Abstract
The cleavage of peptide bonds by metallopeptidases (MPs) is essential for life. These ubiquitous enzymes participate in all major physiological processes, and so their deregulation leads to diseases ranging from cancer and metastasis, inflammation, and microbial infection to neurological insults and cardiovascular disorders. MPs cleave their substrates without a covalent intermediate in a single-step reaction involving a solvent molecule, a general base/acid, and a mono-or dinuclear catalytic metal site. Most monometallic MPs comprise a short metal-binding motif (HEXXH), which includes two metal-binding histidines and a general base/acid glutamate, and they are grouped into the zincin tribe of MPs. The latter divides mainly into the gluzincin and metzincin clans. Metzincins consist of globular ∼130–270-residue catalytic domains, which are usually preceded by N-terminal pro-segments, typically required for folding and latency maintenance. The catalytic domains are often followed by C-terminal domains for substrate recognition and other protein–protein interactions, anchoring to membranes, oligomerization, and compartmentalization. Metzincin catalytic domains consist of a structurally conserved N-terminal subdomain spanning a five-stranded β-sheet, a backing helix, and an active-site helix. The latter contains most of the metal-binding motif, which is here characteristically extended to HEXXHXXGXX(H,D). Downstream C-terminal subdomains are generally shorter, differ more among metzincins, and mainly share a conserved loop—the Met-turn—and a C-terminal helix. The accumulated structural data from more than 300 deposited structures of the 12 currently characterized metzincin families reviewed here provide detailed knowledge of the molecular features of their catalytic domains, help in our understanding of their working mechanisms, and form the basis for the design of novel drugs.
Keywords: structural biochemistry, metzincin clan, catalytic domains, active-site cleft, hydrolytic enzymes, matrix metalloproteases, astacins, ADAM, adamalysins, serralysins, metalloprotease, metalloproteinase
Peptidases
The term “peptidase” is currently recommended for proteolytic enzymes, proteases, and proteinases in general.1 Peptidases are distributed among all kingdoms of life.2–3 They target peptide bonds of proteins and/or peptides and some catalyze extensive protein-processing events such as the digestion or degradation of intake proteins and the development, maintenance, and remodeling of tissues. Other peptidases catalyze limited, specific scission of a small number of peptide bonds, which results in the activation or deactivation of themselves and other (pro)enzymes, bioactive peptides, and DNA repressors.2 In this way, peptidases participate in regulatory mechanisms, which are required in physiological processes such as blood-pressure control, hormone homeostasis, regulation of signal-transduction pathways, and modulation of protein–protein and cell–cell interactions.4 Such limited proteolysis through “sheddases” produces soluble forms from membrane-anchored precursors, thus decreasing protein concentration at cell surfaces and increasing it in the circulation.5,6
Peptidases have traditionally been classified into serine, cysteine, aspartic, and metallopeptidases (MPs) based on elements key for catalysis.8,9 More recently, three categories have been added: N-terminal threonine peptidases, glutamate peptidases, and asparagine peptidases.1,11 In addition, peptidases are divided into endopeptidases and exopeptidases, which cut in the middle or at the ends of substrates, respectively.13 The latter, in turn, can be further classified into aminopeptidases and carboxypeptidases, which cleave off N-terminal and C-terminal amino acids, respectively.1 Overall, exquisite regulation of any proteolytic enzyme is essential for proper functioning and to prevent misdirected temporal and spatial proteolytic activity. Control may be exerted at the transcriptional level or via post-translational modifications. Other regulatory mechanisms include biosynthesis in the form of zymogens that require activation,14 co-localization of enzyme and substrate, cofactor binding and substrate accessibility and specificity, as well as the presence of physiological protein inhibitors.15–16 Failure of regulatory mechanisms can give rise to pathologies such as inflammation, tissue destruction (as in arthritis and fibrotic diseases), neurological diseases (Alzheimer's disease, meningitis and multiple sclerosis), and cardiovascular disorders (stroke, hypertension, thrombosis, bleeding, and myocardial infarct).17,18 Even in tumorigenesis and tumor-progression events, such as angiogenesis, tissue invasion, and metastasis, peptidases play a major role.20–21 A particular group of peptidases are exogenic virulence factors that lead to the depletion of host defenses and tissue destruction.22,23 This is observed during microbial infections that cause inflammation, tetanus, pneumonia, tuberculosis, AIDS, malaria, botulism, gas gangrene, bacterial meningitis, anthrax, etc. but also after poisoning after snake bites. Accordingly, such diverse physiopathological implications make these proteins promising drug targets,25 and their structural studies are thus essential to the understanding of their functional determinants and the design of novel therapeutic agents, now grouped under the term “degradomics.”4,26–29
Here we review the catalytic domains of MPs, in particular from the metzincin clan.28,30–37 To date, structures are available for 12 metzincin families: astacins, adamalysins/ADAMs, serralysins, matrixins (alias matrix metalloproteinases and MMPs), leishmanolysins, snapalysins, pappalysins, archaemetzincins, fragilysins, cholerilysins, toxilysins, and igalysins (for the first collective mention of the first 10 family names, see Ref. 28).
Classification and Cleavage Mechanism of Metallopeptidases
One widely accepted classification of MPs and proteolytic enzymes in general is provided by the MEROPS database, which classifies homologous sets of peptidases and protein inhibitors hierarchically into protein species, families, and clans based on sequence similarity and evolutionary distances (http://merops.sanger.ac.uk;38). This scheme is also followed in a comprehensive study—the Handbook of Proteolytic Enzymes.39 Here, we apply a different approach based on active-site architecture and overall fold similarity (see also Refs. 28, 30, 35, and 40–43). According to this scheme, MPs are first divided into those that have a single catalytic metal and those that have two. Dimetalate MPs mainly include exopeptidases and these are reviewed elsewhere,44,45 so here we focus on structurally characterized members of the subclass of mononuclear MPs (Fig. 1). In addition, isopeptidases such as Jab1/MPN47–48 and AMSH-LP deubiquitinases,49 which cleave amide bonds that are not peptide bonds, and peptide deformylase, which removes N-terminal formyl groups from N-terminal methionine residues,50–51 have also been omitted. This classification is based on recent structural data and thus supersedes previous reports by our group.28,43
Figure 1.

Classification of mononuclear MPs. Within the MP class, mononuclear MPs are a subclass that is divided into tribes currently characterized at the structural level: inverzincins, zincins, relatives of hydrogenase maturating factor (HybD), LAS MPs, and αβα-exopeptidases. These tribes subdivide into clans, which in turn give rise to families. The metzincins are depicted and framed in blue. Metal-binding residues are shown in green, general base/acid residues in magenta, and residues/molecules occupying the position of the Met-turn or Ser/Gly-turn beneath the metal site are in orange. Other residues engaged in substrate binding, stabilization of the reaction intermediate, and/or catalysis are further shown in black, except for X, which stands for any residue and is here only used as a spacer within motifs. Within gluzincins, Z = A/F/S/G/T. Variability within motifs was considered if present only in more than one case. *Leishmanolysins have an extra domain inserted between the glycine and the third histidine metal ligand of the metzincin extended zinc-binding signature. **Igalysins have an extra glycine residue inserted just before the glycine of the metzincin signature. ***Dipeptidyl peptidase III has an extra residue inserted between the general base/acid glutamate and the second metal-binding histidine. This classification supersedes previous schemes.28,43
The active site of MPs accommodates a catalytic divalent metal ion, mostly zinc but also sometimes cobalt, manganese or nickel, which is anchored at the bottom of the active-site cleft of the enzyme by protein residue side chains.41–53 Here it might be appropriate to clarify commonly used terms that tend to be misused in the literature. The intact, active, metal-bound protein is described as the “enzyme” or the “holoenzyme.” If the enzyme has a ligand bound, it can be described as an enzyme–ligand complex. The enzyme lacking a metal ion indispensable for catalysis is described as an “apoenzyme.” Metal-binding residues are mostly histidines, aspartates, and glutamates, which are included in zinc-binding motifs that are characteristic of particular MP tribes, clans, and families (Fig. 1, residues in green). Tribes include αβα-exopeptidases, which comprise the funnelin clan;43–52 LAS MPs;54,55 relatives of hydrogenase maturating factor, HybD, a proven natural nickel-dependent MP;57–58 zincins;30,41 and inverzincins.11,60,61 In all cases, a general/base acid, mostly a glutamate, is further found close to the catalytic metal and is required for catalysis (see the section on “Classification and cleavage mechanism of metallopeptidases” and Fig. 1, residues in magenta).
In general, peptide-bond hydrolysis through monometallic MPs is an ordered single-displacement reaction that follows simple Michaelis-Menten kinetics and occurs optimally at neutral pH. A solvent molecule and a peptide substrate are bound by the active-site cleft and the catalytic metal ion to render the Michaelis complex.63–64 In most cases, the overall active-site environment is in a competent conformation for catalysis in the mature enzyme before recruiting the peptidic substrate, which binds in extended conformation in such a way that substrate side chains upstream (termed P1, P2, P3, etc.) and downstream (P1', P2', P3', etc.) of the scissile bond nestle into active-site cleft subsites S1, S2, S3, etc. and S1', S2', S3', etc. of the enzyme, respectively.52–65 In general, substrate specificity is exerted in MPs through S1'.52 Cleavage can also occur within protein segments adopting compact tertiary structures, even helical conformations, in the unbound state,66 as interaction and binding to the peptidase can energetically compensate for substrate unwinding to yield an extended conformation suitable for catalysis.67 A consensus mechanism for catalysis has been derived after decades of research, mainly on thermolysin from Bacillus thermoproteolyticus and bovine carboxypeptidase A,53–70 which were the first two MPs to be solved for their crystal structures in the late 1960s and early 1970s,71–72 and more recently on MMPs.73–74 This mechanism entails that the solvent molecule is bound first—perhaps it is already part of the mature functional enzyme prior to catalysis as found in the structures of competent MPs in the absence of inhibitors or substrates74–77—by the catalytic metal ion and the general base/acid glutamate, which polarizes the solvent and enhances its nucleophilicity (Fig. 2, I). Once the substrate to be cleaved is bound to the cleft, the scissile carbonyl oxygen is bound and polarized by the catalytic metal, and so the carbonyl carbon is more susceptible to nucleophilic attack by the solvent molecule, which simultaneously transfers a proton to the general base glutamate (Fig. 2, II). The catalytic metal thus serves as a dual oxyanion hole and activates both substrates for the reaction. The attack gives rise to a gem-diolate tetrahedral reaction intermediate (Fig. 2, III), which is stabilized not only by the metal but also by other protein residues such as vicinal histidines, arginines, and tyrosines43,53 (Fig. 1; see also the section on “Common structural features of metzincins”). Subsequently, the intermediate resolves to the double-product complex through scissile bond cleavage and double proton transfer to the new α-amino terminus (Fig. 2, IV). Finally, the two products dissociate from the enzyme and leave the active-site cleft (the nonprimed product possibly first, see Ref. 53), which is replenished with another solvent molecule and ready for a new round of catalysis.
Figure 2.
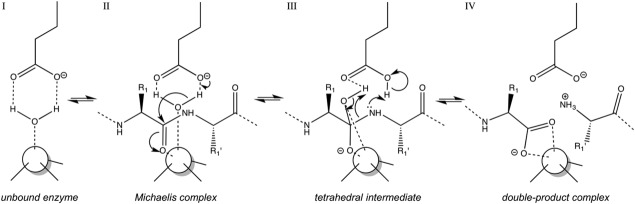
Generally accepted catalytic mechanism of monometallic MPs. The catalytic solvent molecule is bound first to the catalytic metal ion (white sphere) and the general base/acid in the active site in the absence of a peptidic substrate (I). Once the substrate is accommodated in the cleft and the Michaelis complex is formed (II), the polarized solvent molecule attacks the scissile carbonyl group, which leads to the tetrahedral reaction intermediate (III). The latter resolves in scissile bond breakage and double proton transfer to the newly formed α-amino group to render a double-product complex (IV).
Zincins Split into Gluzincins and Metzincins
The zincins were named in 1993 by Bode and colleagues.30 They constitute the most extensive tribe of MPs, with >100 coding genes in humans alone.59 They have a characteristic short metal-binding amino-acid consensus sequence, HEXXH,30,41 first identified by McKerrow in MMPs and serralysins in 1987,79 which is provided by an “active-site helix” and supplies two metal-binding histidine ligands and the general base/acid glutamate (see the section on “Classification and cleavage mechanism of metallopeptidases”). In addition, zincins are divided by a horizontal active-site cleft into an upper N-terminal subdomain (NTS) above the cleft and a lower C-terminal subdomain (CTS) below the cleft when viewed in the standard orientation of MPs, i.e., with the peptide substrate running from left to right.52 The NTS has a helix termed “backing helix” in addition to the active-site helix and a β-sheet of at least three strands, which are mixed parallel/antiparallel. The lowermost strand horizontally frames the top of the active-site cleft, runs antiparallel to a substrate when bound to the cleft, and anchors the latter through inter-main-chain interactions mimicking a β-ribbon. The CTS varies greatly among zincins and is mainly characterized by the presence of a residue (Fig. 1, orange residues) contained in a loop structure providing a basement for the metal-binding site. The two main zincin clans are the gluzincins and the metzincins, whose loop structures are termed, respectively, “Ser/Gly-turn” and “Met-turn” (see below and the section on “Common structural features of metzincins”). Three other zincin clans have an aspartate as third zinc ligand: (i) The S2P-zincins, which include one of the only two integral-membrane MP families structurally characterized to date (those completely imbedded in membranes and not merely type-I or type-II membrane-anchored species); their fold is different from any other zincin, although they have the active-site helix and the short consensus sequence,52–80 and they show a proline below the metal. (ii) The aspzincins, which were named in 1999 by Ichishima and coworkers81 in line with the terms gluzincins and metzincins (see below) due to the aspartate metal ligand81,82 and have an alanine below the metal. (iii) The FtsH-related enzymes, which do not have a protein residue below the metal but rather a solvent molecule.84–87
Gluzincins were so named because of a glutamate found as third metal-binding residue, as observed in archetypal thermolysin.72 The clan name was coined by Hooper in 199441 to distinguish the latter enzyme and related structures from the metzincins, which have a histidine instead of the glutamate (see below) and had been named in 1993 by Bode and colleagues based on structural similarities between astacin, adamalysin II and the serralysin Pseudomonas aeruginosa alkaline proteinase.30 Since then, a number of novel MP structures have become available, which has led us to suggest a redefinition of the common structural features of gluzincins: they are zincins, which in addition to their common structural elements contain a “glutamate helix” below the active-site helix, which runs parallel to it in a horizontal projection but is rotated backward in a vertical projection (see Fig. 3 in Ref. 43). This helix—or a short loop segment immediately preceding it—provides the third metal-binding residue, mostly a glutamate but sometimes also a histidine as in minigluzincins.88 This residue is three positions upstream of the residue of the Ser/Gly-turn providing the basement of the metal-binding site43 (Fig. 1, orange residues). The latter residue varies within gluzincins: it is a phenylalanine in minigluzincins;88 a threonine in clostridial neurotoxins;89–90 a serine in thermolysins;63–93 and a serine or a glycine in cowrins.43 However, the most widespread residue among gluzincin families is an alanine as found in anthrax lethal factor and related proteins such as Escherichia coli titration factor Mlc (Mtfa/Yeel);94–95 in MEROPS M48/M56-family MPs,38–88 which with human FACE1 and yeast Ste24p are the only other integral-membrane MPs in addition to S2P-zincins structurally analyzed to date;96–97 in some members—others have a threonine—of the family encompassing leukotriene A4 hydrolase, tricorn interacting factor F3, and MEROPS M1-family aminopeptidases;98,99 in bacterial collagenases such as those from Clostridium spp.;101–102 in neprilysins;103–104 and in dipeptidyl peptidase III and its structural relatives, which uniquely have an additional residue inserted between the signature glutamate and the second histidine, thus resulting in an exceptional HEXXXH signature.105 This is accounted for by a wider helix turn that allows the second histidine to be suitably positioned to bind the metal ion. Overall, the general hallmark of gluzincins would thus consist of a double consensus motif, HEXXH+(E,H)XX(A,F,T,S,G) (Fig. 1), as well as the minimal structural requirements of zincins plus the presence of a glutamate helix.
