Abstract
Parent-of-origin effects occur when the phenotypic effect of an allele depends on whether it is inherited from an individual’s mother or father. Several phenomena can cause parent-of-origin effects, with the best characterized being parent-of-origin dependent gene expression associated with genomic imprinting. Imprinting plays a critical role in a diversity of biological processes and in certain contexts it structures epigenetic relationships between DNA sequence and phenotypic variation. The development of new mapping approaches applied to the growing abundance of genomic data has demonstrated that imprinted genes can be important contributors to complex trait variation. Therefore, to understand the genetic architecture and evolution of complex traits, including complex diseases and traits of agricultural importance, it is crucial to account for these parent-of-origin effects. Here we discuss patterns of phenotypic variation associated with imprinting, evidence supporting its role in complex trait variation, and approaches for identifying its molecular signatures.
Introduction
Parent-of-origin effects are epigenetic phenomena that appear as phenotypic differences between heterozygotes depending on allelic parent-of-origin. Genomic imprinting occurs when two alleles at a locus are not functionally equivalent and is considered the primary epigenetic phenomenon that can lead to the manifestation of parent-of-origin effects1. Imprinted loci show parent-of-origin dependent gene expression and have been observed in mammals, flowering plants and insects2. However, the taxonomic distribution and the breadth of genomic imprinting (hereafter referred to as ‘imprinting’) are open questions. Imprinting appears to play an important role in modulating sets of complex traits, notably in early development (including embryonic, placental and seed development) and behavior (especially social behavior)1, 3–5. Much of our understanding of the phenotypic consequences of imprinting comes from gross genetic anomalies such as uniparental disomies, translocations, loss of function mutations, and loss of imprinting epimutations, some of which are associated with complex disorders (e.g., Prader-Willi, Beckwith-Weidemann, and Angelman syndromes)1. Genes (or more generally, loci) associated with these disorders show the signature of imprinting manifested as parent-of-origin dependent effects6, with the anomalous phenotype depending on which parent the causal allele(s) is inherited from, rather than an individual’s diploid genotype.
Parent-of-origin effects are often considered synonymous with imprinting but there are other scenarios that can lead to the appearance of a parent-of-origin effect in the absence of imprinting (see below). Here we review recent developments in understanding the role of imprinting as a parent-of-origin effect underlying complex trait variation and provide a primer on approaches that can be used to identify and examine the contribution of imprinted loci to aspects of genetic architecture. Studies suggest that imprinted loci may be important contributors to phenotypic variation7–10, despite the fact that imprinting per se has been confirmed in a relatively small proportion of all genes (<1% in humans or mice11 and an even smaller number in plants12). However, most studies of complex traits have not implemented models that allow for the non-equivalence of parental alleles (i.e., allow for a parent-of-origin effect), thus the number and effects of imprinted genes remain important open questions. Studies that consider genetic and epigenetic variation at imprinted loci as a source of natural variation in complex traits can not only potentially identify additional imprinted genes, but can also reveal an important component of heritable variation that remains ‘hidden’ in traditional complex trait studies.
Other parent-of-origin effects
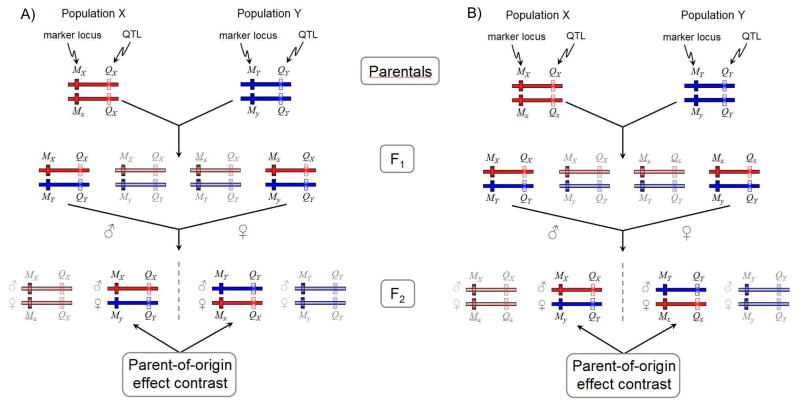
In this review we focus on imprinting, so first it is useful to consider other scenarios that can lead to the appearance of a parent-of-origin effect in the absence of imprinting. One scenario is the possibility that the reciprocal heterozygotes actually have a genetic difference. For example, gene-specific tri-nucleotide expansions can have sex-specific biases in occurrence and therefore transmission, and parent-of-origin effects resulting from such expansions have been associated with disorders such as myotonic dystrophy type-113, 14. Genetic differences between reciprocal heterozygotes are particularly problematic for discovery research using a “line-cross design”15 where individuals from two variable parental populations are intercrossed to produce an experimental population16, 17, as illustrated in Figure 1. In this scenario, spurious imprinting effects can arise when the assumption that the parental strains are fixed for QTL differences but have segregating marker variation is violated. Most problematically, the same conditions making a marker locus informative to detect a parent-of-origin effect (segregating variation at marker loci in parental lines) are the same conditions that can lead to spurious results (segregating variation at linked quantitative trait loci, QTL, in parental lines). The assumptions of the line-cross design are unlikely to be made in studies of natural variation (such as most human studies), and hence the problem of spurious results produced by this phenomenon are unlikely to apply to most approaches used to study parent-of-origin effects. Another confounder is parental genetic effects18. In mammals, studies of parental effects have focused on maternal-effects, however paternal-effects are equally plausible (but presumably less common). From a single locus perspective, parental genetic effects occur when a locus expressed in mothers (or fathers) has some causal influence on the phenotype of her (his) offspring19. For example, maternal effects have been observed in a mouse model of anxiety, where offspring born to mothers that were heterozygote for a knockout of the serotonin A1 receptor, Htr1a, but who did not inherit the mutation themselves, displayed an anxiety-like phenotype20. Maternal effects can lead to the appearance of a parent-of-origin effect when mothers that are homozygous for different alleles have distinct phenotypic effects on their offspring. Because these homozygous mothers can each only produce one type of reciprocal heterozygote, such a maternal effect is expected to lead to a difference in the average phenotypes of the reciprocal heterozygote offspring. Other phenomena that can result in differences between reciprocal heterozygotes include random monoallelic expression and environmentally mediated epigenetic silencing. However, these processes are not expected to produce parent-of-origin dependent patterns and are not considered here.
Figure 1.
