Abstract
Small heat shock proteins (sHSPs) are ubiquitous molecular chaperones that prevent the aggregation of various non-native proteins and play crucial roles for protein quality control in cells. It is poorly understood what natural substrate proteins, with respect to structural characteristics, are preferentially bound by sHSPs in cells. Here we compared the structural characteristics for the natural substrate proteins of Escherichia coli IbpB and Deinococcus radiodurans Hsp20.2 with the respective bacterial proteome at multiple levels, mainly by using bioinformatics analysis. Data indicate that both IbpB and Hsp20.2 preferentially bind to substrates of high molecular weight or moderate acidity. Surprisingly, their substrates contain abundant charged residues but not abundant hydrophobic residues, thus strongly indicating that ionic interactions other than hydrophobic interactions also play crucial roles for the substrate recognition and binding of sHSPs. Further, secondary structure prediction analysis indicates that the substrates of low percentage of β-sheets or coils but high percentage of α-helices are un-favored by both IbpB and Hsp20.2. In addition, IbpB preferentially interacts with multi-domain proteins but unfavorably with α + β proteins as revealed by SCOP analysis. Together, our data suggest that bacterial sHSPs, though having broad substrate spectrums, selectively bind to substrates of certain structural features. These structural characteristic elements may substantially participate in the sHSP–substrate interaction and/or increase the aggregation tendency of the substrates, thus making the substrates more preferentially bound by sHSPs.
Keywords: molecular chaperone, small heat shock protein, protein aggregation, substrate proteins, IbpB, Hsp20.2
Introduction
Small heat shock proteins (sHSPs), as ubiquitous molecular chaperones present in all forms of life, play crucial roles for protein quality control in cells.1 They were able to suppress protein aggregation in an ATP-independent manner2–3 and stabilize stress-damaged cell membranes.4–5 Under in vitro conditions, sHSPs are known to effectively interact with unfolded model substrate proteins and keep them in a folding-competent state for subsequent refolding that is facilitated by such ATP-dependent chaperones as Hsp70s and Hsp100s.6,7 As such, the heterologous over-expression of sHSPs was repeatedly found to increase the tolerance of host cells against various stresses.9,10 Physiologically, sHSPs have been linked to cell differentiation,12 apoptosis,13 animal longevity,14 and dysfunction of them has been related to such diseases15 as cancer development,16 cardiovascular diseases,17 cataracts,18 myopathy,19 and neuron diseases.20–21
One intriguing question regarding the functions of sHSPs is why they are able to protect a great diversity of model or natural substrate proteins.2–26 Whereas characterization of the structures of sHSPs27–28 and determination of their substrate-binding sites24–29 may be helpful in clarifying this question, defining the common structural features among different substrate proteins is also of significance. However, it is hard to determine the high-resolution structures of the substrate proteins due to the structural heterogeneity of sHSP-substrate complexes. Numerous studies using fluorescence spectroscopy, NMR, CD spectroscopy and spin labeling revealed that the substrates are characterized by native-like secondary structures but compromised tertiary structures and somehow exist in molten globule states.30–34 It appears that the substrate proteins, when complexed with sHSPs, are neither folded nor fully unfolded. Another commonly observed structural feature for the substrate proteins is that they are prone to aggregate30–40 in a nucleation-dependent manner.41 Additionally, Carver et al. reported that a disordered intermediate of α-lactalbumin was more efficiently bound by α-crystallin than a less disordered one,35 indicating that structural disorder might also be one of structural features of the sHSP substrates.
The structural features of sHSP substrates had been investigated on a few model substrates,30–42 but not yet on the natural substrates, and this kind of study, if possible, would be more biologically relevant. We have been attempting to investigate the chaperone activity and mechanism of representative sHSPs under both in vitro43–47 and in vivo conditions24–26 for years. Here we attempted to probe the common structural features of natural substrate proteins of sHSPs based on two studies recently contributed by us26 and others,22 in which around 100 natural substrate proteins of two bacterial sHSPs (Escherichia coli IbpB and Deinococcus radiodurans Hsp20.2) have been identified. We systematically compared these substrate proteins with the respective bacterial proteome. Results indicate that bacterial sHSPs selectively bind to substrates of certain structural features.
Results and Discussion
Both IbpB and Hsp20.2 preferentially bind to substrate proteins of high molecular weight or moderate acidity
So far the natural substrate proteins of a few sHSPs have been identified. For instance, a total of 13 and 37 proteins were identified as the substrates of bacterium Synechocystis Hsp16.623 and E. coli IbpA48 in cells, respectively, and a total of 89 proteins present in cell extract were identified as the substrates of bacterium D. radiodurans Hsp20.2 during thermal stress.22 In particular, we recently identified a total of 110 proteins as natural substrates of E. coli IbpB in living cells by using in vivo photo-crosslinking.26 To unravel the structural features of in vivo substrate proteins of sHSPs with statistic significance, we choose IbpB substrate proteins as the subject and compared them with the proteome of E. coli, which comprises 2709 proteins that have been demonstrated at evidence of protein levels in this model organism and are deposited in the UnitProtKB database. It should be pointed out, as also revealed in our earlier study,26 that protein abundance is not the determining factor for a protein to be bound with IbpB. Rather, the principal factor determining such interaction is most likely the aggregation tendency of the protein during its folding/unfolding. In parallel, we also compared Hsp20.2 substrate proteins with the genome-predicted proteome (3167 proteins) of D. radiodurans, and the use of predicted proteome is due to the lack of sufficient experimental data on the proteome.
