Abstract
Apolipoprotein AI (apoA-I) is the principal acceptor of lipids from ATP-binding cassette transporter A1, a process that yields nascent high density lipoproteins. Analysis of lipidated apoA-I conformation yields a belt or twisted belt in which two strands of apoA-I lie antiparallel to one another. In contrast, biophysical studies have suggested that a part of lipid-free apoA-I was arranged in a 4-helix bundle. To understand how lipid-free apoA-I opens from a bundle to a belt while accepting lipid it was necessary to have a more refined model for the conformation of lipid-free apoA-I. This study reports the conformation of lipid-free human apoA-I using lysine-to-lysine chemical cross-linking in conjunction with disulfide cross-linking achieved using selective cysteine mutations. After proteolysis cross-linked peptides were verified by sequencing using tandem mass spectrometry. The resulting structure is compact with roughly 4 helical regions, amino acids 44 through 186, bundled together. C- and N-terminal ends, amino acids 1-43 and 187-243, respectively, are folded such that they lie close to one another. An unusual feature of the molecule is the high degree of connectivity of lysine40 with 6 other lysines, lysines that are close, e.g., lysine59, to distant lysines, e.g., lysine239, that are at the opposite end of the primary sequence. These results are compared and contrasted with other reported conformations for lipid-free human apoA-I and an NMR study of mouse apoA-I.
Cholesterol efflux from cells is mediated by ATP-binding cassette transporter A1 (ABCA1) (1, 2). The primary recipient of the cholesterol and phospholipid is apolipoprotein A-I (apoA-I), which is converted into nascent HDL (nHDL). This lipid transfer process is the first step in reverse cholesterol transport (RCT) (3-5) through which cholesterol in peripheral tissues is transported to the liver for catabolism. ApoA-I has other, essential anti-inflammatory properties and may participate in modulating lipid raft levels on the cell membrane. Various apoA-I entities, e.g., lipid-free apoA-I, nHDL, and mature HDL, are affected with a relatively high degree of specificity by different enzymes, transporters and receptors at various stages of the RCT cycle. Therefore, it is likely that at each step apoA-I reorganizes so that it is uniquely recognized by modifying protein at different points in the RCT cycle.
ApoA-I is a well studied protein that has resisted crystallization. It is composed of 243 amino acids, which, after the first 43 amino acids, called the N-terminal segment, is divided into ten amphipathic helical regions, labeled 1 through 10. There are two 11-amino acid and eight 22-amino acid helices. Numerous studies of the biophysical characteristics of lipid-free apoA-I have suggested that it assumes a compact structure with the helices folded-back along one another. A recent NMR analysis of lipid-free mouse apoA-I yielded a structure having a four-helix bundle composed of what, in human apoA-I, would include the N-terminal end through helix 7 (6). In 1997 Borhani et al. (7) were able to crystallize an N-terminal truncated form of lipid-free apoA-I. The Borhani structure was roughly saddle shaped having many characteristics similar to those proposed for apoA-I on recombinant HDL (rHDL) particles. Smaller segments that often contain proline, located between helical regions, are bent or kinked (7). These kinks may help apoA-I assume a curved conformation. The article stirred new interest in the conformation of lipid-bound apoA-I. There was less interest in the in-solution conformation of intact, lipid-free apoA-I, but two papers were published in 2005 and 2006. The validity of the later paper has been questioned (8). However, the lipid-free solution structure proposed by Silva et al. (9) using chemical cross-linking combined with mass spectrometry and sequence threading has many of the features suggested by other biophysical studies.
Early studies of LCAT-deficient HDL suggested that the first-formed HDL, nascent HDL or nHDL, was a lipid disc (10) that carried two molecules of apoA-I. Jonas’s group perfected the synthesis of synthetic particles using cholate dialysis (11) and the properties of these particles have been studied for many years. Early models agreed that apoA-I was located on the edge of the disk, but disagreed on the conformation of apoA-I (12)(13). After Borhani et al. (7) showed the circular structure for crystalline apoA-I other studies began to confirm this arrangement in a non-crystalline matrix (14-19). One of the first lipidated apoA-Is studied by chemical cross-linking was synthetic, rHDL (rHDL) prepared from lipid-free apoA-I and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) (16, 17, 20). The two strands were anti-parallel, but paired in the 5,5’-region giving a conformation that is best described as a band or belt although one formulation has the belt twisted (21). In contrast the principle lipid-carrying particle formed by ABCA1 is an nHDL with three strands of apoA-I in a belt conformation with the 5,5’,5”-regions adjacent (22, 23). The composition of nHDL is unique in that it carries most of the cholesterol as free cholesterol, e.g., unesterified, like HDL from patients suffering from familial lecithin-cholesterol acyltransferase (LCAT) disease (24-26). In plasma carrying native LCAT nHDL is processed into mature HDL by conversion of free-cholesterol to cholesteryl ester by LCAT. Further modification takes place by the actions of cholesterol ester transfer protein, ATP-binding cassette transporter G1 (ABCG1), hepatic lipase, etc.
Because lipidated apoA-I on nHDL has a much different shape compared to lipid-free apoA-I the lipid-free protein must undergo a major conformational reorganization to accommodate lipid. This reorganization may well be an essential component of the lipidation process. Therefore, to understand how apoA-I changes shape to accept lipid the lipid-free conformation must be identified. In this report we document structural constraints that were obtained using 1) chemical cross-linking combined with mass spectrometry and 2) selective incorporation of cysteine residues using mutagenesis. We incorporated results from other biophysical studies that did not conflict with constraints imposed by cross-linking. Our results are compared to several in-solution structures proposed for lipid-free apoA-I.
Materials and Methods
Materials
d0-Bis(sulfosuccinimidyl)suberate (d0-BS3) and d4-bis(sulfosuccinimidyl)suberate (d4-BS3) were from ThermoFisher. Me2SO, formic acid and guanidine hydrochoride were from Sigma-Aldrich. Sequencing grade modified porcine trypsin and restriction enzymes were from Promega. RapiGest SF™ was obtained from Waters Inc. Potassium chloride, optima grade methanol, chloroform, acetonitrile, ampicillin and glacial acetic acid were from Fisher Scientific. Mark 12 molecular weight standards, SimplyBlue□ SafeStain and isopropylthio-β-D-galactoside were from Invitrogen. Ultrafree-15 centrifuge filters and Biomax□ 10 membranes were from Millipore Corp. 1,4-dithiothreitol was from Soltec Ventures. The IMPACT™ protein expression system, including E. coli strain ER2566, pTYB11 plasmid vector and chitin beads was from New England Biolabs. PCR primers were synthesized by International DNA Technologies. Taq DNA polymerase was from Roche. ApoA-I cDNA constructs were produced by Custom DNA Constructs. DNase I was from Worthington Biochemical. Solvents used in MS and LC-MS were “B&J GC2” grade from Burdick and Jackson. All other solvents and routine reagents were from the highest available commercial grades.
Preparation of Lipid-free Human ApoA-I
ApoA-I was purified from human plasma by sequential ultracentrifugation as previously described (20). The apoA-I was lyophilized to dryness, dissolved in 6 M guanidine hydrochloride and then refolded by dialyzing exhaustively against 10 mM ammonium bicarbonate, pH 7.4. Mass spectrometry and 12% SDS PAGE were used to ensure that there were no contaminating protein and that apoA-I methionines were not oxidized to the sulfoxide form, a common contaminant in apoA-I preparations. Protein concentration was determined using the Lowry assay (27).
Cloning, Protein Expression and Purification
The coding sequence for wild-type and mutant apoA-I was cloned from the CMV5 vector (28) by Custom DNA Constructs (see Supplemental Material), amplified by PCR as described (29) and inserted into the pTYB11 vector (30). Expression and purification of the cysteine containing apoA-I mutants from inclusion bodies were conducted as previously described (28). The protein purity and molecular weight were determined as previously reported (31).
Formation of Intramolecular Disulfide Bonds
To promote the formation of disulfide bonds between specific residues the mutant apoA-I protein folding conditions were optimized to favor intramolecular disulfide bond formation over intermolecular homo-dimerization using a dilute protein concentration of 0.02 μg/μL. Each mutant was denatured with 6M guanidine hydrochloride, reduced with 500 mM dithiothreitol and diluted accordingly. The proteins were then dialyzed extensively with 10 mM ammonium bicarbonate, lyophilized and then refolded by denaturing in a final concentration of 6M guanidine hydrochloride followed by exhaustive dialysis. The disulfide coupling was verified using mass spectrometry after in-gel trypsin digestion. To prove the correct cysteine substitution for the mutants 500 mM 1,4-dithiothreitol was used to reduce disulfide bridges then cysteine residues were alkylated in with 1M iodoacetamide. The position and extent of alkylation was verified using mass spectrometry after in-gel trypsin digestion.
