Abstract
Mammaglobin-A (MGBA), a 10-kD protein, is over expressed in 80% of primary and metastatic human breast cancers. Breast cancer patients demonstrate high frequencies of CD8+ cytotoxic T lymphocytes (CTL) specific to MGBA. Defining CD8+ CTL responses to HLA class I-restricted MGBA-derived epitopes assumes significance in the context of our ongoing efforts to clinically translate vaccine strategies targeting MGBA for prevention and/or treatment of human breast cancers. In this study, we define the CD8+ CTL response to MGBA-derived candidate epitopes presented in the context of HLA-B7, which has a frequency of 17.7% in Caucasian and 15.5% in African American populations. We identified seven MGBA-derived candidate epitopes with high predicted binding scores for HLA-B7 using a computer algorithm. Membrane stabilization studies with TAP-deficient T2 cells transfected with HLA-B7 indicated that MGBA B7.3 (VSKTEYKEL), B7.6 (KLLMVLMLA), B7.7 (NPQVSKTEY), and B7.1 (YAGSGCPLL) have the highest HLA-B7 binding affinities. Further, two CD8+ CTL cell lines generated in vitro against T2.B7 cells individually loaded with MGBA-derived candidate epitopes showed significant cytotoxic activity against MGBA B7.1, B7.3, B7.6, and B7.7. In addition, the same CD8+ CTL lines lysed the HLA-B7+/MGBA+ human breast cancer cell line DU-4475 but had no significant cytotoxicity against HLA-B7− or MGBA− breast cancer cell lines. Cold-target inhibition studies strongly suggest that MGBA B7.3 is an immunodominant epitope. In summary, our results define HLA-B7-restriced, MGBA-derived, CD8+ CTL epitopes with all of the necessary features for developing novel vaccine strategies against HLA-B7 expressing breast cancer patients.
Keywords: Vaccine, CD8 T cell, Mammaglobin-A
Introduction
Mammaglobin-A (MGBA) is a novel breast cancer-associated antigen that was initially identified using a differential screening approach [1]. Subsequently, it has been demonstrated that MGBA has several unique properties which make it a promising target for immune therapy [2, 3]. Gene expression analysis of over 20 different human tissues demonstrated that MGBA is expressed almost exclusively in normal breast epithelium and breast cancer [4]. MGBA is over expressed in up to 80% of primary and metastatic breast cancers [1–3, 5–7]. Near-universal expression of MGBA in breast cancer indicates that the majority of breast cancer patients are likely to be candidates for therapies targeting MGBA, in contrast to Her2/ neu, which is overexpressed in only 30% of breast cancer patients [8]. In addition, the overexpression of MGBA appears to be consistent in all stages of disease [6]. MGBA overexpression is evident in noninvasive breast cancer, well differentiated, and poorly differentiated primary breast cancer, and metastatic disease. Although the functional biology of this molecule remains to be defined, the universal expression of MGBA in breast cancer tissues confirms that it is an attractive target for vaccine therapy.
DNA vaccination is a strategy to induce protective immune responses to infectious disease and cancer. DNA vaccination offers several potential advantages including ease of manufacture and high stability relative to proteins and other biologic agents [9, 10]. In clinical trials, DNA vaccine strategies have been shown to be safe and effective in developing immune responses to malaria [11], HIV [12, 13], and prostate cancer [14]. Vaccine strategies capable of generating immune responses against a broadly expressed tumor antigen such as MGBA are likely to have a significant therapeutic impact in patients with breast cancer. We have previously demonstrated that passive transfer of immune cells from HLA-A2/human CD8 transgenic mice vaccinated with a MGBA DNA vaccine results in breast cancer immunity in a pre-clinical model of human breast cancer [15]. Vaccinated mice demonstrated specific lysis of MGBA+ human breast cancers both in vitro and in vivo. We have previously identified epitopes for HLA-A2 and HLA-A3, the most common HLA class I alleles in the Caucasian population [16–18]. In this communication, we have identified four candidate MGBA-derived epitopes that bind to HLA-B7. CD8+ CTL generated in vitro to these epitopes can specifically lyse human breast cancer cells in a MGBA-specific and HLA-B7-restricted manner.
Materials and methods
Study subjects
Two HLA-B7+ healthy female volunteers were enrolled in this study after obtaining informed consent. HLA typing was performed using sequence-specific oligonucleotide probes that provide low-medium resolution for HLA-B alleles (Dynal Biotech, Lafayette Hill, PA).
Breast cancer cell lines
Breast cancer cell lines were obtained from the American Type Culture Collection (Manassas, VA) (Table 1). HLA typing of the breast cancer cell lines was performed as described above. MGBA expression was determined by reverse transcriptase–polymerase chain reaction and western blotting as previously described [15, 19].
Table 1.
Breast cancer cell lines
Peptides
MGBA peptides that bind the HLA-B7 molecule were identified using the HLA class I peptide binding computer algorithm from the Bioinformatics & Molecular Analysis Section of the National Institutes of Health at http://www-bimas.cit.nih.gov/molbio/hla_bind. Seven MGBA-derived peptides with varying levels of binding affinity for the HLA-B7 molecule were synthesized (Table 2). The EBV peptide RPPIFIRRL, known to bind to the HLA-B7 allele [20], was used as a positive control. Peptides were synthesized by Sigma Genosys. The purity and identity of the peptides were confirmed by high-performance liquid chromatography and mass spectrometry.
Table 2.
HLA-B7-restricted MGBA-derived candidate epitopes
| Peptide | Start position |
Peptide sequence |
HLA-B7 binding scorea |
|---|---|---|---|
| MGBA B7.1 | 17 | YAGSGCPLL | 12.000 |
| MGBA B7.2 | 54 | ATTNAIDEL | 12.000 |
| MGBA B7.3 | 37 | VSKTEYKEL | 4.000 |
| MGBA B7.4 | 4 | LMVLMLAAL | 4.000 |
| MGBA B7.5 | 75 | SNVEVFMQL | 4.000 |
| MGBA B7.6 | 2 | KLLMVLMLA | 0.100 |
| MGBA B7.7 | 34 | NPQVSKTEY | 0.400 |
Estimate of half time of HLA-B7-peptide dissociation based on HLA-peptide binding prediction software from the Bioinformatics and Molecular Analysis Section of the National Institutes of Health at http://www-bimas.cit.nih.gov/molbio/hla_bind
HLA-B7/β2 microglobulin membrane stabilization assay
HLA-B7 membrane stabilization assays were performed in TAP-deficient T2 cells transfected with HLA-B*0702 (T2.B7) [21]. T2.B7 was a gift from Dr. Charles Lutz, University of Kentucky. T2.B7 cells (1 × 106/ml) were incubated in flat-bottom 96-well plates at 26 °C in the presence of each peptide (40 µg/ml) in 200 µl of RPMI-1640 medium (Gibco, Grand Island, NY) supplemented with 10% defined fetal bovine serum (HyClone, Logan, UT), 100 µM non-essential amino acids, 2 mM l-glutamine, 25 mM HEPES, 1 mM sodium pyruvate, 100 units/ ml penicillin, and 100 g/ml streptomycin, and 3 µg/ml of human β2m (Sigma). After 16 h of incubation, the T2.B7 cells were washed three times, and the levels of HLA-B7 expression were determined by flow cytometry. Briefly, the T2.B7 cells were incubated for 30 min at 4°C with the anti-HLA B7 IgG1 mAb, ME1 (1:10 dilution) which was derived from the HB-119 hybridoma cell line (ATCC, VA) and resuspended in phosphate-buffered saline (PBS) supplemented with 2% bovine serum albumin (BSA), 25 mM HEPES, and 0.02% sodium azide. The cells were then washed three times and incubated for 30 min at 4°C with FITC-conjugated goat anti-mouse IgG (10 µg/ml, Becton–Dickinson, Franklin Lakes, NJ). The cells were then washed three times, fixed in 1% paraformaldehyde, and analyzed by single-color flow cytometry using a FACScan flow cytometer (Becton–Dickinson). T2.B7 cells incubated in the presence of the EBV-binding peptide RPPIFIRRL were used as a positive control. Results were expressed as the mean fluorescence shift, and corresponded to the difference between the mean fluorescence obtained with T2.B7 cells incubated in the presence of the peptide (experimental) and the mean fluorescence obtained with “unloaded” T2.B7 cells cultured in the absence of peptides (negative control). No mean fluorescence shift was observed with the MOPC-21 mAb (data not shown).
