Abstract
MicroRNAs (miRNAs) are small, non-coding RNAs that negatively regulate gene expression and have emerged as potential biomarkers in radiation response to human cancer. Only a few miRNAs have been identified in radiation response to prostate cancer and the involvement of the radiation-associated miRNA machinery in the response of prostate cancer cells to radiation is not thoroughly understood. Therefore, the purpose of the present study was to comprehensively investigate the expression levels, arm selection preference and isomiRs of radiation-response miRNAs in radiation-treated PC3 cells using a next-generation sequencing (NGS) approach. Our data revealed that the arm selection preference and 3′ modification of miRNAs may be altered in prostate cancer after radiation exposure. In addition, the proportion of AA dinucleotide modifications at the end of the read gradually increased in a time-dependent manner after PC3 radiation treatment. We also identified 6 miRNAs whose expression increased and 16 miRNAs whose expression decreased after exposure to 10 Gy of radiation. A pathway enrichment analysis revealed that the target genes of these radiation-induced miRNAs significantly co-modulated the radiation response pathway, including the mitogen-activated protein kinase (MAPK), Wnt, transforming growth factor-β (TGF-β) and ErbB signaling pathways. Furthermore, analysis of The Cancer Genome Atlas (TCGA) database revealed that the expression of these radiation-induced miRNAs was frequently dysregulated in prostate cancer. Our study identified radiation-induced miRNA candidates which may contribute to radiosensitivity and can be used as biomarkers for radiotherapy.
Keywords: microRNA, prostate cancer, next-generation sequencing, pathway enrichment analysis, radiation, The Cancer Genome Atlas
Introduction
Prostate carcinoma is the most frequently diagnosed visceral cancer in men worldwide. An increasing prevalence has been reported in recent decades (1). Radiation therapy is one of the primary modalities in prostate cancer treatment. Ionizing radiation damages cells through free radicals from the radiolysis of water that cause DNA double-strand breaks. However, the efficacy of the radiotherapy may be affected by the cellular response to radiation. Radiotherapy is highly effective in treating radiosensitive tumors and enhancing the therapeutic efficacy can increase the overall survival rate. However, the presence of radioresistant tumors leads to cancer relapse and metastasis. Understanding the tumor-radiation-related genes to predict the tumor response to radiotherapy may potentially modulate the treatment outcome for prostate cancer patients.
MicroRNAs (miRNAs) are a family of small, non-coding, single-stranded RNAs composed of ~22 nucleotides (nt) that negatively regulate protein expression at the post-transcriptional level (2). They function as gene regulators by binding to partially complementary sites of mRNAs and cause translation inhibition or direct degradation of the target mRNA. It has been suggested that miRNAs are responsible for controlling ~50% of all protein-coding genes (3). The widespread regulation of protein levels has been studied in cellular models (4). Previous studies have demonstrated that the expression of miRNAs is clearly involved in cancer development, and the deregulation of several miRNAs has been observed in various types of cancer, including prostate cancer. Porkka et al (5) was the first to identify a miRNA signature specific for prostate cancer by systematically profiling prostate cancer cell lines. Numerous studies have identified many dysfunctional miRNAs by using a high-throughput approach, which contributed to prostate cancer progression, including the let-7 family, miR-1, -20a, -21,-34a, -106b, -125b, -205 and -521 (6–13). Although several studies have investigated the role of these dysfunctional miRNAs to develop prostate cancer therapy, few studies have determined the roles of miRNAs in radiation response in prostate cancer. The upregulation of miR-521 reduces the response to radiation damage by specifically targeting a DNA repair protein, the Cockayne syndrome protein A (13). Li et al (14) found that miR-106b was dysregulated after radiation treatment and suppressed radiation-induced p21 activation, suggesting it may override radiation-induced cell cycle arrest and cell growth inhibition. Radiation delivered in daily fractions altered a greater number of miRNAs compared with single-dose radiation, and involved the upregulation of miR-34a and let-7 miRNAs (15).
Next-generation sequencing (NGS) is a high-throughput screening technology, and NGS data can be applied in investigating miRNA expression, miRNA isoforms (isomiRs) and the arm selection preferences of miRNAs. Therefore, the purpose of the present study was to comprehensively investigate the distribution of miRNAs after radiation treatment in PC3 cells by using an NGS approach. Furthermore, we explored the function of radiation-associated miRNA by conducting an in silico analysis.
Materials and methods
Cell culture and radiation treatment
A PC3 cell line was obtained from the American Type Culture Collection and was maintained in RPMI-1640 and supplemented with 10% inactivated fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA). The cells were exposed to various radiation dosages (0, 2, 6, 10, 14 and 18 Gy) and were subsequently cultured in fresh medium. The total RNA was obtained at various time points (0, 5, 15 and 40 h after treatment) by using TRIzol (Invitrogen) according to the manufacturer’s instructions. The concentration, purity and amount of total RNA were determined using a NanoDrop 1000 spectrophotometer (NanoDrop Technologies, Inc., USA).
Collection and preprocessing of sequence reads
PC3 cells were exposed to 10 Gy of radiation. After radiation treatment, the cells were lysed at various time points (0, 5, 15 and 40 h) for RNA extraction. The RNA samples were prepared using an Illumina small RNA preparation kit, and were subsequently sequenced using the Illumina HiSeq platform. The generated sequence reads were first subjected to quality control to remove low-quality reads. The sequence reads were then subjected to 3′ adaptor trimming to generate clean reads, as previously described (15,16). To attain a high confidence level, only the clean reads with a read count ≥2 and with a length ranging from 15 to 27 nt were included in further analyses.
Mapping clean reads to pre-miRNAs
To investigate miRNA expression profiles in different libraries, we mapped the qualified clean reads back to human pre-miRNAs (miRBase 19). To eliminate ambiguous multiple hits during the mapping procedure, no mismatch was allowed. Previous studies reported that, when mapped back to pre-miRNAs, sequence reads usually carried mismatches preferentially located at their terminal 3′ ends (17–20). This mismatch was named the 3′ end modification. To determine whether the 3′ end modification patterns differed among libraries, as described in our previous studies (21), we trimmed and collected the terminal 3′ end mismatches one by one. In addition, the remaining perfect match reads had to be at least 18 nt in length. As a result, we kept reads with no less than 18-nt perfect alignment and 3′ end modification patterns.
Classifying non-miRNA reads into different data sets
The sequence reads that may not be mapped back to pre-miRNAs were classified into classes by mapping to acquire different data sets with Bowtie (22) and allowing a single nucleotide variation. The sequences of mRNAs and other ncRNAs were derived from the NCBI RefSeq 47 (23). The tRNA sequences were downloaded from the Genomic tRNA database (24) and the rRNA sequences were downloaded from the SILVA database (25). The snoRNA, scaRNA and snRNA sequences were all downloaded from NONCODE (26). The sequence reads not belonging to any of the described RNA classes were uploaded to the RepeatMasker to identify repeat elements, which were classified as unknown.
miRNA expression level according to The Cancer Genome Atlas (TCGA) data
TCGA project collects both cancer and corresponding normal tissues from hundreds of prostate cancer patients. We downloaded all level-3 miRNA expression data of prostate adenocarcinoma from the TCGA Data Portal (https://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm). These level-3 data included calculated expressions for each miRNA derived from the Illumina HiSeq sequencing results. A total of 198 tumor samples and 50 normal samples were found at the time the data were downloaded. We kept only the expression data of 50 participants who had both miRNA expression levels from both tumor and normal tissues. Normalized quantification expression levels for these 50 participants were further examined for each investigated miRNA.