The line cross-design and the appearance of pseudo-imprinted loci. Genetically variable individuals from two parental populations, X and Y, are intercrossed to produce an F2 population. Haplotypes are composed of a marker locus (M) and a linked QTL (Q). Haplotypes originating from population X appear in red and from population Y in blue. The marker locus has two alleles in each population, with markers M1 and M2 from population X and M3 and M4 from population Y. The F1 intercross contains four possible unordered genotypes. The F2 population resulting from a random intercross of these F1 genotypes would produce 16 possible ordered genotypes, but for simplicity only the cross between the M1M3 and M2M4 F1 genotypes are illustrated. This cross produces four ordered F2 genotypes, with the paternally inherited allele appearing above the maternally inherited allele. The two genotypes that contain a marker allele from the X and Y parental populations (M1M4and M3M2) contribute to the parent-of-origin effect contrast. A) In the first scenario the parental populations are fixed for alternative QTL alleles (Q1 and Q3). The parent-of-origin effect contrast therefore represents a comparison between the phenotypes of the Q1Q3 and Q3Q1 genotypes that are genetically equivalent at the QTL but differ in the parent-of-origin of alleles. B) In the second scenario population X has segregating variation at the QTL locus, with alleles Q1and Q2, which are linked to markers M1 and M2 respectively. As a result, the parent-of-origin effect contrast represents a comparison between the phenotypes of the Q1Q3 and Q3Q2 genotypes that are not genetically equivalent and hence the contrast confounds parent-of-origin of alleles and allelic differences at the QTL locus.
Identifying phenotypic signatures of imprinted loci
Our understanding of the genotype-phenotype relationship is largely conceptualized through the use of a single locus with two alleles. Within this framework, there are three genotype classes, with the reciprocal heterozygotes grouped into a single class because they are genetically equivalent. However, to understand the contribution of imprinting to the genotype-phenotype relationship we need to characterize the genetically equivalent, but potentially phenotypically non-equivalent, reciprocal heterozygotes as distinct genotype classes7, 21. This increase in the number of genotype classes provides the critical extra degree of freedom required to test for the presence of imprinting. If a locus is imprinted, we expect these two classes to express different alleles (Box 1). Imprinting will manifest as genotype classes that vary phenotypically according to allelic parent-of-origin, forming the foundation of studies aimed at identifying imprinting effects on complex traits (Box 2).
BOX 1. Classification of imprinting patterns.
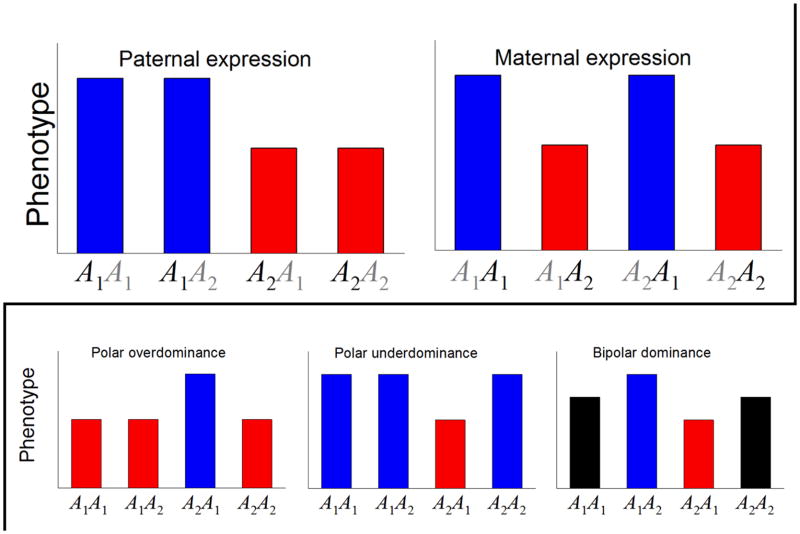
We expect that parent-of-origin dependent monoallelic expression of a single gene will produce a pattern of phenotypic variation in which the phenotypic effect of a locus is determined entirely by the single allele that is expressed (i.e., by the paternally inherited allele for a paternally expressed locus and the maternally inherited allele for a maternally expressed locus). Thus, monoallelic parent-of-origin dependent expression leads to what has been called ‘parental imprinting’8, 9, where the canonical patterns of maternal versus paternal expression depend on whether genotypes group by the maternally versus the paternally inherited allele.
The patterns of phenotypic variation expected for paternal and maternal expression are illustrated in the top two figures, which show the expected phenotypic value for the four possible ordered genotypes at the A locus (with the first allele listed being inherited from the father and the second from the mother). In both cases the A1 allele leads to a larger phenotypic value than the A2 allele and one allele is silenced (appears faded to grey). In each case genotypes group phenotypically by allelic parent of origin, as indicated by the shared color of their shading.
Although most studies have constrained their analysis to parental forms of imprinting, those that have not have generally identified loci showing ‘dominance’ imprinting patterns8–10, 78, where the pattern of effect depends on the combination of alleles. Dominance imprinting occurs in the polar overdominance phenotype associated with the callipyge mutation in sheep49 and has also been observed in humans51 and mice32, 33, 78. Polar overdominance shows the signature of an imprinted locus manifested as a difference between the phenotypes of the reciprocal heterozygotes, but lacks the expected difference between the two homozygotes that should occur under parent-of-origin dependent monoallelic expression. With polar overdominance, the phenotype of the heterozygote is larger than that of the other genotypes, but there is ‘polarity’ because the dominance only appears in one of the two heterozygote configurations (see figure). By analogy, it is also possible for a locus to show a pattern of polar underdominance where one of the heterozygotes has a smaller phenotypic value than the other genotypes (see figure). For both polar over- and underdominance the two homozygotes group phenotypically with one of the heterozygotes (emphasized by the color shading in the figure), but the phenomena differ in the pattern of the grouping.
Finally, it is possible for a locus to show both under- and overdominance at the same time, with one heterozygote having a phenotypic value larger than the two homozygotes and the other heterozygote having a value that is smaller (see figure). This pattern of ‘bipolar dominance’8 reflects the opposing polarity of the heterozygotes.
BOX 2. Genetic effects and mapping models.
There are several different statistical frameworks used to identify imprinting effects, but the vast majority are built on the approach pioneered by Knott et al.21 (which was formalized by Mantey et al. and refined by others 7, 8, 22). This framework is an extension of the single-locus two-allele model underlying most mapping studies. Using unordered genotypes, the simplest mapping model is a regression model built on the classic quantitative genetics model with additive and dominance effects. The additive effect (a) corresponds to a contrast between the two homozygotes, while the dominance effect (d) measures the deviation of the heterozygote from the midpoint (unweighted average) of the two homozygotes79. This model can be expressed as a linear equation wherein the mean phenotypes of the three genotypes at a locus (indicated by the genotype ID with the overbar)22 are decomposed into additive and dominance effects:
where r is the reference point (intercept) for the model (in this case, the midpoint between the two homozygotes, ). Under this model, the additive effect estimated corresponds to half the difference between the means of the two homozygotes ( ) and the dominance effect corresponds to the deviation of the mean heterozygote phenotype from the midpoint between the two homozygotes ( ). To estimate imprinting effects this model uses ordered genotypes21, allowing the estimation of an additional parameter, the imprinting effect (i)8, 22.
This model has the same reference point (intercept) and yields identical definitions of the additive and dominance effects as the unordered genotype model (except that the mean heterozygote in the unordered model is replaced by the mean of the reciprocal heterozygotes, ). Under this model, the imprinting effect is defined as half the difference in the mean phenotypes of the reciprocal heterozygotes ( ). If there is complete silencing of an allele we expect a locus showing paternal expression to have a = i, while maternal expression would correspond to a = −i.