Generation of MGBA-reactive HLA-B7-restricted CD8+ CTL lines
[17, 18]We generated two CD8+ CTL cell lines against HLA-B7-restricted MGBA epitopes in vitro as described previously [18]. Briefly, peripheral blood lymphocytes (2 × 106) were cultured in 2 ml of RPMI-1640 medium, supplemented as described above, in 24-well plates in the presence of a pool of irradiated (10,000 rad) T2.B7 cells (1 × 106) loaded with MGBA B7.1-7 or control EBV peptide RPPIFIRRL. β2m (3 µg/ml) and CD28.2 anti-CD28 mAb (500 ng/ml, BD Biosciences) were added to the cultures. In addition, recombinant human IL-2 (20 U/ml, Chiron, Emeryville, CA) and 5% T cell growth factor prepared from pooled human splenocyte supernatant were added to the cultures after 24 h. PBLs (2 × 105) were re-stimulated every 8–10 days with irradiated, peptideloaded T2.B7 cells (1 × 106) in the presence of irradiated (3,000 rad) autologous peripheral blood lymphocytes (2 × 106) in 24-well plates in 2 ml of culture medium supplemented with IL-2, β2m, and anti-CD28. After stimulation the CD8+ CTL were purified by negative selection (Human CD8+ enrichment kit, Stem Cell, Canada). The purity of resulting CD8+ T cells obtained by this protocol was >95% (data not shown).The MGBA peptide-specific cytotoxic activities of the resulting CD8+ CTL lines were analyzed 7 days after four stimulations. In parallel experiments, PBLs (2 × 106 cells) from the same individuals were stimulated with irradiated (6000 rad) DU-4475 (1 × 106 cells) in 2 ml of complete RPMI-1640 supplemented with β2m (3 µg/ml) and CD28.2 anti-CD28 mAb (500 ng/ml), recombinant human IL-2 (20 U/ml), and 5% human splenocyte supernatant were added to the cultures after 24 h as described above. PBLs (2 × 105) were re-stimulated every 8–10 days with irradiated, DU-4475(1 × 106) in the presence of irradiated (3,000 rad) autologous peripheral blood lymphocytes (2 × 106) in 24-well plates in 2 ml of culture medium supplemented with IL-2, β2m, and anti-CD28. At the end of four stimulations, CD8+ CTLs were isolated by immunomagnetic separation (Stem Cell) and the resulting purity was verified to be >95% (data not shown).
Cytotoxic T lymphocyte assay
Peptide-loaded T2.B7 cells or breast cancer cells were labeled with 51Cr (250 µCi, ICN Pharmaceuticals, Costa Mesa, CA) in 100 µl of complete medium. After 1 h, 5 × 103 labeled cells were plated in triplicate cultures in round bottom 96-well plates in the presence of varying numbers of CD8+ T cells and incubated at 37° C in a humidified 5% CO2 incubator for 4 h. K562, an HLA class I-negative cell line, was used to test non-specific lysis by NK cells. Control wells for determining spontaneous 51Cr release contained labeled target cells alone. Maximal release was determined by adding Triton X-100 (1%) to the target cells. The percent specific lysis was calculated as follows: (experimental 51Cr release − spontaneous 51Cr release)/(maximum 51Cr release − spontaneous 51Cr release)] × 100. Epitope specificity of breast cancer lysis by CD8+ CTLs was further determined in a cold-target inhibition assay by analyzing the capacity of peptide-loaded unlabeled T2.B7 cells to block the lysis of breast cancer cells at an inhibitor to target ratio of 50:1.
Results
Identification of candidate MGBA-derived HLA-B7 epitopes
MGBA-derived epitopes for the HLA-B7 molecule were identified using the HLA class I peptide binding computer algorithm at the Bioinformatics & Molecular Analysis Section of the National Institutes of Health. Peptides with varying levels of binding affinity for the HLA-B7 molecule were identified and synthesized by Sigma Genosys. The EBV peptide sequence RPPIFIRRL was also synthesized and used as positive control. Peptide sequences and their predicted score for binding to HLA-B7 are shown in Table 1.
Confirmation of HLA-B7 specificity of the mAb ME1
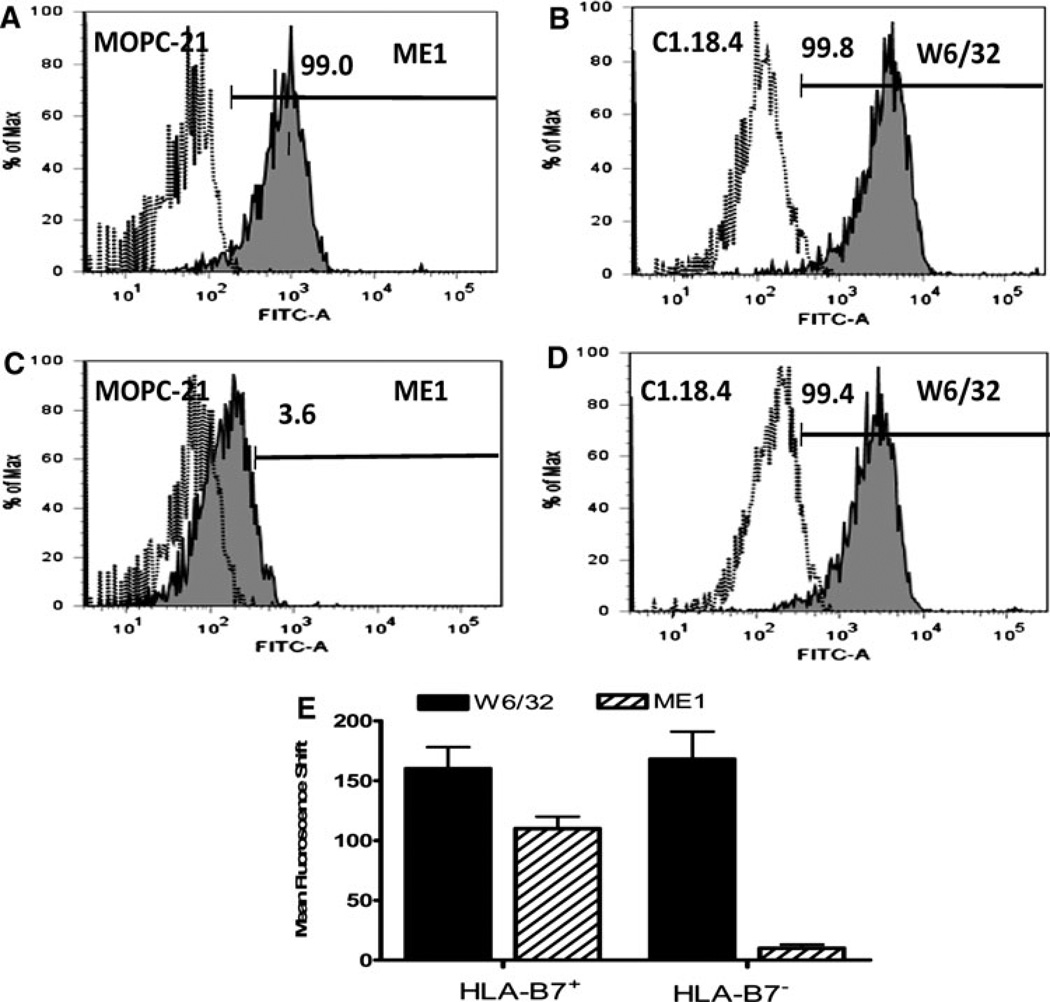
The mAb ME1 (IgG1) was harvested from HB-119 hybridoma culture supernatants. Specific binding of ME1 to HLA-B7 was determined using HLA B7+ (n = 3) and HLA-B7− (n = 3) PBMC from healthy donors by flow cytometry (Fig. 1). Figure 1 is a representative plot obtained from one healthy individual demonstrating binding of ME1 to HLA-B7+ PBMC(A). No binding is seen with HLA-B7− PBMC(C). A pan-specific anti-HLA Class I binding antibody, W6/32, was used as a positive control to demonstrate binding against HLA-B7+ PBMC(B) and HLA-B7− PBMC(D). Isotype control antibodies to ME1, MOPC-21 (IgG1) and W6/32, C1.18.4 (IgG2a) gave negative results, as expected.