Figure 3.
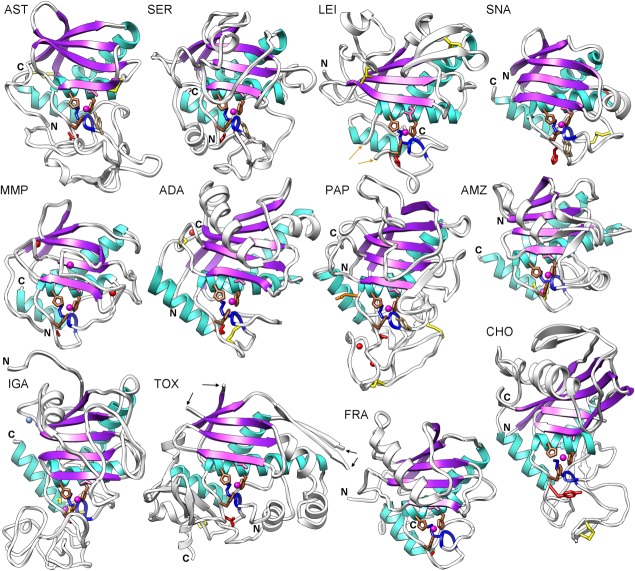
Structure of metzincin catalytic domains. Ribbon-type Richardson plots of a representative member of each of the 12 structurally characterized metzincin families (see the sections on “Common structural features of metzincins” and “Metzincin family-specific features,” and Fig. 1) in standard orientation.52 Depicted are astacin (AST; PDB 1AST; 200 residues), aeruginolysin (SER; PDB 1KAP; 220 residues), leishmanolysin (LEI; PDB 1LML; 213 residues), snapalysin (SNA; PDB 1C7K; 132 residues), human neutrophil collagenase (MMP; PDB 1JAN; 164 residues), adamalysin II (ADA; PDB 1IAG; 203 residues), ulilysin (PAP; PDB 2CKI; 262 residues), Methanopyrus kandleri archaemetzincin AmzA (AMZ; PDB 2X7M; 173 residues), igalysin BACOVA_0063 (IGA; PDB 3P1V; 272 residues), toxilysin EcxA (TOX; PDB 4L63; 258 residues), fragilysin-3 (FRA; PDB 3P24; 188 residues), and cholerilysin StcE (CHO; PDB 3UJZ; 258 residues). The common β-strands and α-helices are shown in purple and cyan, respectively (see Fig. 4 for their nomenclature). Unique regular secondary structure elements of each family are in white. The N-and C-termini are labeled and the side chains of the zinc-binding histidines/aspartates (brown), general base/acid glutamates (pink), Met-turn methionines (blue), disulfide-linked cysteines (yellow), family-specific residues (red), and zinc-binding or substrate-stabilizing tyrosines (where present; in tan) are displayed as sticks. Preceding, inserted, and following (sub-)domains have been omitted for clarity. Catalytic metal ions—mostly zinc—are shown as magenta spheres, calcium cations as red spheres, and the potassium cation of IGA is in blue. Orange arrows pinpoint the anchor points for a domain inserted in LEI between the glycine of the extended consensus sequence and the third metal-binding histidine, PAP is unique in having an asparagine instead of the glycine. Black arrows pinpoint two disordered loops in TOX.
In addition to the families reported in Figure 1, other structures show similarities with gluzincins but are probably unable to cleave peptides. Three similar proteins from Haemophilus influenza,106 Thermotoga maritime,107 and Aquifex aeolicus108 possess a gluzincin-like fold but have the catalytic glutamate replaced by a glycine. The structures reported have a third potential histidine metal-ligand but lack a metal at the potential metal-binding site, and no function or catalytic competence has been reported for any of them. By contrast, an endonuclease function in 70S ribosome quality control and 16S rRNA maturation was reported for Escherichia coli protein YbeY.109 This protein shows a structure halfway between a gluzincin and a metzincin despite lacking the general base/acid glutamate—likewise replaced here by a glycine—and may thus be unable to cleave proteins.110 It bears the extended signature of metzincins (see the section on “Common structural features of metzincins”; the glutamate is replaced by glycine though) and a methionine below the metal-binding site as metzincins do (see the section on “Common structural features of metzincins”), but it also has a glutamate helix as in gluzincins. Actually, YbeY was already identified as a close structural homolog of minigluzincins,88 placing the latter family also at the interface between gluzincins and metzincins, with a likewise bulky hydrophobic residue in the Ser/Gly-turn, a phenylalanine, and a histidine as third metal ligand instead of the usual glutamate.
Finally, inverzincins,11,60,61 whose name was also proposed by Hooper in 1994,41 possess the sequence HXXEH, which is the inverted zincin motif. This led them to be considered a distinct MP tribe (Fig. 1). However, like zincins, they have the motif imbedded within an active-site helix, which runs here characteristically in the opposite direction to the equivalent helix in zincins. In addition, they possess a glutamate helix below the active-site helix, in the same relative position and direction as gluzincins. They have an EXXV motif encompassing the third glutamate metal-binding residue and a valine as the Ser/Ala-turn residue, as well as a mixed β-sheet of at least three strands equivalent to zincins. Furthermore, there is likewise a backing helix in the NTS. Accordingly, although inverzincins are considered a separate tribe from the zincins (Fig. 1), they could also be classified as a special case of gluzincins like the relatives of dipeptidyl peptidase III (see above).
Common Structural Features of Metzincins
Metzincins28,30–37 are present in all the kingdoms of life and more than 300 structures comprising at least the catalytic domain have been deposited with the Protein Data Bank (PDB; Table1). In the present review, 12 catalytic domain lead structures, one for each of the families depicted in Figure 1, will be discussed (Figs. 3 and 4): Astacus astacus crayfish astacin (hereafter AST), Pseudomonas aeruginosa alkaline proteinase (alias aeruginolysin; SER), Leishmania major leishmanolysin (LEI), Streptomyces caespitosus neutral protease (alias ScNP or snapalysin; SNA), human neutrophil collagenase (alias matrix metalloproteinase 8; MMP), Crotalus adamanteus snake venom adamalysin II (ADA), the pappalysin ulilysin from Methanosarcina acetivorans (PAP), the archaemetzincin AmzA from Methanopyrus kandleri (AMZ), the unpublished igalysin “putative MP BACOVA_0063” from Bacteroides ovatus (IGA), the toxilysin EcxA from Escherichia coli (TOX), fragilysin-3 from Bacteroides fragilis (FRA), and the cholerilysin StcE from Escherichia coli (CHO)75–121. These prototypes represent archaea, bacteria, protozoa/protists, and eukaryotes. The structures are ∼130–270-residue globular moieties that share a common scaffold and active-site environment, with the common elements described for zincins in general (see the section on “Zincins split into gluzincins and metzincins”), but each family has distinguishing structural elements (see the section on “Metzincin family-specific features”).
Table 1.
Protein Data Bank Entries Comprising at Least a Metzincin Catalytic Domain (http://www.pdb.org; Search Completed on October 14, 2013)
| Peptidase name | Organism | PDB access code |
|---|---|---|
| ASTACINS | ||
| Astacin | Astacus astacus | 1ast 1iaa 1iab 1iac 1iad 1iae 1qji 1qjj 3lq0 |
| Tolloid-like protease 1 | Homo sapiens | 3edi |
| Bone morphogenic protein 1 | H. sapiens | 3edh 3edg |
| Meprin β | H. sapiens | 4gwm 4gwn |
| HCE-1(high choriolyt. enzyme-1) | Oryzias latipes | 3vtg |
| ZHE1 | Danio rerio | 3lqb |
| SERRALYSINS | ||
| Aeruginolysin | Pseudomonas aeruginosa | 1kap 1jiw 1akl 3vi1 |
| Psychrophilic alkaline protease | Pseudomonas TACII 18 | 1g9k 1h71 1omj 1om6 1om7 1om8 1o0t 1o0q |
| Serralysin | Serratia marcescens/Serratia sp. E-15 | 1af0 1sat 1srp 1smp |
| PrtC | Erwinia chrysanthemi | 1go7 1go8 1k7g 1k7i 1k7q 3hb2 3hbu 3hbv 3hda |
| Psychrophilic marine protease | Flavobacterium sp. YS-80-122 | 3u1r |
| ADAMALYSINS/ADAMs | ||
| Adamalysin II | Crotalus adamanteus | 1iag 2aig 3aig 4aig |
| Atrolysin-C (Ht-d/c) | Crotalus atrox | 1atl 1dth 1htd |
| Bothropasin | Bothrops jararaca | 3dsl |
| Taiwan habu MP | Trimeresurus mucrosquamatus | 1kuf 1kug 1kui 1kuk |
| H2-proteinase | Trimeresurus flavoviridis | 1wni |
| Acutolysin-C | Agkistrodon acutus | 1qua |
| Acutolysin-A | A. acutus | 1bsw 1bud |
| BaP1 | Bothrops asper | 1nd1 2w12 2w13 2w14 2w15 |
| Vascular apoptosis-inducing protein1 VAP1 | C. atrox | 2ero 2erp 2erq |
| Fibrin(ogen)olytic MP | A. acutus | 1yp1 |
| Catrocollastatin/VAP2B | C. atrox | 2dw0 2dw1 2dw2 |
| Russell's viper venom MP | Daboia russelli siamensis | 2e3x |
| BmooMPα-1 | Bothrops moojeni | 3gbo |
| AaHIV | A. acutus | 3hdb |
| Atragin | Naja atra | 3k7l |
| K-like MP | N. atra | 3k7n |
| TM-1 MP | T. mucrosquamatus | 4j4m |
| TNF-α converting enzyme(ADAM-17) | H. sapiens | 1bkc 3edz 3b92 3cki 2oi0 2i47 2fv9 2fv5 2ddf 2a8h 1zxc 3e8r 3ewj 3g42 3kmc 3kme 3l0t 3l0v 3le9 3lea 3lgp 3o64 |
| ADAM-8 | H. sapiens | 4dd8 |
| ADAM-33 | H. sapiens | 1r54 1r55 |
| ADAMTS-1 | H. sapiens | 2jih 2v4b 3q2g 3q2h |
| ADAMTS-4 | H. sapiens | 2rjp 3b2z |
| ADAMTS-5 (alias ADAMTS-11) | H. sapiens | 3b8z 2rjq 3hy7 3hy9 3hyg 3ljt |
| MATRIXINS/MMPs | ||
| Fibroblast collagenase (MMP-1) | H. sapiens | 966c 1ayk 1cge 1cgf 1cgl 1hfc 2ayk 2tcl 3ayk 4ayk 1su3 2j0t 2clt 3shi 4auo |
| Sus scrofa | 1fbl | |
| Gelatinase A (MMP-2) | H. sapiens | 1ck7 1hov 1qib 1gxd 1eak 3ayu |
| Stromelysin-1 (MMP-3) | H. sapiens | 1b3d 1b8y 1biw 1bm6 1bqo 1c3i 1caq 1ciz 1cqr 3usn 1uea 2d1o 1c8t 2jt6 2jt5 2jnp 1d5j 1d7x 1d8f 1d8m 1g05 1g49 1g4k 1hfs 1hy7 1sln 1slm 1qia 1qic 1ums 1umt 1usn 2srt 2usn 1oo9 3ohl 3oho 4dpe 4g9l 4ja1 |
| Matrilysin (MMP-7) | H. sapiens | 1mmp 1mmq 1mmr 2ddy 2y6c 2y6d |
| Neutrophil collagenase (MMP-8) | H. sapiens | 1jan 1a85 1a86 1bzs 1i73 1i76 1jao 1jap 1jaq 1jh1 1jj9 1kbc 1mmb 1mnc 1zvx 2oy4 1zs0 1zp5 2oy2 3dng 3dpe 3dpf 3tt4 |
| Gelatinase B (MMP-9) | H. sapiens | 1gkc 1gkd 1l6j 2ow0 2ow1 2ow2 2ovz 2ovx 4h1q 4h2e 4h3x 4h82 4hma |
| Stromelysin-2 (MMP-10) | H. sapiens | 1q3a 3v96 |
| Stromelysin-3 (MMP-11) | Mus musculus | 1hv5 |
| Macrophage elastase (MMP-12) | H. sapiens | 1jiz 1jk3 2z2d 2k2g 3ba0 2oxu 2oxz 2oxw 2hu6 1z3j 1ycm 1y93 1rmz 1os9 1os2 2poj 1utz 1utt 1ros 2k9c 2krj 2w0d 2wo8 2wo9 2woa 3ehx 3ehy 3f15 3f16 3f17 3f18 3f19 3f1a 3lik 3lil 3lir 3ljg 3lk8 3lka 3n2u 3n2v 3nx7 3rts 3rtt 3ts4 3tsk 3uvc 4efs 4gql 4gr0 4gr3 4gr8 4guy 4h30 4h49 4h76 4h84 4i03 4ijo |
| Collagenase-3 (MMP-13) | H. sapiens | 456c 830c 1eub 1fls 1fm1 2pjt 2ozr 2ow9 2e2d 2d1n 1ztq 1you 1xur 1xud 1xuc 2yig 3elm 3i7g 3i7i 3kec 3kej 3kek 3kry 3ljz 3o2x 3tvc 3zxh 4a7b 4fu4 4fvl 4g0d |
| M. musculus | 1cxv | |
| MT1-MMP (MMP-14) | H. sapiens | 1bqq 1buv 3ma2 |
| MT3-MMP (MMP-16) | H. sapiens | 1rm8 |
| Enamelysin (MMP-20) | H. sapiens | 2jsd |
| Karilysin | Tannerella forsythia | 2xs3 2xs4 4in9 |
| SNAPALYSINS | ||
| Neutral protease | Streptomyces caespitosus | 1c7k 1kuh 4hx3 |
| LEISHMANOLYSINS | ||
| Leishmanolysin | Leishmania major | 1lml |
| PAPPALYSINS | ||
| Ulilysin | Methanosarcina acetivorans | 2cki 2j83 3lum 3lun |
| ARCHAEMETZINCINS | ||
| AmzA | Methanopyrus kandleri | 2x7m |
| Archaeglobus fulgidus | 3zvs 4a3w 4axq | |
| Putative zinc-dependent peptidase | Methanocorpusculum labreanum | 3lmc |
| FRAGILYSINS | ||
| Fragilysin-3 (profragilysin-3) | Bacteroides fragilis | 3p24 |
| CHOLERILYSINS | ||
| StcE | Escherichia coli O157:H7 | 3ujz |
| TOXILYSINS | ||
| EcxAB toxin | E. coli | 4l63 4l6t |
| IGALYSINS | ||
| Putative peptidase BF3526 | B. fragilis | 4df9 |
| Putative metalloendopeptidase BACOVA_0063 | B. ovatus | 3p1v |
The primary reference of each structure may be retrieved from the PDB with the respective access code except for coordinates deposited by structural genomics consortia but not published (denoted as “putative”).