Cross-linking of Lipid-Free ApoA-1 with a Mixture of d0/d4-BS3
The cross-linkers d0-BS3 and d4-BS3 were each added at a molar ratio of 10:1 cross-linker:ApoA-1. For a total starting mass of 140 μg ApoA-1 (5×10−9 moles), 5×10−8 moles of both d0-BS3 and d4-BS3 were used. ApoA-I on ice was diluted to 694 μL in PBS (pH 7.4). 3 μL each of BS3-d0 and BS3-d4 (5×10−8 moles each) were added to the tube, mix by gentle pipetting and then incubated at 37°C. After 5 min the reaction was quenched by adding 7 μL of cold 1M Tris-HCl (pH 7.4) with gentle mixing and cooled on ice for 10 min. Sample volume was adjusted to at least 500 μL with H2O and the sample dialyzed against 2L of 10 mM ammonium bicarbonate (pH 7.4) with at least 3 changes at 1 h intervals. After dialysis samples were stored at -80°C until in-gel trypsin digest.
SDS PAGE and In-gel Trypsin Digest
Products from CCL lipid-free apoA-I were separated on 12% SDS-PAGE. Digestion was accomplished as previously reported (16, 20, 22, 32). Briefly, protein bands from monomeric and dimeric apoA-I were excised from the gel, minced and repeatedly dehydrated with acetonitrile. The gel pieces were rehydrated with a cold, freshly prepared solution containing 20 ng/μL trypsin in 10 mM ammonium bicarbonate, pH 7.8, 0.1 % (w/v) RapiGest SF™ and 1 mM CaCl2. The final trypsin to apoA-I mass ratio was 1:20. After incubating on ice for 10 min the digests were incubated for 18 h at 37 °C.
Peptide Isolation and ES/Q-TOF Mass Spectrometry
Extraction of peptides was accomplished as previously reported (16, 20, 22, 32). Briefly, the digestion solution was removed and gel pieces covered with 200 μL of acetonitrile/formic acid/water, v/v/v, 50:5:45. After sitting for 10 min the solvent was transferred to a fresh tube. The extraction was repeated and the combined aliquots acidified to an HCl:apoA-I ratio of 1:10 (v/v) using 500 mM HCl. After incubating the acidified solution for 35 min at 37°C the sample was centrifuged for 10 min at 13,000 rpm. The supernatant was transferred to a fresh tube before mass spectrometry.
Survey scans were performed on each peptide mixture using a Waters Q-TOF API-US mass spectrometer equipped with a Waters CapLC and Advion Nanomate source. Acquisition was controlled by Mass-Lynx™ 4.0 software. Peptides were loaded onto a PLRP-S trapping column, 0.5 mm diameter × 2.0 mm length, containing 3 micron diameter particles with a pore diameter of 100 Å. Peptides were loaded onto the column in water/acetonitrile/formic acid (97:3:0.2) at 500 nL/min then separated by gradient elution: solvent A (25 mM formic acid in 97% water and 3% acetonitrile) and solvent B solvent B (25 mM formic acid in 3% water 97% acetonitrile). The gradient profile was: 2% solvent B for 3 min, then a linear increase to 40% B at 90 min and then to 80% B in 5 min. At 95 min the composition was ramped to 2% B over 5 min and then equilibrated with 2% B for 30 min. Peptides were eluted at 470 nL/min. Positive ion survey scans were recorded in the continuum mode with a scan window of 300 to 1500 m/z for 2s. The source temperature was 80°C. The cone and capillary voltages were 45 V and 3.5 kV, respectively. Experimental m/z was corrected using apoA-I tryptic fragment 7, m/z = 806.8969, and for the +1 charge state using tryptic fragment 12, m/z = 831.4365. Ions within ±0.051 m/z of the theoretical ions were sequenced. Product ion MS/MS spectra were acquired in the continuum mode from 50 to 1600 m/z using a data directed charge-state selective collision energy and an accumulation time of 2s. Sequence analysis of the MS/MS spectra was performed with a fragment ion tolerance of ± 0.05 m/z.
Circular Dichroism
Wild-Type or cysteine-containing apoA-I mutants were dissolved in 1X phosphate buffered saline (pH 7.1) at 0.06–0.1 mg/mL to prevent protein self-association. The circular dichroism (CD) spectra were recorded with a Jasco J-715 spectropolarimeter (Jasco Corp., Tokyo, Japan) at room temperature using a 0.2 cm quartz cuvette. The spectra were read over wavelengths of 250–190 nm and analyzed as previously described (33).
Molecular Modeling
A composite 3-D structure, based on our own cross-linking and mass spectra data and the data from others (9, 34), was constructed to predict placement of cysteine mutations. The strategy to test apoA-I helical participation involved substituting residues within ~5Å with cysteines to form disulfide bonds between specific helices. Residues selected were chosen if they were ~5Å apart from one another. Cysteine mutations not within 5Å were also designed to determine the accuracy of our structural model. The molecular modeling for apoA-IWT has been described in previous publications (16, 20, 22, 32) using the coordinates for lipid-free △43-apoA-I (7) that were joined with 1-43 amino acids of the N-terminal end. Unmodified, lipid-free apoA-I has been crystallized, but its conformation has been extensively studied. When this study started there was only one creditable solution structure for lipid-free apoA-I by Silva et al. (9). The cross-linked positions were oriented to their correct distances based on the Silva model. To do this we used the maximum distance of Cα–Lysine-(cross-linker)-Cα–Lysine of 26.0 Å for BS3. As required, apoA-I was bent at the proline or glycine-glycine sites between the amphipathic segments of apoA-I. Tools available in Swiss-PdbViewer OSX v4.1 (http://www.expasy.org/spdbv/) were used to optimize the conformations while pdb files were manipulated using PyMOL version 1.5 (http:/www.pymol.org). Swiss-PdbViewer, PyMOL along with Visual Molecular Dynamics (VMD) for Mac OSX, version 1.9.1 (35), were used to generate the molecular figures shown in the manuscript.
Results
Lipid-free human apoA-I was prepared by standard techniques and exhaustively dialyzed to refold the protein into the most stable conformation. Because apoA-I self-associates at concentrations greater than 0.1 mg/mL (36-38) these studies were carried out at 0.2 mg/mL and 1.0 mg/mL. At the lower concentration apoA-I is predominantly a monomer while at the higher concentration favored dimeric association of apoA-I. For these studies a 1:1 mixture of d0-BS3 and d4-BS3 was used to improve detection and identification of cross-linked peptides. After treatment for 5 min with 20 to 1 lysine specific BS3 cross-linker to apoA-I excess cross-linker was deactivated by adding Tris buffer. After concentration samples were separated by SDS-PAGE into monomer and dimer bands formed by chemical cross-linking. Each of these bands was excised and digested with trypsin then subject to LC/MS analysis. In the search for cross-links the results were compared to a table having the masses of all possible interpeptide Lys to Lys cross-linked peptide from d0-BS3 and a second table having all possible intrapeptide Lys to Lys cross-linked peptides from d0-BS3. Each hit was investigated to determine if there is a complimentary isotope series from d0-BS3 substituted with 4 deuterium atoms, d4-BS3. Peptides showing both the d0- and d4-isotopic series were sequenced using MS/MS analysis.
Cross-linking times are kept short, 5 min, and the overall BS3 concentration low. Therefore, multiple cross-links to a single lysine take place because only a small fraction of the total lysines react in a given experiment. The primary products detected in the total ion chromatogram were the unmodified peptides anticipated from tryptic digestion of apoA-I. Another set of products found at low concentration were peptides that had added a single BS3 (data not shown). Because BS3 added to lysine these peptides had 1 missed cut site. The lowest concentration set of products were those in which BS3 coupled two apoA-I tryptic peptides, each of which consisted of a tryptic fragment with 1 missed cut site whether designated as intrapeptide or interpeptide cross-links.
Table 1 shows a list of intrapeptide cross-linked peptides. These are from adjacent lysines cross-linked by BS3 from the monomer bands of 0.2 mg/mL and 1.0 mg/mL cross-linking experiments.
Table 1.
Intrapeptide Peptides
| Theoretical MH+ | d0-m/z | d4-m/Z | positive charge |
Sequenced d0- m/z |
Exp MH+ | Δ m/z | Intensity | Lys1 | Lys2 |
|---|---|---|---|---|---|---|---|---|---|
| 2303.2070 | 768.41 | 769.74 | 3 | 768.41 | 2303.1965 | 0.0105 | 139 | 94 | 96 |
| 2303.2069 | 768.41 | 769.74 | 3 | 768.43 | 2303.1946 | 0.0123 | 1030 | 133 | 140 |
| 2347.2509 | 783.09 | 784.42 | 3 | 783.06 | 2347.2312 | 0.0197 | 230 | 206 | 208 |
| 1672.8467 | 836.93* | 2 | 836.93 | 1672.7819 | 0.0648 | 59 | 88 | 94 | |
| 1717.9165 | 859.46 | 861.46 | 2 | 859.45 | 1717.9421 | −0.0256 | 87 | 96 | 106 |
| 2616.4037 | 872.81 | 874.14 | 3 | 872.79 | 2616.3601 | 0.0436 | 157 | 45 | 59 |
| 3728.8699 | 932.97 | 933.97 | 4 | 932.97 | 3728.8325 | ro.0374 | 70 | 40 | 45 |
| 2016.1017 | 1008.55 | 1010.55 | 2 | 1008.52 | 2016.0919 | 0.0098 | 1360 | 12 | 23 |
| 2109.1120 | 1055.06 | 1057.06 | 2 | 1055.06 | 2109.1335 | −0.0215 | 162 | 238 | 239 |
Table 1 is a compilation of the intrapeptide cross-linked peptides. These peptides were found in both 0.2 mg/mL and 1.0 mg/mL apoA-l. The theoretical and experimental m/z for the intact cross-linked peptide are given, along with m/z for the charge state sequenced and its theoretical m/z. Δ m/z is the difference between the theoretical and experimental m/z for the protonated, intact cross-linked peptide. Maximum ion intensity and amino acid position are listed in the last three rows.