Pathway enrichment analysis
We attempted to determine the functions of the miRNA target genes by investigating the pathways with which the miRNA target genes were involved. Therefore, we first downloaded the target genes of differentially expressed miRNAs from TargetScan 6.0, and then mapped the target genes onto the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways based on the Enzyme Commission (EC) numbers by using the R package SubPathwayMiner v.3.1 (27). Subsequently, the hypergeometric test was performed to identify significantly enriched pathways and calculate the false positive discovery rate in the FDR-corrected q-value.
Results
miRNA profiling of radiation-treated prostate cancer cells
To characterize the mechanism involved in the radiation response of prostate cancer, we used NGS to comprehensively analyze the distribution of miRNAs after radiation treatment in PC3 cells. As indicated in Fig. 1A, PC3 cells were exposed to various dosages of radiation (0, 2, 4, 6, 8, 10, 12, 14, 16 and 18 Gy) and then subjected to a fresh culture medium for an additional 4 days. We found that the growth of the PC3 cells obviously decreased when exposed to 10 Gy of radiation. Therefore, we collected cell RNA at various times (0, 5, 15 and 40 h) following the 10-Gy radiation treatment. We confirmed the expression levels of Cox-2 and p21, which may be induced by radiation at 24 h according to previous studies (16,28). The expression levels of Cox-2 and p21 may be upregulated by radiation treatment in PC3 cells (Fig. 1B). We then performed the comprehensive miRNA profile at various time-points in radiation-treated PC3 cells by using the Illumina HiSeq platform.
Figure 1.
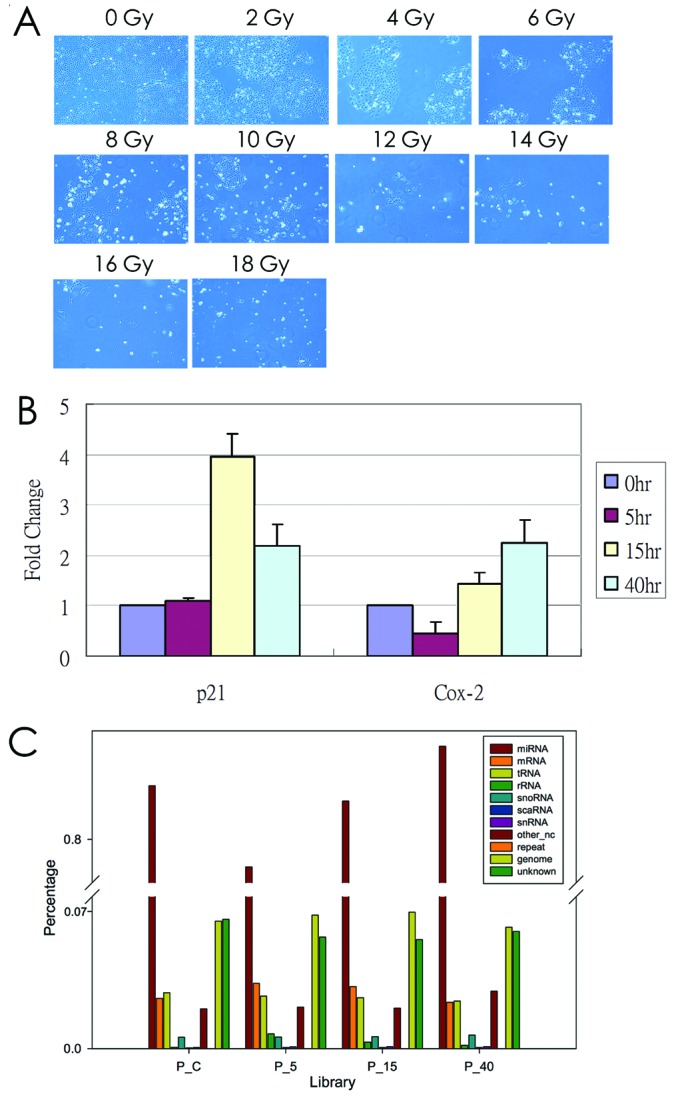
Radiation treatment of human prostate cancer cells, PC3. (A) PC3 cells were treated with various radiation doses (0, 2, 4, 6, 8, 10, 12, 14, 16 and 18 Gy) and were subsequently subjected to fresh culture medium. After culturing for an additional 4 days, the morphology was observed using light microscopy (x40 magnification). (B) The expression pattern of COX-2 and p21 in radiation-treated PC3 cells was examined using a real-time PCR method. S26 was used as an internal control. (C) The distribution of small RNA reads in 11 categories was classified.
Analysis of miRNA sequence reads
Once the samples were sequenced, we collected >9 million clean reads in all libraries (Table I). In addition to miRNA, we also determined which molecules were the remaining non-miRNA reads. By mapping the non-miRNA reads back to a different data set, we classified the reads into 11 categories. Fig. 1C demonstrates that miRNA accounted for 80% of all clean reads in the prostate cell libraries. Other categories accounted for relatively low proportions, which indicated the high performance of the sample preparation protocol. In addition, the proportions of the categories were considerably similar among libraries, indicating that radiation treatment did not alter the composition of RNA samples in the prostate cell libraries.
Table I.
Summary of sequence reads and the detected miRNAs.
| Library | Clean read (n) | miRNA read (%) | pre-miRNA (n) |
miRNA (n) |
|---|---|---|---|---|
| P_C | 9,482,400 | 80.49 | 693 | 916 |
| P_5 | 9,748,570 | 79.75 | 687 | 915 |
| P_15 | 9,589,440 | 80.35 | 712 | 933 |
| P_40 | 10,589,934 | 80.86 | 739 | 964 |
P_C, P_5, P_15 and P_40 are prostate cancer cell lines with different radiation treatment. By mapping the clean sequence reads back to pre-miRNAs, we can quantify how many pre-miRNAs and mature miRNAs were detected.