Figure 4.
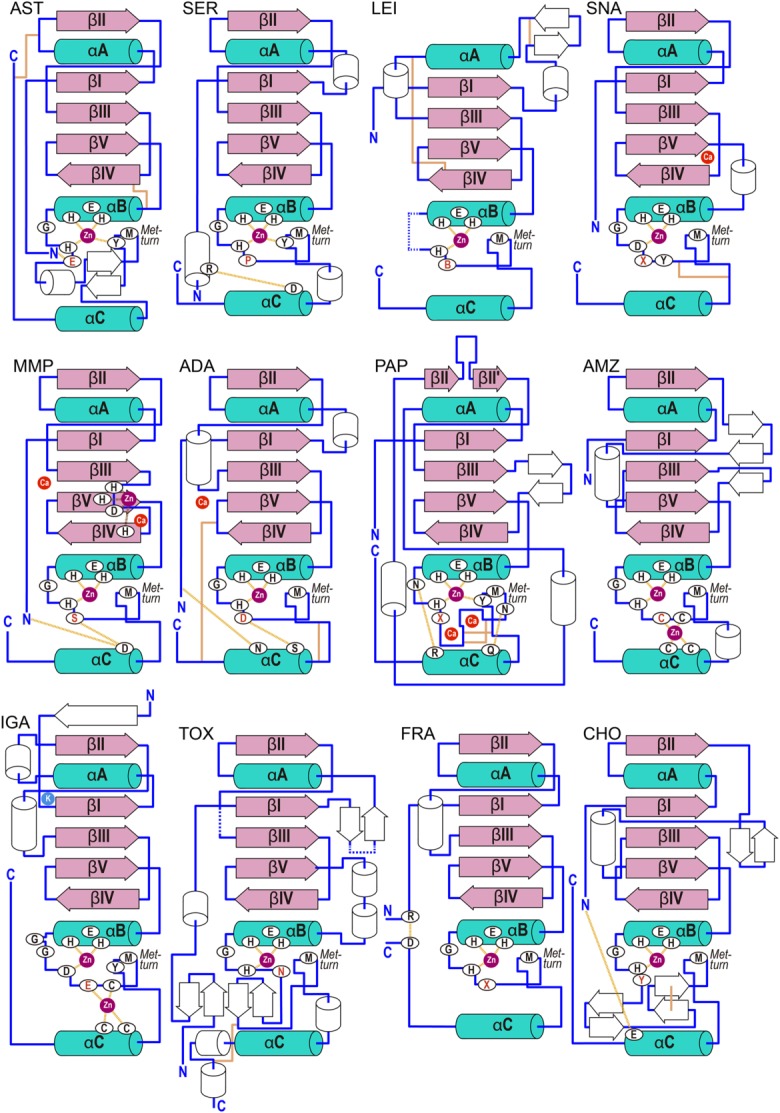
Topology of metzincin catalytic domains. Scheme showing the regular secondary structure elements (helices as rods, strands as arrows) of each metzincin prototype depicted for its structure in Figure 3. The common elements are in pink (strands) and turquoise (helices) and labeled (βI-βV and αA-αC). Hydrogen bonds and metal–ligand bonds are shown as dashed lines, disulfide bonds as orange solid lines. Disordered segments or points of insertion of extra domains are characterized by blue dashes. Family-specific residues are in red, B stands for bulky, hydrophobic residues, X for any residue.
The consensus NTS of metzincins includes a five-stranded twisted β-sheet on the top (Figs. 5). All strands (βI to βV) except the fourth are parallel to each other and to any substrate that is bound in the cleft [Fig. 5(a)]. The antiparallel strand, βIV, forms the lower edge of this sub-domain and creates an upper rim or northern wall of the active-site crevice.122 As discussed for zincins under the section “Zincins split into gluzincins and metzincins,” this strand binds a substrate in an antiparallel manner, mainly on its nonprimed side. The loop segment connecting strands βIII and βIV (LβIIIβIV), the “bulge-edge segment,” generates protruding structural elements, which mainly affect subsites S1' and S2'. This gives rise to extensive variations in enzyme–substrate interactions on the primed side of the active-site clefts. The NTS also contains a backing helix (αA) and an active-site helix (αB), which are arranged on the concave side of the β-sheet identically in all metzincin structures (Figs. 5). Helix αB superimposes well in all structures [Fig. 5(b,c)] and encompasses the first half of the zinc-binding motif, which in metzincins is expanded from HEXXH to HEXXHXXGXX(H,D) and includes the first two zinc-binding histidine residues. At the end of αB, the polypeptide chain takes a sharp, downward turn, mediated by the glycine of the consensus sequence. The main-chain angles of this residue in the lead structures indicate that any other residue would be in a high-energy conformation and thus disfavored. Pappalysins provide the only notable exception, with an asparagine at this position (see the section on “Pappalysins”). The CSD starts right after this glycine and the chain leads to the third zinc ligand, a histidine or an aspartate, which is the last residue of the metzincin zinc-binding motif and approaches the metal from below [Fig. 5(b,c)]. The residue immediately downstream of the latter is termed the “family-specific residue” and is characteristic for each family. The CSD contains few regular secondary-structure elements, mainly a C-terminal helix (αC) at the end of the polypeptide chain. Helices αB and αC are connected by structures that vary in both length and conformation. However, all structures coincide at a conserved 1,4-β-turn containing a strictly invariant methionine at position three, the Met-turn, which is separated from the third zinc-binding histidine by “connecting segments” ranging from six to 77 amino acids. The methionine is superimposable—including the conformation of its side chain [see Fig. 5(b,c)]—and is positioned underneath the catalytic metal, forming a hydrophobic pillow. However, no contact with the metal is observed. Mutation studies indicated a role for this methionine in the folding and stability of the catalytic domains, although the strict conservation of this residue is still a matter of debate.123–129 The S1' pocket of metzincins is the main determinant of substrate specificity and is shaped at the top by the bulge-edge segment and at the bottom by a “wall-forming segment” made up of residues connecting the Met-turn and the C-terminal helix αC.122 This segment also varies in structure and length, ranging from 9 to 37 residues in the reference structures.
Figure 5.
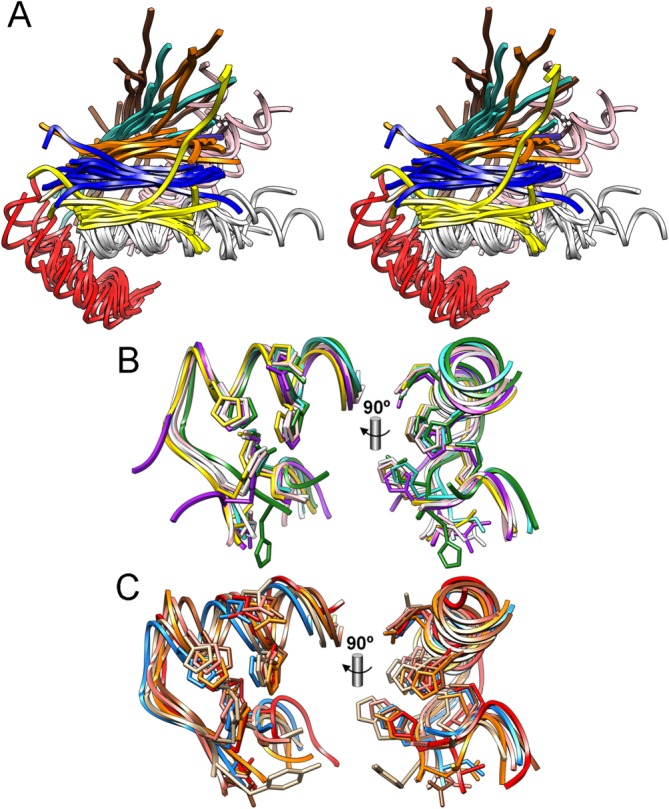
Superposition of common structural elements. (A) Stereo image depicting the superposition of the 12 reference structures only around the common structural elements, β-sheet strands βI (turquoise), βII (brown), βIII (orange), βIV (yellow), and βV (blue), as well as backing helices αA (pink), active-site helices αB (white), and C-terminal helices αC (red). (B) Superposition of the active-site helix and the downstream chain segment until the family-specific residue plus the Met-turn of AST (white), ADA (gold), SER (pink), LEI (purple), MMP (turquoise), and SNA (green) in two orthogonal views. The catalytic metal ions have been omitted for clarity. (C) Same as (B) but showing AMZ (blue), FRA (red), IGA (orange), CHO (tan), PAP (salmon), and TOX (sienna).
The catalytic zinc ion lies roughly at half width of the active-site cleft at its base. It is coordinated by the Nɛ2 atoms of the three consensus histidines or two histidine Nɛ2 atoms and an aspartate Oδ1/2 atom (in SNA and IGA) and the catalytic solvent molecule in an approximately tetrahedral or trigonal pyramidal manner. The solvent molecule is replaced by other ligands in enzyme+inhibitor/product complexes. Some metzincins display a further protein ligand at a slightly greater distance from the catalytic cation in the form of a tyrosine Oη atom, as seen in unbound AST75 and SER111 and suggested for unbound PAP.130 This tyrosine is found two positions downstream of the Met-turn methionine. In SNA and IGA, a tyrosine is found two positions downstream of the metal-binding aspartate, though no longer within binding distance of the catalytic metal ion. The same is observed for the tyrosine found as the family-specific residue in CHO. All these tyrosines may flip back and forth during substrate anchoring, cleavage, and product release, in a motion referred to as the “tyrosine switch” in the particular cases of astacins and serralysins.131,132 In this, they may play a role in substrate and catalytic-solvent binding and stabilization of the tetrahedral intermediate or the newly created α-amino group of the downstream product.28–133 Other, nonconserved residues take over this role in tyrosine-lacking metzincins.
Metzincin Family-Specific Features
Astacins
The first member of the astacin family and first metzincin to be structurally analyzed, in 1992, was the 200-residue digestive crayfish enzyme AST.75–134 Astacins have disparate biological roles in digestion, development, and tissue remodeling and differentiation—by promoting cartilage and bone formation and collagen biosynthesis—exerted through protein degradation, growth-factor activation, extracellular matrix turnover, and extracellular coat degradation (hatching). Six orthologs are found in humans.135 The catalytic domain of mature AST is a packman-like ellipsoid and has two sub-domains roughly equal in size (Fig. 3). While the gene of this peptidase encodes only a short pro-peptide, which shields the active-site cleft in the zymogen following an “aspartate-switch” mechanism,136 and a catalytic MP domain, other family members have additional C-terminal SHK, CUB, LC, TSP, EGF, MAM, TRAF, inserted, transmembrane and cytosolic domains. The astacin family has the longest—together with the families represented by IGA, PAP and CHO—and most sequence-conserved connecting segment among metzincins (see Table1 in Ref. 28). The protein scaffold is crosslinked in AST by four conserved cysteine residues forming two disulfide bridges (Fig. 3). The first two residues of the mature enzyme are buried in an internal cavity in the CSD and the N-terminal α-amino group binds the family-conserved residue, a glutamate in astacins. The unbound coordination of the catalytic zinc is trigonal-bipyramidal due to the presence of a fourth invariant, though more distal, tyrosine-switch zinc ligand and the bound (putatively) catalytic solvent molecule (see the section on “Common structural features of metzincins” and Fig. 3). In addition to AST, the structures of mature human tolloid-like protease 1, bone morphogenetic protein 1, meprin β, high choriolytic enzyme 1, and zebrafish hatching enzyme 1, as well as zymogen structures of AST and meprin β, have been reported, totaling 16 structures (see Table1). Reviews and a monograph on the family have been published.40–142
Serralysins
Pathogenic γ-class proteobacterial genera such as Pseudomonas, Erwinia, Serratia, Pectobacterium, Photorhabdus, Morganella, Preoteus, Xenorhabdus, Dickeya, and Yersinia produce ∼50-kDa bacterial virulence factors, the serralysins, which are secreted as autoactivatable zymogens.143–144 Several of these bacteria infect humans and cause—mostly hospital-acquired—infections such as meningitis, endocarditis, pyelonephritis, plague, dermatitis, soft-tissue infections, septicemia, melioidosis, pneumonia, and other respiratory and urinary tract infections. Serralysins are possibly part of the bacterial armamentarium required for such infections, and they have been shown to be active against coagulation factors and defense-oriented proteins, protease inhibitors, lysozyme, and transferrin. They may also trigger an anaphylactic response. The first serralysin to be biochemically and structurally characterized was SER.111,145 Its mature catalytic domain spans 220-residues (Figs. 3 and 4), lacks disulfide bonds, and is followed by a C-terminal β-roll domain stabilized by calcium ions. As in AST, the two SER subdomains are similar in size, and the enzyme starts with a family-specific α-helix in the CSD that is connected with the molecular moiety by a conserved salt bridge with the C-terminal helix αC (Figs. 3 and 4). An elongated LβIαA gives rise to a flap that runs across the convex surface of the β-sheet and differs among serralysins, thus distinctly conditioning substrate binding. The CSD of SER bears an extra α-helix within the wall-forming segment and a second flap shaped by residues of the connecting segment. These elements likewise modulate substrate binding. Comparison of the structures of tetrapeptide-bound SER and that of the unbound ortholog of Serratia marcescens reveals that the unliganded zinc coordination is similar to astacin (bipyramidal-trigonal) under inclusion of a tyrosine-switch tyrosine.147 Further serralysin structures have been reported from Erwinia chrysanthemi and Flavobacterium sp., totaling 26 (see Table1). Two structures correspond to complexes with cognate protein inhibitors.148–149
Leishmanolysins
Leishmanolysins were originally identified as protozoan cell-surface proteins GP63 in trypanosomes, ciliates, and sarcocysts such as those found in the genera Trypanosoma, Leishmania, Crithidia, Naegleria, Tetrahymena, and Sarcocystis. Collectively, these organisms are associated with major human infections such as African trypanosomiasis, meningoencephalitis, Chagas' disease, and leishmaniasis. Leishmanolysins are the major component of the promastigote surface, and they cleave host surface substrates such as CD4 molecules on human T cells. They further protect protozoan promastigotes from inactivation by complement proteins, thus playing a role as virulence factors. Similar sequences to the protozoan forms have been found in mammals (here called invadolysins), insects, plants, fishes, parasitic worms, sponges, mosses, and some bacteria.28–150 The only structurally analyzed family member is LEI114(Table1). It is synthesized as a 602-residue inactive precursor in the endoplasmic reticulum provided with a signal peptide and a 100-residue pro-domain, which includes a highly conserved cysteine residue potentially acting as a “cysteine switch” (see the section on “Matrixins”). Maturation produces an MP of 276 residues, which includes a 63-residue insertion domain between the glycine and the third metal-binding histidine of the metzincin consensus motif (Figs. 3 and 4), followed by a ∼200-residue C-terminal domain. The MP domain is the most asymmetric among metzincins, with a 171-residue NSD and a 42-amino acid CSD, and its N-terminus is found on the back left surface. The NSD has a β-sheet that lacks strand βII (Figs. 3 and 4) and two unique ∼40-residue inserted flaps preceding and following backing helix αA, which are linked to the molecular moiety through respective disulfide bonds. A slightly prominent bulge-edge segment preceding strand βIV lies on top of the shallow, medium-sized S1' pocket, which is framed by the wall-forming segment and active-site helix αB. At the end of the CSD, αC is followed by a segment in extended conformation, which runs parallel across the back surface (Fig. 3 in Ref. 114).