Fig. 1.
ME1 is specific for HLA-B7. ME1 was harvested from HB-119 hybridoma culture supernatants. ME1 specificity was tested using HLA B7+ and HLA-B7− PBMCs from three healthy donors in each group by flow cytometry. a and c is a representative plot derived from one healthy individual in each group demonstrating specific binding of ME1 (gray filled plots) to HLA-B7+ PBMC obtained (a) but not against HLA-B7− PBMC (c). A panspecific anti-HLA class I binding Ab, W6/32 (gray filled plots), was used as a positive control (b, d). Isotype control Abs to W6/32 (C1.18.4) and ME1 (MOPC-21) (open plots) gave negative results as expected. e Indicates the mean fluorescence shift obtained with W6/32 and ME1 mAbs against HLA-B7+ (n = 3) and HLA-B7 − (n = 3) PBMCs
Binding affinity of candidate MGBA-derived epitopes to HLA-B7 by membrane stabilization assay
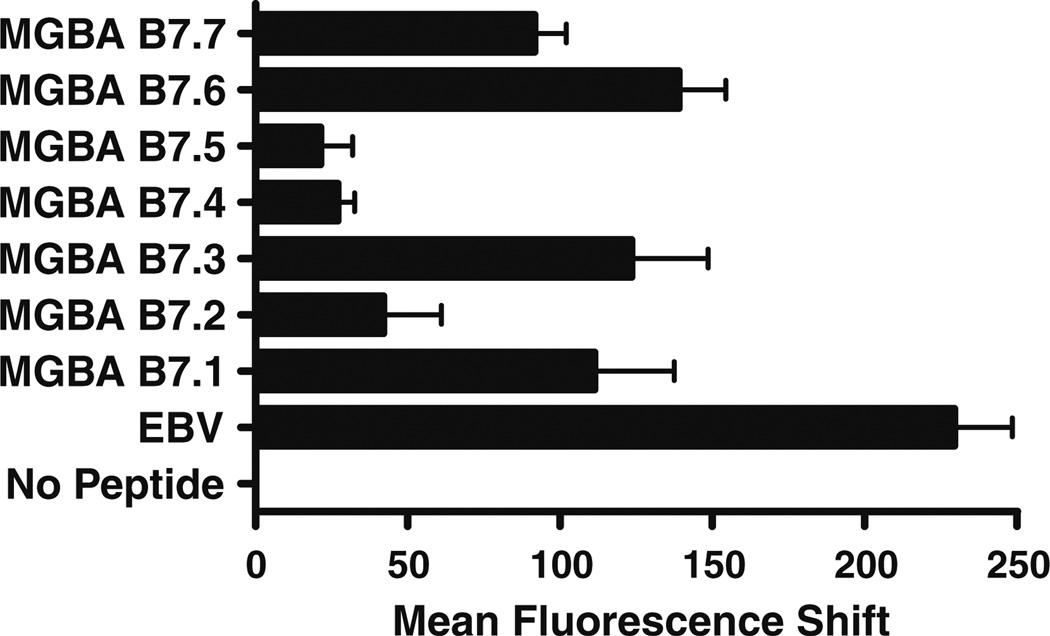
Based on the computer algorithm, we synthesized and tested the MGBA 9-mer peptides with the highest predicted binding affinity for HLA-B7. Seven MGBA peptides were analyzed (Table 2). The actual binding affinity of the candidate epitopes to the HLA-B7 molecule was determined by membrane stabilization assay using TAP-deficient T2 cells transfected with HLA-B*0702 (T2.B7). As shown in Fig. 2, the peptides MGBA B7.6 (KLLMVLMLA), MGBA B7.3 (VSKTEYKEL), MGBA B7.1 (YAGSGCPLL), and MGBA B7.7 (NPQVSKTEY) displayed high affinity for the HLA-B7 molecule, comparable to the binding affinity of the EBV-derived peptide RPPIFIRRL. In contrast, the peptides MGBA B7.2 (ATTNAIDEL), MGBA B7.4 (LMVLMLAAL), and MGBA B7.5 (SNVEVFMQL) demonstrated significantly lower binding. These results clearly demonstrate a discrepancy between the algorithm predictions and the actual binding capacity of the candidate MGBA-derived epitopes. This is expected since the binding affinity for a given epitope in the MHC class I groove is determined by both its amino acid sequence as well as the three-dimensional structure of the binding motif [22, 23]. Several studies have documented this phenomenon between the predicted and the experimental binding affinity for MHC class I epitopes [24, 25].
Fig. 2.