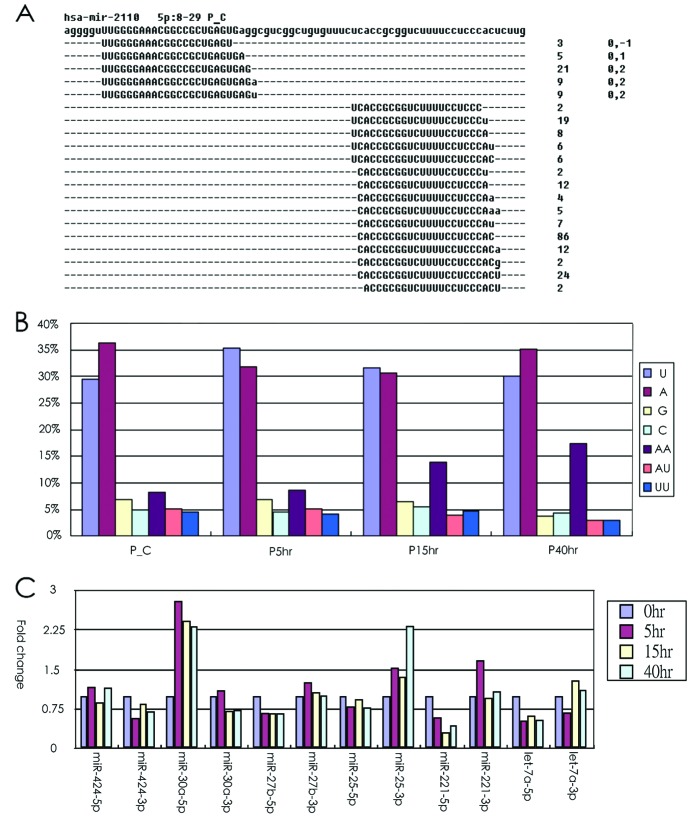
After mapping the clean reads to the genome, most of the miRNA reads tend to exist as isomiR. As demonstrated in Fig. 2A, hsa-miR-2110-5p had 5 isomers, whereas the opposite-arm miRNA-3p had 15 isomiRs, which demonstrated that abundant miRNAs tend to have more isomiRs. Our data revealed that the isomiR quantity was highly correlated with miRNA abundance (Pearson’s correlation coefficient, 0.91). In addition, we observed that the modified nucleotides were preferentially located at the 3′ end of the sequence read (presented in lower case in Fig. 2A). The data indicated that one A nucleotide or one U nucleotide was frequently added at the end of the read. Notably, we found that the proportion of AA dinucleotides modified at the end of the read was gradually increased in a time-dependent manner after the PC3 cells were treated with radiation, which indicated that the 3′ end modification may be altered by radiation treatment in PC3 cells. Our previous studies indicated that the use of miR-5p and -3p may be altered in human cancer (29–31). In the present study, our data indicated that arm selection preference was consistent across nearly all libraries. Only a few cases were observed in which the use of -5p and -3p arm selection had different preferences at various time-points after radiation treatment (Fig. 2C). Further research is required to support these findings.
Figure 2.
The distribution of isomiRs and -5p/-3p arm selection in PC3 radiation treatment. (A) Mapping results of hsa-miR-2110. As shown in 5p:8–29, hsa-miR-2110 encodes mature miRNA at only its 5 p arm, and miRNA spans from nucleotide 8 to nucleotide 29 of the hairpin. The integer values on the left denote the read count of each isomiR. The comma-separated values denote the position shift in the isomiR relative to the miRBase annotated positions (8 to 29). The nucleotides in lowercase type denote the sequence fragments originating from the 3′ modification event. (B) The proportion of 3′ modifications at end of reads at different time-points after PC3 radiation treatment as observed from NGS data. (C) Fold-change of the -5p/-3p arm of miRNA at different time points after PC3 radiation treatment as observed from NGS data. isomiRs, miRNA isoforms; miRNA, microRNA; NGS, next-generation sequencing.
Radiation-response miRNAs in prostate cancer
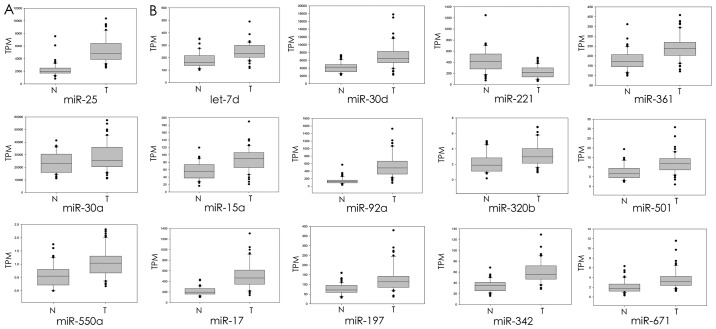
By summarizing the read count of all the isomiRs that belonged to the same mature miRNAs, we quantified the miRNA expression abundances, and presented the result in transcript per million (TPM). Twenty-two miRNAs were selected and are presented in Table II, demonstrating that their expression levels were altered >2-fold after being subjected to radiation exposure (expression of 6 miRNAs increased, and expression of 16 miRNAs decreased). To explore the putative role contributing to prostate cancer progression, we examined the effect of expression levels of radiation-associated miRNAs on prostate cancer from the available TCGA dataset by using an in silico analysis. We downloaded 100 miRNA expression profiles from 50 prostate cancer patients, including 50 cancer lesion and 50 corresponding normal tissues. As demonstrated in Fig. 3A, the expression levels of radiation-induced miRNAs, miR-25, miR-30a and miR-550a, were significantly upregulated in the prostate cancer cells compared with the corresponding normal tissue cells. Twelve radiation-suppressed miRNAs were identified, i.e. let-7d, miR-15a, miR-17, miR-30d, miR-92a, miR-197, miR-221, miR-320b, miR-342, miR-361, miR-501 and miR-671, and a significantly different expression between prostate cancer and the corresponding adjacent part was found, including 11 upregulated and 1 downregulated (Fig. 3B). Overall, the data indicated that most of the radiation-response miRNAs were identified as dysregulated in prostate cancer according to an in silico analysis (15/22; 1 downregulated, 14 upregulated and the rest demonstrated no change in expression in prostate cancer).
Table II.
miRNAs with altered expression in response to radiation in PC3 cells using next-generation sequencing.
| 0 h | 5 h | 15 h | 40 h | Expression data for TCGA | |
|---|---|---|---|---|---|
| Upregulationa | |||||
| hsa-miR-9-5p | 1 | 2.82 | 0.90 | 2.63 | |
| hsa-miR-22-3p | 1 | 1.60 | 2.85 | 2.73 | |
| hsa-miR-25-3p | 1 | 1.54 | 1.39 | 2.33 | Upregulationd |
| hsa-miR-30a-5p | 1 | 2.81 | 2.43 | 2.33 | Upregulationc |
| hsa-miR-550a-3p | 1 | 1.88 | 1.46 | 2.09 | Upregulationd |
| hsa-miR-548h-5p | 1 | 0.50 | 0.44 | 2.56 | |
| Downregulationb | |||||
| hsa-let-7c | 1 | 0.93 | 0.78 | 0.45 | |
| hsa-let-7d-5p | 1 | 0.30 | 0.11 | 0.40 | Upregulationd |
| hsa-let-7e-5p | 1 | 0.58 | 0.55 | 0.40 | |
| hsa-miR-15a-5p | 1 | 0.78 | 0.67 | 0.45 | Upregulationd |
| hsa-miR-17-3p | 1 | 0.52 | 0.49 | 0.47 | Upregulationd |
| hsa-miR-30d-3p | 1 | 0.92 | 0.75 | 0.41 | Upregulationd |
| hsa-miR-92a-5p | 1 | 0.65 | 0.52 | 0.50 | Upregulationd |
| hsa-miR-125a-3p | 1 | 0.42 | 0.42 | 0.32 | |
| hsa-miR-197-3p | 1 | 0.77 | 0.79 | 0.44 | Upregulationd |
| hsa-miR-221-5p | 1 | 0.59 | 0.31 | 0.44 | Downregulationd |
| hsa-miR-320b | 1 | 0.65 | 0.41 | 0.32 | Upregulationd |
| hsa-miR-342-5p | 1 | 0.59 | 0.63 | 0.47 | Upregulationd |
| hsa-miR-361-3p | 1 | 0.45 | 0.53 | 0.40 | Upregulationd |
| hsa-miR-374a-5p | 1 | 1.04 | 0.93 | 0.47 | |
| hsa-miR-501-3p | 1 | 0.77 | 0.48 | 0.45 | Upregulationd |
| hsa-miR-671-3p | 1 | 0.77 | 0.62 | 0.41 | Upregulationd |
Expression levels of miRNA were inducted >2-fold change after PC3 radiation treatment with 10 Gy for 40 h.