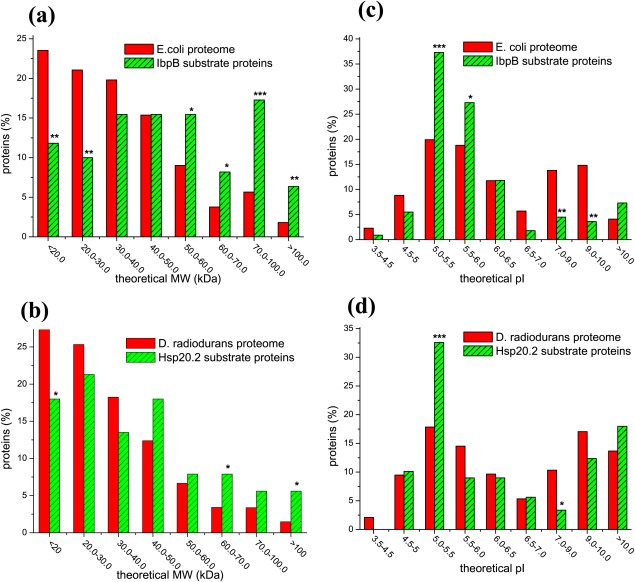
In retrospect, it was reported previously49 that GroEL, an essential molecular chaperone of E. coli, shows certain preference to substrate proteins of middle molecular weight (MW). Prompted by this, we first performed parallel statistic analysis on the MW of the IbpB substrate proteins and E. coli proteome, as well as of the Hsp20.2 substrate proteins and D. radiodurans proteome. Results displayed in Figure 1(a) demonstrate that IbpB preferentially binds to large proteins. Specifically, the proteins of 20–30 kDa represent the mostly un-favored (P < 0.01), somehow similarly to that of GroEL.49 Strikingly, the proteins of >70 kDa represent the most strongly favored for IbpB (P < 0.001) and such MW is beyond the upper limit of MW (60 kDa) for GroEL-encapsulated substrates50 and is also apparently higher than that of GroEL-bound natural substrates.49 In this respect, IbpB and GroEL, respectively as a member of sHSP and Hsp60 families,51 may not be functionally overlapped in protecting the large proteins in E. coli cells. In support of this, IbpB was repeatedly observed to functionally cooperate with DnaK (Hsp70 family) and ClpB (Hsp100 family) by transferring substrates to these ATP-dependent chaperones6–56 but not to GroEL/GroES.6
Figure 1.
IbpB and Hsp20.2 preferentially bind to proteins of high molecular weight or moderate acidity. Percentage of proteins plotted against the indicated molecular weight (Panels a and b) or isoelectric point (Panels c and d) for the E. coli proteome and the 110 substrate proteins of IbpB (Panels a and c), as well as for the D. radiodurans proteome and the 89 substrate proteins of Hsp20.2 ((Panels b and d). The levels of statistical significance are indicated by “*” (P < 0.05), “**” (P < 0.01), and “***” (P < 0.001) in all the figures of the paper.
Similarly, Hsp20.2 also preferentially bind to large proteins but unfavorably to small proteins [Fig. 1(b)], albeit at lower significant levels than does IbpB. Together, such favorable binding to large proteins but unfavorable to small proteins for both IbpB and Hsp20.2 may reflect the general observation that large proteins are more difficult to fold and more prone to misfolding/aggregation during folding/unfolding than small proteins (as reviewed in the Ref.51), thus need more protection by sHSPs.
GroEL was reported to have no any preference to substrates with respect to the isoelectric point (pI).49 By contrast, the pI distribution analysis revealed that IbpB preferentially binds to moderate acidic proteins with a pI of 5.0–5.5 [P < 0.001, Fig. 1(c)], and with a less degree to the proteins with a pI of 5.5–6.0 (P < 0.05). In comparison, moderate basic proteins with a pI of 7.0–10.0 are strongly un-favored for IbpB (P < 0.01). These observations, together with the difference in the MW distribution of their substrates, suggest that GroEL and IbpB are truly not functionally overlapped. Similarly with IbpB, Hsp20.2 shows a significant preference to substrates with a pI of 5.0–5.5 (P < 0.05) but unfavorable binding to substrates with a pI of 7.0–9.0 [Fig. 1(d)].
Both IbpB and Hsp20.2 exhibit preferences to substrate proteins of abundant charged residues rather than abundant hydrophobic residues
The favorable binding to the negatively charged acidic proteins for both IbpB and Hsp20.2, as revealed in Figures 1(c,d), is well consistent with the results of amino acid composition analysis [Fig. 2(a,b)]. Specifically, both IbpB and Hsp20.2 show significant preferential binding to substrates with a middle-to-high percentage of negatively charged residues (10–14%) but unfavorable to those with a low percentage (<8%) of these residues. Interestingly, we also found that both IbpB and Hsp20.2 exhibit significant unfavorable binding to proteins with a low percentage (<10%) of positively charged residues [P < 0.001; Fig. 2(c,d)]. In addition, Hsp20.2 exhibits a remarkable preference to proteins with a high percentage (18–22%, P < 0.001; >22%, P < 0.01) of positively charged residues [Fig. 2(d)], which should be attributed, at least partially, to the enrichment of ribosomal proteins in the substrate pool of Hsp20.2 (referring to the Ref.22). IbpB shows certain preference to proteins with a middle percentage (11–13% and 14–15%) of positively charged residues [Fig. 2(c)].