HLA-B7 membrane stabilization by MGBA-derived candidate epitopes is a surrogate for binding affinity. Membrane stabilization assays were performed in TAP-deficient T2.B7 cells incubated in the presence of candidate epitopes for 16 h. Surface expression of HLAB7 was determined by flow cytometry using the ME1 mAb. T2.B7 cells incubated in the presence of RPPIFIRRL, EBV-derived epitope known to bind HLA-B7, were used as a positive control. Results are expressed as mean fluorescence shift ± standard deviation, and represent the mean fluorescence intensity obtained with T2.B7 cells in the presence of the indicated peptides and the mean fluorescence intensity obtained with T2.B7 cells cultured in the absence of peptides. The results indicate that MGBA B7.6, B7.3, B7.1 and B7.7 are the epitopes with the highest HLA-B7 specific binding affinity
CD8+ CTL response to HLA-B7-restricted, MGBA-derived epitopes
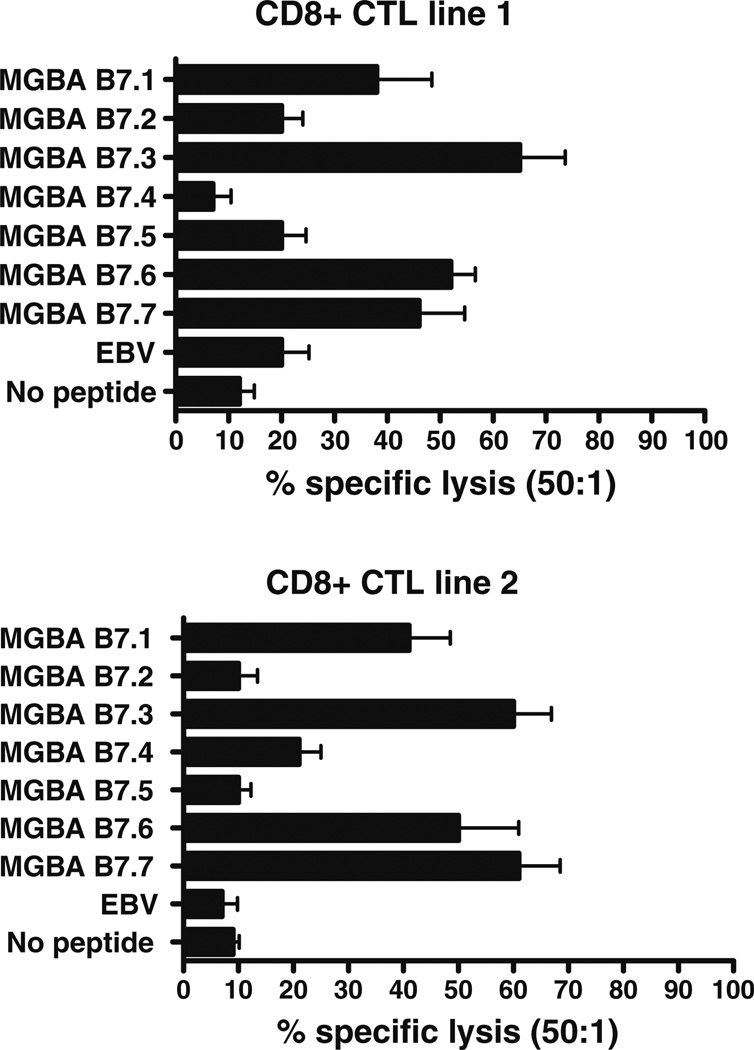
In order to determine whether the HLA-B7-restricted, MGBA-derived, epitopes identified above could induce CD8+ CTL, PBL from two HLA-B7+ healthy female individuals were stimulated in vitro in the presence of a pool of T2.B7 cells, individually loaded with all of the candidate HLA-B7-binding MGBA-derived peptides. Since the affinity of a given peptide to a particular MHC class I molecule does not necessarily correlate with its ability to generate CD8+ CTL responses, we used all the candidate epitopes to examine the profile of CD8+ CTL reactivity. After four stimulations, the cytotoxic activity of the resulting CD8+ CTL lines and their epitope specificity was evaluated using standard 51Cr release assays. The composition of cultures after four stimulations prior to CD8+ negative selection in both CD8+ CTL lines 1 and 2 were: CD3+CD8+: 48.7 ± 18.5%, CD3+CD4+: 31.6 ± 15.2%, CD19+: 3.7 ± 2.9%, CD56+ : 1.5 ± 2.2%. T2 cells loaded with EBV-derived epitope RPPIFIRRL and empty T2.B7 (no peptide) were used as negative controls. The optimal loading dose for cytotoxicity was titrated at varying peptide concentrations (1, 5, 10, 20, 40, 80, 160 µg/ml) following HLA-B7/β2 membrane stabilization assay and was determined to be 40 µg/ml for individual peptides. However, no significant differences in cytotoxicity were noted with varying doses of individual peptides. The results shown in Fig. 3 demonstrate that both CD8+ CTL lines showed significant cytotoxic activity against MGBA-derived epitopes B7.1, B7.3, B7.6, and B7.7 but showed minimal cytotoxic activity against B7.2, B7.4 and B7.5.CD8+ CTLs from both cell lines demonstrated a similar profile of cytotoxicity up to the lower ratio of 12.5:1 (No peptide—8 ± 3, EBV—13 ± 6, MGBA 7.1—24 ± 7, MGBA 7.2—14, MGBA 7.3—49 ± 18, MGBA 7.4—5 ± 3, MGBA 7.5—14 ± 9, MGBA 7.6— 41 ± 16, and MGBA 7.7—37 ± 14).