Snapalysins
Streptomyces are predominantly soil bacteria, which collectively produce over two-thirds of the clinically relevant antibiotics in nature. Only few species are pathogenic to humans and may cause mycetoma. SNA is a secreted neutral protease from Streptomyces caespitosus, which slowly hydrolyses milk. Similar sequences to SNA have been reported from other Streptomyces species and closely related actinomycetales only. The MP consists of a 132-residue catalytic domain preceded by an alanine-rich 100-amino acid N-terminal extension encompassing a signal-peptide and a pro-domain. SNA thus has the shortest metzincin catalytic domain reported to date and the only three reported PDB entries of the family correspond to S. caespitosus, one of them to a heterotetrameric 2 + 2 complex with the dimeric cognate bifunctional protein inhibitor, sermetstatin.151 The SNA structure is a flattened ellipsoid and is divided into two asymmetric sub-domains.113–152 It displays all the characteristic metzincin features, connected by short loops, and could therefore represent a minimal metzincin. Particular distinguishing elements are a small LβIIβIII protruding from the upper sheet within the NSD, a small bulge on top of the primed side of the active-site crevice, a short helix in LβVαB, and a calcium-binding site (Figs. 3 and 4). In addition, an aspartate is the third metal-binding protein ligand and, two positions ahead in the sequence, a conserved tyrosine approaches the metal but does not bind it.
Matrixins
Matrixins alias MMPs are secreted or membrane-bound MPs discovered 50 years ago as responsible for tadpole tail resorption during frog metamorphosis.153 They are found in higher mammals and in animals, plants, and viruses but not in fungi and only in some bacteria, where they do not follow the Darwinian tree-based pathway but may be the result of eukaryotic-to-prokaryotic horizontal gene transfer.154–155 MMPs exert both broad-spectrum turnover and limited proteolysis of extracellular proteins, which includes ectodomain shedding at the plasma membrane in a complementary fashion to adamalysins/ADAMs (see the section on “Adamalysins/ADAMS”). Substrates include extracellular matrix proteins, peptidases and zymogens, inhibitors, clotting factors, antimicrobial peptides, chemotactic and adhesion molecules, growth factors, hormones, and cytokines, as well as their respective receptors.156 In these functions, they contribute to physiological actions ranging from tissue processing during embryogenesis and development, morphogenesis, signaling, and angiogenesis to apoptosis and intestinal defense-protein activation. As a result of this broad spectrum of functions, they play also key roles in pathologies such as inflammation, ulcer, rheumatoid and osteoarthritis, periodontitis, heart failure, atherosclerosis, stroke and cardiovascular disease, fibrosis, emphysema, multiple sclerosis, bacterial meningitis, Alzheimer's disease, HIV-associated dementia, and cancer and metastasis.157
Vertebrate MMPs are multimodular proteins consisting of a ∼20-residue secretory signal peptide, an ∼80-residue pro-peptide, a 160-to-170-residue zinc-and calcium-dependent catalytic domain, a linker region, and a fourfold-propeller hemopexin-like C-terminal domain.158 Further possible modules include fibronectin type-II-related domains, collagen type-V-like and vitronectin-like insertion domains, cysteine-rich, proline-rich, and interleukin-1 receptor-like domains, immunoglobulin-like domains, glycosyl phosphatidylinositol linkage sequences, membrane anchors and cytoplasmic tails; 23 MMP proteins encoded by 24 genes are found in humans termed MMP-1 to MMP-28 (some numbers are unassigned).159 Those MMPs encompassing a membrane anchor gave rise to the MT-MMP subfamily.122–157 MMPs are produced as zymogens that are activated by proteolytic cleavage following a “cysteine switch” or “velcro” mechanism, in which a pro-segment shields access of substrates to the active-site cleft, and a conserved cysteine binds the catalytic metal ion.160–161 The mature catalytic domain of human neutrophil collagenase (MMP112) is taken here as a prototype (Figs. 3 and 4). Like its orthologs and paralogs, it has a shallow active-site cavity, which separates a larger NSD (∼120 residues) from a smaller CSD (∼40 amino acids), and a deep hydrophobic S1' pocket. No disulfide bonds are present in the structure. Characteristically, the N-terminal α-amino group of the mature MP is salt-bridged to the first of two conserved aspartates within helix αC, which also hydrogen-bonds the MMP family-specific residue, a serine or threonine. The NSD displays an S-shaped double loop connecting strands βIII and βIV, which embraces structural zinc and calcium cations. The downstream residues of this segment form a prominent bulge that protrudes into the active-site groove. In addition, LβIVβV and LβIIβIII contribute to a second calcium-binding site on top of the NSD β-sheet (Figs. 3 and 4). In the CSD, MMPs possess the shortest connecting segment within metzincins, with just six to eight residues. MMPs are the most thoroughly studied metzincin family in structural terms, with over 200 structures reported, mostly of MMP-3 and MMP-12 (Table1). These include isolated catalytic domains, multimodular structures and even full-length structures, as well as complexes with their endogenous tissue inhibitors of metalloproteinases (TIMPs), zymogenic structures, and numerous complexes with small-molecule inhibitors. Numerous reviews and monographs have been published and only a few are cited here.157,162–170
Adamalysins/ADAMS
The family, termed adamalysins, ADAMs, or reprolysins, has also been the object of extensive studies:7,171–182 it can be divided into three subgroups, the snake venom MPs, the mammalian ADAMs (19 paralogs in humans), and the likewise mammalian ADAMTSs (19 paralogs in humans). A separate chapter could be dedicated to ADAMDEC1 alias decysin, which has a unique domain structure,183 and the nine testases, which are ADAMs found only in rodents.184 The proteins from snake venom cause destruction of extracellular proteins and hemorrhage following bites, while ADAMs were originally discovered in mammalian reproductive tracts engaged in fertilization and sperm function. They are involved in myogenesis, development and neurogenesis, differentiation of osteoblastic cells, cell-migration modulation, and muscle fusion. Due to these critical roles in physiology, they are also related to human disorders such as asthma, cardiac hypertrophy, obesity-associated adipogenesis and cachexia, rheumatoid arthritis, endotoxic shock, inflammation, and Alzheimer's disease. Together with MMPs, (the section on “Matrixins”), they are major contributors to protein ectodomain shedding. Finally, the ADAMTSs disable cell adhesion by binding to integrins. They also participate in gonad formation, embryonic development and angiogenesis, and procollagen activation, as well as in inflammatory processes, cartilage (aggrecan) degradation in arthritic diseases, bleeding disorders, and glioma-tumor invasion.185
The family members are extracellular multidomain proteins containing a zinc-and calcium-dependent MP domain. In addition, they can display a pro-domain, C-terminal disintegrin-like, cysteine-rich, C-type lectin, EGF-like, thrombospondin 1-like, and/or transmembrane domains, as well as a cytoplasmic domain. In particular, all mammalian ADAMs are type-I membrane-anchored bitopic proteins, while ADAMTSs lack the transmembrane domain and contain multiple copies of a thrombospondin 1-like repeat and a CUB domain instead, thus being soluble extracellular proteases. Zymogenicity is exerted by pro-domains and activation occurs probably following a cysteine switch-like mechanism as in MMPs (see the section on “Matrixins”).28,160 The first structure of a catalytic domain to be solved was that of ADA.76–187 This is a compact ellipsoidal 203-residue molecule with a relatively flat active-site cleft, which separates a 148-residue NSD from a 54-residue CSD (Figs. 3 and 4). Both the N-and the C-termini are surface located; the former is linked by a hydrogen bond to the C-terminal helix αC, which also binds the family-specific residue, a glutamate (Fig. 4). ADA deviates from the metzincin consensus due to two additional helices—the one inserted between βII and βIII and running front-to-back is termed “adamalysin helix”—within the NSD. In the CSD, two disulfide-bonds crosslink the irregular connecting segment and attach αC to the NSD, respectively. A calcium ion is located on the surface, opposite the active site and close to the C-terminus (Figs. 3 and 4). The S1' pocket, characterized by a pronounced bulge-edge segment, is hydrophobic and deep, reminiscent of MMPs.188–189 In addition to ADA, a number of snake venom MPs, ADAM-8, ADAM-17, ADAM-33, ADAMTS-1, -4, and -5 have been structurally analyzed to date (Table1). These structures include one complex of ADAM-17 with the only endogenous ADAM protein inhibitor reported, TIMP-3.190
Pappalysins
This family was named after human PAPP-A alias pappalysin-1, a heavily-glycosylated 170-kDa multidomain protein specifically cleaving insulin-like growth factor binding proteins,124 and its paralog pappalysin-2. Closely related sequences are present in mammals, birds, fishes, amphibians, and echinoderms, and somewhat more distant sequences belong to fungi, bacteria, and archaea.116 Among the latter is 38-kDa PAP from Methanosarcina acetivorans, which encompasses only the pro-domain and the catalytic domain.191 Activation of the PAP zymogen may occur through cysteine-switch as in MMPs (see the section on “Matrixins”) as suggested by the presence of a conserved cysteine in the pro-domain. In vitro, activation occurs autolytically in the presence of calcium. With 262 residues, mature PAP is the second largest metzincin-prototype catalytic domain and the only family member to be structurally analyzed to date (Table1). As characteristic structural elements, it has a loop dividing strand βII into two substrands (βII and βII'; Figs. 3 and 4), and a β-ribbon inserted within LβIIIβIV that protrudes from the molecular surface and frames the active site on its primed side. The segment connecting αA with βII covers the back of the molecule from the NSD to the CSD in a cape-like fashion and includes two unique α-helices. The glycine of the metzincin zinc-binding motif is exceptionally replaced in PAP and a subset of pappalysins by an asparagine, which binds an arginine within helix αC (Fig. 4). This replacement occurs under slight variation of the main-chain angles, which do not correspond here to a high-energy conformation (see the section on “Common structural features of metzincins”) but rather to a left-handed α-helix, although overall, the chain trace is indistinguishable from other metzincins [Fig. 5(b,c)]. The CSD evinces two disulfide bonds and a unique two-calcium site. This site is a molecular switch for activity, as the proteinase can be reversibly inhibited through calcium chelators.116,130 Finally, in the absence of an unbound structure, PAP may possess a fifth zinc-binding tyrosine ligand provided by the Met-turn that is swung out upon substrate binding.
Archaemetzincins
The family name was coined in 200328 and the first members to be studied at the biochemical level were the two human orthologs, archaemetzincin-1 and 2, which were reported to hydrolyze synthetic substrates and bioactive peptides.115 The structures of three archaeal orthologs from Methanopyrus kandleri, Archaeoglobus fulgidus, and Methanocorpusculum labreanum have been reported118–119 (the latter was only deposited by a structural genomics consortium; see Table1), which however corresponded to inactive species. The family has an extended consensus sequence immediately after the third metal-binding histidine, CX4CXMX17CXXC. This sequence encompasses the Met-turn methionine and four cysteines, which bind a second, structural zinc ion in the CTS below the Met-turn (Figs. 3 and 4) in an element termed the “Cys4 zinc finger”.118–119 The cysteines are, respectively, in the position of the family-specific residue; two residues ahead of the Met-turn methionine; and in the first and fourth position of the C-terminal helix αC. Moreover, as found in the 175-residue M. kandleri protein, AMZ, additional family-characteristic structural elements are an adamalysin helix running front-to-back, two short 310-helices, a small three-stranded antiparallel β-sheet on the convex side of the NTS β-sheet, and an extra helix within the wall-forming segment in the CTS (Figs. 3 and 4).
Igalysins
This family was not identified in previous sequence-based studies because it does not contain the intact metzincin zinc-binding consensus sequence but has uniquely an additional glycine preceding the signature glcyine and an aspartate as third zinc ligand (HEXXHXXGGXXD) as found in snapalysins (see the section on “Snapalysins”). Only the deposition—but not publication—by a structural genomics consortium of two proteins from Bacteroides fragilis and Bacteroides ovatus (see Table1), which are thus reported here for the first time, allowed their structural analysis and ascription to the metzincins. These proteins evince significant sequence similarity (BLAST E-values <10−57 at http://www.uniprot.org) with MPs from MEROPS family M64, termed relatives of IgA-peptidase of Clostridium ramosum, which cleave prolyl bonds within an O-glycosylated, tandemly duplicated octapeptide in the hinge region of human immunoglobulin IgA1.193–194 These peptidases should not be confused with those of family M26, which groups MPs represented by IgA1-specific metallopeptidase from Streptococcus sanguinis.38,195 Accordingly, the term “igalysins” will be used hereafter to refer to the novel metzincin family of M64 peptidases.
Detailed inspection of B. ovatus igalysin (IGA) reveals that, with 272 residues, the structure is the largest reported metzincin catalytic domain, and all the characteristic structural elements for metzincins are observed (Figs. 3 and 4). It shows an asymmetric NTS-CTS tandem (166 and 106 residues, respectively) and, within NTS, on top of the β-sheet, it has an extra sixth β-strand inserted before βI, which runs parallel to the upper-rim strand and antiparallel to the other four. In addition, an adamalysin-helix is inserted between βII and βIII and an extra helix at LαAβII (Fig. 4). The insertion of the extra glycine within the consensus sequence occurs at the end of the active-site helix and is compensated for by the extension of the helix by an additional half turn so that, overall, the chain trace hardly differs from that of the other metzincins [Fig. 5(c)]. As to the CTS, it bears the largest connecting segment (77 residues) but the shortest wall-forming segment (nine residues) within metzincins. As in snapalysin, a tyrosine is found two positions downstream of the third zinc-binding residue—likewise an aspartate—but not within binding distance to the metal. However, the most striking feature is the presence of a second structural metal-binding site within the CTS, which coincides in space with the one found in archaemetzincins (see the section on “Pappalysins”) despite completely different chain traces and lengths of the respective connecting segments and wall-forming segments. In IGA, this Cys4 zinc finger is bound, as in archaemetzincin, by the family-specific residue, which is a glutamate instead of a cysteine; by the first and fourth cysteine residues of the C-terminal helix; and by a cysteine two residues downstream of the Met-turn methionine.