Assigning parent-of-origin to alleles
The critical first step in analyzing imprinting effects is assigning parent-of-origin to alleles. The earliest studies used the “line-cross design”15 based on F2 intercross populations in which non-inbred parental lines were crossed17,7, 17, 21, 22. Parent-of-origin of marker alleles is assigned by identifying the grandparent-of-origin of an allele (which requires genotyping founders). This approach has been used to identify imprinting effects on body composition in pigs, but it has been criticized because it can lead to the appearance of imprinting when there are QTL segregating in the parental strains (Figure 1)16. Further, this approach cannot be used to study imprinting effects using biallelic loci, so many genomic regions are uninformative21.
Studies in mice have used a backcross design23 where F1 heterozygotes from an inbred line cross (which have the unordered genotype A1A2) are backcrossed to either parental strain. The parent-of-origin of alleles in all heterozygotes produced in each backcross can be directly inferred. For example, if an A1A2 male is backcrossed to a female from the A1A1 parental line, then all heterozygous offspring will have received the A1 allele from their mother and A2 from their father. Such a design, while intuitive, fully confounds maternal effects with imprinting effects18 and restricts patterns of variation across the genome (since backcrossed populations are necessarily missing subsets of possible multilocus allelic combinations).
Other studies have used an F2 generation of intercrosses between inbred strains, where individuals are produced by genetically identical F1 parents (and hence the pedigree contains no information about allelic parent-of-origin)23. Allelic parent-of-origin in such a population can be inferred if there are sex differences in recombination rates and sufficient marker information to determine the number of recombination events on each chromosomal haplotype. This approach has been used in mice, relying on the fact that females have higher recombination rates than males, which is common in mammals 24. However, this approach can only be implemented in systems where there is a large sex difference in recombination rates and where it is possible to accurately determine the number of recombination events present on each haplotype. This approach lacks power due to high error rates because sex differences in recombination rates can be small.
In samples where parents are genetically variable (such as an advanced intercross), one can simply genotype parents and their offspring and then directly infer allelic inheritance. Allelic parent-of-origin can be determined for all heterozygous offspring produced by all matings between a heterozygote and a homozygote parent or between two opposite homozygote parents. In a population with genotypes in Hardy-Weinberg proportions, this approach can be used to assign the parent-of-origin to alleles in at least three-quarters of all heterozygotes (the proportion of uninformative heterozygotes under random mating is approximately pq, where p and q are the frequencies of the two alleles at a locus). The only families in which parent of origin of alleles cannot be directly inferred are those families where both parents are heterozygotes at the locus in question. Studies in human populations have used family-based genotype information (parent/ offspring trios) to assign parent-of-origin to offspring alleles, and Transmission Disequilibrium Tests (TDT) can be used to identify biased transmission of the parental alleles 25, 26. TDT methods are robust to the confounding effects of population admixture and stratification, however they are generally underpowered because they do not account for between family variation in human samples 27.
Although matings between two heterozygous parents do not allow direct inference of parent-of-origin of alleles, it is possible to use linkage information to infer allelic parent-of-origin in some or all cases (depending on marker density). That is, if allelic parent-of-origin at a locus cannot be determined directly, but the locus is linked to informative loci, the linked marker information can be used to infer allelic parent-of-origin at the ambiguous locus (or assign a conditional probability that each allele came from each parent7, 17). This process can be efficiently achieved through haplotype reconstruction approaches wherein entire chromosomal haplotypes are assigned a parent-of-origin based on algorithms that determine the most likely haplotype configuration in a population 8, 28, or through approaches that more generally use linkage information to assign parent-of-origin probabilities to alleles. Recently, extended pedigree information was used to assign parent-of-origin of haplotypes using a likelihood based framework in >38,000 Icelanders 29.
Imprinting effects on complex traits
Do imprinted QTL map to known imprinted regions?
Studies assigning parent-of-origin to alleles and subsequently mapping QTL with parent-of-origin dependent effects in model organisms have had mixed success in linking loci to known imprinted regions. For example, results from one of the first analyses of imprinted QTL (using a porcine intercross for body composition7) found that three of the four imprinted QTL identified fell outside known imprinted regions. Another study8 mapping body weight and growth in a murine intercross found little overlap between known imprinted genes and imprinted QTL (only 2 of 10 loci overlapped confirmed imprinted genes) but all imprinted QTL contained multiple genes predicted to be imprinted by bioinformatic approaches (discussed below in the Identifying molecular signatures of imprinted loci section)30. Similar patterns, where most QTL map to regions that do not contain known imprinted genes but do contain bioinformatically predicted imprinted genes, were found in a study of bovine growth and body composition 31.
As discussed above, most known imprinted genes are associated with gross genetic anomalies and QTL studies identify genomic regions associated with normally distributed phenotypic variation. While some candidate imprinted QTL may be due to other parent-of-origin effects (see Introduction), these mapping results suggest that there may be more imprinted genes than have been characterized to date, and that imprinted genes are likely associated with normal phenotypic variation. Indeed, recent studies from an F16 generation of a randomly-mated advanced intercross of the LG/J and SM/J inbred mouse lines found imprinted genetic effects to be almost as prevalent as additive genetic effects for multiple metabolic traits: 40 QTL were found associated with variation in adiposity32, 64 with variation in diabetes-related traits33, and 23 with variation in serum lipid levels34; almost all of these QTL have additive effects and about 60% have imprinted effects. Although these candidate imprinted QTL have yet to be validated, simulation studies in an earlier generation of this intercross indicate the distribution of false positives for imprinting effects is the same as that for additive and/or dominance effects8 (i.e., there is no bias for the appearance of parent-of-origin effects) Thus, as with all QTL, loci showing imprinted genetic effects should be treated as candidates that require validation. Such caution is especially critical for QTL mapping to regions with no known imprinted genes.
Studies analyzing known imprinted genes for association with phenotypic variation
When studies have specifically targeted known imprinted genes for association with normal variation rather than with gross genetic anomalies, results indicate that these genes play important roles in complex traits. For example, a study in cattle35 targeted a series of SNPs in eight candidate imprinted genes (CALCR, GRB10, PEG3, PHLDA2, RASGRF1, TSPAN32, ZIM2 and ZNF215) and found six had significant associations with a variety of traits. However, it should be noted that only PEG3 has been shown to be imprinted in cattle, and the associations were not examined with regard to allelic parent-of-origin.
Other studies have honed in on the contribution of IGF2 to traits such as variation in meat quality characteristics in pigs after a QTL mapping study identified a locus containing this gene was strongly associated with variation in muscle mass36–39. This paternally expressed locus was found to have major effects on lean meat content in ham, accounting for 30% of the phenotypic variance in this trait38. The QTL was fine-mapped 36 and an intronic SNP affecting IGF2 expression in postnatal muscle was identified39. This SNP was found to abate ZBED6 repressor binding, leading to an increase in IGF2 expression and elevated muscle mass40. Other SNPs in IGF2 have been associated with milk quality traits in dairy cows41. It is hypothesized that breeding schemes focusing intensive selection on males could potentially favor such variation at paternally expressed loci38.