Expression levels of miRNA were repressed >2-fold change after PC3 radiation treatment with 10 Gy for 40 h.
The difference was indicated to be significant with p-value less than 0.01 or 0.001.
Figure 3.
(A) Expression levels of radiation-induced miRNAs in prostate cancer. (B) Expression levels of radiation-suppressed miRNAs in prostate cancer. Expression levels of miRNAs between tumor and corresponding normal tissues from 50 prostate cancer patients were analyzed using TCGA dataset. The expression levels of miRNAs were presented in transcript per million (TPM). The expression level between tumor and normal cells was evaluated by conducting paired t-tests (P<0.05 was considered significant; NS, non-significant. *P<0.05, **P<0.01, ***P<0.001). TCGA, The Cancer Genome Atlas.
Pathway enrichment analysis of miRNAs
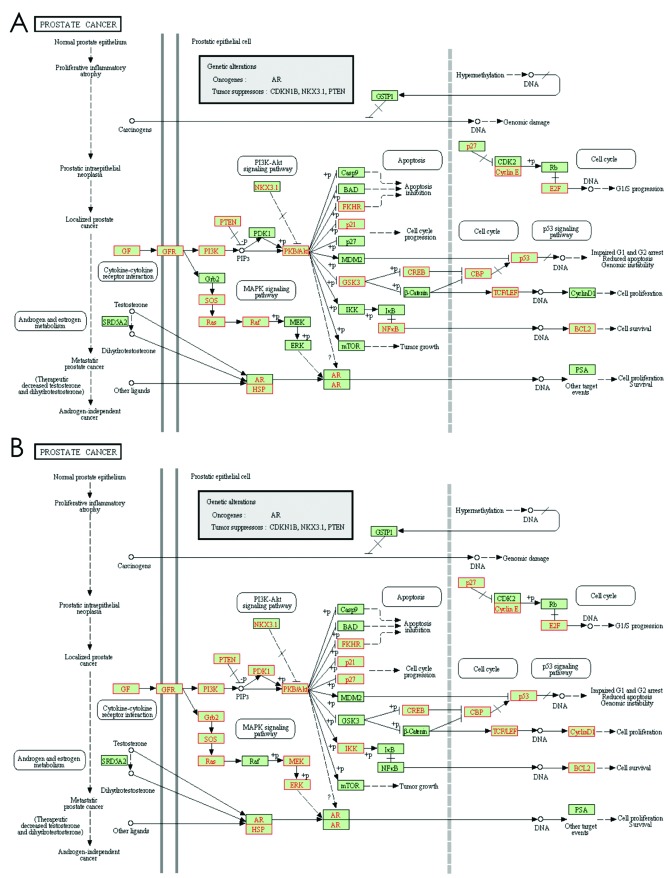
miRNAs can function as either oncogenes or tumor suppressors depending on their target genes. Therefore, identifying a target can facilitate elucidating the role of miRNAs in prostate cancer treatment radiation (32,33). Typically, one miRNA tends to have hundreds of target genes and a group of miRNAs co-modulated as a biological function involved in the regulation of a signaling pathway. Therefore, we further explored the biological function of radiation-response miRNAs by conducting a pathway-enrichment analysis. The putative target genes of miRNAs were obtained from TargetScan 6.0; subsequently, these target genes of the individual miRNAs were mapped onto KEGG pathways. Our data indicated that the target genes of radiation-response miRNAs were frequently significantly enriched in several cancer- or radiation-related pathways, including the mitogen-activated protein kinase (MAPK), ErbB, p53, Wnt, transforming growth factor-β (TGF-β) and mTOR signaling pathways with an FDR <0.05 (Table III). We also subjected the target genes of the 2-gene set, upregulated miRNA and downregulated miRNAs, to pathway-enrichment analysis. Similar results were observed; their targets were significantly enriched in the prostate cancer pathway (Fig. 4) and radiation-related pathways, including the MAPK, ErbB, Wnt and TGF-β signaling pathways (Tables IV and V).
Table III.
The enriched pathways of radiation-induced miRNA target genes.
| microRNA | Cancer-relative pathway (FDR<0.05) |
|---|---|
| Upregulation | |
| hsa-miR-9-5p | Focal adhesion, pathways in cancer, ErbB signaling pathway, MAPK signaling pathway, prostate cancer |
| hsa-miR-22-3p | Chronic myeloid leukemia, MAPK signaling pathway, ErbB signaling pathway, pathways in cancer, glioma, prostate cancer, phosphatidylinositol signaling system, colorectal cancer |
| hsa-miR-25-3p | N.D |
| hsa-miR-30a-5p | N.D |
| hsa-miR-550a-3p | N.D |
| hsa-miR-548h-5p | N.D |
| Downregulation | |
| hsa-let-7c | MAPK signaling pathway, pathways in cancer, p53 signaling pathway, melanoma, chronic myeloid leukemia, glioma, pancreatic cancer, focal adhesion, small cell lung cancer, bladder cancer, prostate cancer |
| hsa-let-7d-5p | MAPK signaling pathway, pathways in cancer, p53 signaling pathway, melanoma, chronic myeloid leukemia, glioma, pancreatic cancer, focal adhesion, small cell lung cancer, bladder cancer, prostate cancer |
| hsa-let-7e-5p | MAPK signaling pathway, pathways in cancer, p53 signaling pathway, melanoma, chronic myeloid leukemia, glioma, pancreatic cancer, focal adhesion, small cell lung cancer, bladder cancer, prostate cancer |
| hsa-miR-15a-5p | Pathways in cancer, regulation of actin cytoskeleton, renal cell carcinoma, MAPK signaling pathway, focal adhesion, melanoma, prostate cancer, Wnt signaling pathway, p53 signaling pathway, mTOR signaling pathway, non-small cell lung cancer, pancreatic cancer, cell cycle |
| hsa-miR-17-3p | MAPK signaling pathway, pathways in cancer, chronic myeloid leukemia, pancreatic cancer, melanoma, bladder cancer, TGF-β signaling pathway, prostate cancer, mTOR signaling pathway, non-small cell lung cancer, renal cell carcinoma, cell cycle, p53 signaling pathway |
| hsa-miR-30d-3p | N.D |
| hsa-miR-92a-5p | N.D |
| hsa-miR-125a-3p | MAPK signaling pathway, adherens junction, pancreatic cancer, TGF-β signaling pathway |
| hsa-miR-197-3p | N.D |
| hsa-miR-221-5p | Wnt signaling pathway, ErbB signaling pathway hsa-miR-320b Chronic myeloid leukemia, non-small cell lung cancer, glioma, pathways in cancer, focal adhesion, pancreatic cancer, melanoma, ErbB signaling pathway, colorectal cancer, TGF-β signaling pathway, prostate cancer, MAPK signaling pathway, mTOR signaling pathway |
| hsa-miR-342-5p | N.D |
| hsa-miR-361-3p | Pathways in cancer, mTOR signaling pathway, melanogenesis, renal cell carcinoma, nucleotide excision repair |
| hsa-miR-374a-5p | Pathways in cancer, prostate cancer, TGF-β signaling pathway, endometrial cancer, non-small cell lung cancer, basal cell carcinoma, MAPK signaling pathway |
| hsa-miR-501-3p | N.D |
| hsa-miR-671-3p | N.D |
Figure 4.