Toxilysins
This is the most recent family, which was discovered while studying bacterial AB5 toxins.121 These are virulence factors of pathogenic bacteria that consist of a pentameric B subunit for host-cell invasion and an A subunit that subverts intracellular functions.197–198 In contrast to previously studied related toxins, EcxAB from E. coli and CfxAB from Citrobacter freundii exhibit the metzincin consensus sequence within their A subunits, and the crystal structure of the former reveals that they fulfill the structural requirements for a novel metzincin family, the toxilysins.121–199 The structure of EcxA, the only one reported to date for the family (Table1), is a compact disk of 258 residues (Figs. 3 and 4). The N-terminal segment is extended by ∼30 residues preceding NTS strand βI and starts within the CTS. There it contributes with two short strands to a four-stranded antiparallel β-sheet. The N-terminal extension is connected with the Met-turn through a double hydrogen bond formed by the family-specific asparagine with both structural elements. Before entering strand βI and already within the NTS, the polypeptide chain also includes an extra helix. In addition, the NTS bears a β-ribbon within LβIαA and two consecutive helices within LβVαB. Within the CTS, the connecting segment provides two additional strands to the aforementioned extra β-sheet and the wall-forming segment contains an extra helix as found in SER and AMZ (see the sections on “Serralysins” and “Pappalysins”). Finally, the C-terminal segment is also extended beyond αC, adopts a helical conformation in two short stretches, and is linked through a disulfide bond to the protein moiety. The final helix interacts with the pentameric B subunit to yield the AB5 complex.
Fragilysins
This is the smallest family, with just one member occurring as three closely related isoforms—fragilysin-1, −2, and −3—with 93–96% sequence identity in enteropathogenic Bacteroides fragilis (ETBF) only. It was first reported in 1984 as a factor potentially causing acute diarrheal disease in newborn lambs.200 The chromosomal pathogenicity islet that encodes fragilysins and is absent in nonenterotoxigenic strains contains a second gene, mpII, which is countertranscribed and encodes a potential MP of similar size and moderate sequence identity (28–30%) to the three fragilysins. It has recently been shown to be an active MP with a specificity for cleavage between flanking arginines.201 Fragilysins are the only proven virulence factors of ETBF and target E-cadherin from the intestine of infected mammalian hosts. The structural and function analysis of FRA revealed a 194-residue pro-domain—the largest among metzincins—with unique fold, which plays a major role in protein folding and inhibits the mature 188-residue MP moiety through an aspartate-switch mechanism similarly to astacins (see the section on “Astacins”).117–202 As distinguishing structural features, FRA has an adamalysin helix within the NTS and a characteristic salt bridge that links the N-and the C-terminus at the back of the molecule (Figs. 3 and 4). Overall, the FRA catalytic domain shows remarkable structural similarity with adamalysins/ADAMs despite just ∼15% sequence identity (see Supporting Information and Fig. S3b in Ref. 117). Taken together with its restriction to just one protein found in just one organism, this indicates that it may be the result of horizontal gene transfer of an ADAM gene from a mammalian host and subsequent evolution in a bacterial environment, thus giving rise to a unique pro-domain, small structural changes, and a different protein sequence expressed as three isoforms.117–202
Cholerilysins
In pathogenic Vibrio cholerae, the transmembrane DNA-binding protein ToxR coordinates the expression of essential virulence genes encoded by the Vibrio pathogenicity island, including those encoding virulence factors such as the toxin-coregulated type-IV pilus. Among them, ToxR-activated gene A (TagA) lipoprotein has been described.203 Together with a paralog, another protein from Burholderia, and protein StcE (secreted protease of C1-esterase inhibitor; CHO), they were proposed in 2003 to constitute the cholerilysin family,28 which coincides with MEROPS family M6638 and the SLiMe family.204–205 CHO is an 898-residue virulence factor secreted by enterohemorrhagic E. coli. It acts as a mucinase hydrolyzing glycoproteins of the mucosal barrier that lines and protects the mammalian gastrointestinal tract, thus contributing to bacterial penetration during infection.206 CHO includes a central 258-residue catalytic domain flanked upstream by two immunoglobulin-like domains and an insertion region and downstream by two disordered regions and a C-terminal domain.120 This is the only structure reported for the family (Table1): it is shaped like a banana, while comprising all the structural hallmarks of metzincins. However, in comparison with other metzincins, CHO shows an unusually open active-site cleft, which is compatible with binding of large O-glycosylated substrates.120 In addition, it features a helix on the convex face of the NTS β-sheet, in a similar position though different orientation to the adamalysin helix of other metzincins. Furthermore, an active-site flap with a short β-ribbon is inserted after strand βII, which replaces the bulge-edge segment in other metzincins in shaping the active-site cleft top on its primed side. Between the family-specific tyrosine, which points toward the bulk solvent in the reported structure but could belong to a tyrosine switch (see the section on “Common structural features of metzincins”), and the Met-turn, two additional β-ribbons are found within the CTS; the first contains a conserved sequence, WGWD,205 and the second is crosslinked by a disulfide bond. Finally, the N-terminal α-amino nitrogen is linked through a salt bridge with a glutamate in the penultimate position of the C-terminal helix αC, five residues upstream of the catalytic domain C-terminus.
Concluding Remarks
The metzincins are ubiquitous MPs that participate in pathophysiological metabolic processes. Twelve metzincin families have been characterized at the structural level, some are restricted to certain taxa, others span several kingdoms, and even others are most likely the product of horizontal gene transfer. In addition to those structurally characterized, several more families may enlarge the clan based on the presence of the characteristic extended zinc-binding consensus sequence pattern and its small variants as found in some current family members. The structural evidence, accumulated over the past 21 years, has enabled us to confirm the hypothesis proposed in 1993 that they share a set of characteristic structural elements and a catalytic metal environment despite negligible overall sequence similarity. Accordingly, this MP clan can be envisaged as an example of divergent evolution since the onset of life—they are present in archaea—from an archetypal minimal urmetzincin, which may have spanned ∼100–110 residues and may not have looked very different from the smallest current member, 132-residue snapalysin. Within this initial scaffold, functional requirements and the associated evolutionary processes would have led to the introduction of specific structural elements, which eventually resulted in the distinct current families. The structural information reviewed here may help to ascribe the function of proteins encoded by newly discovered gene sequences and in the elucidation of common catalytic mechanisms but also of their detailed differences, which may enable the design of specific and selective inhibitors and activators to modulate their activity.
Acknowledgments
Ulrich Baumann and Johann (Hans) Brandstetter are warmly thanked for critical reading of the manuscript, as is Robin Rycroft. This study was supported in part by grants from European, Spanish, and Catalan agencies.
References
- Barrett AJ, Rawlings ND, Salvesen G, Woessner JF. In: Handbook of proteolytic enzymes. Rawlings ND, Salvesen G, editors. Oxford (UK): Academic Press; 2013. pp. Ii–Iiv. [Google Scholar]
- Neurath H. Limited proteolysis and zymogen activation. In: Reich E, Rifkins DB, Shaw E, editors. Proteases and biological control. New York: Cold Spring Harbor Laboratory Press; 1975. pp. 51–64. [Google Scholar]
- Neurath H, Walsh KA. Role of proteolytic enzymes in biological regulation. Proc Natl Acad Sci USA. 1976;73:3825–3832. doi: 10.1073/pnas.73.11.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C, Overall CM. Protease degradomics: a new challenge for proteomics. Nat Rev Mol Cell Biol. 2002;3:509–519. doi: 10.1038/nrm858. [DOI] [PubMed] [Google Scholar]
- Kheradmand F, Werb Z. Shedding light on sheddases: role in growth and development. BioEssays. 2002;24:8–12. doi: 10.1002/bies.10037. [DOI] [PubMed] [Google Scholar]
- Arribas J, Merlos-Suárez A. Shedding of plasma membrane proteins. Curr Top Dev Biol. 2003;54:125–144. doi: 10.1016/s0070-2153(03)54007-8. [DOI] [PubMed] [Google Scholar]
- Blobel CP. ADAMs: key components in EGFR signalling and development. Nat Rev Mol Cell Biol. 2005;6:32–43. doi: 10.1038/nrm1548. [DOI] [PubMed] [Google Scholar]
- Hartley BS. Chemical nature of the active centres of enzymes. Ann Repts Prog Chem. 1955;51:303–311. [Google Scholar]
- Hartley BS. Proteolytic enzymes. Ann Rev Biochem. 1960;29:45–72. doi: 10.1146/annurev.bi.29.070160.000401. [DOI] [PubMed] [Google Scholar]
- Barrett AJ, McDonald JK. Nomenclature: protease, proteinase and peptidase. Biochem J. 1986;237:935–935. doi: 10.1042/bj2370935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Bateman A. MEROPS: the peptidase database. Nucleic Acids Res. 2010;38:D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Bateman A. Asparagine peptide lyases: a seventh catalytic type of proteolytic enzymes. J Biol Chem. 2011;286:38321–38328. doi: 10.1074/jbc.M111.260026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett AJ, Rawlings ND, Woessner JF. Handbook of proteolytic enzymes. Amsterdam: Elsevier; 2004. [Google Scholar]
- Stroud RM, Kossiakoff AA, Chambers JL. Mechanisms of zymogen activation. Annu Rev Biophys Bioeng. 1977;6:177–193. doi: 10.1146/annurev.bb.06.060177.001141. [DOI] [PubMed] [Google Scholar]
- Holzer H, Heinrich PC. Control of proteolysis. Annu Rev Biochem. 1980;49:63–91. doi: 10.1146/annurev.bi.49.070180.000431. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Tolle DP, Barrett AJ. Evolutionary families of peptidase inhibitors. Biochem J. 2004;378:705–716. doi: 10.1042/BJ20031825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollery CM, Libby P. Atherosclerosis and proteinase activation. Cardiovasc Res. 2006;69:625–635. doi: 10.1016/j.cardiores.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Murphy G, Nagase H. Reappraising metalloproteinases in rheumatoid arthritis and osteoarthritis: destruction or repair? Nat Clin Pract Rheumatol. 2008;4:128–135. doi: 10.1038/ncprheum0727. [DOI] [PubMed] [Google Scholar]
- Nalivaeva NN, Fisk LR, Belyaev ND, Turner AJ. Amyloid-degrading enzymes as therapeutic targets in Alzheimer's disease. Curr Alzheimer Res. 2008;5:212–224. doi: 10.2174/156720508783954785. [DOI] [PubMed] [Google Scholar]
- Chang C, Werb Z. The many faces of metalloproteases: cell growth, invasion, angiogenesis and metastasis. Trends Cell Biol. 2001;11:S37–43. doi: 10.1016/s0962-8924(01)02122-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C, Matrisian LM. Emerging roles of proteases in tumour suppression. Nat Rev Cancer. 2007;7:800–808. doi: 10.1038/nrc2228. [DOI] [PubMed] [Google Scholar]
- Miyoshi S, Shinoda S. Microbial metalloproteases and pathogenesis. Microbes Infect. 2000;2:91–98. doi: 10.1016/s1286-4579(00)00280-x. [DOI] [PubMed] [Google Scholar]
- Taylor LH, Latham SM, Woolhouse ME. Risk factors for human disease emergence. Phil Trans Royal Soc London B. 2001;356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potempa J, Pike RN. Bacterial peptidases. Contrib Microbiol. 2005;12:132–180. doi: 10.1159/000081693. [DOI] [PubMed] [Google Scholar]
- Turk B. Targeting proteases: successes, failures and future prospects. Nat Rev Drug Discov. 2006;5:785–799. doi: 10.1038/nrd2092. [DOI] [PubMed] [Google Scholar]
- Bjarnason JB, Fox JB. Hemorrhagic metalloproteinases from snake venoms. Pharmac Ther. 1994;62:325–372. doi: 10.1016/0163-7258(94)90049-3. [DOI] [PubMed] [Google Scholar]
- Tonello F, Morante S, Rossetto O, Schiavo G, Montecucco C. Tetanus and botulism neurotoxins: a novel group of zinc-endopeptidases. Adv Exp Med Biol. 1996;389:251–260. [PubMed] [Google Scholar]
- Gomis-Rüth FX. Structural aspects of the metzincin clan of metalloendopeptidases. Mol Biotech. 2003;24:157–202. doi: 10.1385/MB:24:2:157. [DOI] [PubMed] [Google Scholar]
- Quesada V, Ordoñez GR, Sánchez LM, Puente XS, López-Otn C. The Degradome database: mammalian proteases and diseases of proteolysis. Nucleic Acids Res. 2009;37:D239–D243. doi: 10.1093/nar/gkn570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode W, Gomis-Rüth FX, Stöcker W. Astacins, serralysins, snake venom and matrix metalloproteinases exhibit identical zinc-binding environments (HEXXHXXGXXH and Met-turn) and topologies and should be grouped into a common family, the 'metzincins'. FEBS Lett. 1993;331:134–140. doi: 10.1016/0014-5793(93)80312-i. [DOI] [PubMed] [Google Scholar]
- Stöcker W, Bode W. Structural features of a superfamily of zinc-endopeptidases: the metzincins. Curr Opin Struct Biol. 1995;5:383–390. doi: 10.1016/0959-440x(95)80101-4. [DOI] [PubMed] [Google Scholar]