Fig. 3.
Defining the CD8+ CTL response to candidate MGBA-derived HLA-B7-restricted epitopes. Two CD8+ CTL lines were generated from individual HLA-B7+ healthy individuals by in vitro stimulations with pooled T2.B7 cells individually loaded with candidate MGBA-derived epitopes. The epitope specificity of the CD8+ CTL lines was then evaluated using T2.B7 cell lines individually loaded with candidate epitopes by means of a standard 51Cr release assay. T2 cells loaded with RPPIFIRRL (EBV-derived epitope) and empty T2.B7 cells (no peptide) were used as negative controls. The assay was performed at E:T ratios of 50:1, 25:1, 12.5:1 and 6.25:1 in triplicate cultures. Results obtained at E:T ratios of 50:1 are presented as mean ± standard deviation
Specific lysis of human breast cancer cells by CD8+ CTL generated against MGBA-derived epitopes
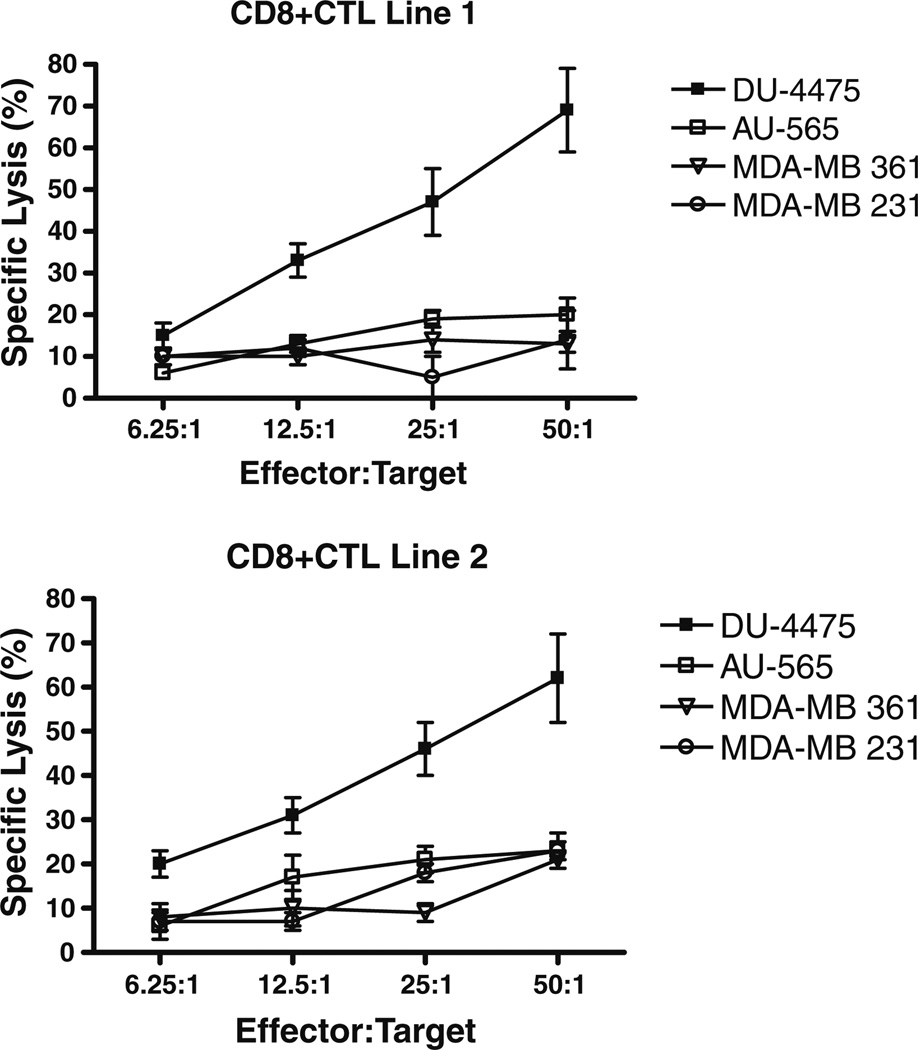
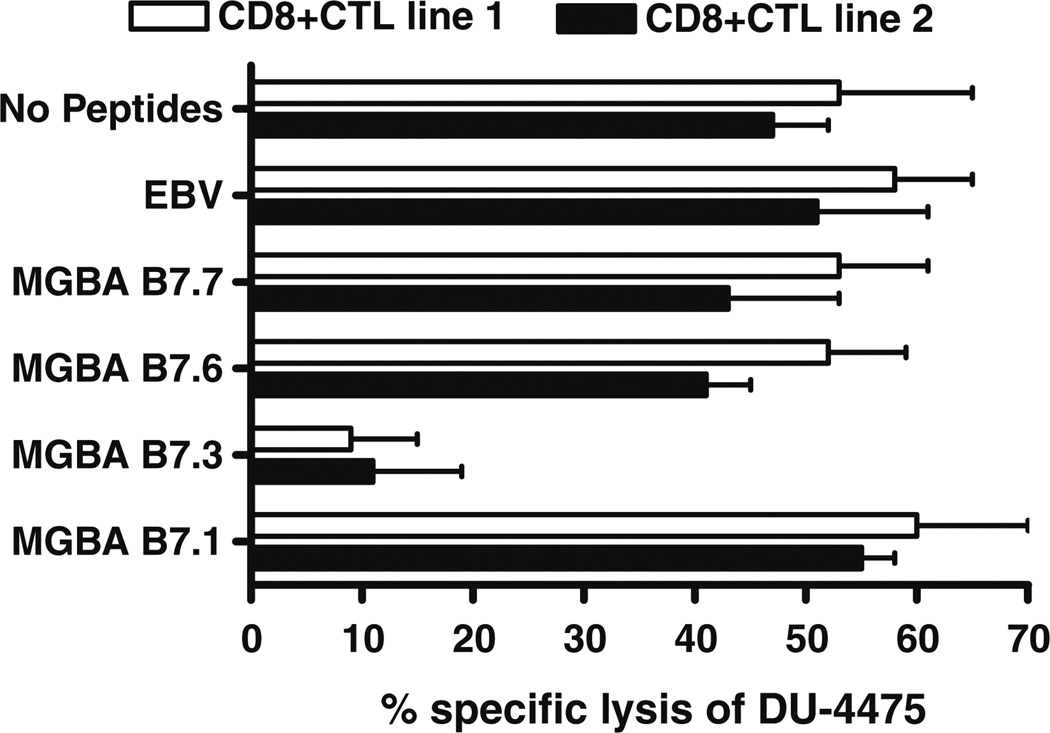
In order to determine the cytotoxic reactivity of HLA-B7-restricted CD8+ CTL against human breast cancer cell lines that endogenously express HLA-B7 and MGBA, we determined the specific lysis of four different breast cancer cell lines by CD8+ CTL lines generated in vitro. The human breast cancer cell lines used for this assay, their HLA phenotypes, as well as the status of MGBA expression are given in Table 1. The CD8+ CTL generated from two HLA-B7+ healthy donors were stimulated by pooled T2.B7 cells loaded with all seven peptides (B7.1–7.7) derived from MGBA. As demonstrated in Fig. 4, both CD8+ CTL lines demonstrated significant cytotoxicity against the breast cancer cell line DU-4475, which expresses both HLA-B7 and MGBA but not against other breast cancer cell lines AU-565 (HLA-B7−/MGBA+)[26], MDA-361 (HLA-B7−/MGBA+) [27], and MDA-231 (HLA-B7−/MGBA−) [28]. More importantly, no cytotoxicity was observed against K562, which is sensitive to NK cell-mediated lysis (CTL line 1: 1.1 ± 0.4%; CTL line 2: 1.5 ± 0.3% at E:T ratio 50:1). At the lower E:T ratios (12.5:1 and 6.25:1) the CTL cell lines 1 and 2 showed significant lysis of DU-4475 but not AU-565, MDA-361, and MDA-231. However, at higher E:T ratios (50:1), a minimal cytotoxicity (AU-565: 18 ± 5, MDA-361: 11 ± 4, MDA-231:12 ± 7) with either HLA-B7- or MGBA-cell lines were noted which we consider as nonspecific lysis. These results demonstrate that MGBA is processed by breast cancer cells, and that candidate HLAB7-binding epitopes are presented by HLA-B7.
Fig. 4.
