Abstract
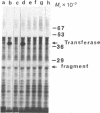
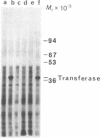
In Saccharomyces cerevisiae, utilization of galactose requires four inducible enzyme activities. Three of these activities (galactose-1-phosphate uridyl transferase, EC 2.7.7.10; uridine diphosphogalactose 4-epimerase, EC 5.1.3.2; and galactokinase, EC 2.7.1.6) are specified by three tightly linked genes (GAL7, GAL10, and GAL1, respectively) on chromosome II, whereas the fourth, galactose transport, is specified by a gene (GAL2) located on chromosome XII. Although classic genetic analysis has revealed both positive and negative regulatory genes that coordinately affect the appearance of all four enzyme activities, neither the basic events leading to the appearance of enzyme activities nor the roles of the regulatory genes have yet been determined. Regulation of inducible enzyme activity could be mediated by events related to transcription, translation, or enzyme activation. For the purpose of studying galactose pathway induction and its regulation, we have developed an immunoprecipitation assay that enables us to detect the GAL7 specified uridyl transferase polypeptide in yeast extracts and among the polypeptides synthesized in an RNA-dependent in vitro translation system. Use of this immunoprecipitation assay in conjunction with in vivo labeling experiments demonstrates the presence of [3H]leucine-labeled transferase in extracts prepared from cells grown in galactose but not from cells grown in glucose. This galactose-specific induction of transferase polypeptide is mediated by the de novo appearance of a functional mRNA species whose synthetic capacity is detectable by the combination of in vitro translation and immunoprecipitation. The appearance of functional transferase mRNA depends on wild-type expression of the positive regulatory gene, GAL4. Cells carrying a nonsense (amber) mutation in the GAL4 gene fail to produce the transferase mRNA, whereas a nonsense suppressor of the GAL4 amber mutant regains the galactose-specific mRNA response. Our results establish that the induction of the GAL7 specified uridyl transferase activity is mediated by de novo appearance of a functional mRNA and that this galactose-specific response is dependent on a wild-type GAL4 gene product.
Keywords: in vitro translation, immunoprecipitation, GAL gene cluster, positive regulation
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bassel J., Mortimer R. Genetic order of the galactose structural genes in Saccharomyces cerevisiae. J Bacteriol. 1971 Oct;108(1):179–183. doi: 10.1128/jb.108.1.179-183.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan P., Douglas H. C. Genetic control of phosphoglucomutase variants in Saccharomyces cerevisiae. J Bacteriol. 1969 May;98(2):532–535. doi: 10.1128/jb.98.2.532-535.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo V. P. Galactose transport in Saccharomyces cerevisiae. I. Nonmetabolized sugars as substrates and inducers of the galactose transport system. J Bacteriol. 1968 May;95(5):1727–1731. doi: 10.1128/jb.95.5.1727-1731.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOUGLAS H. C. A mutation in saccharomyces that affects phosphoglucomutase activity and galactose utilization. Biochim Biophys Acta. 1961 Sep 2;52:209–211. doi: 10.1016/0006-3002(61)90924-6. [DOI] [PubMed] [Google Scholar]
- DOUGLAS H. C., CONDIE F. The genetic control of galactose utilization in Saccharomyces. J Bacteriol. 1954 Dec;68(6):662–670. doi: 10.1128/jb.68.6.662-670.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOUGLAS H. C., HAWTHORNE D. C. ENZYMATIC EXPRESSION AND GENETIC LINKAGE OF GENES CONTROLLING GALACTOSE UTILIZATION IN SACCHAROMYCES. Genetics. 1964 May;49:837–844. doi: 10.1093/genetics/49.5.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas H. C., Hawthorne C. D. Uninducible mutants in the gal i locus of Saccharomyces cerevisiae. J Bacteriol. 1972 Mar;109(3):1139–1143. doi: 10.1128/jb.109.3.1139-1143.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas H. C., Hawthorne D. C. Regulation of genes controlling synthesis of the galactose pathway enzymes in yeast. Genetics. 1966 Sep;54(3):911–916. doi: 10.1093/genetics/54.3.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firtel R. A., Lodish H. F. A small nuclear precursor of messenger RNA in the cellular slime mold Dictyostelium discoideum. J Mol Biol. 1973 Sep 15;79(2):295–314. doi: 10.1016/0022-2836(73)90007-7. [DOI] [PubMed] [Google Scholar]