- Stöcker W, Grams F, Baumann U, Reinemer P, Gomis-Rüth FX, McKay DB, Bode W. The metzincins—Topological and sequential relations between the astacins, adamalysins, serralysins, and matrixins (collagenases) define a superfamily of zinc-peptidases. Prot Sci. 1995;4:823–840. doi: 10.1002/pro.5560040502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode W, Grams F, Reinemer P, Gomis-Rüth FX, Baumann U, McKay DB, Stöcker W. The metzincin-superfamily of zinc-peptidases. Adv Exp Med Biol. 1996;389:1–11. doi: 10.1007/978-1-4613-0335-0_1. [DOI] [PubMed] [Google Scholar]
- Sterchi EE. Special issue: metzincin metalloproteinases. Mol Aspects Med. 2008;29:255–257. doi: 10.1016/j.mam.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Gomis-Rüth FX. Catalytic domain architecture of metzincin metalloproteases. J Biol Chem. 2009;284:15353–15357. doi: 10.1074/jbc.R800069200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera S, Khrestchatisky M, Kaczmarek L, Rosenberg GA, Jaworski DM. Metzincin proteases and their inhibitors: foes or friends in nervous system physiology? J Neurosci. 2010;30:15337–15357. doi: 10.1523/JNEUROSCI.3467-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban NP, Rudakova NL, Sharipova MR. Structural and functional characteristics and properties of metzincins. Biochem Biokhim. 2012;77:119–127. doi: 10.1134/S0006297912020010. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40:D343–D350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Salvesen G. Handbook of proteolytic enzymes. Oxford (UK): Academic Press; 2013. p. 4094. [Google Scholar]
- Jiang W, Bond JS. Families of metalloendopeptidases and their relationships. FEBS Lett. 1992;312:110–114. doi: 10.1016/0014-5793(92)80916-5. [DOI] [PubMed] [Google Scholar]
- Hooper NM. Families of zinc metalloproteases. FEBS Lett. 1994;354:1–6. doi: 10.1016/0014-5793(94)01079-x. [DOI] [PubMed] [Google Scholar]
- Lewis AP, Thomas PJ. A novel clan of zinc metallopeptidases with possible intramembrane cleavage properties. Prot Sci. 1999;8:439–442. doi: 10.1110/ps.8.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis-Rüth FX. Structure and mechanism of metallocarboxypeptidases. Crit Rev Biochem Mol Biol. 2008;43:319–345. doi: 10.1080/10409230802376375. [DOI] [PubMed] [Google Scholar]
- Lowther WT, Matthews BW. Metalloaminopeptidases: common functional themes in disparate structural surroundings. Chem Rev. 2002;102:4581–4608. doi: 10.1021/cr0101757. [DOI] [PubMed] [Google Scholar]
- Seibert CM, Raushel FM. Structural and catalytic diversity within the amidohydrolase superfamily. Biochemistry. 2005;44:6383–6391. doi: 10.1021/bi047326v. [DOI] [PubMed] [Google Scholar]
- Botelho TO, Guevara T, Marrero A, Arêde P, Fluxa VS, Reymond JL, Oliveira DC, Gomis-Rüth FX. Structural and functional analyses reveal that Staphylococcus aureus antibiotic resistance factor HmrA is a zinc-dependent endopeptidase. J Biol Chem. 2011;286:25697–25709. doi: 10.1074/jbc.M111.247437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran HJ, Allen MD, Lowe J, Bycroft M. Structure of the Jab1/MPN domain and its implications for proteasome function. Biochemistry. 2003;42:11460–11465. doi: 10.1021/bi035033g. [DOI] [PubMed] [Google Scholar]
- Ambroggio XI, Rees DC, Deshaies RJ. JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2004;2:E2. doi: 10.1371/journal.pbio.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, Ookata K, Nureki O, Iwai K, Komada M, Fukai S. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;455:358–362. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- Meinnel T, Blanquet S, Dardel F. A new subclass of the zinc metalloproteases superfamily revealed by the solution structure of peptide deformylase. J Mol Biol. 1996;262:375–386. doi: 10.1006/jmbi.1996.0521. [DOI] [PubMed] [Google Scholar]
- Chan MK, Gong W, Rajagopalan PT, Hao B, Tsai CM, Pei D. Crystal structure of the Escherichia coli peptide deformylase. Biochemistry. 1997;36:13904–13909. doi: 10.1021/bi9711543. [DOI] [PubMed] [Google Scholar]
- Gomis-Rüth FX, Botelho TO, Bode W. A standard orientation for metallopeptidases. Biochim Biophys Acta. 2012;1824:157–163. doi: 10.1016/j.bbapap.2011.04.014. [DOI] [PubMed] [Google Scholar]
- Auld DS. Chapter 78—Catalytic mechanism for metallopeptidases. In: Rawlings ND, Salvesen G, editors. Handbook of proteolytic enzymes. Oxford: Academic Press; 2013. pp. 370–396. [Google Scholar]
- Bochtler M, Odintsov SG, Marcyjaniak M, Sabala I. Similar active sites in lysostaphins and D-Ala-D-Ala metallopeptidases. Prot Sci. 2004;13:854–861. doi: 10.1110/ps.03515704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcyjaniak M, Odintsov SG, Sabala I, Bochtler M. Peptidoglycan amidase MepA is a LAS metallopeptidase. J Biol Chem. 2004;279:43982–43989. doi: 10.1074/jbc.M406735200. [DOI] [PubMed] [Google Scholar]
- van Heijenoort J. Peptidoglycan hydrolases of Escherichia coli. Microbiol Mol Biol Rev: MMBR. 2011;75:636–663. doi: 10.1128/MMBR.00022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche E, Paschos A, Beisel HG, Bock A, Huber R. Crystal structure of the hydrogenase maturating endopeptidase HYBD from Escherichia coli. J Mol Biol. 1999;288:989–998. doi: 10.1006/jmbi.1999.2719. [DOI] [PubMed] [Google Scholar]
- Theodoratou E, Huber R, Bock A. [NiFe]-Hydrogenase maturation endopeptidase: structure and function. Biochem Soc Trans. 2005;33:108–111. doi: 10.1042/BST0330108. [DOI] [PubMed] [Google Scholar]
- Lenart A, Dudkiewicz M, Grynberg M, Pawlowski K. CLCAs—A family of metalloproteases of intriguing phylogenetic distribution and with cases of substituted catalytic sites. PloS One. 2013;8:e62272. doi: 10.1371/journal.pone.0062272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ. Homologues of insulinase, a new superfamily of metalloendopeptidases. Biochem J. 1991;275:389–391. doi: 10.1042/bj2750389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker AB, Roth RA. An unusual active site identified in a family of zinc metalloendopeptidases. Proc Natl Acad Sci USA. 1992;89:3835–3839. doi: 10.1073/pnas.89.9.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama Y, Chuma A, Mikami B, Hashimoto W, Murata K. Heterosubunit composition and crystal structures of a novel bacterial M16B metallopeptidase. J Mol Biol. 2011;407:180–192. doi: 10.1016/j.jmb.2011.01.038. [DOI] [PubMed] [Google Scholar]
- Matthews BW. Structural basis of the action of thermolysin and related zinc peptidases. Acc Chem Res. 1988;21:333–340. [Google Scholar]
- Polgár L. Basic kinetic mechanisms of proteolytic enzymes. In: Sterchi EE, Stöcker W, editors. Proteolytic enzymes—Tools and targets. Berlin/Heidelberg: Springer Verlag; 1999. pp. 148–166. [Google Scholar]
- Schechter I, Berger A. On the size of active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- Timmer JC, Zhu W, Pop C, Regan T, Snipas SJ, Eroshkin AM, Riedl SJ, Salvesen GS. Structural and kinetic determinants of protease substrates. Nat Struct Mol Biol. 2009;16:1101–1108. doi: 10.1038/nsmb.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madala PK, Tyndall JDA, Nall T, Fairlie DP. Update 1: proteases universally recognize beta strands in their active sites. Chem Rev. 2010;110:PR1–PR31. doi: 10.1021/cr900368a. [DOI] [PubMed] [Google Scholar]
- Vallee BL, Neurath H. Carboxypeptidase, a zinc metalloprotein. J Am Chem Soc. 1954;76:5006. [Google Scholar]
- Vallee BL, Auld DS. Zinc coordination, function, and structure of zinc enzymes and other proteins. Biochemistry. 1990;29:5647–5659. doi: 10.1021/bi00476a001. [DOI] [PubMed] [Google Scholar]
- Vallee BL, Auld DS. Active-site zinc ligands and activated H2O of zinc enzymes. Proc Natl Acad Sci USA. 1990;87:220–224. doi: 10.1073/pnas.87.1.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscomb WN, Hartsuck JA, Reeke GN, Jr, Quiocho FA, Bethge PH, Ludwig ML, Steitz TA, Muirhead H, Coppola JC. The structure of carboxypeptidase A. VII. The 2.0-Å resolution studies of the enzyme and of its complex with glycyltyrosine, and mechanistic deductions. Brookhaven Symp Biol. 1968;21:24–90. [PubMed] [Google Scholar]
- Matthews BW, Jansonius JN, Colman PM, Schoenborn BP, Dupourque D. Three-dimensional structure of thermolysin. Nature. 1972;238:37–41. doi: 10.1038/newbio238037a0. [DOI] [PubMed] [Google Scholar]
- Pelmenschikov V, Siegbahn PEM. Catalytic mechanism of matrix metalloproteinases: two-layered ONIOM study. Inorg Chem. 2002;41:5659–5666. doi: 10.1021/ic0255656. [DOI] [PubMed] [Google Scholar]
- Bertini I, Calderone V, Fragai M, Luchinat C, Maletta M, Yeo KJ. Snapshots of the reaction mechanism of matrix metalloproteinases. Angew Chem Int Ed Engl. 2006;45:7952–7955. doi: 10.1002/anie.200603100. [DOI] [PubMed] [Google Scholar]
- Bode W, Gomis-Rüth FX, Huber R, Zwilling R, Stöcker W. Structure of astacin and implications for activation of astacins and zinc-ligation of collagenases. Nature. 1992;358:164–167. doi: 10.1038/358164a0. [DOI] [PubMed] [Google Scholar]
- Gomis-Rüth FX, Kress LF, Bode W. First structure of a snake venom metalloproteinase: a prototype for matrix metalloproteinases/collagenases. EMBO J. 1993;12:4151–4157. doi: 10.1002/j.1460-2075.1993.tb06099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilshtain-Vardi A, Glick M, Greenblatt HM, Goldblum A, Shoham G. Refined structure of bovine carboxypeptidase A at 1.25 Å resolution. Acta Cryst. 2003;59:323–333. doi: 10.1107/s0907444902015706. [DOI] [PubMed] [Google Scholar]
- Jongeneel CV, Bouvier J, Bairoch A. A unique signature identifies a family of zinc-dependent metallopeptidases. FEBS Lett. 1989;242:211–214. doi: 10.1016/0014-5793(89)80471-5. [DOI] [PubMed] [Google Scholar]
- McKerrow JH. Human fibroblast collagenase contains an amino acid sequence homologous to the zinc-binding site of Serratia protease. J Biol Chem. 1987;262:5943–5943. [PubMed] [Google Scholar]
- Feng L, Yan H, Wu Z, Yan N, Wang Z, Jeffrey PD, Shi Y. Structure of a site-2 protease family intramembrane metalloprotease. Science. 2007;318:1608–1612. doi: 10.1126/science.1150755. [DOI] [PubMed] [Google Scholar]
- Fushimi N, Ee CE, Nakajima T, Ichishima E. Aspzincin, a family of metalloendopeptidases with a new zinc-binding motif. Identification of new zinc-binding sites (His(128), His(132), and Asp(164)) and three catalytically crucial residues (Glu(129), Asp(143), and Tyr(106)) of deuterolysin from Aspergillus oryzae by site-directed mutagenesis. J Biol Chem. 1999;274:24195–24201. doi: 10.1074/jbc.274.34.24195. [DOI] [PubMed] [Google Scholar]
- Hori T, Kumasaka T, Yamamoto M, Nonaka N, Tanaka N, Hashimoto Y, Ueki U, Takio K. Structure of a new ‘aspzincin’ metalloendopeptidase from Grifola frondosa: implications for the catalytic mechanism and substrate specificity based on several different crystal forms. Acta Cryst. 2001;57:361–368. doi: 10.1107/s0907444900019740. [DOI] [PubMed] [Google Scholar]
- McAuley KE, Jia-Xing Y, Dodson EJ, Lehmbeck J, Ostergaard PR, Wilson KS. A quick solution: ab initio structure determination of a 19 kDa metalloproteinase using ACORN. Acta Cryst. 2001;57:1571–1578. doi: 10.1107/s090744490101335x. [DOI] [PubMed] [Google Scholar]
- Bieniossek C, Schalch T, Bumann M, Meister M, Meier R, Baumann U. The molecular architecture of the metalloprotease FtsH. Proc Natl Acad Sci USA. 2006;103:3066–3071. doi: 10.1073/pnas.0600031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suno R, Niwa H, Tsuchiya D, Zhang X, Yoshida M, Morikawa K. Structure of the whole cytosolic region of ATP-dependent protease FtsH. Mol Cell. 2006;22:575–585. doi: 10.1016/j.molcel.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Bieniossek C, Niederhauser B, Baumann UM. The crystal structure of apo-FtsH reveals domain movements necessary for substrate unfolding and translocation. Proc Natl Acad Sci USA. 2009;106:21579–21584. doi: 10.1073/pnas.0910708106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langklotz S, Baumann U, Narberhaus F. Structure and function of the bacterial AAA protease FtsH. Biochim Biophys Acta. 2012;1823:40–48. doi: 10.1016/j.bbamcr.2011.08.015. [DOI] [PubMed] [Google Scholar]
- López-Pelegrín M, Cerdà-Costa N, Martínez-Jiménez F, Cintas-Pedrola A, Canals A, Peinado JR, Martí-Renom MA, López-Otín C, Arolas JL, Gomis-Rüth FX. A novel family of soluble minimal scaffolds provides structural insight into the catalytic domains of integral-membrane metallopeptidases. J Biol Chem. 2013;288:21279–21294. doi: 10.1074/jbc.M113.476580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecucco C, Schiavo G. Tetanus and botulism neurotoxins: a new group of zinc proteases. Trends Biochem Sci. 1993;18:324–327. doi: 10.1016/0968-0004(93)90065-u. [DOI] [PubMed] [Google Scholar]
- Swaminathan S. Molecular structures and functional relationships in clostridial neurotoxins. FEBS J. 2011;278:4467–4485. doi: 10.1111/j.1742-4658.2011.08183.x. [DOI] [PubMed] [Google Scholar]
- Tronrud DE, Roderick SL, Matthews BW. Structural basis for the action of thermolysin. Matrix. 1992;1:107–111. [PubMed] [Google Scholar]
- Adekoya OA, Sylte I. The thermolysin family (M4) of enzymes: therapeutic and biotechnological potential. Chem Biol Drug Des. 2009;73:7–16. doi: 10.1111/j.1747-0285.2008.00757.x. [DOI] [PubMed] [Google Scholar]
- Eijsink VG, Matthews BW, Vriend G. The role of calcium ions in the stability and instability of a thermolysin-like protease. Prot Sci. 2011;20:1346–1355. doi: 10.1002/pro.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannifer AD, Wong TY, Schwarzenbacher R, Renatus M, Petosa C, Bienkowska J, Lacy DB, Collier RJ, Park S, Leppla SH, Hanna P, Liddington RC. Crystal structure of the anthrax lethal factor. Nature. 2001;414:229–233. doi: 10.1038/n35101998. [DOI] [PubMed] [Google Scholar]