Figure 2.
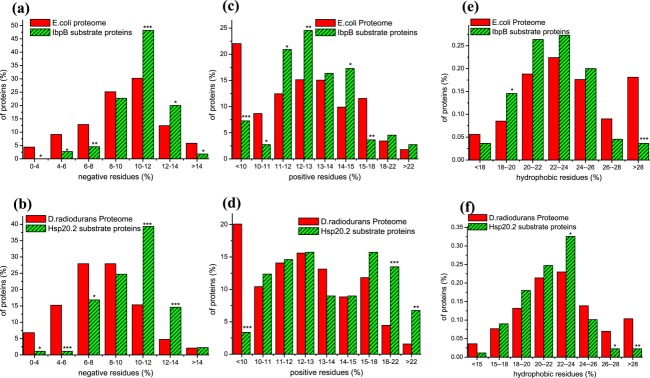
The substrates of IbpB and Hsp20.2 contain abundant charged residues but not hydrophobic residues Percentage of proteins plotted against the indicated content of negatively charged (Asp and Glu; Panels a and b), positively charged (Lys, Arg and His; Panels c and d) and aliphatic hydrophobic residues (Val, Leu, Ile; Panels e and f) for the E. coli proteome and IbpB substrate proteins (Panels a, c and e), as well as for the D. radiodurans proteome and Hsp20.2 substrate proteins ((Panels b, d, and f).
In light of these observations, together with the pI values of IbpB and Hsp20.2 themselves being respectively 5.2 and 5.0, we suggest that the negatively and positively charged residues abundantly present in the substrates of IbpB and Hsp20.2 may directly participate in the sHSP-substrate interactions through ionic interactions under neutral pH conditions. In support of this, we recently revealed the significantly enriched presence of the charged substrate-binding residues in IbpB, that is, among the 48 substrate-binding residues of IbpB, nine are positively charged and four negatively charged.24 In addition, sHSPs were reported to be unable to suppress the thermal protein aggregations under acidic pH,57–58 which apparently reflects the bleaching of ionic interactions between sHSPs and substrates under such acidic pH conditions.
Solvent-exposure of non-native hydrophobic surfaces in proteins is widely considered as a critical factor for protein aggregation.59 In line with this, hydrophobic interactions are considered to dominantly participate in the sHSP-substrate interaction as revealed by using hydrophobic probes under in vitro conditions.45–64 However, we unexpectedly found that both IbpB and Hsp20.2 do not prefer for substrate proteins with a high percentage of hydrophobic residues, including aliphatic residues [Leu, Ile, and Val; Fig. 2(e,f)] and aromatic residues [Trp, Phe, and Tyr; Fig. S1]. These observations thus indicate that the hydrophobic interactions between sHSPs and substrates may not be dominant as generally believed.45–63 Rather, other types of non-covalent interactions (e.g., ionic interactions as described above) also play crucial roles.
Other notable observations
Secondary structure elements
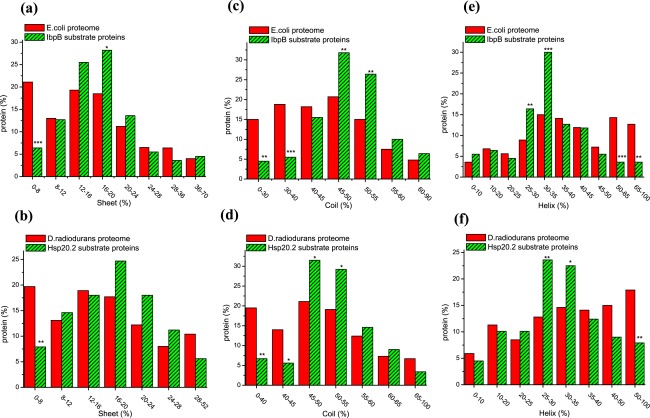
It would be of significance to unravel the features of the substrate proteins at the level of secondary structures, given that they are characterized by native-like secondary structures when complexed with sHSPs.30–34 For this purpose, secondary structure predictions were performed on the E. coli and D. radiodurans proteomes using the algorithm of PSIPRED. It should be pointed out that such prediction is not as precise as the calculation of molecular weight and amino acid composition of proteins as describe above, and that the prediction results would somehow depend on the prediction algorithm that we used. Data indicate that both IbpB and Hsp20.2 unfavorably bind to substrate proteins with a low percentage of β-sheets [Fig. 3(a,b)] or coils [Fig. 3(c,d)], or those with a high percentage of α-helixes [Fig. 3(e,f)].
Figure 3.
Both IbpB and Hsp20.2 unfavorably bind to substrate proteins of low percentage of β-sheets or coils but to those of high percentage of α-helixes Percentage of proteins plotted against the indicated content of secondary structural elements (β-sheet, coil and α-helix) for the E. coli proteome and IbpB substrate proteins (Panels a, c, and e), as well as for the D. radiodurans proteins and Hsp20.2 substrate proteins (Panels b, d, and f).
