis thaliana cDNA by complementation of a yeast abc1 deletion mutant deficient in complex III respiratory activity. Gene. 1998;221:117–125. doi: 10.1016/s0378-1119(98)00417-x. [DOI] [PubMed] [Google Scholar]
- 43.Friso G, et al. Reconstruction of metabolic pathways, protein expression, and homeostasis machineries across maize bundle sheath and mesophyll chloroplasts: large-scale quantitative proteomics using the first maize genome assembly. Plant Physiol. 2010;152:1219–1250. doi: 10.1104/pp.109.152694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Majeran M, et al. Structural and metabolic transitions of C4 leaf development and differentiation defined by microscopy and quantitative proteomics. The Plant Cell. 2010;22:3509–3542. doi: 10.1105/tpc.110.079764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Majeran W, et al. Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: a new conceptual framework for nucleoid functions. Plant Physiol. 2012;158:156–189. doi: 10.1104/pp.111.188474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emanuelsson O, et al. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 47.Bousquet I, et al. ABC1, a novel yeast nuclear gene has a dual function in mitochondria: it suppresses a cytochrome b mRNA translation defect and is essential for the electron transfer in the bc 1 complex. Embo J. 1991;10:2023–2031. doi: 10.1002/j.1460-2075.1991.tb07732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Do TQ, et al. A Defect in Coenzyme Q Biosynthesis Is Responsible for the Respiratory Deficiency in Saccharomyces cerevisiae abc1 Mutants. J. Biol. Chem. 2001;276:18161–18168. doi: 10.1074/jbc.M100952200. [DOI] [PubMed] [Google Scholar]
- 49.Brasseur G, et al. The nuclear ABC1 gene is essential for the correct conformation and functioning of the cytochrome bc1 complex and the neighbouring complexes II and IV in the mitochondrial respiratory chain. Eur J Biochem. 1997;246:103–111. doi: 10.1111/j.1432-1033.1997.t01-1-00103.x. [DOI] [PubMed] [Google Scholar]
- 50.Hsieh EJ, et al. A tRNA(TRP) gene mediates the suppression of cbs2-223 previously attributed to ABC1/COQ8. Biochem Biophys Res Commun. 2004;317:648–653. doi: 10.1016/j.bbrc.2004.03.096. [DOI] [PubMed] [Google Scholar]
- 51.Poon WW, et al. Identification of Escherichia coli ubiB, a gene required for the first monooxygenase step in ubiquinone biosynthesis. J Bacteriol. 2000;182:5139–5146. doi: 10.1128/jb.182.18.5139-5146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mollet J, et al. CABC1 Gene Mutations Cause Ubiquinone Deficiency with Cerebellar Ataxia and Seizures. The American Journal of Human Genetics. 2008;82:623–630. doi: 10.1016/j.ajhg.2007.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lagier-Tourenne C, et al. ADCK3, an Ancestral Kinase, Is Mutated in a Form of Recessive Ataxia Associated with Coenzyme Q10 Deficiency. The American Journal of Human Genetics. 2008;82:661–672. doi: 10.1016/j.ajhg.2007.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nowicka B, Kruk J. Occurrence, biosynthesis and function of isoprenoid quinones. Biochim Biophys Acta. 2010;1797:1587–1605. doi: 10.1016/j.bbabio.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 55.Price DC, et al. Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science. 2012;335:843–847. doi: 10.1126/science.1213561. [DOI] [PubMed] [Google Scholar]
- 56.Tran UC, Clarke CF. Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion The Role of Coenzyme Q in Cellular Metabolism: Current Biological and Clinical Aspects. 2007;7(Supplement):S62–S71. doi: 10.1016/j.mito.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gray MW, et al. Mitochondrial Evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 58.Esser C, et al. The origin of mitochondria in light of a fluid prokaryotic chromosome model. Biology Letters. 2007;3:180–184. doi: 10.1098/rsbl.2006.0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Collins MD, Jones D. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol Rev. 1981;45:316–354. doi: 10.1128/mr.45.2.316-354.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sadre R, et al. Plastoquinone-9 biosynthesis in cyanobacteria differs from that in plants and involves a novel 4-hydroxybenzoate solanesyltransferase. Biochem J. 2012 doi: 10.1042/BJ20111796. [DOI] [PubMed] [Google Scholar]
- 61.Leonard CJ, et al. Novel families of putative protein kinases in bacteria and archaea: evolution of the "eukaryotic" protein kinase superfamily. Genome Res. 1998;8:1038–1047. doi: 10.1101/gr.8.10.1038. [DOI] [PubMed] [Google Scholar]
- 62.Delumeau O, et al. Functional and Structural Characterization of RsbU, a Stress Signaling Protein Phosphatase 2C. Journal of Biological Chemistry. 2004;279:40927–40937. doi: 10.1074/jbc.M405464200. [DOI] [PubMed] [Google Scholar]
- 63.Locke JC, et al. Stochastic pulse regulation in bacterial stress response. Science. 2011;334:366–369. doi: 10.1126/science.1208144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liobikas JB, Danas, Stanys Vidmantas, Toleikis Adolfas, Eriksson Ove. Identification and analysis of a novel serine beta-lactamase-like plant protein by a bioinformatic approach. Biologija. 2006:126–129. [Google Scholar]
- 65.Mustafiz A, et al. Functional & Integrative Genomics. Vol. 11. Springer Berlin: Heidelberg; 2011. Genome-wide analysis of rice and Arabidopsis identifies two glyoxalase genes that are highly expressed in abiotic stresses; pp. 293–305. [DOI] [PubMed] [Google Scholar]
- 66.Waterworth WM, et al. A plant DNA ligase is an important determinant of seed longevity. The Plant Journal. 2010;63:848–860. doi: 10.1111/j.1365-313X.2010.04285.x. [DOI] [PubMed] [Google Scholar]
- 67.Tauche A, et al. Ubiquinone biosynthesis in Saccharomyces cerevisiae: the molecular organization of O-methylase Coq3p depends on Abc1p/Coq8p. FEMS Yeast Research. 2008;8:1263–1275. doi: 10.1111/j.1567-1364.2008.00436.x. [DOI] [PubMed] [Google Scholar]
- 68.Merchant SS, et al. The Chlamydomonas Genome Reveals the Evolution of Key Animal and Plant Functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fusco N, et al. Identification of cadmium-regulated genes by cDNA-AFLP in the heavy metal accumulator Brassica juncea L. Journal of Experimental Botany. 2005;56:3017–3027. doi: 10.1093/jxb/eri299. [DOI] [PubMed] [Google Scholar]
- 70.Gao Q-S, et al. Cloning of an ABC1-like Gene ZmABC1-10 and Its Responses to Cadmium and Other Abiotic Stresses in Maize (Zea mays L.) Acta Agronomica Sinica. 2010;36:2073–2083. [Google Scholar]
- 71.Wang C, et al. TaABC1, a member of the activity of bc1 complex protein kinase family from common wheat, confers enhanced tolerance to abiotic stresses in Arabidopsis. Journal of Experimental Botany. 2010;63:1299–1311. doi: 10.1093/jxb/erq377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gao Q-s, et al. Systematic Identification of Rice ABC1 Gene Family and Its Response to Abiotic Stress. Rice Science. 2011;18:167–177. [Google Scholar]
- 73.Grobei MA, et al. Deterministic protein inference for shotgun proteomics data provides new insights into Arabidopsis pollen development and function. Genome Res. 2009;19:1786–1800. doi: 10.1101/gr.089060.108. [DOI] [PMC free article] [PubMed] [Google Scholar]