Sequenced from experiments that used only d0-BS3.
Table 2 shows a list of mostly interpeptide cross-linked peptides. All peptides had both the d0 and d4 isotope series and gave the correct amino acid sequences by MS/MS analysis. These peptides are formed by BS3 cross-linking of lysines close in space on the monomer, but not necessarily close in the primary sequence. Two bolded peptides, m/z 2233.12 and 2358.22, were found only in the dimer band from 1.0 mg/mL apoA-I cross-linking experiments.
Table 2.
Interpeptide Linked Peptides
| Theoretical MH+ |
d0-m/z | d4-m/Z | positive charge |
Sequenced d0- m/z |
Exp MH+ | Δ m/z | Intensity | Lys1 | Lys2 |
|---|---|---|---|---|---|---|---|---|---|
| 2159.2150 | 720.41 | 721.74 | 3 | 720.42 | 2159.2395 | −0.0245 | 54 | 118 | 133 |
| 2233.1217 | 745.05 | 746.39 | 3 | 745.05 | 2233.1458 | −0.0241 | 51 | lα | 118 |
| 2358.2185 | 786.74 | 788.09 | 3 | 786.72 | 2358.2163 | 0.0022 | 44 | 88 | 118 |
| 1671.9138 | 836.46 | 838.46 | 2 | 836.46 | 1671.9401 | −0.0263 | 98 | 94 | 239 |
| 3627.9461 | 907.74 | 908.74 | 4 | 907.74 | 3627.8796 | 0.0665 | 90 | 12 | 195 |
| 2737.4365 | 913.15 | 914.48 | 3 | 913.15 | 2737.4180 | 0.0185 | 124 | 40 | 239 |
| 3669.8953 | 918.23 | 919.23 | 4 | 918.24 | 3669.8948 | 0.0005 | 68 | 23 | 59 |
| 2826.4517 | 942.82 | 944.16 | 3 | 942.80 | 2826.3882 | 0.0635 | 1180 | 1α | 12 |
| 2915.5614 | 972.53 | 973.86 | 3 | 972.52 | 2915.5664 | −0.0050 | 145 | 118 | 140 |
| 3003.6103 | 1001.84 | 1003.18 | 3 | 1001.88 | 3003.6216 | −0.0113 | 42 | 40 | 118 |
| 4016.0589 | 1004.77 | 1005.77 | 4 | 1004.77 | 4016.0642 | −0.0053 | 73 | 40 | 59 |
| 4043.0731 | 1011.52 | 1012.52 | 4 | 1011.52 | 4043.0825 | −0.0094 | 141 | 40 | 140 |
| 3245.5703 | 1082.53 | 1083.86 | 3 | 1082.53 | 3245.5815 | −0.0112 | 44 | 1α | 59 |
| 3286.7267 | 1096.25 | 1097.58 | 3 | 1096.24 | 3286.7563 | −0.0296 | 172 | 40 | 133 |
| 3291.7169 | 1097.91 | 1099.25 | 3 | 1097.90 | 3291.7061 | 0.0108 | 39 | 40 | 182 |
Table 2 is a compilation of the interpeptide cross-linked peptides. Peptides in regular type were found in both 0.2 mg/mL and 1.0 mg/mL apoA-l. Bold type indicate peptides detected only at an apoA-l concentration of 1.0 mg/mL. The theoretical and experimental m/z for the intact cross-linked peptide are given, along with m/z for the charge state sequenced and its theoretical m/z. Δ m/z is the difference between the theoretical and experimental m/z for the protonated, intact cross-linked peptide. Maximum ion intensity and amino acid position are listed in the last three rows.
For the initial phase of the analyses we compared the cross-links with the several available models. Chemical cross-links were oriented to the correct distances based on whether these cross-links were intrapeptide or interpeptide. We used the maximum distance of Cα–Lysine-(BS3)-Cα–Lysine at 26Å to establish the appropriate separations.
The structure for lipid-free apoA-I by Gangani et al. (9) was used as the preliminary model for these studies. After repeated analysis of lipid-free apoA-I conformation using chemical cross-linking the structure of Gangani et al. was adjusted to reflect new constraints. Based on the revised structure a series of mutants each having 2 cysteines were prepared to further refine the conformation of apoA-I. A separation of around 5Å was assumed necessary for disulfide bond formation. Initial efforts centered on searching the model for pairs of amino acids adjacent to one another, but located in different helical regions. For example, F104 was close to H162. These were mutated and found to readily form a disulfide bond. It was first assumed that the region 165 to 160 was helical and that position R160 would be on the opposite face of the helix, compared to H162. When we made the F104C, R160C mutation set they also formed a disulfide bond. The interpretation was that both R160 and H162 faced position F104 and the model was adjusted. Position M148 appeared to be well separated from F104 in the model so this double Cys mutation was prepared to test whether there was random disulfide formation when apoA-I was refolding. This Cys-substitution pair did not form a disulfide bond. Table 3 shows the mutant apoA-Is having 2 cysteine substitutions, whether the cysteines formed a disulfide bond and details about the ions monitored. If the mutants formed disulfides the structure was adjusted to accommodate these additional constraints. Figure 1 shows three stereoscopic views for lipid-free apoA-I.
Table 3.
Mutant ApoA-Is with 2 Cysteine Substitutions
| Cysteine Substitutions |
Disulfide Tryptic Peptides (m/z)+charge |
Locked | CAM- Peptide (m/z)+charge |
CAM-Peptide (m/z)+charge |
|---|---|---|---|---|
| F104C-H162C* | T14-T26 (820.05)+3 |
Yes | 625.81+2 | 662.81+2 |
| D103C-R177C | T14-T28 (733.04)+3 |
Yes | 649.32+2 | 494.78+2 |
| F104C-M148C | T14-T22 (737.01)+3 |
No | 633.28+2 | 530.77+2 |
| F104C-R160C | T14-T25 (644.10)+5 |
Yes | 633.28+2 | 517.77+4 |
| D157C-L178C | T25-T29 (665.34)+2 |
No | 413.76+2 | 620.36+1 |
| V53C-R123C | T7-T18 (758.35)+4 |
Yes | 837.32+2 | 736.35+2 |
| D13C-V67C | T3-T9 (790.14)+4 |
No | 640.87+2 | 997.51+2 |
| G26CK59C | T4-T7 (778.69)+3 |
No | 537.26+1* | 957.43+2 |
| L200C-L233C | T32-T35 (645.55)+4 |
Yes | 421.53+3 | 717.34+3 |
The term Locked indicates that a disulfide bond was identified between the two Cys using mass spectrometry after trypsin proteolysis.
Analysis was performed on a protein that had a substitution Q→L at position 98.
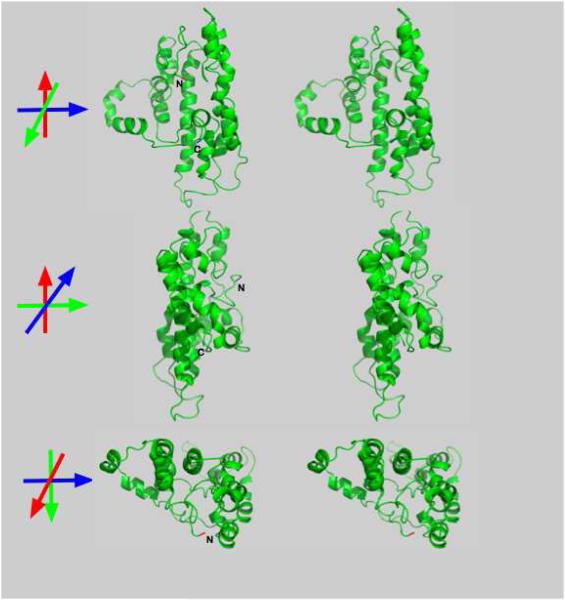
Figure 1. Lipid-free apoA-I.
Three orthogonal, cross-eye stereo views of the conformation proposed for lipid-free apoA-I. C- and N-terminal ends of the molecule are associated. The C- and N-terminal AA shown in blue and red, respectively. A four-helix bundle that comprises most of the molecule.