These observations may be explained as follows. First, the proteins of low percentage of coils may be more likely structurally folded than those of high percentage of coils, and are thus unfavorably bound by sHSPs. Second, the enrichment of β-sheets in both sHSPs27–28 and the substrates suggests a possibility that, intermolecular β-sheets may be prevalently formed between sHSPs and substrates and even play crucial for the substrate recognition and binding of sHSPs. In support of this, sHSPs were repeatedly reported to suppress the fibril formation of amyloid proteins that are enriched in β-sheets (as extensively reviewed65). It follows that the proteins of low percentage of β-sheets are unfavorably bound by sHSPs, as we observed [Fig. 3(a,b)]. Third, the apparent unfavorable binding of sHSPs to substrates of high percentage of α-helixes may not be biologically relevant. Rather, it is just numerically obtained during prediction using the PSIPRED for secondary structure prediction, i.e., the combined content of α-helixes, coils and β-sheets in a protein is always taken as 100%.
SCOP
Given the preference of IbpB and Hsp20.2 for substrate proteins with characteristic primary and secondary structures, we thought to examine whether these substrates contain common folds. We focused on the IbpB substrates with known 3D structures or with homologues of known structures, using the protein domain-classification databases SCOP.66 The structural classification of proteins in the E. coli proteome was directly adopted from the SCOP database. Data as displayed in Figure 4 indicate that IbpB significantly preferentially binds to substrate proteins of multi-domain (P < 0.05), which are defined as those with domains of different classes.66 This may reflect that the multi-domain proteins are more prone to aggregate during folding/unfolding than the single-domain proteins. In addition, this finding is in accordance with the substrate preference of IbpB to proteins of high MW (>70 kDa) [Fig. 1(a)]. Notably, IbpB was found to unfavorably bind to substrate proteins of α + β fold, which are defined as those in which α-helices and β-strands are largely segregated.66 Nevertheless, why this protein fold is unfavorably bound by IbpB cannot be explained based on our current understanding on proteins structures.
Figure 4.
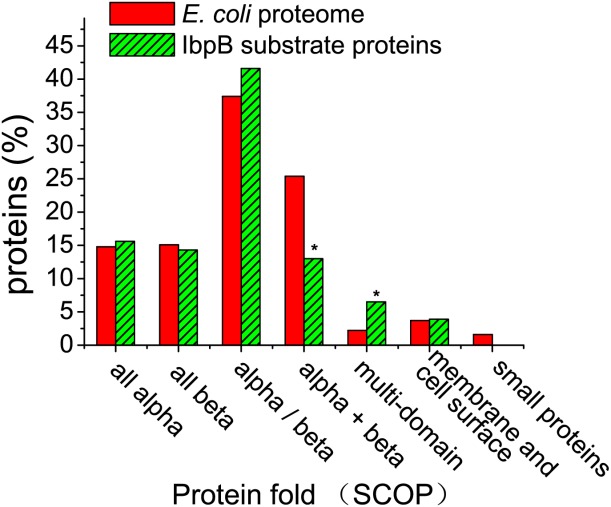
IbpB preferentially binds to substrate proteins of multi-domain percentage of proteins plotted against the indicated classes for the IbpB substrate proteins with known 3-D structures or with homologues of known structures according to the protein domain-classification databases SCOP.66 A total of 73 IbpB substrate proteins and 1274 E. coli proteins were assigned with a SCOP protein class.
Conclusion and Perspective
The nature of sHSP–substrate interactions is still far from clear. Here we report, for the first time, that certain structural characteristic elements are present in the natural substrates of IbpB and Hsp20.2. These elements may directly participate in the sHSP–substrate interaction. Alternatively, they may increase the aggregation tendency of substrates and thus make the substrates more preferentially bound by sHSPs, given that the model substrates of sHSPs were found to be aggregation-prone per se.30–34 Specifically, the preferential binding to proteins of high MW or multi-domain may reflect the fact that these substrates have higher tendency to aggregate than the proteins of low MW or single domain. Further, the presence of abundant charged residues, instead of abundant hydrophobic residues, in the substrates indicates the ionic interactions other than the hydrophobic interactions play crucial roles in the sHSP–substrate interactions. In particular, the remarkable preferential binding of IbpB to acidic proteins with a pI of 5.0–5.5 apparently reflects the ionic interactions formed between the negatively charged residues of the substrates and the positively charged substrate-binding residues in IbpB. Although the genome-predicted proteome for D. radiodurans was used as a reference in comparative studies, the similarity of the 89 Hsp20.2 substrates with the 110 IbpB substrates in terms of multilevel structural characteristics suggests that this compromise does not severely interfere with the analysis.
Since there is no report on the substrate profiling of sHSPs of archaea or eukaryotes at present, whether their natural substrate proteins show similar characteristic structural features, as we observed here on the substrates of bacterial sHSPs, is unknown and merits further explorations. In addition, whether the structural elements at other levels (e.g., the tertiary and quaternary structures) are preferentially or unfavorably present in the substrates of sHSPs also needs to be investigated. On the other hand, characterization of the structural features of the model substrate proteins under in vitro conditions still represents an important way to probe the mechanism of substrate recognition and interaction of sHSPs.
Materials and Methods
Data source
A total of 2709 E. coli proteins, which have been demonstrated to be expressed at protein levels, were downloaded from the UnitProtKB database (date of 2013/7/3). A total of 3167 D. radiodurans proteins (predicted by the genome sequence) were downloaded from the NCBI database. The IbpB substrate proteins were adopted from our earlier study,26 and the Hsp20.2 substrate proteins were from the earlier report22 after removing five potential functional partners from a total of 94 Hsp20.2-interacting proteins.