Human complex traits
In human studies, parent-of-origin effects have been implicated in many complex disorders including: Alzheimer’s disease42, autism43, bipolar disorder and schizophrenia44, 45, cancer29, adiposity and type-2 diabetes29, 46, 47, and type-1 diabetes26. Although most of these results have not been validated, they do indicate that imprinting effects underlie some of the variation observed in these traits. Unfortunately human samples often do not have data available to determine parent-of-origin of alleles, and/or are underpowered to incorporate this information into their analyses due to small sample sizes. However, recent large-scale studies have found interesting parent-of-origin effects associated with variation in multiple complex disorders26, 29, 47, with the implication that incorporating parent-of-origin of alleles into mapping models will increase a study’s power to account for a trait’s heritable variation. Large-scale studies with pedigree information will be important for developing models and tools that can accommodate the extra degrees of freedom resulting from distinguishing among heterozygote classes.
Complex imprinting patterns
In addition to implying that there are more imprinted genes than are currently characterized and that allelic parent-of-origin contributes to complex trait variation, studies that identify imprinted genetic effects have revealed that imprinting patterns can be complex. Complex imprinting patterns (Box 1) can arise when a locus contains multiple genes that can differ in their imprinting status. Imprinted genes tend to have a clustered distribution11, and within imprinted gene clusters there can be both maternally and paternally expressed genes regulated by the same imprint control region (for example at the H19/IGF2 locus associated with Beckwith-Weidemann, H19 is expressed from the maternal allele while IGF2 is expressed from the paternal allele48).
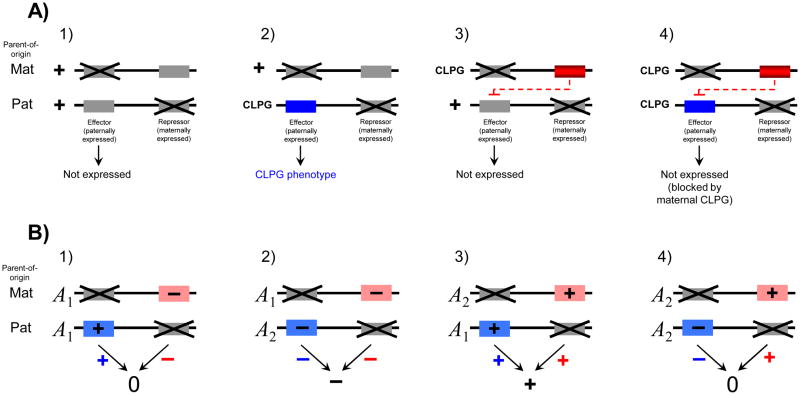
The first example of a locus with a complex imprinting pattern is the callipyge locus (CLPG) in sheep 49, 50, which shows polar overdominance (Box 1). Polar overdominance has also been described at the DLK1 gene in humans which is associated with juvenile obesity51. Early work hypothesized that the complex pattern results from a mutation that switches parent-of-origin specific expression from paternal to maternal (or vice-versa)52, 53. The CLPG mutation is an A→G transition in the intergenic region flanked by the paternally expressed DLK1 protein-coding gene and the maternally expressed GTL2 noncoding RNA gene. Focused studies of the CLPG mutation show it affects molecular marks including local DNA hypomethylation and DNase-1 hypersensitivity, and long-range bidirectional transcription throughout the intergenic region54. Additionally, RNA interference of the paternally expressed PEG11 by miRNAs processed by the maternally expressed antiPEG11 has been described. Both PEG11 and antiPEG11 expression are affected by the CLPG mutation55. Precise details of how the polar overdominance phenotype is achieved remain unclear, however the predominant model is that it is the result of up-regulation of a paternally expressed ‘effector’ and a maternally expressed ‘repressor’ 50, 56, which may be linked to the molecular mechanisms described at the locus (Figure 2.A). We have described an analogous ‘effector/repressor’ model to explain the appearance of bipolar dominance imprinting, where homozygotes are phenotypically identical but heterozygotes have different phenotypes (Box 1; Figure 2.B). An implication of a bipolar dominance effect, in a disease context, is that the same allele can be both protective and a potential risk factor in the heterozygote depending on parent-of-origin. Kong et al. 29 recently identified a pattern consistent with bipolar dominance in a large genotyped human population. Here the same SNP variant associated with type-2 diabetes risk when paternally inherited was found to be protective when maternally inherited.
Figure 2.
Molecular mechanisms that generate complex phenotypic patterns associated with genomic imprinting. In each figure the locus is composed of two imprinted genes, one showing maternal expression and the other paternal expression. The imprinted copies appear with a cross through them. Genes in grey are not expressed, either because they are imprinted or because they are inactive. Genes in blue are paternally expressed while those in red are maternally expressed. A) A working model for polar overdominance following Georges et al.50 (for simplicity the long range control element is not included). A.1) Individuals homozygous for the wild-type allele (W) do not express the effector or repressor and show the wild-type phenotype, A.2) Heterozygotes that inherit the active effector (CLPG allele) from their fathers and the inactive (wild-type) repressor from their mothers manifest the callipyge phenotype, A.3) Heterozygotes that inherit the active repressor (CLPG allele) from their mothers but the inactive effector from their fathers have a wild-type phenotype (the repressor is expressed but has no effect if there is no effector to block in trans). A.4) The CLPG homozygote expresses the effector from the paternally inherited copy and the repressor from the maternally inherited copy. The repressor blocks the effector in trans and results in a wild-type phenotype. B) Hypothetical model for the origin of bipolar dominance imprinting8. The A1 allele has a positive effect on the phenotype when paternally inherited (because of a paternally expressed gene) but a negative effect when maternally inherited (because of a maternally expressed gene) while the A2 allele has the opposite pattern of effect. The effect of the two genes are summed together to determine the influence of the A locus on the phenotype. B.1) In the A1A1 homozygote the paternally inherited positive effect cancels out the maternally inherited negative effect to result in a net effect of zero B.2) in the A2A1 heterozygote both the paternally inherited (A2) and maternally inherited (A1) alleles have a negative effect, resulting in a net negative phenotypic effect, B.3) In the A1A2 heterozygotes both the paternally inherited (A1) and maternally inherited (A2) alleles have a positive effect, resulting in a net positive phenotypic effect, B.4) In the A2A2 homozygote the paternally inherited negative effect cancels out the maternally inherited positive effect to result in a net effect of zero
Context dependent imprinting effects
Studies identifying imprinted genetic effects on complex traits strongly suggest that these effects can be context dependent, and imprinting patterns are not consistent among traits, environments, or between sexes. Our mapping results of multiple metabolic parameters in mice (adiposity, serum lipids, glucose and insulin levels) found context dependency to be prevalent at candidate imprinted QTL57. For example, a locus associated with free-fatty acids levels (DMetS1b) showed imprinted effects in females, but high-fat fed females showed maternal expression while low-fat fed females showed paternal expression57. This result indicates that imprinted genetic effects may occur at many levels. Due to the complexity of the genotype-phenotype relationship, it can be difficult to systematically study these effects in human samples. This constraint may be especially true for more ‘plastic’ complex traits such as obesity where the genetic architecture results from multidimensional interactions among genes and environment as well as from inter-organ cross talk (see Box 3 for a discussion of epistasis and genomic imprinting).