Discussion
Apolipoprotein A-I is the principal protein of HDL, the lipoprotein associated with RCT and essential for maintaining cholesterol balance. It is synthesized as a pre-proprotein, secreted in the pro-form and then converted to mature apoA-I by bone morphogenic protein. An initial step in the formation of HDL is lipidation of lipid-free apoA-I by ABCA1 to form the first lipidated HDL species, nHDL. Because of its central role in cholesterol efflux the structure and conformation has been studied for many years. These analyses employed a combination of biophysical techniques including sedimentation velocity analysis and limited proteolysis (39-41). Limited proteolysis suggested that the C-terminal 53 amino acids were flexible while the 190 N-terminal amino acids were folded into a domain in which helical structure predominated (39). Denaturation studies on a variety of apoA-I variants suggested that the N-terminal and central regions had a folded domain structure and that modifications to the C-terminal end had little effect on guanidine hydrochloride denaturation (42).
Structural attributes of lipid-free apoA-I
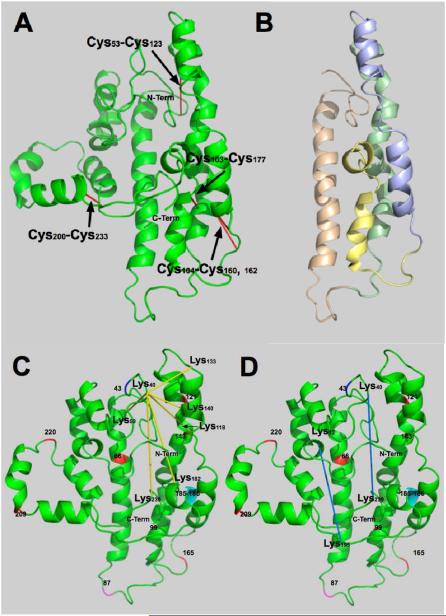
As a starting point for the structure we used the analysis by Silva et al. (9) and reports by Rogers et al. (39-41, 43). It is usually assumed that the secondary structure of the repeating 11- and 22-amino acid was predominantly the α-helix with bends punctuated by prolines or other helix-breaking amino acid combinations. The secondary structure was optimized using secondary structural information identified in other studies (6, 44-48). Figure 1 shows the proposed structure for lipid-free apoA-I based on the constraints imposed by disulfide bond formation and chemical cross-linking with BS3. The lipid-free structure with Cys-Cys bonds in red is shown in Figure 2A. Regions containing a high content of α-helices are color coded in Figure 2B. A series of Cys-disulfide bonds tied together helical regions: A-B, Cys53-Cys123; B-C, Cys104-Cys162, Cys104-Cys160, C-D and B-D, Cys103-Cys177. Because the Cα-Cα Cys-Cys distance has a maximum length of 6.8Å (49) the residues must lie close together. Formation of a Cys200-Cys233 disulfide bond suggests that the C-terminal end is folded back on itself. Figure 2B emphasizes the internal region of lipid-free apoA-I, the four-helix bundle, by showing the central region after the N- and C-termini were removed for clarity. The C-terminal end of the proposed structure is more exposed to the solvent, Figure 2A, suggesting that it would be susceptible to proteolytic digestion, consistent with a structure in which the central regions had considerable helical character and could undergo cooperative unfolding (43, 50, 51).
Figure 2. Disulfide bonds and four-helix bundle.
A. Proposed structure for lipid-free apoA-I showing the positions of disulfide bonds that formed spontaneously. B. Conformation shown in A after removing C- and N-termini for clarity: N-terminal, amino acids (AA) 1-43; tan region, AA 44-92, helices 1 and 2; green region, AA 93-130, helices 3-5; blue region, AA 131-166, helices 5 and 6; yellow region, AA 167-187, helix 7; and C-terminal end, AA 188-243, helices 8-10. C. Proposed structure showing the BS3 cross-links radiating from Lys40. D. BS3 cross-links that show the C- and N- terminal ends are close. Cross-links are shown in yellow. Prolines are shown in red, AA 66, 99, 121, 143, 165, 209 and 220. The end of the N-terminal region is shown at AA 43 in blue. Helical region 2 end, AA 87, is shown in magenta. Glycines between helical regions 8 and 9 are shown in cyan.
Possibly the most unusual feature found in this study is the connectivity of N-terminal region Lys40 with many different domains of lipid-free apoA-I, Figure 2C, including Lys59 (part of Helical region A), Lys118 (Helical region B), Lys133 and Lys140 (Helical region C), Lys182 (Helical region D) and Lys239 (C-terminal region). Extended connectivity of Lys40 suggests that the region around Lys40 is sufficiently unobstructed to accommodate reaction of BS3 with six neighboring Lys and that the C- and N-terminal ends do not cover this region and block access of BS3 to Lys40. These results also suggest that lysines cross-linking with Lys40 have similar degrees of hydrogen bonding and, therefore, similar reactivity with BS3.
Two BS3 cross-links show that the C- and N-terminal ends of the molecule are close to one-another, Figure 2D. These are BS3 cross-links Lys239 and Lys195 to Lys40 and Lys12, respectively. Chemical cross-links with Lys40 and cross-links between Lys12 and Lys59 located in helical repeat 1 suggest that the N-terminal end is closely associated with the core helix bundle. The absence of an extensive set of interpeptide cross-links indicates that the C-terminal end is not as tightly associated with the helix core bundle as is the N-terminal region. Portions of the C-terminal end are sufficiently close that a disulfide bridge formed between Cys200 and Cys233. The C-terminal region has been proposed as the site of initial interaction between apoA-I and lipid (40, 52).
Comparison to other lipid-free structures
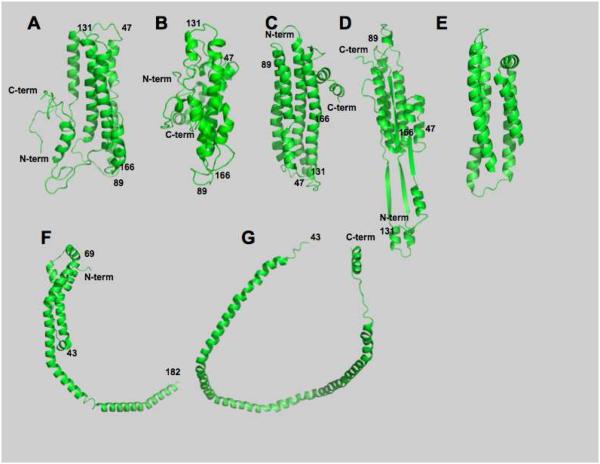
Figure 3 shows published structures that made significant contributions to the understanding of lipid-free apoA-I structure. To compare these structures, distances between Cα, asymmetric backbone carbon, of cross-linked amino acids reported in our study were measured from the model coordinates for each published structure and collected in Table 4. The first set of structures were determined in solution, Figures 3A through 3D. Silva et al. proposed a solution structure using chemical cross-linking combined with mass spectrometry and sequence threading (9) that is shown in Figure 3A. This structure has many of the features suggested by earlier biophysical studies and is similar to the structure proposed herein, Figure 3B. Two other solution structures have been reported. The first employed high-field NMR to deduce the solution structure of a C-terminal truncated mouse apoA-I (6, 53) amino acids 1-216, Figure 3C. Mouse apoA-I and the truncated mutant in particular are more soluble and less subject to aggregation (6). The second study employed a series of full-length, cysteine-substituted apoA-Is to which thio-specific nitroxide spin-labels were coupled through the thiol moiety of the cysteine side chain. Dipolar coupling and solvent accessibility were measured by electron paramagnetic resonance spectroscopy (EPR). Analysis of the EPR data yielded the structure (45) shown in Figure 3D. Common factors in structures A-D are parallel helix bundles that are reminiscent of the LDL receptor-binding domain reported for apolipoprotein (apoE) (54) shown in Figure 3E. Structures A through D are similar to to proposals obtained using other biophysical techniques (37-41, 44, 47, 48).
Figure 3. Comparison of several lipid-free structures for apoA-I.
A, Silva et al. (9); B, Herein; C, Yang, et al. (6); D, Lagerstedt et al. (45); E, LDL receptor-binding domain reported of apoE (54); F, Mei and Atkinson (34); E, Borhani et al (7). C- and N-termini are indicated as are selected amino acid positions on the structures.
Table 4.