CD8+ CTL lines generated in vitro with candidate epitope-pulsed T2.B7 cells recognize breast cancers in a MGBA-specific and HLA-B7-restricted fashion. Two CD8+ CTL lines were generated from HLA-B7+ healthy females by means of four weekly in vitro stimulations with pooled T2.B7 cells individually loaded with MGBA B7.1-7. The MGBA specificity, and HLA-B7-restriction of the resulting CD8+ CTL lines were evaluated against the DU-4475 (HLA-B7+/MGBA+), AU-565 (HLA-B7−/MGBA+), MDA-361 (HLA-B7−/MGBA+), and MDA-231(HLA-B7−/MGBA−) breast cancer cell lines by means of a standard 51Cr release assay. Results obtained in triplicate cultures are expressed as mean ± standard deviation
Defining the epitope specificity of HLA-B7-restricted CD8+ CTL
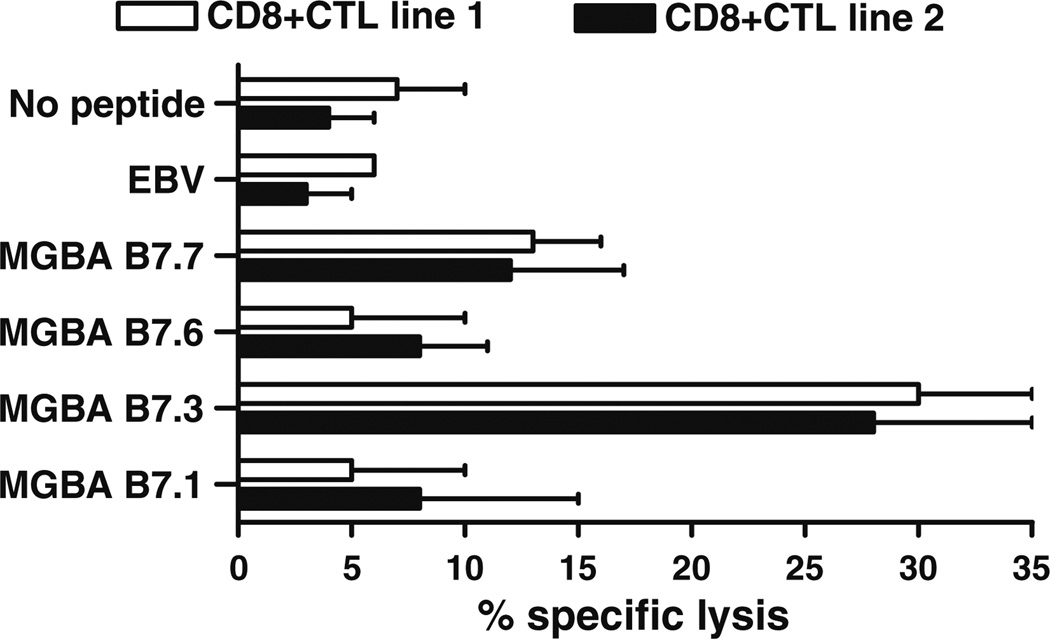
In order to determine the epitope specificity of the HLAB7-restricted CD8+ CTL against the HLA-B7+ MGBA+ human breast cancer cell line, DU-4475, a standard 51Cr release assay at varying E:T ratios (50:1 to 6.25:1) was performed in the presence of 20-fold excess of unlabeled T2.B7 cells individually loaded with either MGBA B7.1, B7.3, B7.6, or B7.7. Unloaded T2.B7 cells or RPPIFIRRL-loaded T2.B7 cells were used as negative controls. As shown in Fig. 5, the lysis of DU-4475 cell line by both CD8+ CTL lines (E:T ratio: 50:1) was inhibited by the addition of unlabeled T2.B7 cells loaded with MGBA B7.3, but not MGBA B7.1, B7.6, B 7.7, RPPIFIRRL or no peptide. To confirm these findings, two additional CD8+ CTL lines were generated from the same HLA-B7+ healthy female individuals in parallel experiments by four weekly in vitro stimulations with DU-4475. The peptide and HLAB7 specificity of the resulting CD8+ CTL cell lines were evaluated using T2.B7 cells individually loaded with MGBA HLA-B7 peptides by a standard 51Cr release assay. T2.B7 cells loaded with RPPIFIRRL, and empty T2.B7 cells (no peptide) were used as negative controls. As presented in Fig. 6, both CD8+ CTL lines generated in vitro demonstrated significant cytotoxicity against T2.B7 cells loaded with MGBA B7.3 but not those loaded with MGBA B7.1, B7.6, or B7.7. K562 cells were not lysed by these cell lines (CTL line 1: 0.8 ± 0.7%; CTL line 2 0.9 ± 0.3% at E:T ratio 50:1). These results demonstrate that breast cancer cell lines express the MGBA epitope B7.3, and support the hypothesis that this naturally-processed epitope is immunodominant.
Fig. 5.
The MGBA B7.3 epitope is the dominant epitope recognized on DU-4475 breast cancer cells by HLA-B7-restricted CD8+ CTL generated in vitro. The peptide specificity of the resulting CD8+ CTL lines generated from HLA-B7+ healthy female individuals by means of four weekly in vitro stimulations with pooled T2.B7 cells individually loaded with MGBA-derived candidate epitopes was evaluated using HLA-B7+/MGBA+ DU-4475 breast cancer cells in standard 51Cr release assays at varying E:T ratios in the presence of 20-fold excess of unlabeled T2.B7 cells individually loaded with MGBA-derived candidate epitopes. T2.B7 cells loaded with the EBV-derived epitope RPPIFIRRL, and empty T2.B7 cells (no peptide) were used as negative controls. The lysis of DU-4475 cells by both CD8+ CTL lines (50:1) was inhibited by the addition of unlabeled T2.B7 cells loaded with MGBA B7.3, but not T2.B7 cells loaded with MGBA B7.1, B7.6, B7.7, RPPIFIRRL, or no peptide. Results obtained in triplicate cultures at an E:T ratio of 50:1 are expressed as mean ± standard deviation
Fig. 6.
Recognition of MGBA B7.3-loaded T2.B7 cells by CD8+ CTL generated in vitro by stimulation with DU-4475 breast cancer cells. Two CD8+ CTL lines were generated from HLA-B7+ healthy female individuals by means of four weekly in vitro stimulations with the HLA-B7+/MGBA+ breast cancer cell line, DU-4475. The peptide specificity of the resulting CD8+ CTL cell lines was evaluated against T2.B7 cells individually loaded with MGBA candidate epitopes by a standard 51Cr release assay. T2.B7 cells loaded with the EBV-derived epitope RPPIFIRRL, and empty T2.B7 cells (no peptide) were used as negative controls. Both CD8+ CTL lines demonstrated significant lysis of T2.B7 cell loaded with MGBA B7.3, but no significant lysis of cells loaded with MGBA B7.1, B7.6, B7.7, EBV or no peptides. Results obtained at an E:T ratio of 50:1 in triplicate cultures are expressed as mean ± standard deviation
Discussion
Near-universal expression of MGBA in both primary and metastatic breast cancers is well documented [4, 29, 30], making MGBA an attractive immunotherapy target for the prevention and treatment of human breast cancer. The identification of MGBA-derived epitopes recognized by CD8+ CTL is critical for the successful clinical development of effective vaccine strategies. We have previously identified MGBA-derived epitopes presented in the context of HLA-A2 and HLA-A3, the most common HLA class I alleles in the general population [17, 18]. In this communication, we describe the identification of HLA-B7-restricted MGBA epitopes. HLA-B7 is also present in the population with high frequency [31], and is present in 17.7% of Caucasian, 15.5% of African American, 11.8% of Hispanic, 9.6% of Japanese, and 6.9% of Chinese populations. Therefore, identification of HLA-B7-restricted MGBA-derived CD8+ cytotoxic T cell epitopes in conjunction with the previously identified HLA-A2- and HLA-A3-restricted epitopes will provide important insights into the MGBA-specific response in the majority of individuals.