BOX 3. Epistasis and genomic imprinting.
Analyses of interactions among imprinted genes suggest that they may be particularly ‘interactive’, being enriched in complex networks that include both imprinted and non-imprinted genes80, 81. Effects of epistatic interactions involving imprinting effects on complex traits occur when: the effect of one locus depends on the parent-of-origin of alleles at another locus; and/or the imprinting effect of a locus changes as a function of the alleles present at another locus (or loci). This latter scenario can potentially include cases where one locus modifies the imprinting status of another locus. Here we briefly discuss the contribution of these types of epistatic interactions involving imprinting effects to variation in complex traits and to the evolution of imprinting patterns. We provide a formal framework for dissecting epistatic interactions involving imprinting effects elsewhere81. See also Li et al.82 for methods to identify epistatic interactions involving imprinted genes in a Bayesian framework.
From a statistical perspective, epistatic interactions involving imprinting effects appear logically as interaction terms in the multi-locus extension to the mapping model framework presented in Box 281. For example, there may be a statistical interaction between the additive effect of one locus and the imprinting effect of another locus (‘ additive-by-imprinting’ epistasis), meaning that the additive effect of one locus depends on the type of heterozygote present at another locus, while the imprinting effect of that other locus depends on the type of homozygote present at the first locus. More generally, epistasis involving imprinting occurs whenever one must consider the parent-of-origin of alleles to understand how effects at one locus change as a function of the genotype present at another locus (and vice versa). In some scenarios, the change in the effect of a locus as a function of the genetic background provided by another locus can correspond to a change in the pattern of imprinting. For example, a locus may show a pattern of maternal expression on one genetic background and a pattern of paternal expression on a different genetic background. Consequently, the status of imprinting at the locus could evolve as genetic background changes.
Few studies have considered epistatic interactions between imprinted genes in the context of quantitative genetic variation, but there have been studies that have shown that epistatic interactions in the broad sense can involve imprinted genes. For example, Reilly et al.83 found that the development of neural tumors in a mouse model is influenced by epistatic interactions involving an imprinted locus and tumor suppressor genes. Studies of the effects of various combinations of uniparental disomies containing imprinted regions in mice have shown that the combinations often have non-additive effects on developmental traits84. Studies have also shown that trans acting factors can change the imprinting status of a locus. For example, imprinting of the gene MEDEA in Arabidopsis endosperm is controlled by antagonism between at least two other genes and hence changes at those other genes can disrupt the presence of imprinting at MEDEA.
Further confounding is the fact that the trait being studied may be a composite that combines tissues or developmental stages that include both imprinted and non-imprinted expression. For example, many genes are imprinted only in the placenta58, likewise UBE3a in Angelman syndrome is biallelically expressed in most tissues but is maternally expressed in most neurons59 and the imprinted IGF2 gene is biallelically expressed in some tissues11. This can result in a phenotypic difference between reciprocal heterozygotes, but the difference is not as pronounced as the difference between the homozygotes. This scenario is called partial imprinting and has been observed both in the phenotypic manifestation at imprinted QTL8, 16, 23 and in gene expression differences60.
Thus animal models can be important complements to human studies because developmental stage, genetics and environment can be controlled and monitored. Imprinted patterns and genes identified in animal studies can be translated to human studies and the unit of translation is the imprinted ‘locus’ or the pathway containing the imprinted gene(s). It is unclear how often genes imprinted in one species are also imprinted in another. Studies comparing sets of predicted imprinted genes in humans and mice have suggested from 32%61 to 87%62 of imprinted genes are imprinted in both species. However, much work is required to validate such estimates, let alone to determine if conserved imprinted genes are also conserved in their phenotypic effects and/or underlying molecular mechanisms (not to mention tissue, developmental or environmental contexts)63, 64.
Identifying molecular signatures of imprinted loci
The ultimate support for an imprinted effect comes from molecular characterization of a locus. Such support is especially important when imprinted genetic effects are mapped to genomic regions that do not contain confirmed imprinted genes. Advances in whole-genome sequencing technologies can facilitate molecular characterization of candidate loci using DNA sequence features that, in some contexts, distinguish imprinted from non-imprinted genes. Features including the concentration and orientation of SINE repetitive elements and local CG content in conjunction with epigenetic features such as histone modification sites have been used to train algorithms that bioinformatically predict imprinted loci from whole-genome sequence65. Some of these predictions have been integrated with data from genes with known parent-of-origin allelic expression biases and used not only to further classify a predicted imprinted gene as maternally or paternally expressed, but also to identify potential patterns (and hence molecular mechanisms) that may distinguish between parent-of-origin of alleles61, 66. Using such computational predictions, Luedi et al.61 identified two novel imprinted genes in humans, KCNK9, which is maternally expressed in fetal brain and DLGAP2, which is paternally expressed in testes. It has been hypothesized that different mechanisms control maternal versus paternal expression biases67, 68, and identification of patterns associated with allele-specific regulation can provide a framework for clarifying DNA sequence/ gene expression relationships underlying the phenotypic signatures of imprinting.
Phenotypic variation at imprinted loci can be directly linked to genomic variation through analysis of parent-of-origin dependent gene expression. RNA-sequencing is the gold standard for quantifying whole-genome allele-specific biases in transcription (in which there is an unequal number of transcripts from the maternally and paternally derived alleles of a gene) and several studies have made use of this technology in an effort to identify novel imprinted genes in reciprocal crossings of inbred model organisms69–71. Inconsistent criteria for ascertaining parent-of-origin dependent biases has led to substantial discrepancies among results and failure to validate most predictions72. Further confounding factors include evidence that parent-of-origin dependent effects are context dependent, as discussed above.
A potential mechanism underlying allele-specific expression is DNA methylation. The addition of a methyl group to DNA nucleotides can occur in an allele-specific manner, and allele-specific methylation in imprint control regions (referred to as differentially methylated regions, DMR) is associated with parent-of-origin dependent gene expression. For example at the DMR at the H19/IGF2 locus, the maternal allele is unmethylated allowing CTCF transcription factor binding and preventing IGF2 promoter activation. The methylated paternal allele prevents CTCF binding and the downstream enhancer is able to activate IGF2 transcription73, 74. Next-generation sequencing technologies that specifically target methylated DNA have been used to identify DMRs that may associate with imprinted genes75–77. A promising avenue of research is to integrate RNA sequencing data with whole-genome methylation data and identify regions where both allele-specific expression and methylation correlate in a parent-of-origin dependent manner that associates with phenotypic variation. In addition to DMRs in imprint control regions, imprinted gene clusters often contain non-coding RNAs that have regulatory roles. Hence the phenotypic manifestation of a particular ‘locus’ can be determined by the joint action of multiple imprinted (coding) genes and non-coding elements. Thus characterizing genomic context, such as identifying clusters of non-coding RNA elements that may affect local gene expression in potentially complex interacting combinations, can be a tool for identifying loci that that show complex imprinting patterns such as polar overdominance or bipolar dominance.