Comparison of Distances Between Cross-United Peptides Reported in This Study with Other Published Models for Lipid-Free ApoA-l
| Lys1 | Lys2 | Lagerstedt(45) | Yang(6) | Mei(34) | Borhani(7) | Silva(9) | Pollard |
|---|---|---|---|---|---|---|---|
| 1α | 12 | 12.9 | 16.1 | 18.5*2 | - | 26.1 | 20.0 |
| 1α | 59 | 39.6 | 57.4 | 25.2*5 | - | 44.2 | 25.1 |
| 12 | 195 | 82.0 | 13.8 | - | - | 34.5 | 25.9 |
| 23 | 59 | 10.5 | 28.7 | 15.6 | - | 16.4 | 13.2 |
| 40 | 59 | 18.4 | 4.5 | 35.5 | - | 22.3 | 20.0 |
| 40 | 118 | 27.3 | 24.8 | 16.8 | 104.3*3 | 21.2 | 20.1 |
| 40 | 133 | 62.2 | 22.1 | 29.5 | 106.6*6 | 19.3 | 11.9 |
| 40 | 140 | 70.0 | 17.5 | 38.4 | 109.0*2 | 23.2 | 12.5 |
| 40 | 182 | 16.1 | 53.4 | 79.7*7 | 79.9*8 | 41.0 | 23.2 |
| 40 | 239 | 27.0 | - | - | 25.0** | 31.5 | 26.0 |
| 94 | 239 | 19.8 | - | - | 92.4 | 38.2 | 26.0 |
| 118 | 133 | 41.3 | 25.5 | 24.1 | 24.7 | 16.6 | 20.9 |
| 118 | 140 | 51.4 | 32.4 | 35.3 | 34.9 | 10.8 | 11.4 |
| 104 | 162 | 23.9 | 22.9 | 77.2 | 72.6 | 8.2 | 9.7 |
| 103 | 177 | 11.7 | 6.1 | 93.9 | 75.7 | 24.8 | 6.7 |
| 104 | 160 | 22.9 | 28.3 | 75.7 | 69.0 | 9.2 | 8.3 |
| 53 | 123 | 19.4 | 27.3 | 42.9 | 84.9 | 11.3 | 11.4 |
| 200 | 233 | 31.1 | - | - | 54.7 | 35.0 | 6.4 |
For some of the models the primary sequence was truncated. Where there is an amino acid (AA) close to the position in full-length apoA-l it is used and listed as following:
AA 3-12;
AA 4-59;
AA 43-118;
AA 43-133;
AA 43-140;
AA 40-181.
AA 43-182;
AA 43-239.
The last two published models were derived from X-ray crystallographic analysis of truncated apoA-I molecules. Atkinson’s group recently analyzed the structure of a C-terminal truncation mutant composed of amino acids 1-184. The resulting structure showed how the N-terminal end of apoA-I is folded (34, 55), Figure 3F, as was suggested from cross-linking studies of lipidated apoA-I (16, 20). The complimentary structure that rekindled interest in apoA-I structure, N-terminal truncated lipid-free apoA-I having amino acids 44-243, called △43 apoA-I (7), is shown in Figure 3G. Both X-ray crystallographic structures were similar in that they had a more-or-less circular, belt-like conformation with hydrophobic amino acids facing the interior and are reminiscent of structures proposed for lipidated apoA-I (16, 17, 20, 56) and not the structures of lipid-free apoA-I in solution obtained using chemical cross-linking and from other biophysical studies.
Structure 3E shows a “classic” apolipoprotein 4-helix bundle, the LDL receptor-binding domain of apoE (54). Structures 3A, 3B and 3C have a 4-helix bundle as the core of the molecule. The bundle structure of 3B can be seen more easily when the C- and N-terminal ends are hidden, Figure 2B. Truncated mouse apoA-I, 3C, has considerably more helical motif toward the N-terminus than does human apoA-I. Attempts to increase the helical content of the N-terminal end of human apoA-I did not yield a structure with the identified cross-links and the N-terminal region covered the Lys40 region that displayed extensive connectivity to other parts of the molecule. Human and mouse apoA-I have only about 63% homology suggesting that the differences between their respective structures may be due to differences in primary sequence (6).
This study found 8 of the 10 cross-links reported by Silva et al. (9) and identified intrapeptide cross-link, Lys45 to Lys59, not reported in the earlier study. However, there was a much greater difference in the number of identified interpeptide cross-links between the two studies. Only two interpeptide cross-links were common to both studies: Lys23 to Lys59 and Lys118 to Lys140. This study identified 11 cross-links not reported by Silva et al. (9) and did not find 5 other cross-links. Similarities and differences are listed in Table 5. A greater degree of confidence is assigned to the peptides reported herein because each was validated by MS/MS sequencing.
Table 5.
| Intermolecular Cross-links | Intramolecular Cross-links | ||||
|---|---|---|---|---|---|
| Theoretical MH+ |
Lysl | Lys2 | Theoretical MH+ |
Lysl | Lys2 |
| 2826.45 | 1α | 12 | 2016.10 | 12 | 23 |
| 3245.57 | 1α | 59 | 3728.87 | 40 | 45 |
| 2814.35 | 1α | 96 | 2616.40 | 45 | 59 |
| 2233.12 | lα | 118 | 1672.85 | 88 | 94 |
| 3627.95 | 12 | 195 | 2303.21 | 94 | 96 |
| 3669.90 | 23 | 59 | 1717.92 | 96 | 106 |
| 4016.06 | 40 | 59 | 2782.44 | 106 | 107 |
| 3003.61 3286.73 |
40 40 |
118 133 |
2303.21 2347.25 |
133 206 |
140 208 |
| 4043.07 3291.72 |
40 40 |
140 182 |
2863.63 2109.11 |
226 238 |
238 239 |
| 2737.44 2358.22 |
40 88 |
239 118 |
|||
| 2080.14 | 94 | 208 | |||
| 1671.91 | 94 | 239 | |||
| 3615.85 | 96 | 195 | |||
| 2630.30 | 96 | 208 | |||
| 4186.21 | 96 | 226 | |||
| 2159.21 | 118 | 133 | |||
| 2915.56 | 118 | 140 | |||
Table 5 shows cross-links reported in this study and that by Silva et al. (9). A white background indicates that it is only from this study. A light gray background is for cross-linked peptides found in both studies. The dark gray background indicates that the cross-linked peptide was found only in (9).
Cross-links Lys40 to Lys133 and Lys182 to Lys239 rule out extended structures including both structures derived by X-ray crystallography, Figures 3F and 3G, and the one derived using EPR, Figure 3D. Structure 3G led to reinvestigation of apoA-I conformation on phospholipid discs with the conclusion that the two apoA-I molecules on 9.6 and 7.8 nm diameter synthetic rHDL particles, rHDL, have an antiparallel arrangement with the 5,5’-helical regions adjacent. The “Belt-buckle” hypothesis for apoA-I conformation suggests that the N- and C-terminal regions fold back onto the body of the chain, with the fold-back more extensive for the N-terminal end. Structure 3D has both the helix bundle along with the extended form that is similar to that proposed by Rogers et al. (40, 41). This structure does not fit the cross-link constraints identified in this study, but has many similarities, and it suggests that lipid-free apoA-I may open to an extended conformation that is in equilibrium with the compact conformation.
The structure compared to other biophysical studies
The structure of apoA-I has been investigated for at least 40 years and the proposed structure is in general agreement with previous studies suggesting that C- and N-terminal regions are located close together and that it has a helix-bundle core. Several studies showed that apoA-I self associates, but is a monomer below 0.1 mg/mL (38, 57, 58), the concentration of lipid-free apoA-I in human plasma (59, 60). Davidson et al. using chemical cross-linking estimated that more than 85% of lipid-free apoA-I was monomeric below 0.1 mg/mL (61). Cross-linking conditions used in this study yielded less than 1 percent dimer formation at 0.2 mg/mL (data not shown). Two additional cross-links in monomer fractions were identified from cross-linking conducted at 1.0 mg/mL compared to 0.2 mg/mL apoA-I. The positioning of these cross-links suggests that association may have compacted the structure.
Sedimentation velocity data showed the monomer had the shape of a prolate ellipsoid with a major axis of 75Å and a minor axis of 12.5Å (38) or dimensions of 150Å × 25Å. Minor axis dimensions showed that there was sufficient volume for adjacent, antiparallel α-helices (38), like the 4-helix bundle reported for crystalline apoE (54). The width to length ratio of the model proposed in this study, about 3.8, indicates a more compact structure than that suggested from other biophysical studies, (38). The proposed structure has dimensions of roughly 69Å × 47Å × 18Å. Based on the experimental specific volume of 0.74 cm3/g (62) the calculated volume of the proposed structure was 3.4 × 104 Å3 similar to volumes calculated using programs VADAR (63) and 3V (64). However, by simply swinging the C- and N-termini onto the major axis the length of the molecule was extended to about 161Å. Thermal unfolding studies in low ionic strength buffers indicated that apoA-I transitioned to a state with defined secondary structure having lax tertiary structure (65), the definition of the molten globular state. This state may be easily accessible and a prerequisite for opening the structure to open up to bind lipid.
Confounding factors to the interpretation of the cross-linking results
There are usually a sufficient number of available lysine pairs to obtain a reasonable number of constraints. However, steric hinderance, hydrogen bonding, or proximity to other lysines moderate the reactivity with cross-linkers. In earlier studies there was concern that differences in lysine reactivity could be overcome by using higher concentrations of cross-linker. Therefore, in our previous studies we compared cross-linking with different ratios of cross-linker to apoA-I (16, 20). Repeated experiments showed that the same set of cross-linked peptides were found for cross-linker to apoA-I ratios of 2:1 to 20:1. Ratios of 10:1 to 20:1 increased the yield of cross-linked peptides and improved the S/N for MS/MS sequencing. Tables 1 and 2 include the ion intensity of cross-linked peptides and these give a rough estimate of their concentration. A caveat to increasing the cross-linking apoA-I ratio or any process that increases the yield of BS3 to lysine coupling is that a large fraction of available lysines will react. There can be two consequences, the first is that there will be a substantial reduction in tryptic cut sites and the second is that there could be a lower yield of cross-links from less reactive lysine pairs. To mitigate these outcomes analyses were performed for a short time and the reaction quenched with Tris to prevent continued reaction. Analysis of the tryptic digests showed that the majority of the tryptic peptides had not been modified by BS3 (data not shown).
