Abstract
Objective
Easy tool for newborn screening of Gaucher and Hurler diseases.
Methods
Method comparison between fluorometric enzymatic activity assay on a digital microfluidic platform and micro-titer plate bench assay was performed on normal (n=100), Gaucher (n=10) and Hurler (n=7) dried blood spot samples.
Results
Enzymatic activity analysis of glucocerebrosidase (Gaucher) and α-L-iduronidase (Hurler) revealed similar discrimination between normal and affected samples on both platforms.
Conclusions
Digital microfluidics is suitable for Gaucher and Hurler newborn screening.
Keywords: Newborn screening, lysosomal storage disease, digital microfluidics, dried blood spot, Gaucher disease, Hurler disease
1. Introduction
Enzyme replacement therapies are now available for certain lysosomal storage diseases (LSD) [1], fueling the development of rapid and inexpensive multiplex technologies for newborn screening (NBS) using a single dried blood spot (DBS) [2]. Current technologies for LSD screening include micro-titer plate fluorometry [3–5], tandem mass spectrometry [6], and immunoassays [7].
We published a novel method for screening Pompe, Fabry [8] and Hunter diseases [9] from DBS using digital microfluidic fluorometry (DMF) with comparable results to standard micro-titer plate bench fluorometry (MTPF). Here, we present demonstration of assays for two additional LSDs: Gaucher (glucocerebrosidase (GBA) deficiency) and Hurler diseases (mucopolysaccharidosis type I, α-iduronidase (IDU) deficiency) [10]. We demonstrate performance of a digital microfluidic system for LSD NBS through a method comparison for GBA (EC 3.2.1.21) and IDU (EC 3.2.1.76) enzyme activities with “gold standard” micro-titer plate fluorometric assays performed at Duke University Biochemical Genetics Laboratory (BGL). The method comparison studies illustrate the potential of the DMF platform to screen for LSDs; a pilot study conducted using an early prototype of a digital microfluidic cartridge designed by Advanced Liquid Logic (ALL) for a tri-plex assay (Pompe, Fabry, and Gaucher) resulted in two confirmed cases of Gaucher from a total of 8,012 DBS samples [11].
2. Methods
2.1 Micro-Titer Plate Fluorometric Enzyme Assay Protocols
2.1.1 Gaucher
The protocol to determine enzymatic activity for GBA using the MTPF method was originally developed by Chamoles et al. [3] and later modified [5]. Assays were performed on individual 3 mm punches from normal and affected DBS at Duke BGL essentially as described [5]. Briefly, the DBS was reconstituted in 200 µL extraction buffer for one hour at room temperature (see Supplemental Material Table 1). Then, 40 µL of DBS extract were added to 80 µL of substrate buffer containing 4-methylumbelliferyl β-D-glucopyranoside (4-MU-β-Gluc), both in the presence and absence of the β-glucosidase inhibitor conduritol B epoxide. Reactions were stopped by mixing with 100 µL stop buffer after incubation for 20 hours at 37 °C, and end point fluorescence was measured. Enzymatic activity was reported by subtracting inhibited activity from total enzymatic activity.
2.1.2 Hurler
The MTPF method to determine enzymatic activity of IDU was developed by Chamoles et al. [4]. Assays were performed on individual 3 mm punches from normal and affected DBS at Duke BGL, in the presence of the β-glucuronidase inhibitor D-saccharic acid 1,4 lactone. Briefly, the DBS was reconstituted in 160 µL extraction buffer for 30 minutes at room temperature (Supplemental Material Table 1). After centrifugation at 5,000 rpm for 1 minute, 20 µL of DBS extract were added to 10 µL of substrate buffer containing 4-methylumbelliferyl α-L-iduronide (4-MU- α-IDU), mixed, and incubated for 20 hours at 37 °C. Stop buffer (100 µl) was added, and end point fluorescence was measured. Enzymatic activities for GBA and IDU were calculated and reported as pmol/punch/hour of incubation, which was converted to μmol/L of blood/hour of incubation by dividing with the volume of blood (3.1 μL) in one DBS punch.