Using an HLA class I peptide-binding algorithm, seven MGBA-derived peptides (MGBA B7.1–7.7) with a range of predicted binding affinity for the HLA-B7 molecule were identified (Table 2). The actual binding affinity of the identified epitopes to HLA-B7 molecule was determined by membrane stabilization assay using TAP-deficient T2 cells transfected with HLA-B*0702. The peptides MGBA B7.3 (VSKTEYKEL), B7.6 (KLLMVLMLA), B7.7 (NPQVSKTEY), and B7.1 (YAGSGCPLL) displayed a high affinity for HLA-B7 molecule comparable to the binding affinity of EBV-derived peptide RPPIFIRRL. In contrast, the peptides MGBA B7.2 (ATTNAIDEL), B7.4 (LMVLMLAAL), B7.5 (SNVEVFMQL) demonstrated significantly low affinity. This discrepancy between the computer-predicted and experimental binding affinity for a given epitope in the HLA class I groove has been documented by previous studies in our laboratory [17, 18] and others as well [24, 25]. This is most likely explained by the findings that binding affinity for an individual epitope to each HLA class I molecule is determined by both its amino acid sequence and the three-dimensional structure of the binding motif within the HLA class I groove [22, 23]. To determine whether CD8+ CTL generated against MGBA-derived epitopes presented by HLA-B7 were able to recognize human breast cancer cells, we generated two CD8+ CTL lines from two HLA-B7+ healthy individuals using pooled T2.B7 cells individually pulsed with MGBA-derived peptides B7.1-7.7. These CD8+ CTL cell lines generated in vitro recognized MGBA B7.1, B7.3, B7.6, and B7.7 epitopes but not MGBA B7.2, B7.4, and B7.5. We propose that this is due to the significantly lower binding affinity of peptides MGBA B7.2, B7.4, and B7.5 for the HLA-B7 molecule as compared to the other peptides. The resulting CD8+ CTL lines generated in vitro displayed significant cytotoxic activity against DU-4475, a HLA-B7+ MGBA+ breast cancer cell line [19]. The lack of reactivity against AU-565 (HLA-B7− MGBA+), MDA-361 (HLA-B7− MGBA+), MDA-231 (HLA-B7− MGBA−) clearly demonstrate that the generated CD8+ CTL lines exhibited the cytotoxic reactivity against human breast cancer cells in a MGBAspecific and HLA-B7-restricted manner (Fig. 4). We also determined that CD8+ CTL recognized only a single epitope, MGBA B7.3, on the DU-4475 cell line (Figs. 5, 6). Cancers are known to have heterogenous expression of tumor-associated antigens due to differential cleavage by the proteasome of endogenous proteins [32]. In this regard, a potential limitation of our study is that we only analyzed a single breast cancer cell line, which cannot fully evaluate the heterogeneity of breast cancer-associated tumor antigens in vivo. At the same time, our results demonstrate the feasibility of four MGBA-derived epitopes, MGBA B7.1, 7.3, 7.6, and 7.7 to induce CD8+ CTL responses specifically against HLA-B7+ MGBA+ breast cancer cells. Of the epitopes identified, we provide evidence for MGBA B7.3 (VSKTEYKEL) as the immunodominant epitope which displays the capacity to induce significant lysis of a HLAB7 + MGBA+ breast cancer cell line tested in vitro. These results will provide the opportunity to design and test epitope-specific vaccination strategies such as peptide vaccines, or single chain trimer DNA vaccines. Furthermore, CD8 + CTLs have been shown to recycle and lyse targets sequentially for two or more cycles without further reactivation [33].
In conclusion, we identified four MGBA-derived epitopes, MGBA B7.1, MGBA B7.3, MGBA B7.6, and MGBA B7.7 presented in the context of the HLA-B7 molecule which can be used to generate CD8+ CTL in vitro. The CD8+ CTL demonstrate the ability to effectively lyse breast cancer cells, confirming that breast cancers process and present MGBA-derived epitopes in an HLA-B7 restricted fashion. Results presented in this study with MGBA epitopes for HLA-B7, along with MGBA epitopes previously identified for HLA-A2 and A3, provide the opportunity for efficient breast vaccine development. Further, the identified peptide sequences could be used in future epitope-specific vaccine strategies including peptide vaccine strategies, single chain trimer DNA vaccine strategies, and for developing immune-monitoring reagents such as tetramers for staining HLA-B7-resticted, MGBA-responsive cytotoxic T-cells following MGBA vaccination.
Acknowledgments
This work was supported in part by the Department of Defense Breast Cancer Research Program (W81XWH-06-1-0677), and BJC Foundation (TM). The authors would like to thank Billie Glasscock for her help in submitting this manuscript.
Contributor Information
Haseeb Ilias Basha, Department of Surgery, Washington University School of Medicine, Box 8109, 3328 CSRB, 660 South Euclid Avenue, Saint Louis, MO 63110, USA.
Venkataswarup Tiriveedhi, Department of Surgery, Washington University School of Medicine, Box 8109, 3328 CSRB, 660 South Euclid Avenue, Saint Louis, MO 63110, USA.
Timothy P. Fleming, Department of Surgery, Washington University School of Medicine, Box 8109, 3328 CSRB, 660 South Euclid Avenue, Saint Louis, MO 63110, USA
William E. Gillanders, Department of Surgery, Washington University School of Medicine, Box 8109, 3328 CSRB, 660 South Euclid Avenue, Saint Louis, MO 63110, USA
T. Mohanakumar, Email: kumart@wustl.edu, Department of Surgery, Washington University School of Medicine, Box 8109, 3328 CSRB, 660 South Euclid Avenue, Saint Louis, MO 63110, USA; Department Pathology and Immunology, Washington University School of Medicine, Saint Louis, MO, USA.
References
- 1.Watson MA, Fleming TP. Isolation of differentially expressed sequence tags from human breast cancer. Cancer Res. 1994;54(17):4598–4602. [PubMed] [Google Scholar]
- 2.Fleming TP, Watson MA. Mammaglobin, a breast-specific gene, and its utility as a marker for breast cancer. Ann N Y Acad Sci. 2000;923:78–89. doi: 10.1111/j.1749-6632.2000.tb05521.x. [DOI] [PubMed] [Google Scholar]
- 3.Goedegebuure PS, Watson MA, Viehl CT, Fleming TP. Mammaglobin-based strategies for treatment of breast cancer. Curr Cancer Drug Targets. 2004;4(6):531–542. doi: 10.2174/1568009043332862. [DOI] [PubMed] [Google Scholar]
- 4.Watson MA, Fleming TP. Mammaglobin, a mammary-specific member of the uteroglobin gene family, is overexpressed in human breast cancer. Cancer Res. 1996;56(4):860–865. [PubMed] [Google Scholar]
- 5.Mikhitarian K, Gillanders WE, Almeida JS, Hebert Martin R, Varela JC, Metcalf JS, Cole DJ, Mitas M. An innovative microarray strategy identities informative molecular markers for the detection of micrometastatic breast cancer. Clin Cancer Res. 2005;11(10):3697–3704. doi: 10.1158/1078-0432.CCR-04-2164. [DOI] [PubMed] [Google Scholar]
- 6.Watson MA, Dintzis S, Darrow CM, Voss LE, DiPersio J, Jensen R, Fleming TP. Mammaglobin expression in primary, metastatic, and occult breast cancer. Cancer Res. 1999;59(13):3028–3031. [PubMed] [Google Scholar]