Concluding remarks
Recent empirical research indicates that parent-of-origin effects are an important component of the genetic architecture of complex traits, and that complex patterns of imprinted genetic effects are prevalent. In animal research, there is a need to develop and incorporate models that consider developmental stage, tissue-specificity and context dependency into models of discovery research. In human association studies, there is a need to develop and incorporate models that allow parental alleles in heterozygotes to be functionally non-equivalent. Most studies of complex traits in both model organisms and human samples are underpowered (or the data are just not available), and there is currently no way to predict these complicated epigenetic effects from DNA sequence alone. What is lacking is a framework of DNA sequence–imprinted function relationships. There is a clear need for further research integrating complex trait mapping results with next-generation sequencing data to understand how imprinted genes contribute to the patterns of phenotypic variation seen in both natural and experimental populations. Studies that consider how and when imprinted genetic effects are conserved among species and/or are modified by environmental factors or genetic background will be particularly relevant for advancing the field of complex trait research.
Online Key Points.
Parent-of-origin effects likely contribute to the genetic architecture of complex traits, yet they are rarely included in studies of genetic architecture
It is critical to distinguish between reciprocal heterozygotes when identifying parent-of-origin effects, but several phenomena besides genomic imprinting can potentially produce phenotypic differences between reciprocal heterozygotes.
In human studies, large-scale samples that incorporate pedigree information will be important for developing models and tools that can accommodate parent-of-origin effects into analyses.
Animal studies will be essential for developing a framework of DNA sequence–imprinted–function relationships, particularly because parent-of-origin effects can be context dependent.
Research that integrates complex trait mapping results with next-generation sequencing data to identify patterns that have predictive power will be essential to advance the field.
Acknowledgments
Heather A Lawson is supported by National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health awards K01DK095003 to HAL and P30DK056341 to the Washington University School of Medicine Nutrition and Obesity Research Center.
Glossary
- Parental Effects
Occur when genes expressed in the mother or father have a causal influence on their offspring’s phenotype
- Quantitative Trait Locus (QTL)
A region of the genome in which genetic variation at a marker locus is significantly correlated with phenotypic variation for a complex trait
- Parental Imprinting
Occurs when either only the maternally or only the paternally inherited allele affects the phenotype. In a two-allele system, genotypes will group into two phenotypic classes based on the maternally or paternally expressed allele (Box 1)
- Dominance Imprinting
A complex imprinting pattern where parent-of-origin of alleles affects dominance at a locus. For example, bi-polar dominance imprinting occurs when one heterozygote shows overdominance and the reciprocal shows underdominance (Box 1)
- Advanced Intercross
The result of continued random mating of a population derived from a cross between inbred lines. Advanced intercrosses provide higher resolution for QTL than traditional (e.g., F2) mapping approaches because of the accumulation of recombination through each generation of random mating
- Allele-Specific Bias
Occurs when the two alleles in a heterozygote are not functionally equivalent. Can arise from expression bias wherein one allele is expressed at a higher rate than the other (the null expectation being that both alleles will be expressed at approximately the same rate). There can also be methylation biases, wherein one allele is preferentially methylated (or unmethylated), which can underlie allele-specific expression biases
- Epigenetic
A difference in phenotype resulting from variations in DNA chemistry rather than DNA sequence. Epigenetic changes can be cell specific, modified by environmental factors, affect gene expression, and may underlie some parent-of-origin effects on complex traits
- Complex Trait
A quantitative trait that is influenced by many genetic, epigenetic, and environmental factors and their interactions
- Differentially Methylated Region (DMR)
Genomic region where the pattern of methylation (the ratio of methylated to unmethylated sequence) is different between two alleles at the same locus
- Line-cross design
An approach to QTL mapping in which two non-inbred lines are crossed to produce a mapping population. The approach assumes that the two lines are fixed for different QTL alleles, but there is variation at marker loci segregating within lines (Figure 1)
Biographies
Heather A. Lawson is faculty in the Departments of Genetics and Medicine at Washington University School of Medicine, St Louis, USA. She received her PhD in anthropology from The Pennsylvania State University, University Park, USA, under the mentorships of Kenneth Weiss and Webb Miller. She subsequently completed a postdoc with Dr James Cheverud at Washington University. Her research is focused on molecular mechanisms underlying the genetic architecture of metabolic traits in different environments. She is interested in translating genetic and epigenetic molecular and analytical observations to physiological endpoints that are relevant to clinical manifestations of metabolic disease.
James M. Cheverud is Chair of the Department of Biology at Loyola University in Chicago, USA, and Professor Emeritus at Washington University, St Louis, USA. He received his BA from Northwestern University, Evanston, USA, and his MS and PhD from the University of Wisconsin-Madison, USA, under the mentorship of Kenneth Bennett. His research focuses on quantitative genetics and evolutionary diversification in primates, the genetic architecture of complex traits and the evolution of gene effects through epistatic interactions.
Jason B. Wolf is a Senior Lecturer in the Department of Biology & Biochemistry at the University of Bath, UK. He received his PhD from the University of Kentucky, under the mentorship of Allen Moore, Edmund D. Brodie III and Andy Sih. His research integrates theoretical, experimental and computational approaches to understand the evolutionary genetics of complex traits. He is particularly interested in non-Mendelian inheritance, including genomic imprinting, maternal effects, and social effects.