Bioinformatics analysis
The MW and pI of each protein were calculated using the Compute pI/Mw software (http://web.expasy.org/compute_pi/). Amino acid composition analysis for all proteins was performed using the BioEdit software. Secondary structure prediction was performed using PSIPRED67 by subjecting the amino acid sequences to the algorithm installed in a local super-computer. The protein class of E. coli proteome was adopted directly from the SCOP database (http://scop.mrc-lmb.cam.ac.uk/scop/) and that of IbpB substrate proteins was referred to the records of each protein deposited in the UnitProtKB database.
Statistics
Statistics was performed in Microsoft Excel software using the binominal distribution or u-test. Significance levels (with P values being less than 0.05, 0.01, or 0.001) were indicated in Figures 1, 2, 3, 4, and Supporting Information Figure S1.
Glossary
- sHSPs
small heat shock proteins; MW, molecular weight; pI, isoelectric point.
Additional Supporting Information may be found in the online version of this article.
References
- de Jong WW, Leunissen JA, Voorter CE. Evolution of the alpha-crystallin/small heat-shock protein family. Mol Biol Evol. 1993;10:103–126. doi: 10.1093/oxfordjournals.molbev.a039992. [DOI] [PubMed] [Google Scholar]
- Horwitz J. Alpha-crystallin can function as a molecular chaperone. Proc Natl Acad Sci USA. 1992;89:10449–10453. doi: 10.1073/pnas.89.21.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob U, Gaestel M, Engel K, Buchner J. Small heat shock proteins are molecular chaperones. J BIol Chem. 1993;268:1517–1520. [PubMed] [Google Scholar]
- Torok Z, Goloubinoff P, Horvath I, Tsvetkova NM, Glatz A, Balogh G, Varvasovszki V, Los DA, Vierling E, Crowe JH, Vigh L. Synechocystis HSP17 is an amphitropic protein that stabilizes heat-stressed membranes and binds denatured proteins for subsequent chaperone-mediated refolding. Proc Natl Acad Sci USA. 2001;98:3098–3103. doi: 10.1073/pnas.051619498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkova NM, Horvath I, Torok Z, Wolkers WF, Balogi Z, Shigapova N, Crowe LM, Tablin F, Vierling E, Crowe JH, Vigh L. Small heat-shock proteins regulate membrane lipid polymorphism. Proc Natl Acad Sci USA. 2002;99:13504–13509. doi: 10.1073/pnas.192468399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veinger L, Diamant S, Buchner J, Goloubinoff P. The small heat-shock protein IbpB from Escherichia coli stabilizes stress-denatured proteins for subsequent refolding by a multichaperone network. J Biol Chem. 1998;273:11032–11037. doi: 10.1074/jbc.273.18.11032. [DOI] [PubMed] [Google Scholar]
- Lee GJ, Roseman AM, Saibil HR, Vierling E. A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J. 1997;16:659–671. doi: 10.1093/emboj/16.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrnsperger M, Graber S, Gaestel M, Buchner J. Binding of non-native protein to Hsp25 during heat shock creates a reservoir of folding intermediates for reactivation. EMBO J. 1997;16:221–229. doi: 10.1093/emboj/16.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh CH, Chang PF, Yeh KW, Lin WC, Chen YM, Lin CY. Expression of a gene encoding a 16.9-kDa heat-shock protein, Oshsp16.9, in Escherichia coli enhances thermotolerance. Proc Natl Acad Sci USA. 1997;94:10967–10972. doi: 10.1073/pnas.94.20.10967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Matsumura Y, Tsuchido T. Small heat shock proteins, IbpA and IbpB, are involved in resistances to heat and superoxide stresses in Escherichia coli. FEMS Microbiol Lett. 2000;184:165–171. doi: 10.1111/j.1574-6968.2000.tb09009.x. [DOI] [PubMed] [Google Scholar]
- Landry J, Chretien P, Lambert H, Hickey E, Weber LA. Heat shock resistance conferred by expression of the human HSP27 gene in rodent cells. J Cell Biol. 1989;109:7–15. doi: 10.1083/jcb.109.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favet N, Duverger O, Loones MT, Poliard A, Kellermann O, Morange M. Overexpression of murine small heat shock protein HSP25 interferes with chondrocyte differentiation and decreases cell adhesion. Cell Death Differ. 2001;8:603–613. doi: 10.1038/sj.cdd.4400847. [DOI] [PubMed] [Google Scholar]
- Salinthone S, Ba M, Hanson L, Martin JL, Halayko AJ, Gerthoffer WT. Overexpression of human Hsp27 inhibits serum-induced proliferation in airway smooth muscle myocytes and confers resistance to hydrogen peroxide cytotoxicity. Am J Physiol Lung Cell Mol Physiol. 2007;293:L1194–L1207. doi: 10.1152/ajplung.00453.2006. [DOI] [PubMed] [Google Scholar]