- Xu Q, Gohler AK, Kosfeld A, Carlton D, Chiu HJ, Klock HE, Knuth MW, Miller MD, Elsliger MA, Deacon AM, Godzik A, Lesley SA, Jahreis K, Wilson IA. The structure of Mlc titration factor A (MtfA/YeeI) reveals a prototypical zinc metallopeptidase related to anthrax lethal factor. J Bacteriol. 2012;194:2987–2999. doi: 10.1128/JB.00038-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor EE, Jr, Horanyi PS, Clark KM, Fedoriw N, Connelly SM, Koszelak-Rosenblum M, Zhu G, Malkowski MG, Wiener MC, Dumont ME. Structure of the integral membrane protein CAAX protease Ste24p. Science. 2013;339:1600–1604. doi: 10.1126/science.1232048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley A, Dong YY, Pike AC, Dong L, Shrestha L, Berridge G, Stansfeld PJ, Sansom MS, Edwards AM, Bountra C, von Delft F, Bullock AN, Burgess-Brown NA, Carpenter EP. The structural basis of ZMPSTE24-dependent laminopathies. Science. 2013;339:1604–1607. doi: 10.1126/science.1231513. [DOI] [PubMed] [Google Scholar]
- Kyrieleis OJ, Goettig P, Kiefersauer R, Huber R, Brandstetter H. Crystal structures of the tricorn interacting factor F3 from Thermoplasma acidophilum, a zinc aminopeptidase in three different conformations. J Mol Biol. 2005;349:787–800. doi: 10.1016/j.jmb.2005.03.070. [DOI] [PubMed] [Google Scholar]
- Addlagatta A, Gay L, Matthews BW. Structure of aminopeptidase N from Escherichia coli suggests a compartmentalized, gated active site. Proc Natl Acad Sci USA. 2006;103:13339–13344. doi: 10.1073/pnas.0606167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholander F, Muroya A, Roques BP, Fournie-Zaluski MC, Thunnissen MM, Haeggstrom JZ. Structure-based dissection of the active site chemistry of leukotriene A4 hydrolase: implications for M1 aminopeptidases and inhibitor design. Chem Biol. 2008;15:920–929. doi: 10.1016/j.chembiol.2008.07.018. [DOI] [PubMed] [Google Scholar]
- Eckhard U, Schonauer E, Nuss D, Brandstetter H. Structure of collagenase G reveals a chew-and-digest mechanism of bacterial collagenolysis. Nat Struct Mol Biol. 2011;18:1109–1114. doi: 10.1038/nsmb.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhard U, Schonauer E, Brandstetter H. Structural basis for activity regulation and substrate preference of clostridial collagenases G, H, and T. J Biol Chem. 2013;288:20184–20194. doi: 10.1074/jbc.M112.448548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oefner C, D'Arcy A, Hennig M, Winkler FK, Dale GE. Structure of human neutral endopeptidase (Neprilysin) complexed with phosphoramidon. J Mol Biol. 2000;296:341–349. doi: 10.1006/jmbi.1999.3492. [DOI] [PubMed] [Google Scholar]
- Bland ND, Pinney JW, Thomas JE, Turner AJ, Isaac RE. Bioinformatic analysis of the neprilysin (M13) family of peptidases reveals complex evolutionary and functional relationships. BMC Evol Biol. 2008;8:16. doi: 10.1186/1471-2148-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baral PK, Jajcanin-Jozic N, Deller S, Macheroux P, Abramic M, Gruber K. The first structure of dipeptidyl-peptidase III provides insight into the catalytic mechanism and mode of substrate binding. J Biol Chem. 2008;283:22316–22324. doi: 10.1074/jbc.M803522200. [DOI] [PubMed] [Google Scholar]
- Yeh DC, Parsons LM, Parsons JF, Liu F, Eisenstein E, Orban J. NMR structure of HI0004, a putative essential gene product from Haemophilus influenzae, and comparison with the X-ray structure of an Aquifex aeolicus homolog. Prot Sci. 2005;14:424–430. doi: 10.1110/ps.041096705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penhoat CH, Li Z, Atreya HS, Kim S, Yee A, Xiao R, Murray D, Arrowsmith CH, Szyperski T. NMR solution structure of Thermotoga maritima protein TM1509 reveals a Zn-metalloprotease-like tertiary structure. J Struct Funct Genom. 2005;6:51–62. doi: 10.1007/s10969-005-5277-z. [DOI] [PubMed] [Google Scholar]
- Oganesyan V, Busso D, Brandsen J, Chen S, Jancarik J, Kim R, Kim SH. Structure of the hypothetical protein AQ_1354 from Aquifex aeolicus. Acta Cryst. 2003;59:1219–1223. doi: 10.1107/s0907444903011028. [DOI] [PubMed] [Google Scholar]
- Jacob AI, Kohrer C, Davies BW, RajBhandary UL, Walker GC. Conserved bacterial RNase YbeY plays key roles in 70S ribosome quality control and 16S rRNA maturation. Mol Cell. 2013;49:427–438. doi: 10.1016/j.molcel.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan C, Fedorov EV, Shi W, Ramagopal UA, Thirumuruhan R, Manjasetty BA, Almo SC, Fiser A, Chance MR, Fedorov AA. The ybeY protein from Escherichia coli is a metalloprotein. Acta Cryst. 2005;61:959–963. doi: 10.1107/S1744309105031131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann U, Wu S, Flaherty KM, McKay DB. Three-dimensional structure of the alkaline protease of Pseudomonas aeruginosa: A two-domain protein with a calcium binding parallel beta roll motif. EMBO J. 1993;12:3357–3364. doi: 10.1002/j.1460-2075.1993.tb06009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode W, Reinemer P, Huber R, Kleine T, Schnierer S, Tschesche H. The X-ray crystal structure of the catalytic domain of human neutrophil collagenase inhibited by a substrate analogue reveals the essentials for catalysis and specificity. EMBO J. 1994;13:1263–1269. doi: 10.1002/j.1460-2075.1994.tb06378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurisu G, Kinoshita T, Sugimoto A, Nagara A, Kai Y, Kasai N, Harada S. Structure of the zinc endoprotease from Streptomyces caespitosus. J Biochem. 1997;121:304–308. doi: 10.1093/oxfordjournals.jbchem.a021587. [DOI] [PubMed] [Google Scholar]
- Schlagenhauf E, Etges R, Metcalf P. The crystal structure of the Leishmania major surface proteinase leishmanolysin (gp63) Structure. 1998;6:1035–1046. doi: 10.1016/s0969-2126(98)00104-x. [DOI] [PubMed] [Google Scholar]
- Díaz-Perales A, Quesada V, Peinado JR, Ugalde AP, Alvarez J, Suarez MF, Gomis-Rüth FX, López-Otín C. Identification and characterization of human archaemetzincin-1 and −2, two novel members of a family of metalloproteases widely distributed in Archaea. J Biol Chem. 2005;280:30367–30375. doi: 10.1074/jbc.M504533200. [DOI] [PubMed] [Google Scholar]
- Tallant C, García-Castellanos R, Seco J, Baumann U, Gomis-Rüth FX. Molecular analysis of ulilysin, the structural prototype of a new family of metzincin metalloproteases. J Biol Chem. 2006;281:17920–17928. doi: 10.1074/jbc.M600907200. [DOI] [PubMed] [Google Scholar]
- Goulas T, Arolas JL, Gomis-Rüth FX. Structure, function and latency regulation of a bacterial enterotoxin potentially derived from a mammalian adamalysin/ADAM xenolog. Proc Natl Acad Sci USA. 2010;108:1856–1861. doi: 10.1073/pnas.1012173108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltersperger S, Widmer C, Wang M, Baumann U. Crystal structure of archaemetzincin AmzA from Methanopyrus kandleri at 1.5 Å resolution. Proteins. 2010;78:2720–2723. doi: 10.1002/prot.22777. [DOI] [PubMed] [Google Scholar]
- Graef C, Schacherl M, Waltersperger S, Baumann U. Crystal structures of archaemetzincin reveal a moldable substrate-binding site. PloS One. 2012;7:e43863. doi: 10.1371/journal.pone.0043863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu AC, Worrall LJ, Strynadka NC. Structural insight into the bacterial mucinase StcE essential to adhesion and immune evasion during enterohemorrhagic E. Coli infection. Structure. 2012;20:707–717. doi: 10.1016/j.str.2012.02.015. [DOI] [PubMed] [Google Scholar]
- Ng NM, Littler DR, Paton AW, Le Nours J, Rossjohn J, Paton JC, Beddoe T. EcxAB is a founding member of a new family of metalloprotease AB toxins with a hybrid cholera-like B subunit. Structure. 2013;21:2003–2013. doi: 10.1016/j.str.2013.08.024. [DOI] [PubMed] [Google Scholar]
- Maskos K, Bode W. Structural basis of matrix metalloproteinases and tissue inhibitors of metalloproteinases. Mol Biotechnol. 2003;25:241–266. doi: 10.1385/MB:25:3:241. [DOI] [PubMed] [Google Scholar]
- Pieper M, Betz M, Budiša N, Gomis-Rüth FX, Bode W, Tschesche H. Expression, purification, characterization, and X-ray analysis of selenomethionine 215 variant of leukocyte collagenase. J Prot Chem. 1997;16:637–650. doi: 10.1023/a:1026327125333. [DOI] [PubMed] [Google Scholar]
- Boldt HB, Overgaard MT, Laursen LS, Weyer K, Sottrup-Jensen L, Oxvig C. Mutational analysis of the proteolytic domain of pregnancy-associated plasma protein-A (PAPP-A): classification as a metzincin. Biochem J. 2001;358:359–367. doi: 10.1042/0264-6021:3580359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hege T, Baumann U. The conserved methionine residue of the metzincins: a site-directed mutagenesis study. J Mol Biol. 2001;314:181–186. doi: 10.1006/jmbi.2001.5125. [DOI] [PubMed] [Google Scholar]
- Butler GS, Tam EM, Overall CM. The canonical methionine 392 of matrix metalloproteinase 2 (gelatinase A) is not required for catalytic efficiency or structural integrity: probing the role of the methionine-turn in the metzincin metalloprotease superfamily. J Biol Chem. 2004;279:15615–15620. doi: 10.1074/jbc.M312727200. [DOI] [PubMed] [Google Scholar]
- Perez L, Kerrigan JE, Li X, Fan H. Substitution of methionine 435 with leucine, isoleucine, and serine in tumor necrosis factor α converting enzyme inactivates ectodomain shedding activity. Biochem Cell Biol. 2007;85:141–149. doi: 10.1139/o06-179. [DOI] [PubMed] [Google Scholar]
- Oberholzer AE, Bumann M, Hege T, Russo S, Baumann U. The metzincin's canonical methionine is responsible for the structural integrity of the zinc-binding site. Biol Chem. 2009;390:875–881. doi: 10.1515/BC.2009.100. [DOI] [PubMed] [Google Scholar]
- Tallant C, García-Castellanos R, Baumann U, Gomis-Rüth FX. On the relevance of the Met-turn methionine in metzincins. J Biol Chem. 2010;285:13951–13957. doi: 10.1074/jbc.M109.083378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallant C, García-Castellanos R, Marrero A, Canals F, Yang Y, Reymond JL, Solà M, Baumann U, Gomis-Rüth FX. Activity of ulilysin, an archaeal PAPP-A-related gelatinase and IGFBP protease. Biol Chem. 2007;388:1243–1253. doi: 10.1515/BC.2007.143. [DOI] [PubMed] [Google Scholar]
- Grams F, Dive V, Yiotakis A, Yiallouros I, Vassiliou S, Zwilling R, Bode W, Stöcker W. Structure of astacin with a transition-state analogue inhibitor. Nat Struct Biol. 1996;3:671–675. doi: 10.1038/nsb0896-671. [DOI] [PubMed] [Google Scholar]
- Yiallouros I, Grosse-Berkhoff E, Stöcker W. The roles of Glu93 and Tyr149 in astacin-like zinc peptidases. FEBS Lett. 2000;484:224–228. doi: 10.1016/s0014-5793(00)02163-3. [DOI] [PubMed] [Google Scholar]
- Hege T, Baumann U. Protease C of Erwinia chrysanthemi: the crystal structure and role of amino acids Y228 and E189. J Mol Biol. 2001;314:187–193. doi: 10.1006/jmbi.2001.5124. [DOI] [PubMed] [Google Scholar]
- Gomis-Rüth FX, Stöcker W, Huber R, Zwilling R, Bode W. Refined 1.8 Å X-ray crystal structure of astacin, a zinc-endopeptidase from the crayfish Astacus astacus L. Structure determination, refinement, molecular structure and comparison with thermolysin. J Mol Biol. 1993;229:945–968. doi: 10.1006/jmbi.1993.1098. [DOI] [PubMed] [Google Scholar]
- Gomis-Rüth FX, Trillo-Muyo S, Stöcker W. Functional and structural insights into astacin metallopeptidases. Biol Chem. 2012;393:1027–1041. doi: 10.1515/hsz-2012-0149. [DOI] [PubMed] [Google Scholar]
- Guevara T, Yiallouros I, Kappelhoff R, Bissdorf S, Stöcker W, Gomis-Rüth FX. Proenzyme structure and activation of astacin metallopeptidase. J Biol Chem. 2010;285:13958–13965. doi: 10.1074/jbc.M109.097436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöcker W, Gomis-Rüth FX, Bode W, Zwilling R. Implications of the three-dimensional structure of astacin for the structure and function of the astacin-family of zinc-endopeptidases. Eur J Biochem. 1993;214:215–231. doi: 10.1111/j.1432-1033.1993.tb17915.x. [DOI] [PubMed] [Google Scholar]
- Bond JS, Beynon RJ. The astacin family of metalloendopeptidases. Prot Sci. 1995;4:1247–1261. doi: 10.1002/pro.5560040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwilling R, Stöcker W. Naturwissenschaftliche Forschungsergebnisse. Hamburg (Germany): Verlag Dr. Kovač; 1997. The astacins—Structure and function of a new protein family; p. 370. [Google Scholar]
- Villa JP, Bertenshaw GP, Bylander JE, Bond JS. Meprin proteolytic complexes at the cell surface and in extracellular spaces. The Biochemical Society Symposium. 2003:53–63. doi: 10.1042/bss0700053. [DOI] [PubMed] [Google Scholar]
- Semenova SA, Rudenskaia GN. The astacin family of metalloproteinases. Biomeditsinskaia khimiia. 2008;54:531–554. [PubMed] [Google Scholar]
- Sterchi EE, Stöcker W, Bond JS. Meprins, membrane-bound and secreted astacin metalloproteinases. Mol Aspects Med. 2008;29:309–328. doi: 10.1016/j.mam.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Morihara K. Serralysin and related bacterial proteinases. Meth Enzymol. 1995;248:395–413. doi: 10.1016/0076-6879(95)48026-9. [DOI] [PubMed] [Google Scholar]
- Baumann U. Chapter 180—Serralysin and related enzymes. In: Rawlings ND, Salvesen GS, editors. Handbook of proteolytic enzymes. Oxford: Academic Press; 2013. pp. 864–867. [Google Scholar]
- Morihara K. Studies on the protease of Pseudomonas. II. Crystallization of the protease and its physicochemical and general properties. Bull Agric Chem Soc Jpn. 1957;21:11–17. [Google Scholar]
- Morihara K, Homma JY. Pseudomonas proteases. In: Holder IA, editor. Bacterial enzymes and virulence. Boca Ratón, FL: CRC Press; 1985. pp. 41–79. [Google Scholar]
- Baumann U. Crystal structure of the 50 kDa metallo protease from Serratia marcescens. J Mol Biol. 1994;242:244–251. doi: 10.1006/jmbi.1994.1576. [DOI] [PubMed] [Google Scholar]