The enriched pathway of the target gene union of radiation-response miRNAs. (A) The target gene union of radiation-upregulated miRNAs enriched in the prostate cancer pathway (FDR=0.001). (B) The target gene union of radiation-downregulated miRNAs enriched in the prostate cancer pathway (FDR=3.6E-9). The target genes are labeled in red.
Table IV.
The enriched pathways of radiation-upregulated miRNA target genes.
| Pathway Id | Pathway name | FDR |
|---|---|---|
| Path:04360 | Axon guidance | 7.88E-08 |
| Path:04722 | Neurotrophin signaling pathway | 1.11E-07 |
| Path:04010 | MAPK signaling pathway | 1.11E-07 |
| Path:05200 | Pathways in cancer | 3.25E-07 |
| Path:04012 | ErbB signaling pathway | 3.14E-06 |
| Path:04120 | Ubiquitin mediated proteolysis | 8.82E-06 |
| Path:04144 | Endocytosis | 1.24E-05 |
| Path:04520 | Adherens junction | 5.86E-05 |
| Path:05412 | Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 7.26E-05 |
| Path:04510 | Focal adhesion | 0.000105 |
| Path:04916 | Melanogenesis | 0.000245 |
| Path:05220 | Chronic myeloid leukemia | 0.000245 |
| Path:04810 | Regulation of actin cytoskeleton | 0.000261 |
| Path:05214 | Glioma | 0.000261 |
| Path:05414 | Dilated cardiomyopathy | 0.000268 |
| Path:04910 | Insulin signaling pathway | 0.000282 |
| Path:04720 | Long-term potentiation | 0.000294 |
| Path:04070 | Phosphatidylinositol signaling system | 0.000294 |
| Path:05410 | Hypertrophic cardiomyopathy (HCM) | 0.000802 |
| Path:05100 | Bacterial invasion of epithelial cells | 0.001018 |
| Path:05215 | Prostate cancer | 0.001096 |
| Path:04310 | Wnt signaling pathway | 0.001153 |
| Path:04914 | Progesterone-mediated oocyte maturation | 0.002548 |
| Path:05211 | Renal cell carcinoma | 0.003135 |
| Path:04920 | Adipocytokine signaling pathway | 0.003902 |
| Path:04350 | TGF-β signaling pathway | 0.003902 |
| Path:04666 | Fc γ R-mediated phagocytosis | 0.004314 |
| Path:04141 | Protein processing in endoplasmic reticulum | 0.005738 |
| Path:05210 | Colorectal cancer | 0.005747 |
| Path:04130 | SNARE interactions in vesicular transport | 0.007624 |
| Path:05216 | Thyroid cancer | 0.008679 |
| Path:00562 | Inositol phosphate metabolism | 0.008679 |
| Path:00532 | Glycosaminoglycan biosynthesis-chondroitin sulfate | 0.008679 |
| Path:05014 | Amyotrophic lateral sclerosis (ALS) | 0.008679 |
| Path:05212 | Pancreatic cancer | 0.008679 |
| Path:04020 | Calcium signaling pathway | 0.008679 |
| Path:05218 | Melanoma | 0.010147 |
| Path:04930 | Type II diabetes mellitus | 0.018227 |
| Path:04962 | Vasopressin-regulated water reabsorption | 0.018227 |
| Path:04530 | Tight junction | 0.019289 |
| Path:04512 | ECM-receptor interaction | 0.019289 |
| Path:04912 | GnRH signaling pathway | 0.019289 |
| Path:05223 | Non-small cell lung cancer | 0.023349 |
| Path:05222 | Small cell lung cancer | 0.032425 |
| Path:05213 | Endometrial cancer | 0.035464 |
| Path:04662 | B cell receptor signaling pathway | 0.035464 |
| Path:05131 | Shigellosis | 0.035464 |
| Path:05221 | Acute myeloid leukemia | 0.03804 |
| Path:04540 | Gap junction | 0.041068 |
| Path:00250 | Alanine, aspartate and glutamate metabolism | 0.045237 |
| Path:04114 | Oocyte meiosis | 0.049286 |
Table V.
The enriched pathways of radiation-downregulated miRNA target genes.