- Morrow G, Battistini S, Zhang P, Tanguay RM. Decreased lifespan in the absence of expression of the mitochondrial small heat shock protein Hsp22 in Drosophila. J Biol Chem. 2004;279:43382–43385. doi: 10.1074/jbc.C400357200. [DOI] [PubMed] [Google Scholar]
- Laskowska E, Matuszewska E, Kuczynska-Wisnik D. Small heat shock proteins and protein-misfolding diseases. Curr Pharm Biotechnol. 2010;11:146–157. doi: 10.2174/138920110790909669. [DOI] [PubMed] [Google Scholar]
- Richards EH, Hickey E, Weber L, Master JR. Effect of overexpression of the small heat shock protein HSP27 on the heat and drug sensitivities of human testis tumor cells. Cancer Res. 1996;56:2446–2451. [PubMed] [Google Scholar]
- Hollander JM, Martin JL, Belke DD, Scott BT, Swanson E, Krishnamoorthy V, Dillmann WH. Overexpression of wild-type heat shock protein 27 and a nonphosphorylatable heat shock protein 27 mutant protects against ischemia/reperfusion injury in a transgenic mouse model. Circulation. 2004;110:3544–3552. doi: 10.1161/01.CIR.0000148825.99184.50. [DOI] [PubMed] [Google Scholar]
- Litt M, Kramer P, LaMorticella DM, Murphey W, Lovrien EW, Weleber RG. Autosomal dominant congenital cataract associated with a missense mutation in the human alpha crystallin gene CRYAA. Hum Mol Genet. 1998;7:471–474. doi: 10.1093/hmg/7.3.471. [DOI] [PubMed] [Google Scholar]
- Vicart P, Caron A, Guicheney P, Li Z, Prevost MC, Faure A, Chateau D, Chapon F, Tome F, Dupret JM, Paulin D, Fardeau M. A missense mutation in the alphaB-crystallin chaperone gene causes a desmin-related myopathy. Nat Genet. 1998;20:92–95. doi: 10.1038/1765. [DOI] [PubMed] [Google Scholar]
- Irobi J, Van Impe K, Seeman P, Jordanova A, Dierick I, Verpoorten N, Michalik A, De Vriendt E, Jacobs A, Van Gerwen V, Vennekens K, Mazanec R, Tournev I, Hilton-Jones D, Talbot K, Kremensky I, Van Den Bosch L, Robberecht W, Van Vandekerckhove J, Van Broeckhoven C, Gettemans J, De Jonghe P, Timmerman V. Hot-spot residue in small heat-shock protein 22 causes distal motor neuropathy. Nat Genet. 2004;36:597–601. doi: 10.1038/ng1328. [DOI] [PubMed] [Google Scholar]
- Evgrafov OV, Mersiyanova I, Irobi J, Van Den Bosch L, Dierick I, Leung CL, Schagina O, Verpoorten N, Van Impe K, Fedotov V, Dadali E, Auer-Grumbach M, Windpassinger C, Wagner K, Mitrovic Z, Hilton-Jones D, Talbot K, Martin JJ, Vasserman N, Tverskaya S, Polyakov A, Liem RK, Gettemans J, Robberecht W, De Jonghe P, Timmerman V. Mutant small heat-shock protein 27 causes axonal Charcot-Marie-Tooth disease and distal hereditary motor neuropathy. Nat Genet. 2004;36:602–606. doi: 10.1038/ng1354. [DOI] [PubMed] [Google Scholar]
- Bepperling A, Alte F, Kriehuber T, Braun N, Weinkauf S, Groll M, Haslbeck M, Buchner J. Alternative bacterial two-component small heat shock protein systems. Proc Natl Acad Sci USA. 2012;109:20407–20412. doi: 10.1073/pnas.1209565109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basha E, Lee GJ, Breci LA, Hausrath AC, Buan NR, Giese KC, Vierling E. The identity of proteins associated with a small heat shock protein during heat stress in vivo indicates that these chaperones protect a wide range of cellular functions. J Biol Chem. 2004;279:7566–7575. doi: 10.1074/jbc.M310684200. [DOI] [PubMed] [Google Scholar]
- Fu X, Shi X, Yin L, Liu J, Joo K, Lee J, Chang Z. Small heat shock protein IbpB acts as a robust chaperone in living cells by hierarchically activating its multi-type substrate-binding residues. J Biol Chem. 2013;288:11897–11906. doi: 10.1074/jbc.M113.450437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R, Kim KK, Yokota H, Kim SH. Small heat shock protein of Methanococcus jannaschii, a hyperthermophile. Proc Natl Acad Sci USA. 1998;95:9129–9133. doi: 10.1073/pnas.95.16.9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Shi X, Yan L, Zhang H, Chang Z. In vivo substrate diversity and preference of small heat shock protein IbpB as revealed by using a genetically incorporated photo-cross-linker. J Biol Chem. 2013;288:31646–31654. doi: 10.1074/jbc.M113.501817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KK, Kim R, Kim SH. Crystal structure of a small heat-shock protein. Nature. 1998;394:595–599. doi: 10.1038/29106. [DOI] [PubMed] [Google Scholar]
- Van Montfort RL, Basha E, Friedrich KL, Slingsby C, Vierling E. Crystal structure and assembly of a eukaryotic small heat shock protein. Nat Struct Biol. 2001;8:1025–1030. doi: 10.1038/nsb722. [DOI] [PubMed] [Google Scholar]