- Gallis B. M., McDonnell J. P., Hopper J. E., Young E. T. Translation of poly(riboadenylic acid)-enriched messenger RNAs from the yeast, Saccharomyces cerevisiae, in heterologous cell-free systems. Biochemistry. 1975 Mar 11;14(5):1038–1046. doi: 10.1021/bi00676a024. [DOI] [PubMed] [Google Scholar]
- Hopper J. E., Bostian K. A., Rowe L. B., Tipper D. J. Translation of the L-species dsRNA genome of the killer-associated virus-like particles of Saccharomyces cerevisiae. J Biol Chem. 1977 Dec 25;252(24):9010–9017. [PubMed] [Google Scholar]
- Hopper J. E., Ko G., Young E. T. Comparative analysis of the in vivo and in vitro expression of bacteriophage T7 messenger RNAs during infection of Escherichia coli. J Mol Biol. 1975 Jun 5;94(4):539–554. doi: 10.1016/0022-2836(75)90320-4. [DOI] [PubMed] [Google Scholar]
- JOHNSTON F. B., STERN H. Mass isolation of viable wheat embryos. Nature. 1957 Jan 19;179(4551):160–161. doi: 10.1038/179160b0. [DOI] [PubMed] [Google Scholar]
- KALCKAR H. M., BRAGANCA B., MUNCH-PETERSEN H. M. Uridyl transferases and the formation of uridine diphosphogalactose. Nature. 1953 Dec 5;172(4388):1038–1038. [PubMed] [Google Scholar]
- Kew O. M., Douglas H. C. Genetic co-regulation of galactose and melibiose utilization in Saccharomyces. J Bacteriol. 1976 Jan;125(1):33–41. doi: 10.1128/jb.125.1.33-41.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosterlitz H. W. The fermentation of galactose and galactose-1-phosphate. Biochem J. 1943 Sep;37(3):322–326. doi: 10.1042/bj0370322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LELOIR L. F. The enzymatic transformation of uridine diphosphate glucose into a galactose derivative. Arch Biochem Biophys. 1951 Sep;33(2):186–190. doi: 10.1016/0003-9861(51)90096-3. [DOI] [PubMed] [Google Scholar]
- Palacios R., Palmiter R. D., Schimke R. T. Identification and isolation of ovalbumin-synthesizing polysomes. I. Specific binding of 125 I-anti-ovalbumin to polysomes. J Biol Chem. 1972 Apr 25;247(8):2316–2321. [PubMed] [Google Scholar]
- Palmiter R. D. Differential rates of initiation of conalbumin and ovalbumin messenger ribonucleic acid in reticulocyte lysates. J Biol Chem. 1974 Nov 10;249(21):6779–6787. [PubMed] [Google Scholar]
- Palmiter R. D., Oka T., Schimke R. T. Modulation of ovalbumin synthesis by estradiol-17 beta and actinomycin D as studied in explants of chick oviduct in culture. J Biol Chem. 1971 Feb 10;246(3):724–737. [PubMed] [Google Scholar]
- Roberts B. E., Paterson B. M. Efficient translation of tobacco mosaic virus RNA and rabbit globin 9S RNA in a cell-free system from commercial wheat germ. Proc Natl Acad Sci U S A. 1973 Aug;70(8):2330–2334. doi: 10.1073/pnas.70.8.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SPIEGELMAN S., SUSSMAN R. R., PINSKA E. On the cytoplasmic nature of "long-term adaptation" in yeast. Proc Natl Acad Sci U S A. 1950 Nov;36(11):591–606. doi: 10.1073/pnas.36.11.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih D. S., Kaesberg P. Translation of brome mosaic viral ribonucleic acid in a cell-free system derived from wheat embryo. Proc Natl Acad Sci U S A. 1973 Jun;70(6):1799–1803. doi: 10.1073/pnas.70.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W. Analysis of bacteriophage T7 early RNAs and proteins on slab gels. J Mol Biol. 1973 Sep 15;79(2):237–248. doi: 10.1016/0022-2836(73)90003-x. [DOI] [PubMed] [Google Scholar]