- 7.Gillanders WE, Mikhitarian K, Hebert R, Mauldin PD, Palesch Y, Walters C, Urist MM, Mann GB, Doherty G, Herrmann VM, et al. Molecular detection of micrometastatic breast cancer in histopathology-negative axillary lymph nodes correlates with traditional predictors of prognosis: an interim analysis of a prospective multi-institutional cohort study. Ann Surg. 2004;239(6):828–837. doi: 10.1097/01.sla.0000128687.59439.d6. (discussion 837–840) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244(4905):707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 9.Donnelly JJ, Ulmer JB, Shiver JW, Liu MA. DNA vaccines. Annu Rev Immunol. 1997;15:617–648. doi: 10.1146/annurev.immunol.15.1.617. [DOI] [PubMed] [Google Scholar]
- 10.Gurunathan S, Klinman DM, Seder RA. DNA vaccines: immunology, application, and optimization*. Annu Rev Immunol. 2000;18:927–974. doi: 10.1146/annurev.immunol.18.1.927. [DOI] [PubMed] [Google Scholar]
- 11.Wang R, Doolan DL, Le TP, Hedstrom RC, Coonan KM, Charoenvit Y, Jones TR, Hobart P, Margalith M, Ng J, et al. Induction of antigen-specific cytotoxic T lymphocytes in humans by a malaria DNA vaccine. Science. 1998;282(5388):476–480. doi: 10.1126/science.282.5388.476. [DOI] [PubMed] [Google Scholar]
- 12.MacGregor RR, Boyer JD, Ugen KE, Lacy KE, Gluckman SJ, Bagarazzi ML, Chattergoon MA, Baine Y, Higgins TJ, Ciccarelli RB, et al. First human trial of a DNA-based vaccine for treatment of human immunodeficiency virus type 1 infection: safety and host response. J Infect Dis. 1998;178(1):92–100. doi: 10.1086/515613. [DOI] [PubMed] [Google Scholar]
- 13.Ugen KE, Nyland SB, Boyer JD, Vidal C, Lera L, Rasheid S, Chattergoon M, Bagarazzi ML, Ciccarelli R, Higgins T, et al. DNA vaccination with HIV-1 expressing constructs elicits immune responses in humans. Vaccine. 1998;16(19):1818–1821. doi: 10.1016/s0264-410x(98)00180-7. [DOI] [PubMed] [Google Scholar]
- 14.Pavlenko M, Roos AK, Lundqvist A, Palmborg A, Miller AM, Ozenci V, Bergman B, Egevad L, Hellstrom M, Kiessling R, et al. A phase I trial of DNA vaccination with a plasmid expressing prostate-specific antigen in patients with hormone-refractory prostate cancer. Br J Cancer. 2004;91(4):688–694. doi: 10.1038/sj.bjc.6602019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Narayanan K, Jaramillo A, Benshoff ND, Campbell LG, Fleming TP, Dietz JR, Mohanakumar T. Response of established human breast tumors to vaccination with mammaglobin-A cDNA. J Natl Cancer Inst. 2004;96(18):1388–1396. doi: 10.1093/jnci/djh261. [DOI] [PubMed] [Google Scholar]
- 16.Manna PP, Jaramillo A, Majumder K, Campbell LG, Fleming TP, Dietz JR, Dipersio JF, Mohanakumar T. Generation of CD8 + cytotoxic T lymphocytes against breast cancer cells by stimulation with mammaglobin-A-pulsed dendritic cells. Breast Cancer Res Treat. 2003;79(1):133–136. doi: 10.1023/a:1023323509888. [DOI] [PubMed] [Google Scholar]
- 17.Jaramillo A, Majumder K, Manna PP, Fleming TP, Doherty G, Dipersio JF, Mohanakumar T. Identification of HLA-A3-restricted CD8 + T cell epitopes derived from mammaglobin-A, a tumor-associated antigen of human breast cancer. Int J Cancer. 2002;102(5):499–506. doi: 10.1002/ijc.10736. [DOI] [PubMed] [Google Scholar]
- 18.Jaramillo A, Narayanan K, Campbell LG, Benshoff ND, Lybarger L, Hansen TH, Fleming TP, Dietz JR, Mohanakumar T. Recognition of HLA-A2-restricted mammaglobin-A-derived epitopes by CD8 + cytotoxic T lymphocytes from breast cancer patients. Breast Cancer Res Treat. 2004;88(1):29–41. doi: 10.1007/s10549-004-8918-1. [DOI] [PubMed] [Google Scholar]
- 19.Burdall SE, Hanby AM, Lansdown MR, Speirs V. Breast cancer cell lines: friend or foe? Breast Cancer Res. 2003;5(2):89–95. doi: 10.1186/bcr577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gronen F, Ruprecht K, Weissbrich B, Klinker E, Kroner A, Hofstetter HH, Rieckmann P. Frequency analysis of HLA-B7-restricted Epstein-Barr virus-specific cytotoxic T lymphocytes in patients with multiple sclerosis and healthy controls. J Neuroimmunol. 2006;180(1–2):185–192. doi: 10.1016/j.jneuroim.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 21.Smith KD, Lutz CT. Peptide-dependent expression of HLA-B7 on antigen processing-deficient T2 cells. J Immunol. 1996;156(10):3755–3764. [PubMed] [Google Scholar]
- 22.Kuhns JJ, Batalia MA, Yan S, Collins EJ. Poor binding of a HER-2/neu epitope (GP2) to HLA-A2.1 is due to a lack of interactions with the center of the peptide. J Biol Chem. 1999;274(51):36422–36427. doi: 10.1074/jbc.274.51.36422. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka Y, Amos KD, Joo HG, Eberlein TJ, Goedegebuure PS. Modification of the HER2/NEU-derived tumor antigen GP2 improves induction of GP2-reactive cytotoxic T lymphocytes. Int J Cancer. 2001;94(4):540–544. doi: 10.1002/ijc.1508. [DOI] [PubMed] [Google Scholar]
- 24.Flower DR. Towards in silico prediction of immunogenic epitopes. Trends Immunol. 2003;24(12):667–674. doi: 10.1016/j.it.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 25.Liu W, Meng X, Xu Q, Flower DR, Li T. Quantitative prediction of mouse class I MHC peptide binding affinity using support vector machine regression (SVR) models. BMC Bioinformatics. 2006;7:182. doi: 10.1186/1471-2105-7-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Earley S, Plopper GE. Disruption of focal adhesion kinase slows transendothelial migration of AU-565 breast cancer cells. Biochem Biophys Res Commun. 2006;350(2):405–412. doi: 10.1016/j.bbrc.2006.09.056. [DOI] [PubMed] [Google Scholar]
- 27.Ueno NT, Bartholomeusz C, Xia W, Anklesaria P, Bruckheimer EM, Mebel E, Paul R, Li S, Yo GH, Huang L, et al. Systemic gene therapy in human xenograft tumor models by liposomal delivery of the E1A gene. Cancer Res. 2002;62(22):6712–6716. [PubMed] [Google Scholar]
- 28.Lee-Huang S, Huang PL, Sun Y, Chen HC, Kung HF, Huang PL, Murphy WJ. Inhibition of MDA-MB-231 human breast tumor xenografts and HER2 expression by anti-tumor agents GAP31 and MAP30. Anticancer Res. 2000;20(2A):653–659. [PubMed] [Google Scholar]
- 29.Zuo L, Li L, Wang Q, Fleming TP, You S. Mammaglobin as a potential molecular target for breast cancer drug delivery. Cancer Cell Int. 2009;9:8. doi: 10.1186/1475-2867-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watson MA, Darrow C, Zimonjic DB, Popescu NC, Fleming TP. Structure and transcriptional regulation of the human mammaglobin gene, a breast cancer associated member of the uteroglobin gene family localized to chromosome 11q13. Oncogene. 1998;16(6):817–824. doi: 10.1038/sj.onc.1201597. [DOI] [PubMed] [Google Scholar]
- 31.Bodmer JG, Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Charron D, Dupont B, Erlich HA, Fauchet R, Mach B, et al. Nomenclature for factors of the HLA system, 1996. Tissue Antigens. 1997;49(3 Pt 2):297–321. doi: 10.1111/j.1399-0039.1997.tb02759.x. [DOI] [PubMed] [Google Scholar]
- 32.Luckey CJ, Marto JA, Partridge M, Hall E, White FM, Lippolis JD, Shabanowitz J, Hunt DF, Engelhard VH. Differences in the expression of human class I MHC alleles and their associated peptides in the presence of proteasome inhibitors. J Immunol. 2001;167(3):1212–1221. doi: 10.4049/jimmunol.167.3.1212. [DOI] [PubMed] [Google Scholar]
- 33.Berke G. The binding and lysis of target cells by cytotoxic lymphocytes: molecular and cellular aspects. Annu Rev Immunol. 1994;12:735–773. doi: 10.1146/annurev.iy.12.040194.003511. [DOI] [PubMed] [Google Scholar]