- Jaya N, Garcia V, Vierling E. Substrate binding site flexibility of the small heat shock protein molecular chaperones. Proc Natl Acad Sci USA. 2009;106:15604–15609. doi: 10.1073/pnas.0902177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das KP, Choo-Smith LP, Petrash JM, Surewicz WK. Insight into the secondary structure of non-native proteins bound to a molecular chaperone alpha-crystallin. An isotope-edited infrared spectroscopic study. J Biol Chem. 1999;274:33209–33212. doi: 10.1074/jbc.274.47.33209. [DOI] [PubMed] [Google Scholar]
- Das KP, Petrash JM, Surewicz WK. Conformational properties of substrate proteins bound to a molecular chaperone alpha-crystallin. J Biol Chem. 1996;271:10449–10452. doi: 10.1074/jbc.271.18.10449. [DOI] [PubMed] [Google Scholar]
- Lindner RA, Kapur A, Carver JA. The interaction of the molecular chaperone, alpha-crystallin, with molten globule states of bovine alpha-lactalbumin. J Biol Chem. 1997;272:27722–27729. doi: 10.1074/jbc.272.44.27722. [DOI] [PubMed] [Google Scholar]
- Rawat U, Rao M. Interactions of chaperone alpha-crystallin with the molten globule state of xylose reductase. Implications for reconstitution of the active enzyme. J Biol Chem. 1998;273:9415–9423. doi: 10.1074/jbc.273.16.9415. [DOI] [PubMed] [Google Scholar]
- Sathish HA, Stein RA, Yang G, McHaourab HS. Mechanism of chaperone function in small heat-shock proteins. Fluorescence studies of the conformations of T4 lysozyme bound to alphaB-crystallin. J Biol Chem. 2003;278:44214–44221. doi: 10.1074/jbc.M307578200. [DOI] [PubMed] [Google Scholar]
- Carver JA, Lindner RA, Lyon C, Canet D, Hernandez H, Dobson CM, Redfield C. The interaction of the molecular chaperone alpha-crystallin with unfolding alpha-lactalbumin: a structural and kinetic spectroscopic study. J Mol Biol. 2002;318:815–827. doi: 10.1016/S0022-2836(02)00144-4. [DOI] [PubMed] [Google Scholar]
- Carver JA, Lindner RA. NMR spectroscopy of alpha-crystallin. Insights into the structure, interactions and chaperone action of small heat-shock proteins. Int J BIol Macromol. 1998;22:197–209. doi: 10.1016/s0141-8130(98)00017-8. [DOI] [PubMed] [Google Scholar]
- Carver JA, Guerreiro N, Nicholls KA, Truscott RJ. On the interaction of alpha-crystallin with unfolded proteins. Biochim Biophys Acta. 1995;1252:251–260. doi: 10.1016/0167-4838(95)00146-l. [DOI] [PubMed] [Google Scholar]
- Treweek TM, Lindner RA, Mariani M, Carver JA. The small heat-shock chaperone protein, alpha-crystallin, does not recognize stable molten globule states of cytosolic proteins. Biochim Biophys Acta. 2000;1481:175–188. doi: 10.1016/s0167-4838(00)00109-6. [DOI] [PubMed] [Google Scholar]
- Farahbakhsh ZT, Huang QL, Ding LL, Altenbach C, Steinhoff HJ, Horwitz J, Hubbell WL. Interaction of alpha-crystallin with spin-labeled peptides. Biochemistry. 1995;34:509–516. doi: 10.1021/bi00002a015. [DOI] [PubMed] [Google Scholar]
- Lindner RA, Treweek TM, Carver JA. The molecular chaperone alpha-crystallin is in kinetic competition with aggregation to stabilize a monomeric molten-globule form of alpha-lactalbumin. Biochem J. 2001;354:79–87. doi: 10.1042/0264-6021:3540079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin GL, Carver JA, Bottomley SP. The selective inhibition of serpin aggregation by the molecular chaperone, alpha-crystallin, indicates a nucleation-dependent specificity. J Biol Chem. 2003;278:48644–48650. doi: 10.1074/jbc.M308376200. [DOI] [PubMed] [Google Scholar]
- Stengel F, Baldwin AJ, Bush MF, Hilton GR, Lioe H, Basha E, Jaya N, Vierling E, Benesch JL. Dissecting heterogeneous molecular chaperone complexes using a mass spectrum deconvolution approach. Chem Biol. 2012;19:599–607. doi: 10.1016/j.chembiol.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L, Abulimiti A, Li W, Chang Z. Monodisperse Hsp16.3 nonamer exhibits dynamic dissociation and reassociation, with the nonamer dissociation prerequisite for chaperone-like activity. J Mol Biol. 2002;319:517–526. doi: 10.1016/S0022-2836(02)00311-X. [DOI] [PubMed] [Google Scholar]
- Chang Z, Primm TP, Jakana J, Lee IH, Serysheva I, Chiu W, Gilbert HF, Quiocho FA. Mycobacterium tuberculosis 16-kDa antigen (Hsp16.3) functions as an oligomeric structure in vitro to suppress thermal aggregation. J Biol Chem. 1996;271:7218–7223. [PubMed] [Google Scholar]
- Fu X, Zhang H, Zhang X, Cao Y, Jiao W, Liu C, Song Y, Abulimiti A, Chang Z. A dual role for the N-terminal region of Mycobacterium tuberculosis Hsp16.3 in self-oligomerization and binding denaturing substrate proteins. J Biol Chem. 2005;280:6337–6348. doi: 10.1074/jbc.M406319200. [DOI] [PubMed] [Google Scholar]
- Jiao W, Qian M, Li P, Zhao L, Chang Z. The essential role of the flexible termini in the temperature-responsiveness of the oligomeric state and chaperone-like activity for the polydisperse small heat shock protein IbpB from Escherichia coli. J Mol Biol. 2005;347:871–884. doi: 10.1016/j.jmb.2005.01.029. [DOI] [PubMed] [Google Scholar]