| Pathway Id | Pathway name | FDR |
|---|---|---|
| Path:04010 | MAPK signaling pathway | 0 |
| Path:04360 | Axon guidance | 0 |
| Path:05200 | Pathways in cancer | 0 |
| Path:04722 | Neurotrophin signaling pathway | 1.18E-12 |
| Path:04310 | Wnt signaling pathway | 1.50E-10 |
| Path:04510 | Focal adhesion | 4.27E-10 |
| Path:04144 | Endocytosis | 1.40E-09 |
| Path:04810 | Regulation of actin cytoskeleton | 2.34E-09 |
| Path:05215 | Prostate cancer | 3.58E-09 |
| Path:05211 | Renal cell carcinoma | 3.58E-09 |
| Path:04720 | Long-term potentiation | 1.14E-08 |
| Path:05220 | Chronic myeloid leukemia | 3.41E-08 |
| Path:04120 | Ubiquitin mediated proteolysis | 7.40E-08 |
| Path:04020 | Calcium signaling pathway | 1.10E-07 |
| Path:05212 | Pancreatic cancer | 1.57E-07 |
| Path:05214 | Glioma | 1.59E-07 |
| Path:05218 | Melanoma | 2.36E-07 |
| Path:05223 | Non-small cell lung cancer | 3.64E-07 |
| Path:04916 | Melanogenesis | 4.67E-07 |
| Path:04520 | Adherens junction | 4.67E-07 |
| Path:04910 | Insulin signaling pathway | 6.79E-07 |
| Path:04350 | TGF-β signaling pathway | 7.72E-07 |
| Path:05210 | Colorectal cancer | 2.50E-06 |
| Path:04012 | ErbB signaling pathway | 3.89E-06 |
| Path:05222 | Small cell lung cancer | 3.20E-05 |
| Path:04730 | Long-term depression | 5.13E-05 |
| Path:05221 | Acute myeloid leukemia | 7.50E-05 |
| Path:04150 | mTOR signaling pathway | 8.68E-05 |
| Path:05213 | Endometrial cancer | 8.68E-05 |
| Path:05217 | Basal cell carcinoma | 9.44E-05 |
| Path:04710 | Circadian rhythm-mammal | 9.64E-05 |
| Path:04070 | Phosphatidylinositol signaling system | 9.64E-05 |
| Path:04540 | Gap junction | 9.83E-05 |
| Path:04115 | p53 signaling pathway | 0.000113 |
| Path:04141 | Protein processing in endoplasmic reticulum | 0.000137 |
| Path:04114 | Oocyte meiosis | 0.000196 |
| Path:05014 | Amyotrophic lateral sclerosis (ALS) | 0.000353 |
| Path:04970 | Salivary secretion | 0.000414 |
| Path:04660 | T cell receptor signaling pathway | 0.000449 |
| Path:04666 | Fc γ R-mediated phagocytosis | 0.000526 |
| Path:04512 | ECM-receptor interaction | 0.000526 |
| Path:05142 | Chagas disease | 0.000563 |
| Path:04062 | Chemokine signaling pathway | 0.000812 |
| Path:04210 | Apoptosis | 0.000844 |
| Path:04914 | Progesterone-mediated oocyte maturation | 0.001465 |
| Path:04110 | Cell cycle | 0.001527 |
| Path:04662 | B cell receptor signaling pathway | 0.00169 |
| Path:04530 | Tight junction | 0.001692 |
| Path:04930 | Type II diabetes mellitus | 0.002128 |
| Path:05412 | Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 0.002128 |
| Path:04920 | Adipocytokine signaling pathway | 0.002184 |
| Path:04664 | Fc ɛ RI signaling pathway | 0.002621 |
| Path:04960 | Aldosterone-regulated sodium reabsorption | 0.002802 |
| Path:04912 | GnRH signaling pathway | 0.002955 |
| Path:04320 | Dorso-ventral axis formation | 0.00373 |
| Path:05219 | Bladder cancer | 0.00373 |
| Path:04340 | Hedgehog signaling pathway | 0.005218 |
| Path:04971 | Gastric acid secretion | 0.005633 |
| Path:04130 | SNARE interactions in vesicular transport | 0.006926 |
| Path:00532 | Glycosaminoglycan biosynthesis-chondroitin sulfate | 0.006926 |
| Path:05131 | Shigellosis | 0.008185 |
| Path:05160 | Hepatitis C | 0.008185 |
| Path:04630 | Jak-STAT signaling pathway | 0.008368 |
| Path:05410 | Hypertrophic cardiomyopathy (HCM) | 0.009939 |
| Path:05414 | Dilated cardiomyopathy | 0.010733 |
| Path:00534 | Glycosaminoglycan biosynthesis-heparan sulfate | 0.011359 |
| Path:04670 | Leukocyte transendothelial migration | 0.011852 |
| Path:04370 | VEGF signaling pathway | 0.013265 |
| Path:00562 | Inositol phosphate metabolism | 0.013625 |
| Path:04270 | Vascular smooth muscle contraction | 0.014557 |
| Path:00512 | O-Glycan biosynthesis | 0.016472 |
| Path:04330 | Notch signaling pathway | 0.026769 |
| Path:04142 | Lysosome | 0.038882 |
| Path:00533 | Glycosaminoglycan biosynthesis-keratan sulfate | 0.038882 |
| Path:05145 | Toxoplasmosis | 0.048785 |
Discussion
Our previous studies indicated that the distributions of 3′ end modifications and the arm selection preference of miRNAs were different between normal and tumor tissues (29–31). The -5p and -3p of miRNA play a distant role by suppressing the different target genes. It was previously reported that, in contrast to the oncogenic effect of miR-17 (-5p), miR-17*(-3P) plays a tumor suppressive role in prostate cancer (9,34,35). The miR-28-5p and miR-28-3p also play opposite roles in colon cancer cell proliferation and migration (36). In the present study, our data showed that the -5p and -3p of particular miRNAs were differently regulated by radiation (shown in Fig. 2C). Several studies have demonstrated that miRNAs contain various ends, which were caused by either RNA editing or non-template nucleotide additions (17,18,20). These miRNA isoforms (isomiRs) contribute to increased miRNA stability or strengthened miRNA-target gene interaction and are differentially expressed in different cellular conditions, including cancer (16,37,38). Our data revealed that the proportion of AA dinucleotide modifications at the end of the read gradually increased in a time-dependent manner after the PC3 cells were treated with radiation, suggesting that radiation may influence the particular miRNA stability or efficiency of silencing targets by regulating the 3′ end modifications, which warrants further research.
miRNAs are known to function as gene silencers and are involved in modulating biological functions, including cell growth, apoptosis, the cell cycle and the metastasis of cancer (39). Comprehensive miRNA profiling of prostate cancer has indicated that several miRNAs are differentially expressed between prostate cancer and the adjacent normal, which contributes to prostate cancer progression (40–42). In the present study, we analyzed miRNA expression from TCGA database and found that the expression levels of radiation-induced miRNAs were frequently dysregulated in prostate cancer (Fig. 3). Our results are consistent with those of previous studies and demonstrated that miR-25, miR-17, miR-30d and miR-92a are overexpressed, and miR-221 is downregulated in prostate cancer (9,42–44). However, the expression levels of let-7d and miR-15a decreased according to TCGA, which contradicted the results of previous studies (45–47). These dysfunctional miRNAs have potential to be used as biomarkers for prostate cancer prognosis or diagnosis. Therefore, understanding the function of miRNAs may provide practical benefits for clinical applications. Predicting the outcome of cancer treatment is the most promising application of miRNAs. Gonzales et al found miR-141 to be consistent with changes in other conventional biomarkers and to the clinical outcomes, suggesting that miR-141 can be used as a marker for monitoring therapeutic response in prostate cancer patients (48). The prognostic value of miRNA expression profiling in prostate cancer has also been demonstrated by Hulf et al. They demonstrated that DNA methylation and histone H3K9-deacetylation of the miR-205 locus is associated with miRNA silencing and deregulation of MED1, which is predictive of a poor prognosis in localized prostate cancer (49).
Ionizing radiation is one of the 3 primary modalities used in cancer therapy. Radiation induces considerable DNA damages, which, if not repaired, cause cancer cells to progress to apoptosis and cell cycle arrest. Some cancer cells are resistant to radiation treatment due to activation of complex signaling pathways that counteract these damages, including ErbB, nuclear factor κB (NFκB), MAPK, PI3K/AKT and transforming growth factor-β (TGF-β) signaling pathways (50–52). Several radiation-related miRNAs have been identified that contribute to the radiosensitivity of cancer cells by modulating the radiation-response signaling pathway (51,52). Since miRNAs are generally slightly repressed by their target genes, the alteration of an individual miRNA is insufficient for accomplishing a biological function. Previous studies have introduced the concept of miRNA regulatory modules (MRMs), which potentially serve as a model for understanding the detailed influences of miRNAs in cellular biological functions (53–55). Therefore, in the present study, we were particularly interested in the consequences of changes in a group of radiation-induced miRNAs in prostate cancer. Our data indicated that targets of the co-expressed miRNAs were enriched in a radiation-related signaling pathway, suggesting that they co-modulated an abundance of target genes in the same pathway (Table III).
miRNAs regulate various factors in radiation-related biological pathways and may affect the radiosensitivity of tumor cells (51). Radiation-response miRNAs have been identified in prostate cancer by using a microarray approach (13–15). By comparing these data, we identified known and unknown radiation-response miRNAs in prostate cancer by using an NGS approach. Li et al reported that the expression levels of miR-9, miR-22 and miR-30a decreased in radiation-treated PC3 cells (14). Radiation reduced the expression level of an miR-17-92a cluster and the let-7 family in prostate cancer (15). In the present study, we also identified radiation-response miRNAs that had been reported in other types of cancer but not in prostate cancer, such as miR-25, miR-15a, miR-30d, miR-125a, miR-221 and miR-342 (21,56–63). In addition, we identified a group of radiation-response miRNAs that have not been reported in any type of cancer. The pathway-enrichment analysis revealed that their targets are frequently enriched in the radiation-response signaling pathway.