Studies have shown that analysis of cross-linked proteins yields distance constraints that are remarkably similar to those determined by other techniques like X-ray crystallography. Young et al. (66) compared distance constraints for fibroblast growth factor-2 from cross-linking and found them to agree well with results from X-ray crystallography. Similar results have been obtained for bovine (67, 68) and human (69) serum albumin and bovine rhodopsin (70).
Studies that compared the calculated maximum lysineCα-BS3-lysineCα separations, 26Å, to the same separations taken from X-ray crystallographic data showed that the X-ray results often yielded a slightly greater separation, 28Å, a bit longer than the cross-linker arm length (71-74). This is in contrast to other calculations showing that the arm lengths of cross-linkers are shorter (75) than usually assumed from simple conformational analysis. Therefore, in solution, regions of the protein may be slightly closer than anticipated from the crystal structure or alternatively, motions inherent in parts of the protein, particularly the N- and C-termini, may permit cross-links to form that would not be anticipated from a ridged crystal structure. However, because of conformational flexibility BS3 can cross-link lysines that are separated by much less than 26Å. By constraining the maximum lysineCα-BS3-lysineCα distance to 26Å, this study may report a more compact structure for lipid-free apoA-I. Because the largest separations were found between the C- and N-terminal ends, increasing the maximum lysineCα-BS3-lysineCα distance would place the C- and N-termini farther apart than reported herein.
During review the question was ask “does cross-linking at one site result in constraints that make additional cross-linking more or less likely?” An implied question is whether the “first” cross-link may direct subsequent cross-linking to yield an incorrect protein conformation. Given that several studies have reported cross-linking constraints very similar to those obtained with X-ray crystallography, it is likely that cross-linking of two closely associated lysines does not have a profound effect on the structure. However, this observation does not indicate if the frequency of other cross-links were affected.
The frequency of cross-linking depends on the time two lysines remain in proximity. Therefore, the constraints reflect the time-average position of lysines relative to one another. Proteins in solution exhibit more motion than crystallized protein, particularly at the C- and N-termini or at loops, and this mobility may lead to a greater variety of cross-links between the mobile region and more constrained regions. Chemical cross-linking of a pair of lysines depends on several factors including the reactivity of the individual lysines with BS3, the time the lysines remain in proximity, and competition between hydrolysis of BS3 by the solvent rendering the cross-linker incapable of reacting with a second lysine. These arguments suggest that only lysines in close proximity can be coupled by BS3 and argue against apoA-I having an extended conformation with the C- and N-termini extended from the helix bundle core.
Disulfide bond formation provided useful evidence for the structure of lipid-free apoA-I as it did in a study helix of bundle opening when apoE accepts lipids (76). Disulfide bond formation takes place on refolding. If this process does not follow two-state mechanism, involving a molten globule state, apoA-I may explore several different conformations, some of which could permit disulfide formation in a non-Native conformation. Disulfide formation is not instantaneous and requires oxidation of two cysteines in a process involving reduction of dioxygen. To prevent random disulfide formation refolding of lipid-free apoA-I starts in a highly reducing environment with disulfide formation taking place later after a stable conformation is achieved.
In conclusion, these studies suggest that lipid-free apoA-I in solution is a squat, oblate spheroid, with the helical regions 1 through 7 associated in a 4-helix bundle. The N-terminal region, AA 1-43, and the C-terminal amphipathic helices 8-10, are close to one another and associated with the helix bundle. The conformation reported herein should be contrasted with that proposed for rHDL, nHDL and native HDL in which the apoA-I has opened up to a more extended conformation, very similar to the structure reported for crystalline △43 apoA-I, Figure 3G. The model presented here will be useful in predicting how lipid-free apoA-I associates with ABCA1 for subsequent lipidation to generate nHDL.
Acknowledgments
Funding Source Statement. These studies were supported by grants from the National Institutes of Health NHLBI HL-49373 and HL-64163 (MST) and the American Heart Association 09GRNT2280053 (MJT). The Waters Q-TOF mass spectrometer was obtained using funds from NIH Shared Instrumentation Grant 1S10RR17846 (MJT). The MS analyses were performed in the Mass Spectrometer Facility of the Comprehensive Cancer Center of Wake Forest School of Medicine supported in part by NCI center grant 5P30CA12197.
Abbreviations
- ABCA1
ATP-binding cassette transporter A1
- ABCG1
ATP-binding cassette transporter G1
- apoA-I
apolipoprotein A-I
- BS3
Bis[sulfosuccinimidyl] suberate
- HDL
high density lipoprotein
- nHDL
nascent HDL
- LCAT
lecithin-cholesterol acyltransferase
- RCT
reverse cholesterol transport
- rHDL
recombinant HDL
- MS/MS
tandem mass spectrometry
References
- 1.Oram JF, Vaughan AM. ABCA1-mediated transport of cellular cholesterol and phospholipids to HDL apolipoproteins. Current Opinion in Lipidology. 2000;11:253–260. doi: 10.1097/00041433-200006000-00005. [DOI] [PubMed] [Google Scholar]
- 2.Wang N, Silver DL, Costet P, Tall AR. Specific binding of ApoA-I, enhanced cholesterol efflux, and altered plasma membrane morphology in cells expressing ABC1. Journal of Biological Chemistry. 2000;275:33053–33058. doi: 10.1074/jbc.M005438200. [DOI] [PubMed] [Google Scholar]
- 3.Yancey PG, Bortnick AE, Kellner-Weibel G, De La Llera-Moya M, Phillips MC, Rothblat GH. Importance of Different Pathways of Cellular Cholesterol Efflux. Arterioscler Thromb Vasc Biol. 2003;23:712–719. doi: 10.1161/01.ATV.0000057572.97137.DD. [DOI] [PubMed] [Google Scholar]
- 4.Libby P. Managing the risk of atherosclerosis: The role of high-density lipoprotein. Am J Card. 2001;88:3N–8N. doi: 10.1016/s0002-9149(01)02145-2. [DOI] [PubMed] [Google Scholar]
- 5.Rader DJ, Tall AR. The not-so-simple HDL story: Is it time to revise the HDL cholesterol hypothesis? Nat Med. 2012;18:1344–1346. doi: 10.1038/nm.2937. [DOI] [PubMed] [Google Scholar]
- 6.Yang Y, Hoyt D, Wang J. A complete NMR spectral assignment of the lipid-free mouse apolipoprotein A-I (apoAI) C-terminal truncation mutant, apoAI(1-216) Biomol NMR Assign. 2007;1:109–111. doi: 10.1007/s12104-007-9031-2. [DOI] [PubMed] [Google Scholar]
- 7.Borhani DW, Rogers DP, Engler JA, Brouillette CG. Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc Natl Acad Sci U S A. 1997;94:12291–12296. doi: 10.1073/pnas.94.23.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.2013. Multiple. H.M. Krishna Murthy, In Wikipedia.