2.2 Digital Microfluidic Enzyme Assay Protocol (Gaucher and Hurler)
In contrast to MTPF, the extract from a single 3 mm DBS punch was used to perform enzymatic assays for both GBA and IDU on the DMF platform. These punches were taken from the same DBS samples that were analyzed using MTPF. Although DMF assays were performed using essentially similar reagents to those used for MTPF, there were substantial differences in reagent composition and assay conditions (Supplemental Material Table 1). DBS extracts were reconstituted in extraction buffer in standard deep-well 96-well plates and transferred to DMF cartridges using a multi-channel pipette. Hands-on time of less than 12 minutes to set up the DMF assay included time involved in punching and loading samples and reagents onto the cartridge. All subsequent fluid handling operations were automated on the cartridge.
The protocol to perform enzymatic analysis on the DMF cartridge was the same as previously described [8–9]. Briefly, reagent droplets (~100 nL) were dispensed and merged with sample droplets (~100 nL) to form reaction droplets (~200 nL) that were incubated for ~1 hour at 37 °C on a disposable digital microfluidic cartridge. After incubation, stop buffer droplets (~100 nL) were dispensed and merged with the reaction mixture. End point fluorescence was measured at 370 nm excitation and 460 nm emission; enzymatic activity was reported as µmoles of 4-methyl umbelliferone (4-MU) produced per liter of blood per hour of incubation using a 4-MU calibration curve generated on the same cartridge.
3. Results and Discussion
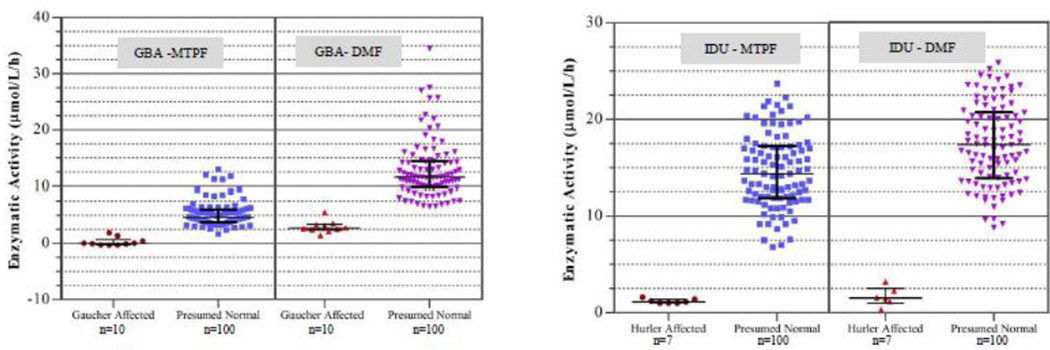
The comparison of assay results for Gaucher and Hurler diseases is summarized in Figure 1, which illustrates plots of enzymatic activity using the MTPF method at Duke BGL and DMF method at ALL for GBA and IDU. The horizontal black line represents the median and error bars represent standard deviation. For IDU, the enzymatic activity for all presumed normal and affected samples matched well between the two methods despite differences in reagents and incubation times. For GBA, the median enzymatic activity value obtained by DMF was two times higher than that obtained by MTPF. The reason for this may be partly due to the lack of an inhibitor in the DMF method and differences in extraction buffer composition. Despite these differences, separation between the highest affected sample and the lowest normal was 0.5 µmol/L/h using MTPF and 1 µmol/L/h using DMF for GBA, while it was 5.2 µmol/L/h and 5.6 µmol/L/h for IDU using MTPF and DMF methods, respectively. For the DMF assay, we were able to utilize a single extraction buffer and a single stop buffer (Supplemental Material Table 1) in order to minimize the number of reagents required for multiplex analysis [9].
Figure 1.
Enzymatic activity comparison for GBA and IDU between micro-titer plate fluorometry (Duke BGL) and digital microfluidics (ALL) methods. The same set of normal (n=100) and affected (GBA: n=10; IDU: n=7) DBS samples were used in both methods. The horizontal black line in the middle represents the median while the error bars represent standard deviation.
4. Conclusions
We successfully demonstrated that enzymatic activity results for GBA and IDU obtained from a DMF assay were comparable to those obtained by a “gold standard” MTPF assay. These results, in conjunction with previous comparison studies for Pompe, Fabry, and Hunter diseases [8–9], provide evidence that digital microfluidic enzymatic assays are a viable method for multiplex screening of lysosomal storage diseases.
Supplementary Material
Highlights.
Digital microfluidic enzymatic analysis of GBA and IDU comparable to micro-titer plate fluorometry.
Multiplex enzymatic assays are completed from a single dried blood spot punch.
Digital microfluidics is suitable for newborn screening of Gaucher and Hurler diseases.