- Jiao W, Hong W, Li P, Sun S, Ma J, Qian M, Hu M, Chang Z. The dramatically increased chaperone activity of small heat-shock protein IbpB is retained for an extended period of time after the stress condition is removed. Biochem J. 2008;410:63–70. doi: 10.1042/BJ20071120. [DOI] [PubMed] [Google Scholar]
- Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, Davey M, Parkinson J, Greenblatt J, Emili A. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- Houry WA, Frishman D, Eckerskorn C, Lottspeich F, Hartl FU. Identification of in vivo substrates of the chaperonin GroEL. Nature. 1999;402:147–154. doi: 10.1038/45977. [DOI] [PubMed] [Google Scholar]
- Brinker A, Pfeifer G, Kerner MJ, Naylor DJ, Hartl FU, Hayer-Hartl M. Dual function of protein confinement in chaperonin-assisted protein folding. Cell. 2001;107:223–233. doi: 10.1016/s0092-8674(01)00517-7. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Mogk A, Deuerling E, Vorderwulbecke S, Vierling E, Bukau B. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol Microbiol. 2003;50:585–595. doi: 10.1046/j.1365-2958.2003.03710.x. [DOI] [PubMed] [Google Scholar]
- Thomas JG, Baneyx F. Roles of the Escherichia coli small heat shock proteins IbpA and IbpB in thermal stress management: comparison with ClpA, ClpB, and HtpG In vivo. J Bacteriol. 1998;180:5165–5172. doi: 10.1128/jb.180.19.5165-5172.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuszewska M, Kuczynska-Wisnik D, Laskowska E, Liberek K. The small heat shock protein IbpA of Escherichia coli cooperates with IbpB in stabilization of thermally aggregated proteins in a disaggregation competent state. J Biol Chem. 2005;280:12292–12298. doi: 10.1074/jbc.M412706200. [DOI] [PubMed] [Google Scholar]
- Ratajczak E, Zietkiewicz S, Liberek K. Distinct activities of Escherichia coli small heat shock proteins IbpA and IbpB promote efficient protein disaggregation. J Mol Biol. 2009;386:178–189. doi: 10.1016/j.jmb.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Cashikar AG, Duennwald M, Lindquist SL. A chaperone pathway in protein disaggregation. Hsp26 alters the nature of protein aggregates to facilitate reactivation by Hsp104. J Biol Chem. 2005;280:23869–23875. doi: 10.1074/jbc.M502854200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitre M, Weidmann S, Rieu A, Fenel D, Schoehn G, Ebel C, Coves J, Guzzo J. The oligomer plasticity of the small heat-shock protein Lo18 from Oenococcus oeni influences its role in both membrane stabilization and protein protection. Biochem J. 2012;444:97–104. doi: 10.1042/BJ20120066. [DOI] [PubMed] [Google Scholar]
- Bukach OV, Seit-Nebi AS, Marston SB, Gusev NB. Some properties of human small heat shock protein Hsp20 (HspB6) Eur J Biochem. 2004;271:291–302. doi: 10.1046/j.1432-1033.2003.03928.x. [DOI] [PubMed] [Google Scholar]
- De Bernardez Clark E, Schwarz E, Rudolph R. Inhibition of aggregation side reactions during in vitro protein folding. Methods Enzymol. 1999;309:217–236. doi: 10.1016/s0076-6879(99)09017-5. [DOI] [PubMed] [Google Scholar]
- Benesch JL, Ayoub M, Robinson CV, Aquilina JA. Small heat shock protein activity is regulated by variable oligomeric substructure. J Biol Chem. 2008;283:28513–28517. doi: 10.1074/jbc.M804729200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs M, Poirier DJ, Seguin SJ, Lambert H, Carra S, Charette SJ, Landry J. Identification of the key structural motifs involved in HspB8/HspB6-Bag3 interaction. Biochem J. 2010;425:245–255. doi: 10.1042/BJ20090907. [DOI] [PubMed] [Google Scholar]
- Sharma KK, Kaur H, Kumar GS, Kester K. Interaction of 1,1'-bi(4-anilino)naphthalene-5,5'-disulfonic acid with alpha-crystallin. J Biol Chem. 1998;273:8965–8970. doi: 10.1074/jbc.273.15.8965. [DOI] [PubMed] [Google Scholar]
- Sharma KK, Kumar GS, Murphy AS, Kester K. Identification of 1,1'-bi(4-anilino)naphthalene-5,5'-disulfonic acid binding sequences in alpha-crystallin. J Biol Chem. 1998;273:15474–15478. doi: 10.1074/jbc.273.25.15474. [DOI] [PubMed] [Google Scholar]
- Fu X, Chang Z. Identification of bis-ANS binding sites in Mycobacterium tuberculosis small heat shock protein Hsp16.3: evidences for a two-step substrate-binding mechanism. Biochem Biophys Res Commun. 2006;349:167–171. doi: 10.1016/j.bbrc.2006.08.036. [DOI] [PubMed] [Google Scholar]
- Ecroyd H, Carver JA. Crystallin proteins and amyloid fibrils. Cell Mol Life Sci. 2009;66:62–81. doi: 10.1007/s00018-008-8327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.