- Baumann U, Bauer M, Letoffe S, Delepelaire P, Wandersman C. Crystal structure of a complex between Serratia marcescens metallo-protease and an inhibitor from Erwinia chrysanthemi. J Mol Biol. 1995;248:653–661. doi: 10.1006/jmbi.1995.0249. [DOI] [PubMed] [Google Scholar]
- Hege T, Feltzer RE, Gray RD, Baumann U. Crystal structure of a complex between Pseudomonas aeruginosa alkaline protease and its cognate inhibitor. Inhibition by a zinc-NH2 coordinative bond. J Biol Chem. 2001;276:35087–35092. doi: 10.1074/jbc.M104020200. [DOI] [PubMed] [Google Scholar]
- McHugh B, Krause SA, Yu B, Deans AM, Heasman S, McLaughlin P, Heck MM. Invadolysin: a novel, conserved metalloprotease links mitotic structural rearrangements with cell migration. J Cell Biol. 2004;167:673–686. doi: 10.1083/jcb.200405155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trillo-Muyo S, Martínez-Rodríguez S, Arolas JL, Gomis-Rüth FX. Mechanism of action of a Janus-faced single-domain protein inhibitor simultaneously targeting two peptidase classes. Chem Sci. 2013;4:791–797. [Google Scholar]
- Kurisu G, Kai Y, Harada S. Structure of the zinc-binding site in the crystal structure of a zinc endoprotease from Streptomyces caespitosus at 1 Å resolution. J Inorg Biochem. 2000;82:225–228. doi: 10.1016/s0162-0134(00)00136-7. [DOI] [PubMed] [Google Scholar]
- Gross J, Lapière CM. Collagenolytic activity in amphibian tissues: a tissue culture assay. Proc Natl Acad Sci USA. 1962;48:1014–1022. doi: 10.1073/pnas.48.6.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdà-Costa N, Guevara T, Karim AY, Ksiazek M, Nguyen KA, Arolas JL, Potempa J, Gomis-Rüth FX. The structure of the catalytic domain of Tannerella forsythia karilysin reveals it is a bacterial xenologue of animal matrix metalloproteinases. Mol Microbiol. 2011;79:119–132. doi: 10.1111/j.1365-2958.2010.07434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevara T, Ksiazek M, Skottrup PD, Cerda-Costa N, Trillo-Muyo S, de Diego I, Riise E, Potempa J, Gomis-Ruth FX. Structure of the catalytic domain of the Tannerella forsythia matrix metallopeptidase karilysin in complex with a tetrapeptidic inhibitor. Acta Cryst. 2013;69:472–476. doi: 10.1107/S1744309113007392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCawley LJ, Matrisian LM. Matrix metalloproteinases: they're not just for matrix anymore! Curr Opin Cell Biol. 2001;13:534–540. doi: 10.1016/s0955-0674(00)00248-9. [DOI] [PubMed] [Google Scholar]
- Nagase H, Jr Woessner JF. Matrix metalloproteinases. J Biol Chem. 1999;274:21491–21494. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- Tallant C, Marrero A, Gomis-Rüth FX. Matrix metalloproteinases: fold and function of their catalytic domains. Biochim Biophys Acta Mol Cell Res. 2010;1803:20–28. doi: 10.1016/j.bbamcr.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Puente XS, Sánchez LM, Overall CM, López-Otín C. Human and mouse proteases: a comparative genomic approach. Nat Rev Genet. 2003;4:544–558. doi: 10.1038/nrg1111. [DOI] [PubMed] [Google Scholar]
- Springman EB, Angleton EL, Birkedal-Hansen H, Van Wart HE. Multiple modes of activation of latent human fibroblast collagenase: evidence for the role of a Cys73 active-site zinc complex in latency and a "cysteine switch" mechanism for activation. Proc Natl Acad Sci USA. 1990;87:364–368. doi: 10.1073/pnas.87.1.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wart HE, Birkedal-Hansen H. The cysteine switch: a principle of regulation of metalloproteinase activity with potential applicability to the entire matrix metalloproteinase gene family. Proc Natl Acad Sci USA. 1990;87:5578–5582. doi: 10.1073/pnas.87.14.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagase H. Activation mechanisms of matrix metalloproteinases. Biol Chem. 1997;378:151–160. [PubMed] [Google Scholar]
- Woessner JF, Jr, Nagase H. Matrix metalloproteinases and TIMPs. In: Sheterline P, editor. Protein profile series. New York: Oxford University Press; 2000. [Google Scholar]
- Clark IM. Matrix metalloproteinase protocols. In: Walker JM, editor. Methods in molecular biology. Totowa, NJ: Humana Press Inc; 2001. p. 545. [Google Scholar]
- Visse R, Nagase H. Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function, and biochemistry. Circ Res. 2003;92:827–839. doi: 10.1161/01.RES.0000070112.80711.3D. [DOI] [PubMed] [Google Scholar]
- Murphy G, Nagase H. Progress in matrix metalloproteinase research. Mol Aspects Med. 2008;29:290–308. doi: 10.1016/j.mam.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy G. Tissue inhibitors of metalloproteinases. Genome Biol. 2011;12:233. doi: 10.1186/gb-2011-12-11-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapinska A, Fields GB. Chemical biology for understanding matrix metalloproteinase function. Chembiochem. 2012;13:2002–2020. doi: 10.1002/cbic.201200298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiro N, Miyagi E. Matrix metalloproteinases—Biology, functions and clinical implications. Protein biochemistry, synthesis, structure abd cellular functions. Hauppauge, NY: Nova Science Publishers Inc; 2012. 241 pp. [Google Scholar]
- Tocchi A, Parks WC. Functional interactions between matrix metalloproteinases and glycosaminoglycans. FEBS J. 2013;280:2332–2341. doi: 10.1111/febs.12198. [DOI] [PubMed] [Google Scholar]
- Fox JW, Bjarnasson JB. Snake venom metalloendopeptidases: reprolysins. Meth Enzymol. 1995;248:345–368. doi: 10.1016/0076-6879(95)48023-4. [DOI] [PubMed] [Google Scholar]
- Schlondorff J, Blobel CP. Metalloprotease-disintegrins: modular proteins capable of promoting cell-cell interactions and triggering signals by protein-ectodomain shedding. J Cell Sci. 1999;112:3603–3617. doi: 10.1242/jcs.112.21.3603. [DOI] [PubMed] [Google Scholar]
- Cal S, Obaya AJ, Llamazares M, Garabaya C, Quesada V, López-Otín C. Cloning, expression analysis, and structural characterization of seven novel human ADAMTSs, a family of metalloproteinases with disintegrin and thrombospondin-1 domains. Gene. 2002;283:49–62. doi: 10.1016/s0378-1119(01)00861-7. [DOI] [PubMed] [Google Scholar]
- Fox JW, Serrano SM. Structural considerations of the snake venom metalloproteinases, key members of the M12 reprolysin family of metalloproteinases. Toxicon. 2005;45:969–985. doi: 10.1016/j.toxicon.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Hooper NM, Lendeckel U. The ADAM family of proteases. Dordrecht, The Netherlands: Springer; 2005. p. 344. [Google Scholar]
- Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Mol Aspects Med. 2008;29:258–289. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy G. The ADAMs: signalling scissors in the tumour microenvironment. Nat Rev Cancer. 2008;8:929–941. doi: 10.1038/nrc2459. [DOI] [PubMed] [Google Scholar]
- Apte SS. A disintegrin-like and metalloprotease (reprolysin-type) with thrombospondin type 1 motif (ADAMTS) superfamily: functions and mechanisms. J Biol Chem. 2009;284:31493–31497. doi: 10.1074/jbc.R109.052340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S. Three-dimensional domain architecture of the ADAM family proteinases. Semin Cell Dev Biol. 2009;20:146–152. doi: 10.1016/j.semcdb.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Klein T, Bischoff R. Active metalloproteases of the A disintegrin and metalloprotease (ADAM) family: Biological function and structure. J Proteome Res. 2011;10:17–33. doi: 10.1021/pr100556z. [DOI] [PubMed] [Google Scholar]
- Le Goff C, Cormier-Daire V. The ADAMTS(L) family and human genetic disorders. Human Mol Genet. 2011;20:R163–167. doi: 10.1093/hmg/ddr361. [DOI] [PubMed] [Google Scholar]
- Weber S, Saftig P. Ectodomain shedding and ADAMs in development. Development. 2012;139:3693–3709. doi: 10.1242/dev.076398. [DOI] [PubMed] [Google Scholar]
- Lund J, Olsen OH, Sorensen ES, Stennicke HR, Petersen HH, Overgaard MT. ADAMDEC1 is a metzincin metalloprotease with dampened proteolytic activity. J Biol Chem. 2013;288:21367–21375. doi: 10.1074/jbc.M113.474536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu GZ, Lin Y, Myles DG, Primakoff P. Identification of four novel ADAMs with potential roles in spermatogenesis and fertilization. Gene. 1999;234:227–237. doi: 10.1016/s0378-1119(99)00208-5. [DOI] [PubMed] [Google Scholar]
- Nagase H, Fushimi K. Elucidating the function of noncatalytic domains of collagenases and aggrecanases. Connect Tissue Res. 2008;49:169–174. doi: 10.1080/03008200802151698. [DOI] [PubMed] [Google Scholar]
- Grams F, Huber R, Kress LF, Moroder L, Bode W. Activation of snake venom metalloproteinases by a cysteine switch-like mechanism. FEBS Lett. 1993;335:76–80. doi: 10.1016/0014-5793(93)80443-x. [DOI] [PubMed] [Google Scholar]
- Gomis-Rüth FX, Kress LF, Kellermann J, Mayr I, Lee X, Huber R, Bode W. Refined 2.0Å X-ray crystal structure of the zinc-endopeptidase adamalysin II. Primary and tertiary structure determination, refinement, molecular structure and comparison with astacin, collagenase and thermolysin. J Mol Biol. 1994;239:513–544. doi: 10.1006/jmbi.1994.1392. [DOI] [PubMed] [Google Scholar]
- Cirilli M, Gallina C, Gavuzzo E, Giordano C, Gomis-Rüth FX, Gorini B, Kress LF, Mazza F, PaglialungaParadisi M, Pochetti G, Politi V. 2-Å X-ray structure of adamalysin II complexed with a peptide phosphonate inhibitor adopting a retro-binding mode. FEBS Lett. 1997;418:319–322. doi: 10.1016/s0014-5793(97)01401-4. [DOI] [PubMed] [Google Scholar]
- Gomis-Rüth FX, Meyer EF, Kress LF, Politi V. Structures of adamalysin II with peptidic inhibitors. Implications for the design of tumor necrosis factor α convertase inhibitors. Prot Sci. 1998;7:283–292. doi: 10.1002/pro.5560070207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska M, Goettig P, Maskos K, Belouski E, Winters D, Hecht R, Black R, Bode W. Structural determinants of the ADAM inhibition by TIMP-3: crystal structure of the TACE-N-TIMP-3 complex. J Mol Biol. 2008;381:1307–1319. doi: 10.1016/j.jmb.2008.06.088. [DOI] [PubMed] [Google Scholar]
- Baumann U, Gomis-Rüth FX. Chapter 270—Ulilysin (Methanosarcina acetivorans. In: Rawlings ND, Salvesen G, editors. Handbook of proteolytic enzymes. Oxford: Academic Press; 2013. pp. 1208–1211. [Google Scholar]
- García-Castellanos R, Tallant C, Marrero A, Solà M, Baumann U, Gomis-Rüth FX. Substrate specificity of a metalloprotease of the pappalysin family revealed by an inhibitor and a product complex. Arch Biochem Biophys. 2007;457:57–72. doi: 10.1016/j.abb.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Kosowska K, Reinholdt J, Rasmussen LK, Sabat A, Potempa J, Kilian M, Poulsen K. The Clostridium ramosum IgA proteinase represents a novel type of metalloendopeptidase. J Biol Chem. 2002;277:11987–11994. doi: 10.1074/jbc.M110883200. [DOI] [PubMed] [Google Scholar]
- Potempa J, Poulsen K. Chapter 274—The IgA protease of Clostridium ramosum. In: Rawlings ND, Salvesen G, editors. Handbook of proteolytic enzymes. Oxford: Academic Press; 2013. pp. 1225–1228. [Google Scholar]
- Mulks MH, Shoberg RJ. Bacterial immunoglobulin A1 proteases. Meth Enzymol. 1994;235:543–554. doi: 10.1016/0076-6879(94)35169-4. [DOI] [PubMed] [Google Scholar]
- Plaut AG, Qiu J. Chapter 278—IgA-specific metalloendopeptidase. In: Rawlings ND, Salvesen G, editors. Handbook of proteolytic enzymes. Oxford: Academic Press; 2013. pp. 1243–1248. [Google Scholar]
- Merritt EA, Hol WG. AB5 toxins. Curr Opin Struct Biol. 1995;5:165–171. doi: 10.1016/0959-440x(95)80071-9. [DOI] [PubMed] [Google Scholar]
- Beddoe T, Paton AW, Le Nours J, Rossjohn J, Paton JC. Structure, biological functions and applications of the AB5 toxins. Trends Biochem Sci. 2010;35:411–418. doi: 10.1016/j.tibs.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis-Rüth FX. A different look for AB5 toxins. Structure. 2013;21:1909–1910. doi: 10.1016/j.str.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Myers LL, Firehammer BD, Shoop DS, Border MM. Bacteroides fragilis: a possible cause of acute diarrheal disease in newborn lambs. Infect Immun. 1984;44:241–244. doi: 10.1128/iai.44.2.241-244.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiryaev SA, Remacle AG, Chernov AV, Golubkov VS, Motamedchaboki K, Muranaka N, Dambacher CM, Capek P, Kukreja M, Kozlov IA, Perucho M, Cieplak P, Strongin AY. Substrate cleavage profiling suggests a distinct function of Bacteroides fragilis metalloproteinases (fragilysin and metalloproteinase II) at the microbiome-inflammation-cancer interface. J Biol Chem. 2013 doi: 10.1074/jbc.M113.516153. doi: 10.1074/jbc.M113.516153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulas T, Gomis-Rüth FX, Fragilysin . In: Handbook of proteolytic enzymes. Rawlings ND, Salvesen GS, editors. Oxford: Academic Press; 2013. pp. 887–891. [Google Scholar]
- Karaolis DKR, Lan R, Kaper JB, Reeves PR. Comparison of Vibrio cholerae pathogenicity islands in sixth and seventh pandemic strains. Infect Immun. 2001;69:1947–1952. doi: 10.1128/IAI.69.3.1947-1952.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabady RL, Yanta JH, Halladin DK, Schofield MJ, Welch RA. TagA is a secreted protease of Vibrio cholerae that specifically cleaves mucin glycoproteins. Microbiology. 2011;157:516–525. doi: 10.1099/mic.0.044529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabady RL, Welch RA. Chapter 286—StcE peptidase and the StcE-like metallopeptidases. In: Rawlings ND, Salvesen G, editors. Handbook of proteolytic enzymes. Oxford: Academic Press; 2013. pp. 1272–1280. [Google Scholar]
- Lathem WW, Grys TE, Witowski SE, Torres AG, Kaper JB, Tarr PI, Welch RA. StcE, a metalloprotease secreted by Escherichia coli O157:H7, specifically cleaves C1 esterase inhibitor. Molecul Microbiol. 2002;45:277–288. doi: 10.1046/j.1365-2958.2002.02997.x. [DOI] [PubMed] [Google Scholar]