Acknowledgements
We acknowledge Dr. Shu Chaing (North Carolina Division of Public Health) for providing de-identified normal DBS. Research reported in this publication was partly supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health under award number R44HD057713. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Non Standard Abbreviations
- LSD
Lysosomal storage diseases
- NBS
Newborn screening
- DBS
Dried blood spot
- DMF
Digital microfluidic platform
- MTPF
Micro-titer plate based fluorometric method
- GBA
Acid β-D-glucosidase
- IDU
Acid α-L-iduronidase
- BGL
Duke Biochemical Genetics Laboratory
- ALL
Advanced Liquid Logic, Inc.
- 4-MU- β-Gluc
4-methylumbelliferyl β-D-glucopyranoside
- 4-MU
4-methyl umbelliferone
- 4-MU- α-IDU
4-methylumbelliferyl α-L-iduronide
- EDTA
Ethylenediaminetetraacetic acid
- DMSO
Dimethyl sulfoxide
- EXT
Extraction buffer
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lim-Melia ER, Kronn DF. Current enzyme replacement therapy for the treatment of lysosomal storage diseases. Pediatr. Ann. 2009;38:448–455. doi: 10.3928/00904481-20090723-09. [DOI] [PubMed] [Google Scholar]
- 2.Zhou H, Fernhoff P, Vogt RF. Newborn bloodspot screening for lysosomal storage disorders. J Pediatr. 2011;159(1):7–13. doi: 10.1016/j.jpeds.2011.02.026. [DOI] [PubMed] [Google Scholar]
- 3.Chamoles NA, Blanco M, Gaggioli D, Casentini C. Gaucher and Niemann-Pick diseases--enzymatic diagnosis in dried blood spots on filter paper: retrospective diagnoses in newbornscreening cards. Clin. Chim. Acta. 2002;317(1–2):191–197. doi: 10.1016/s0009-8981(01)00798-7. [DOI] [PubMed] [Google Scholar]
- 4.Chamoles NA, Blanco MB, Gaggioli D, Casentini C. Hurler-like phenotype: enzymatic diagnosis in dried blood spots on filter paper. Clin. Chem. 2001;4(12):2098–2012. [PubMed] [Google Scholar]
- 5.Olivova P, Cullen E, Titlow M, Kallwass H, Barranger J, Zhang K, Keutzer J. An improved high-throughput dried blood spot screening method for Gaucher disease. Clin. Chim. Acta. 2008;398:163–164. doi: 10.1016/j.cca.2008.08.024. [DOI] [PubMed] [Google Scholar]
- 6.Mechtler TP, Metz TF, Muller HG, Ostermann K, Ratschmann R, De Jesus VR, Shushan B, Di Bussolo JM, Herman JL, Herkner KR, Kasper DC. Short-incubation mass spectrometry assay for lysosomal storage disorders in newborn and high-risk population screening. J. Chromat. B. 2012;908:9–17. doi: 10.1016/j.jchromb.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meikle PJ, Grasby DJ, Dean CJ, Lang DL, Bockmann M, Whittle AM, Fietz MJ, Simonsen H, Fuller M, Brooks DA, Hopwood JJ. Newborn screening for lysosomal storage disorders. Molecular Genetics and Metabolism. 2006;88:307–314. doi: 10.1016/j.ymgme.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Sista RS, Eckhardt AE, Wang T, Graham C, Rouse JL, Norton SM, Srinivasan V, Pollack MG, Tolun AA, Bali D, Millington DS, Pamula VK. Digital microfluidic platform for multiplexing enzyme assays: implications for lysosomal storage disease screening in newborns. Clin. Chem. 2011;57(10):1444–1451. doi: 10.1373/clinchem.2011.163139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sista R, Eckhardt AE, Wang T, Sellos-Moura M, Pamula VK. Rapid, single-step assay for Hunter syndrome in dried blood spots using digital microfluidics. Clin. Chim. Acta. 2011;412(19–20):1895–1897. doi: 10.1016/j.cca.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 10.Marsden D, Levy H. Newborn screening of lysosomal storage disorders. Clin. Chem. 2010;56:1071–1079. doi: 10.1373/clinchem.2009.141622. [DOI] [PubMed] [Google Scholar]
- 11.Burton B, Charrow J, Angle B, Widera S, Waggoner D. A pilot newborn screening program for lysosomal storage disorders (LSD) in Illinois. Molecular Genetics and Metabolism. 2012;105(2):S23–S24. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.