- 9.Silva RA, Hilliard GM, Fang J, Macha S, Davidson WS. A three-dimensional molecular model of lipid-free apolipoprotein A-I determined by cross-linking/mass spectrometry and sequence threading. Biochemistry. 2005;44:2759–2769. doi: 10.1021/bi047717+. [DOI] [PubMed] [Google Scholar]
- 10.Forte T, Norum KR, Glomset JA, Nichols AV. Plasma lipoproteins in familial lecithin: cholesterol acyltransferase deficiency: structure of low and high density lipoproteins as revealed by elctron microscopy. J Clin Invest. 1971;50:1141–1148. doi: 10.1172/JCI106586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matz CE, Jonas A. Micellar complexes of human apolipoprotein A-I with phosphatidylcholines and cholesterol prepared from cholate-lipid dispersions. J Biol Chem. 1982;257:4535–4540. [PubMed] [Google Scholar]
- 12.Segrest JP, Jackson RL, Morrisett JD, Gotto A. M., Jr. A molecular theory of lipid-protein interactions in the plasma lipoproteins. FEBS Lett. 1974;38:247–258. doi: 10.1016/0014-5793(74)80064-5. [DOI] [PubMed] [Google Scholar]
- 13.Wald JH, Coormaghtigh E, De Meutter J, Ruysschaert JM, Jonas A. Investigation of the lipid domains and apolipoprotein orientation in reconstituted high density lipoproteins by fluorescence and IR methods. J Biol Chem. 1990;265:20044–20050. [PubMed] [Google Scholar]
- 14.Koppaka V, Silvestro L, Engler JA, Brouillette CG, Axelsen PH. The structure of human lipoprotein A-I. Evidence for the "belt" model. J Biol Chem. 1999;274:14541–14544. doi: 10.1074/jbc.274.21.14541. [DOI] [PubMed] [Google Scholar]
- 15.Segrest JP, Jones MK, Klon AE, Sheldahl CJ, Hellinger M, De Loof H, Harvey SC. A detailed molecular belt model for apolipoprotein A-I in discoidal high density lipoprotein. J Biol Chem. 1999;274:31755–31758. doi: 10.1074/jbc.274.45.31755. [DOI] [PubMed] [Google Scholar]
- 16.Bhat S, Sorci-Thomas MG, Alexander ET, Samuel MP, Thomas MJ. Intermolecular contact between globular N-terminal fold and C-terminal domain of ApoA-I stabilizes its lipid-bound conformation: studies employing chemical cross-linking and mass spectrometry. J Biol Chem. 2005;280:33015–33025. doi: 10.1074/jbc.M505081200. [DOI] [PubMed] [Google Scholar]
- 17.Davidson WS, Hilliard GM. The spatial organization of apolipoprotein A-I on the edge of discoidal high density lipoprotein particles: a mass specrometry study. J Biol Chem. 2003;278:27199–27207. doi: 10.1074/jbc.M302764200. [DOI] [PubMed] [Google Scholar]
- 18.Tricerri MA, Behling Agree AK, Sanchez SA, Bronski J, Jonas A. Arrangement of apolipoprotein A-I in reconstituted high-density lipoprotein disks: an alternative model based on fluorescence resonance energy transfer experiments. Biochemistry. 2001;40:5065–5074. doi: 10.1021/bi002815q. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Lyles DS, Thomas MJ, Pan W, Sorci-Thomas MG. Structural determination of lipid-bound ApoA-I using fluorescence resonance energy transfer. J Biol Chem. 2000;275:37048–37054. doi: 10.1074/jbc.M005336200. [DOI] [PubMed] [Google Scholar]
- 20.Bhat S, Sorci-Thomas MG, Tuladhar R, Samuel MP, Thomas MJ. Conformational adaptation of apolipoprotein A-I to discretely sized phospholipid complexes. Biochemistry. 2007;46:7811–7821. doi: 10.1021/bi700384t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas MJ, Bhat S, Sorci-Thomas MG. Three-dimensional models of HDL apoA-I: implications for its assembly and function. J Lipid Res. 2008;49:1875–1883. doi: 10.1194/jlr.R800010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sorci-Thomas MG, Owen JS, Fulp B, Bhat S, Zhu X, Parks JS, Shah D, Jerome WG, Gerelus M, Zabalawi M, Thomas MJ. Nascent high density lipoproteins formed by ABCA1 resemble lipid rafts and are structurally organized by three ApoA-I monomers. J Lipid Res. 2012;53:1890–1909. doi: 10.1194/jlr.M026674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gursky O. Crystal structure of Delta(185-243)ApoA-I suggests a mechanistic framework for the protein adaptation to the changing lipid load in good cholesterol: from flatland to sphereland via double belt, belt buckle, double hairpin and trefoil/tetrafoil. J Mol Biol. 2013;425:1–16. doi: 10.1016/j.jmb.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norum KR, Glomset JA, Nichols AV, Forte T. Plasma lipoproteins in familial lecithin: cholesterol acyltransferase deficiency: physical and chemical studies of low and high density lipoproteins. J Clin Invest. 1971;50:1131–1140. doi: 10.1172/JCI106585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchell CD, King WC, Applegate KR, Forte T, Glomset JA, Norum KR, Gjone E. Characterization of apolipoprotein E-rich high density lipoproteins in familial lecithin:cholesterol acyltransferase deficiency. J Lipid Res. 1980;21:625–634. [PubMed] [Google Scholar]
- 26.Glomset JA, Norum KR, King W. Plasma lipoproteins in familial lecithin: cholesterol acyltransferase deficiency: lipid composition and reactivity in vitro. J Clin Invest. 1970;49:1827–1837. doi: 10.1172/JCI106400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lowry OJ, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;19:265–275. [PubMed] [Google Scholar]
- 28.Owen JS, Bharadwaj MS, Thomas MJ, Bhat S, Samuel MP, Sorci-Thomas MG. Ratio determination of plasma wild-type and L159R apoA-I using mass spectrometry: tools for studying apoA-IFin. J Lipid Res. 2007;48:226–234. doi: 10.1194/jlr.D600031-JLR200. [DOI] [PubMed] [Google Scholar]
- 29.Bhat S, Zabalawi M, Willingham MC, Shelness GS, Thomas MJ, Sorci-Thomas MG. Quality control in the apoA-I secretory pathway: deletion of apoA-I helix 6 leads to the formation of cytosolic phospholipid inclusions. J Lipid Res. 2004;45:1207–1220. doi: 10.1194/jlr.M300498-JLR200. [DOI] [PubMed] [Google Scholar]
- 30.Li HH, Thomas MJ, Pan W, Alexander E, Samuel M, Sorci-Thomas MG. Preparation and incorporation of probe-labeled apoA-I for fluorescence resonance energy transfer studies of rHDL. J Lipid Res. 2001;42:2084–2091. [PubMed] [Google Scholar]
- 31.Li HH, Lyles DS, Pan W, Alexander E, Thomas MJ, Sorci-Thomas MG. ApoA-I structure on discs and spheres. Variable helix registry and conformational states. J Biol Chem. 2002;277:39093–39101. doi: 10.1074/jbc.M206770200. [DOI] [PubMed] [Google Scholar]
- 32.Bhat S, Sorci-Thomas MG, Calabresi L, Samuel MP, Thomas MJ. Conformation of dimeric apolipoprotein A-I milano on recombinant lipoprotein particles. Biochemistry. 2010;49:5213–5224. doi: 10.1021/bi1003734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alexander ET, Bhat S, Thomas MJ, Weinberg RB, Cook VR, Bharadwaj MS, Sorci-Thomas M. Apolipoprotein A-I helix 6 negatively charged residues attenuate lecithin-cholesterol acyltransferase (LCAT) reactivity. Biochemistry. 2005;44:5409–5419. doi: 10.1021/bi047412v. [DOI] [PubMed] [Google Scholar]
- 34.Mei X, Atkinson D. Crystal structure of C-terminal truncated apolipoprotein A-I reveals the assembly of high density lipoprotein (HDL) by dimerization. J Biol Chem. 2011;286:38570–38582. doi: 10.1074/jbc.M111.260422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 36.Vitello LB, Scanu AM. Studies on human serum high density lipoproteins. Self-association of apolipoprotein A-I in aqueous solutions. J Biol Chem. 1976;251:1131–1136. [PubMed] [Google Scholar]
- 37.Teng TL, Edelstein C, Barbeau DL, Scanu AM. An ultracentrifugal study of the self-association of canine apolipoprotein A-I in solution. J Biol Chem. 1977;252:8634–8638. [PubMed] [Google Scholar]
- 38.Barbeau DL, Jonas A, Teng T, Scanu AM. Asymmetry of apolipoprotein A-I in solution as assessed from ultracentrifugal, viscometric, and fluorescence polarization studies. Biochemistry. 1979;18:362–369. doi: 10.1021/bi00569a021. [DOI] [PubMed] [Google Scholar]
- 39.Roberts LM, Ray MJ, Shih TW, Hayden E, Reader MM, Brouillette CG. Structural analysis of apolipoprotein A-I: limited proteolysis of methionine-reduced and - oxidized lipid-free and lipid-bound human apo A-I. Biochemistry. 1997;36:7615–7624. doi: 10.1021/bi962952g. [DOI] [PubMed] [Google Scholar]
- 40.Rogers DP, Roberts LM, Lebowitz J, Datta G, Anantharamaiah GM, Engler JA, Brouillette CG. The lipid-free structure of apolipoprotein A-I: effects of amino-terminal deletions. Biochemistry. 1998;37:11714–11725. doi: 10.1021/bi973112k. [DOI] [PubMed] [Google Scholar]
- 41.Rogers DP, Roberts LM, Lebowitz J, Engler JA, Brouillette CG. Structural analysis of apolipoprotein A-I: effects of amino- and carboxy-terminal deletions on the lipid-free structure. Biochemistry. 1998;37:945–955. doi: 10.1021/bi9713512. [DOI] [PubMed] [Google Scholar]
- 42.Saito H, Dhanasekaran P, Nguyen D, Holvoet P, Lund-Katz S, Phillips MC. Domain structure and lipid interaction in human apolipoproteins A-I and E, a general model. J Biol Chem. 2003;278:23227–23232. doi: 10.1074/jbc.M303365200. [DOI] [PubMed] [Google Scholar]