In summary, in the present study, we thoroughly investigated radiation-response miRNAs, which may be involved in the radiosensitivity of prostate cancer, by modulating radiation-related signaling pathways using an NGS approach. These miRNA candidates may be effective targets for improving the efficacy of radiation treatment in future prostate cancer therapy. In addition, we observed that 3′ end modifications and the -5p/-3p arm selection of miRNAs were altered in prostate cancer after radiation treatment. These finding require further research.
Acknowledgements
This study was supported by grants from Kaohsiung Veterans General Hospital (VGHKS 102-005 and VGHKS 102-074). The authors thank Genomics and Proteomics Core Laboratory, Department of Medical Research, Kaohsiung Chang Gung Memorial Hospital, for the assistance with NGS data analysis.
References
- 1.Crawford ED. Epidemiology of prostate cancer. Urology. 2003;62(Suppl 1):3–12. doi: 10.1016/j.urology.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 4.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 5.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 6.Sevli S, Uzumcu A, Solak M, Ittmann M, Ozen M. The function of microRNAs, small but potent molecules, in human prostate cancer. Prostate Cancer Prostatic Dis. 2010;13:208–217. doi: 10.1038/pcan.2010.21. [DOI] [PubMed] [Google Scholar]
- 7.Li T, Li D, Sha J, Sun P, Huang Y. MicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cells. Biochem Biophys Res Commun. 2009;383:280–285. doi: 10.1016/j.bbrc.2009.03.077. [DOI] [PubMed] [Google Scholar]
- 8.Ambs S, Prueitt RL, Yi M, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68:6162–6170. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sylvestre Y, De Guire V, Querido E, et al. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282:2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 10.Fujita Y, Kojima K, Hamada N, et al. Effects of miR-34a on cell growth and chemoresistance in prostate cancer PC3 cells. Biochem Biophys Res Commun. 2008;377:114–119. doi: 10.1016/j.bbrc.2008.09.086. [DOI] [PubMed] [Google Scholar]
- 11.Shi XB, Xue L, Yang J, et al. An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc Natl Acad Sci USA. 2007;104:19983–19988. doi: 10.1073/pnas.0706641104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gandellini P, Folini M, Longoni N, et al. miR-205 exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase Cɛ. Cancer Res. 2009;69:2287–2295. doi: 10.1158/0008-5472.CAN-08-2894. [DOI] [PubMed] [Google Scholar]
- 13.Josson S, Sung SY, Lao K, Chung LW, Johnstone PA. Radiation modulation of microRNA in prostate cancer cell lines. Prostate. 2008;68:1599–1606. doi: 10.1002/pros.20827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li B, Shi XB, Nori D, et al. Down-regulation of microRNA 106b is involved in p21-mediated cell cycle arrest in response to radiation in prostate cancer cells. Prostate. 2011;71:567–574. doi: 10.1002/pros.21272. [DOI] [PubMed] [Google Scholar]
- 15.John-Aryankalayil M, Palayoor ST, Makinde AY, et al. Fractionated radiation alters oncomir and tumor suppressor miRNAs in human prostate cancer cells. Radiat Res. 2012;178:105–117. doi: 10.1667/rr2703.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cloonan N, Wani S, Xu Q, et al. MicroRNAs and their isomiRs function cooperatively to target common biological pathways. Genome Biol. 2011;12:R126. doi: 10.1186/gb-2011-12-12-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ebhardt HA, Tsang HH, Dai DC, Liu Y, Bostan B, Fahlman RP. Meta-analysis of small RNA-sequencing errors reveals ubiquitous post-transcriptional RNA modifications. Nucleic Acids Res. 2009;37:2461–2470. doi: 10.1093/nar/gkp093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reid JG, Nagaraja AK, Lynn FC, et al. Mouse let-7 miRNA populations exhibit RNA editing that is constrained in the 5′-seed/cleavage/anchor regions and stabilize predicted mmu-let-7a:mRNA duplexes. Genome Res. 2008;18:1571–1581. doi: 10.1101/gr.078246.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morin RD, O’Connor MD, Griffith M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaudhry MA, Omaruddin RA, Brumbaugh CD, Tariq MA, Pourmand N. Identification of radiation-induced microRNA transcriptome by next-generation massively parallel sequencing. J Radiat Res. 2013;54:808–822. doi: 10.1093/jrr/rrt014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pruesse E, Quast C, Knittel K, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu C, Bai B, Skogerbø G, et al. NONCODE: an integrated knowledge database of non-coding RNAs. Nucleic Acids Res. 2005;33:D112–D115. doi: 10.1093/nar/gki041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li C, Li X, Miao Y, et al. SubpathwayMiner: a software package for flexible identification of pathways. Nucleic Acids Res. 2009;37:e131. doi: 10.1093/nar/gkp667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.John-Aryankalayil M, Palayoor ST, Cerna D, et al. Fractionated radiation therapy can induce a molecular profile for therapeutic targeting. Radiat Res. 2010;174:446–458. doi: 10.1667/RR2105.1. [DOI] [PubMed] [Google Scholar]
- 29.Chang HT, Li SC, Ho MR, et al. Comprehensive analysis of microRNAs in breast cancer. BMC Genomics. 2012;13(Suppl 7):S18. doi: 10.1186/1471-2164-13-S7-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li SC, Liao YL, Ho MR, Tsai KW, Lai CH, Lin WC. miRNA arm selection and isomiR distribution in gastric cancer. BMC Genomics. 2012;13(Suppl 1):S13. doi: 10.1186/1471-2164-13-S1-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li SC, Tsai KW, Pan HW, Jeng YM, Ho MR, Li WH. MicroRNA 3′ end nucleotide modification patterns and arm selection preference in liver tissues. BMC Syst Biol. 2012;6(Suppl 2):S14. doi: 10.1186/1752-0509-6-S2-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krek A, Grün D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 33.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Y, Fang F, Zhang J, Josson S, St Clair WH, St Clair DK. miR-17*suppresses tumorigenicity of prostate cancer by inhibiting mitochondrial antioxidant enzymes. PLoS One. 2010;5:e14356. doi: 10.1371/journal.pone.0014356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X, Ladd A, Dragoescu E, Budd WT, Ware JL, Zehner ZE. MicroRNA-17-3p is a prostate tumor suppressor in vitro and in vivo, and is decreased in high grade prostate tumors analyzed by laser capture microdissection. Clin Exp Metastasis. 2009;26:965–979. doi: 10.1007/s10585-009-9287-2. [DOI] [PubMed] [Google Scholar]