Weblink: http://www.evolutionarygenetics.org
References
- 1.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nature Reviews Genetics. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 2.Ferguson-Smith A. Genomic imprinting: the emergence of an epigenetic paradigm. Nature Reviews Genetics. 2011;12:565–575. doi: 10.1038/nrg3032. [DOI] [PubMed] [Google Scholar]
- 3.Tycko B, Morison IM. Physiological functions of imprinted genes. Journal of Cellular Physiology. 2002;192:245–258. doi: 10.1002/jcp.10129. [DOI] [PubMed] [Google Scholar]
- 4.Swaney WT, Curley JP, Champagne FA, Keverne EB. The paternally expressed gene Peg3 regulates sexual experience-dependent preferences for estrous odors. Behavioral Neuroscience. 2008;122:963–973. doi: 10.1037/a0012706. [DOI] [PubMed] [Google Scholar]
- 5.Curley JP. Is there a genomically imprinted social brain? BioEssays. 2011;33:662–668. doi: 10.1002/bies.201100060. [DOI] [PubMed] [Google Scholar]
- 6.Wolf JB, Hager R, Cheverud JM. Genomic imprinting effects on complex traits: A phenotype-based perspective. Epigenetics. 2008;3:295–299. doi: 10.4161/epi.3.6.7257. [DOI] [PubMed] [Google Scholar]
- 7.de Koning DJ, et al. Genome-wide scan for body composition in pigs reveals important role of imprinting. Proceedings of the National Academy of Sciences, USA. 2000;97:7947–7950. doi: 10.1073/pnas.140216397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolf JBM, CJ, Roseman C, Hager R. Genome-wide analysis reveals a complex pattern of genomic imprinting in mice. PLoS Genetics. 2008;4:e1000091. doi: 10.1371/journal.pgen.1000091. This study developed the framework in Box 1 to describe the complex patterns of imprinting identified, including first describing the patterns of polar underdominance and bipolar dominance imprinting. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheverud JM, et al. Genomic imprinting effects on adult body composition in mice. Proceedings of the National Academy of Sciences, USA. 2008;105:4253–4258. doi: 10.1073/pnas.0706562105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawson HA, et al. The importance of context to the genetic architecture of diabetes-related traits is revealed in a genome-wide scan of a LG/J 3 SM/J murine model. Mammalian Genome. 2011;22:197–208. doi: 10.1007/s00335-010-9313-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends in Genetics. 2005;21:457–465. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Raissig MT, Baroux C, Grossniklaus U. Regulation and flexibility of genomic imprinting during seed development. The Plant Cell. 2011;23:16–26. doi: 10.1105/tpc.110.081018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pearson CE. Slipping while sleeping? Trinucleotide repeat expansions in germ cells. Trends Mol Med. 2003;9:490–5. doi: 10.1016/j.molmed.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 14.Tome S, et al. Maternal germline-specific effect of DNA ligase I on CTG/CAG instability. Hum Mol Genet. 2011;20:2131–43. doi: 10.1093/hmg/ddr099. [DOI] [PubMed] [Google Scholar]
- 15.Haley CS, Knott SA, Elsen JM. Mapping quantitative trait loci in crosses between outbred lines using least squares. Genetics. 1994;136:1195–1207. doi: 10.1093/genetics/136.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Koning DJ, Bovenhuis H, Van Arendonk JAM. On the detection of imprinted quantitative trait loci in experimental crosses of outbred species. Genetics. 2002;161:931–938. doi: 10.1093/genetics/161.2.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandor C, Georges M. On the detection of imprinted quantitative trait loci in line crosses: effect of linkage disequilibrium. Genetics. 2008;180:1167–1175. doi: 10.1534/genetics.108.092551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hager RM, CJ, Roseman C, Wolf JB. Maternal effects as the cause of parent-of-origin effects that mimic genomic imprinting. Genetics. 2008:178. doi: 10.1534/genetics.107.080697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolf JB, Wade MJ. What are maternal effects (and what are they not)? Philosophical Transactions of the Royal Society of London. Series B, Biological sciences. 2009;364:1107–1115. doi: 10.1098/rstb.2008.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gleason G, et al. The serotonin1A receptor gene as a genetic and prenatal maternal environmental factor in anxiety. Proc Natl Acad Sci U S A. 2010;107:7592–7. doi: 10.1073/pnas.0914805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knott SA, et al. Multiple marker mapping of quantitative trait loci in a cross between outbred wild boar and large white pigs. Genetics. 1998;149:1069–1080. doi: 10.1093/genetics/149.2.1069. This study developed the first model for identifying imprinted loci in QTL analyses. It is also the first application of the line-cross design to identify parent-of-origin effects. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mantey C, Brockmann GA, Kalm E, Reinsch N. Mapping and exclusion mapping of genomic imprinting effects in mouse F2 families. Journal of Heredity. 2005;96:329–338. doi: 10.1093/jhered/esi044. This paper develops a formal framework for decomposing and characterizing the effects of imprinted loci using a linear model (as outlined in Box 2) [DOI] [PubMed] [Google Scholar]
- 23.Cui Y, Cheverud JM, Wu R. A statistical model for dissecting genomic imprinting through genetic mapping. Genetica. 2007;130:227–239. doi: 10.1007/s10709-006-9101-x. [DOI] [PubMed] [Google Scholar]
- 24.Cui Y, Lu Q, Cheverud JM, Littell RC, Wu R. Model for mapping imprinted quantitative trait loci in an inbred F2 design. Genomics. 2006;87:543–551. doi: 10.1016/j.ygeno.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 25.Martin ER, Rampersaud E. Family-based genetic association tests. Cold Spring Harb Protoc 2011. 2011 doi: 10.1101/pdb.top96. pdb top96. [DOI] [PubMed] [Google Scholar]
- 26.Wallace C, et al. The imprinted DLK1-MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. Nat Genet. 2009;42:68–71. doi: 10.1038/ng.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belonogova NM, Axenovich TI, Aulchenko YS. A powerful genome-wide feasible approach to detect parent-of-origin effects in studies of quantitative traits. Eur J Hum Genet. 2010;18:379–84. doi: 10.1038/ejhg.2009.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Jiang T. Efficient inference of haplotypes from genotypes on a pedigree. Journal of Bioinformatics and Computational Biology. 2003;1:41–69. doi: 10.1142/s0219720003000204. [DOI] [PubMed] [Google Scholar]
- 29.Kong A, et al. Parental origin of sequence variants associated with complex diseases. Nature. 2009;462:868–74. doi: 10.1038/nature08625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luedi PP, Hartemink AJ, Jirtle RL. Genome-wide prediction of imprinted murine genes. Genome Research. 2005;15:875–884. doi: 10.1101/gr.3303505. This paper develops and implements a bioinformatic approach to identifying imprinted genes in mice using sequence characteristics to train a statistical model based on a machine learning algorithm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imumorin IG, et al. Genome scan for parent-of-origin QTL effects on bovine growth and carcass traits. Frontiers in Genetics. 2011;2 doi: 10.3389/fgene.2011.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheverud JM, et al. Diet-dependent genetic and genomic imprinting effects on obesity in mice. Obesity (Silver Spring) 2010;19:160–70. doi: 10.1038/oby.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawson HA, et al. The importance of context to the genetic architecture of diabetes-related traits is revealed in a genome-wide scan of a LG/J x SM/J murine model. Mamm Genome. 2011;22:197–208. doi: 10.1007/s00335-010-9313-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawson HA, et al. Genetic, epigenetic, and gene-by-diet interaction effects underlie variation in serum lipids in a LG/JxSM/J murine model. J Lipid Res. 2010;51:2976–84. doi: 10.1194/jlr.M006957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magee DA, et al. DNA sequence polymorphisms in a panel of eight candidate bovine imprinted genes and their association with performance traits in Irish Holstein-Friesian cattle. BMC Genetics. 2010;11 doi: 10.1186/1471-2156-11-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nezer C, et al. Haplotype sharing refines the location of an imprinted quantitative trait locus with major effect on muscle mass to a 250-kb chromosome segment containing the porcine IGF2 gene. Genetics. 2003;165:277–285. doi: 10.1093/genetics/165.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nezer C, et al. An imprinted QTL with major effect on muscle mass and fat deposition maps to the IGF2 locus in pigs. Nature Genetics. 1999;21:155–156. doi: 10.1038/5935. [DOI] [PubMed] [Google Scholar]