- 43.Rogers DP, Brouillette CG, Engler JA, Tendian SW, Roberts L, Mishra VK, Anantharamaiah GM, Lund-Katz S, Phillips MC, Ray MJ. Truncation of the amino terminus of human apolipoprotien A-I substantially alters only the lipid-free conformation. Biochemistry. 1997;36:288–300. doi: 10.1021/bi961876e. [DOI] [PubMed] [Google Scholar]
- 44.Chetty PS, Mayne L, Lund-Katz S, Stranz D, Englander SW, Phillips MC. Helical structure and stability in human apolipoprotein A-I by hydrogen exchange and mass spectrometry. Proc Natl Acad Sci U S A. 2009;106:19005–19010. doi: 10.1073/pnas.0909708106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lagerstedt JO, Budamagunta MS, Liu GS, Devalle NC, Voss JC, Oda MN. The "beta-clasp" model of apolipoprotein A-I - A lipid-free solution structure determined by electron paramagnetic resonance spectroscopy. Biochim Biophys Acta. 2012;1821:448–455. doi: 10.1016/j.bbalip.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brouillette CG, Dong WJ, Yang ZW, Ray MJ, Protasevich II, Cheung HC, Engler JA. Forster resonance energy transfer measurements are consistent with a helical bundle model for lipid-free apolipoprotein A-I. Biochemistry. 2005;44:16413–16425. doi: 10.1021/bi051018v. [DOI] [PubMed] [Google Scholar]
- 47.Wu Z, Wagner MA, Zheng L, Parks JS, Shy JM, 3rd, Smith JD, Gogonea V, Hazen SL. The refined structure of nascent HDL reveals a key functional domain for particle maturation and dysfunction. Nat Struct Mol Biol. 2007;14:861–868. doi: 10.1038/nsmb1284. Erratum: Nat Struct Mol Biol. 2008. 2015: 2330. [DOI] [PubMed] [Google Scholar]
- 48.Mayne L, Kan ZY, Chetty PS, Ricciuti A, Walters BT, Englander SW. Many overlapping peptides for protein hydrogen exchange experiments by the fragment separation-mass spectrometry method. J Am Soc Mass Spectrom. 2011;22:1898–1905. doi: 10.1007/s13361-011-0235-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fass D. Disulfide bonding in protein biophysics. Annu Rev Biophys. 2012;41:63–79. doi: 10.1146/annurev-biophys-050511-102321. [DOI] [PubMed] [Google Scholar]
- 50.Leroy A, Jonas A. Native-like structure and self-association behavior of apolipoprotein A-I in a water/n-propanol solution. Biochim Biophys Acta. 1994;1212:285–294. doi: 10.1016/0005-2760(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 51.Reijngoud DJ, Phillips MC. Mechanism of dissociation of human apolipoprotein A-I from complexes with dimyristoylphosphatidylcholine as studied by guanidine hydrochloride denaturation. Biochemistry. 1982;21:2969–2976. doi: 10.1021/bi00541a026. [DOI] [PubMed] [Google Scholar]
- 52.Saito H, Dhanasekaran P, Nguyen D, Deridder E, Holvoet P, Lund-Katz S, Phillips MC. Alpha-helix formation is required for high affinity binding of human apolipoprotein A-I to lipids. J Biol Chem. 2004;279:20974–20981. doi: 10.1074/jbc.M402043200. [DOI] [PubMed] [Google Scholar]
- 53.Chen J, Wang J, Yang Y. Monomeric mouse apoAI(1-216) In Protein Data Bank. 2012 p 2LEM, RCSB. [Google Scholar]
- 54.Wilson C, Wardell MR, Weisgraber KH, Mahley RW, Agard DA. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991;252:1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- 55.Gursky O, Mei X, Atkinson D. The crystal structure of the C-terminal truncated apolipoprotein a-I sheds new light on amyloid formation by the N-terminal fragment. Biochemistry. 2012;51:10–18. doi: 10.1021/bi2017014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davidson WS, Silva RA. Apolipoprotein structural organization in high density lipoproteins: belts, bundles, hinges and hairpins. Curr Opin Lipidol. 2005;16:295–300. doi: 10.1097/01.mol.0000169349.38321.ad. [DOI] [PubMed] [Google Scholar]
- 57.Sparks DL, Frank PG, Braschi S, Neville TA, Marcel YL. Effect of apolipoprotein A-I lipidation on the formation and function of pre-beta and alpha-migrating LpA-I particles. Biochemistry. 1999;38:1727–1735. doi: 10.1021/bi981945k. [DOI] [PubMed] [Google Scholar]
- 58.Jayaraman S, Abe-Dohmae S, Yokoyama S, Cavigiolio G. Impact of self-association on function of apolipoprotein A-I. J Biol Chem. 2011;286:35610–35623. doi: 10.1074/jbc.M111.262485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Asztalos BF, Roheim PS, Milani RL, Lefevre M, McNamara JR, Horvath KV, Schaefer EJ. Distribution of apoA-I-containing HDL subpopulations in patients with coronary heart disease. Arteriosclerosis, Thrombosis, and Vascular Biology. 2000;20:2670–2676. doi: 10.1161/01.atv.20.12.2670. [DOI] [PubMed] [Google Scholar]
- 60.Schonfeld G, Pfleger B. The structure of human high density lipoprotein and the levels of apolipoprotein A-I in plasma as determined by radioimmunoassay. J Clin Invest. 1974;54:236–246. doi: 10.1172/JCI107758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Davidson WS, Hazlett T, Mantulin WW, Jonas A. The role of apolipoprotein AI domains in lipid binding. Proc Natl Acad Sci U S A. 1996;93:13605–13610. doi: 10.1073/pnas.93.24.13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gwynne J, Brewer B, Jr., Edelhoch H. The molecular properties of ApoA-I from human high density lipoprotein. J Biol Chem. 1974;249:2411–2416. [PubMed] [Google Scholar]
- 63.Willard L, Ranjan A, Zhang H, Monzavi H, Boyko RF, Sykes BD, Wishart DS. VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Res. 2003;31:3316–3319. doi: 10.1093/nar/gkg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Voss NR, Gerstein M. 3V: cavity, channel and cleft volume calculator and extractor. Nucleic Acids Res. 2010;38:W555–562. doi: 10.1093/nar/gkq395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gursky O, Atkinson D. Thermal unfolding of human high-density apolipoprotein A-1: implications for a lipid-free molten globular state. Proc Natl Acad Sci U S A. 1996;93:2991–2995. doi: 10.1073/pnas.93.7.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Young MM, Tang N, Hempel JC, Oshiro CM, Taylor EW, Kuntz ID, Gibson BW, Dollinger G. High throughput protein fold identification by using experimental constraints derived from intramolecular cross-links and mass spectrometry. Proc Natl Acad Sci U S A. 2000;97:5802–5806. doi: 10.1073/pnas.090099097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leitner A, Reischl R, Walzthoeni T, Herzog F, Bohn S, Forster F, Aebersold R. Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.014126. M111 014126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang BX, Kim HY, Dass C. Probing three-dimensional structure of bovine serum albumin by chemical cross-linking and mass spectrometry. J Am Soc Mass Spectrom. 2004;15:1237–1247. doi: 10.1016/j.jasms.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 69.Huang BX, Dass C, Kim HY. Probing conformational changes of human serum albumin due to unsaturated fatty acid binding by chemical cross-linking and mass spectrometry. Biochem J. 2005;387:695–702. doi: 10.1042/BJ20041624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jacobsen RB, Sale KL, Ayson MJ, Novak P, Hong J, Lane P, Wood NL, Kruppa GH, Young MM, Schoeniger JS. Structure and dynamics of dark-state bovine rhodopsin revealed by chemical cross-linking and high-resolution mass spectrometry. Protein Sci. 2006;15:1303–1317. doi: 10.1110/ps.052040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kalisman N, Adams CM, Levitt M. Subunit order of eukaryotic TRiC/CCT chaperonin by cross-linking, mass spectrometry, and combinatorial homology modeling. Proc Natl Acad Sci U S A. 2012;109:2884–2889. doi: 10.1073/pnas.1119472109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, Aebersold R. Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol Cell Proteomics. 2010;9:1634–1649. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen ZA, Jawhari A, Fischer L, Buchen C, Tahir S, Kamenski T, Rasmussen M, Lariviere L, Bukowski-Wills JC, Nilges M, Cramer P, Rappsilber J. Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 2010;29:717–726. doi: 10.1038/emboj.2009.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Seebacher J, Mallick P, Zhang N, Eddes JS, Aebersold R, Gelb MH. Protein cross-linking analysis using mass spectrometry, isotope-coded cross-linkers, and integrated computational data processing. J Proteome Res. 2006;5:2270–2282. doi: 10.1021/pr060154z. [DOI] [PubMed] [Google Scholar]
- 75.Green NS, Reisler E, Houk KN. Quantitative evaluation of the lengths of homobifunctional protein cross-linking reagents used as molecular rulers. Protein Sci. 2001;10:1293–1304. doi: 10.1110/ps.51201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lu B, Morrow JA, Weisgraber KH. Conformational reorganization of the four-helix bundle of human apolipoprotein E in binding to phospholipid. J Biol Chem. 2000;275:20775–20781. doi: 10.1074/jbc.M003508200. [DOI] [PubMed] [Google Scholar]