- 36.Almeida MI, Nicoloso MS, Zeng L, et al. Strand-specific miR-28-5p and miR-28-3p have distinct effects in colorectal cancer cells. Gastroenterology. 2012;142:886–896. doi: 10.1053/j.gastro.2011.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fernandez-Valverde SL, Taft RJ, Mattick JS. Dynamic isomiR regulation in Drosophila development. RNA. 2010;16:1881–1888. doi: 10.1261/rna.2379610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo L, Li H, Liang T, et al. Consistent isomiR expression patterns and 3′ addition events in miRNA gene clusters and families implicate functional and evolutionary relationships. Mol Biol Rep. 2012;39:6699–6706. doi: 10.1007/s11033-012-1493-3. [DOI] [PubMed] [Google Scholar]
- 39.Pan HW, Li SC, Tsai KW. MicroRNA dysregulation in gastric cancer. Curr Pharm Des. 2013;19:1273–1284. doi: 10.2174/138161213804805621. [DOI] [PubMed] [Google Scholar]
- 40.Leite KR, Tomiyama A, Reis ST, et al. MicroRNA expression profiles in the progression of prostate cancer - from high-grade prostate intraepithelial neoplasia to metastasis. Urol Oncol. 2013;31:796–801. doi: 10.1016/j.urolonc.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 41.Schubert M, Spahn M, Kneitz S, et al. Distinct microRNA expression profile in prostate cancer patients with early clinical failure and the impact of let-7 as prognostic marker in high-risk prostate cancer. PLoS One. 2013;8:e65064. doi: 10.1371/journal.pone.0065064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walter BA, Valera VA, Pinto PA, Merino MJ. Comprehensive microRNA Profiling of Prostate Cancer. J Cancer. 2013;4:350–357. doi: 10.7150/jca.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kobayashi N, Uemura H, Nagahama K, et al. Identification of miR-30d as a novel prognostic maker of prostate cancer. Oncotarget. 2012;3:1455–1471. doi: 10.18632/oncotarget.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poliseno L, Salmena L, Riccardi L, et al. Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal. 2010;3:ra29. doi: 10.1126/scisignal.2000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramberg H, Alshbib A, Berge V, Svindland A, Taskén KA. Regulation of PBX3 expression by androgen and Let-7d in prostate cancer. Mol Cancer. 2011;10:50. doi: 10.1186/1476-4598-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bonci D, Coppola V, Musumeci M, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 47.Porkka KP, Ogg EL, Saramaki OR, et al. The miR-15a-miR-16-1 locus is homozygously deleted in a subset of prostate cancers. Genes Chromosomes Cancer. 2011;50:499–509. doi: 10.1002/gcc.20873. [DOI] [PubMed] [Google Scholar]
- 48.Gonzales JC, Fink LM, Goodman OB, Jr, Symanowski JT, Vogelzang NJ, Ward DC. Comparison of circulating microRNA 141 to circulating tumor cells, lactate dehydrogenase, and prostate-specific antigen for determining treatment response in patients with metastatic prostate cancer. Clin Genitourin Cancer. 2011;9:39–45. doi: 10.1016/j.clgc.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 49.Hulf T, Sibbritt T, Wiklund ED, et al. Epigenetic-induced repression of microRNA-205 is associated with MED1 activation and a poorer prognosis in localized prostate cancer. Oncogene. 2013;32:2891–2899. doi: 10.1038/onc.2012.300. [DOI] [PubMed] [Google Scholar]
- 50.Runkle EA, Zhang H, Cai Z, et al. Reversion of the ErbB malignant phenotype and the DNA damage response. Exp Mol Pathol. 2012;93:324–333. doi: 10.1016/j.yexmp.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao L, Bode AM, Cao Y, Dong Z. Regulatory mechanisms and clinical perspectives of miRNA in tumor radiosensitivity. Carcinogenesis. 2012;33:2220–2227. doi: 10.1093/carcin/bgs235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao L, Lu X, Cao Y. MicroRNA and signal transduction pathways in tumor radiation response. Cell Signal. 2013;25:1625–1634. doi: 10.1016/j.cellsig.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joung JG, Hwang KB, Nam JW, Kim SJ, Zhang BT. Discovery of microRNA-mRNA modules via population-based probabilistic learning. Bioinformatics. 2007;23:1141–1147. doi: 10.1093/bioinformatics/btm045. [DOI] [PubMed] [Google Scholar]
- 54.Tran DH, Satou K, Ho TB. Finding microRNA regulatory modules in human genome using rule induction. BMC Bioinformatics. 2008;9(Suppl 12):S5. doi: 10.1186/1471-2105-9-S12-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoon S, De Micheli G. Prediction of regulatory modules comprising microRNAs and target genes. Bioinformatics. 2005;21(Suppl 2):ii93–ii100. doi: 10.1093/bioinformatics/bti1116. [DOI] [PubMed] [Google Scholar]
- 56.Chaudhry MA, Sachdeva H, Omaruddin RA. Radiation-induced micro-RNA modulation in glioblastoma cells differing in DNA-repair pathways. DNA Cell Biol. 2010;29:553–561. doi: 10.1089/dna.2009.0978. [DOI] [PubMed] [Google Scholar]
- 57.Wang Q, Li P, Li A, et al. Plasma specific miRNAs as predictive biomarkers for diagnosis and prognosis of glioma. J Exp Clin Cancer Res. 2012;31:97. doi: 10.1186/1756-9966-31-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chaudhry MA, Omaruddin RA. Differential regulation of microRNA expression in irradiated and bystander cells. Mol Biol. 2012;46:634–643. [PubMed] [Google Scholar]
- 59.Vincenti S, Brillante N, Lanza V, et al. HUVEC respond to radiation by inducing the expression of pro-angiogenic microRNAs. Radiat Res. 2011;175:535–546. doi: 10.1667/RR2200.1. [DOI] [PubMed] [Google Scholar]
- 60.Chun-Zhi Z, Lei H, An-Ling Z, et al. MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer. 2010;10:367. doi: 10.1186/1471-2407-10-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu Y, Zhou B, Wu D, Yin Z, Luo D. Baicalin modulates microRNA expression in UVB irradiated mouse skin. J Biomed Res. 2012;26:125–134. doi: 10.1016/S1674-8301(12)60022-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wagner-Ecker M, Schwager C, Wirkner U, Abdollahi A, Huber PE. MicroRNA expression after ionizing radiation in human endothelial cells. Radiat Oncol. 2010;5:25. doi: 10.1186/1748-717X-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y, Scheiber MN, Neumann C, Calin GA, Zhou D. MicroRNA regulation of ionizing radiation-induced premature senescence. Int J Radiat Oncol Biol Phys. 2011;81:839–848. doi: 10.1016/j.ijrobp.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]