- 38.Jeon JT, et al. A paternally expressed QTL affecting skeletal and cardiac muscle mass in pigs maps to the IGF2 locus. Nature Genetics. 1999;21:157–158. doi: 10.1038/5938. [DOI] [PubMed] [Google Scholar]
- 39.Van Laere A-S, et al. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature. 2003;425:832–836. doi: 10.1038/nature02064. This study demonstrates that a QTL identified in a mapping study is caused by a particular nucleotide substitution in an intron of the imprinted IGF2 gene. [DOI] [PubMed] [Google Scholar]
- 40.Markljung E, et al. ZBED6, a novel transcription factor derived from a domesticated DNA transposon regulates IGF2 expression and muscle growth. PLoS Biol. 2009;7:e1000256. doi: 10.1371/journal.pbio.1000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berkowicz EW, et al. Single nucleotide polymorphisms at the imprinted bovine insulin-like growth factor 2 (IGF2) locus are associated with dairy performance in Irish Holstein-Friesian cattle. Journal of Dairy Research. 2011;78:1–8. doi: 10.1017/S0022029910000567. [DOI] [PubMed] [Google Scholar]
- 42.Bassett SS, Avramopoulos D, Fallin D. Evidence for parent of origin effect in late-onset Alzheimer disease. Am J Med Genet. 2002;114:679–86. doi: 10.1002/ajmg.10648. [DOI] [PubMed] [Google Scholar]
- 43.Lamb JA, et al. Analysis of IMGSAC autism susceptibility loci: evidence for sex limited and parent of origin specific effects. J Med Genet. 2005;42:132–7. doi: 10.1136/jmg.2004.025668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Francks C, et al. Parent-of-origin effects on handedness and schizophrenia susceptibility on chromosome 2p12-q11. Hum Mol Genet. 2003;12:3225–30. doi: 10.1093/hmg/ddg362. [DOI] [PubMed] [Google Scholar]
- 45.Cichon S, et al. A genome screen for genes predisposing to bipolar affective disorder detects a new susceptibility locus on 8q. Hum Mol Genet. 2001;10:2933–44. doi: 10.1093/hmg/10.25.2933. [DOI] [PubMed] [Google Scholar]
- 46.Lindsay RS, Kobes S, Knowler WC, Bennett PH, Hanson RL. Genome-wide linkage analysis assessing parent-of-origin effects in the inheritance of type 2 diabetes and BMI in Pima Indians. Diabetes. 2001;50:2850–7. doi: 10.2337/diabetes.50.12.2850. [DOI] [PubMed] [Google Scholar]
- 47.Small KS, et al. Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat Genet. 2011;43:561–4. doi: 10.1038/ng.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sha K. A mechanistic view of genomic imprinting. Annu Rev Genomics Hum Genet. 2008;9:197–216. doi: 10.1146/annurev.genom.122007.110031. [DOI] [PubMed] [Google Scholar]
- 49.Cockett NE, et al. Polar overdominance at the ovine callipyge locus. Science. 1996;273:236–238. doi: 10.1126/science.273.5272.236. This paper was the first to identify an imprinted locus affecting a quantitative trait and was also the first to identify a locus showing a pattern of dominance imprinting. [DOI] [PubMed] [Google Scholar]
- 50.Georges M, Charlier C, Cockett N. The callipyge locus: evidence for the trans interaction of reciprocally imprinted genes. Trends in Genetics. 2003;19:248–252. doi: 10.1016/S0168-9525(03)00082-9. [DOI] [PubMed] [Google Scholar]
- 51.Wermter AK, et al. Preferential reciprocal transfer of paternal/maternal DLK1 alleles to obese children: first evidence of polar overdominance in humans. Eur J Hum Genet. 2008;16:1126–34. doi: 10.1038/ejhg.2008.64. [DOI] [PubMed] [Google Scholar]
- 52.Sapienza C, Paquette J, Pannunzio P, Albrechtson S, Morgan K. The polar-lethal Ovum mutant gene maps to the distal portion of mouse chromosome 11. Genetics. 1992;132:241–6. doi: 10.1093/genetics/132.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cockett NE, et al. Polar overdominance at the ovine callipyge locus. Science. 1996;273:236–8. doi: 10.1126/science.273.5272.236. [DOI] [PubMed] [Google Scholar]
- 54.Takeda H, et al. The callipyge mutation enhances bidirectional long-range DLK1-GTL2 intergenic transcription in cis. Proc Natl Acad Sci U S A. 2006;103:8119–24. doi: 10.1073/pnas.0602844103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davis E, et al. RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr Biol. 2005;15:743–9. doi: 10.1016/j.cub.2005.02.060. [DOI] [PubMed] [Google Scholar]
- 56.Bidwell CA, et al. In: Livestock Epigenetics. Khatib H, editor. Wiley-Blackwell; Chichester, UK: 2012. [Google Scholar]
- 57.Lawson HA, et al. Genetic effects at pleiotropic loci are context-dependent with consequences for the maintenance of genetic variation in populations. PLoS Genet. 2011;7:e1002256. doi: 10.1371/journal.pgen.1002256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wagschal A, Feil R. Genomic imprinting in the placenta. Cytogenetics and Genome Research. 2006;113:90–98. doi: 10.1159/000090819. [DOI] [PubMed] [Google Scholar]
- 59.Mabb AM, Judson MC, Zylka MJ, Philpot BD. Angelman syndrome: insights into genomic imprinting and neurodevelopmental phenotypes. Trends in Neuroscience. 2011;34:293–303. doi: 10.1016/j.tins.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marcos L, et al. Genome-wide assessment of imprinted expression in human cells. Genome Biology. 2011;12:R25. doi: 10.1186/gb-2011-12-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luedi PP, et al. Computational and experimental identification of novel human imprinted genes. Genome Res. 2007;17:1723–30. doi: 10.1101/gr.6584707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steinhoff C, Paulsen M, Kielbasa S, Walter J, Vingron M. Expression profile and transcription factor binding site exploration of imprinted genes in human and mouse. BMC Genomics. 2009;10:144. doi: 10.1186/1471-2164-10-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Monk D, et al. Limited evolutionary conservation of imprinting in the human placenta. Proc Natl Acad Sci U S A. 2006;103:6623–8. doi: 10.1073/pnas.0511031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frost JM, Moore GE. The importance of imprinting in the human placenta. PLoS Genet. 2010;6:e1001015. doi: 10.1371/journal.pgen.1001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brideau CM, Eilertson KE, Hagarman JA, Bustamante CD, Soloway PD. Successful computational prediction of novel imprinted genes from epigenomic features. Mol Cell Biol. 2010;30:3357–70. doi: 10.1128/MCB.01355-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luedi PP, Hartemink AJ, Jirtle RL. Genome-wide prediction of imprinted murine genes. Genome Res. 2005;15:875–84. doi: 10.1101/gr.3303505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mancini-Dinardo D, Steele SJ, Levorse JM, Ingram RS, Tilghman SM. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20:1268–82. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends Genet. 2005;21:457–65. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 69.Waters AJ, et al. Parent-of-origin effects on gene expression and DNA methylation in the maize endosperm. Plant Cell. 2011;23:4221–33. doi: 10.1105/tpc.111.092668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gregg C, et al. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science. 2010;329:643–8. doi: 10.1126/science.1190830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang X, et al. Transcriptome-wide identification of novel imprinted genes in neonatal mouse brain. PLoS One. 2008;3:e3839. doi: 10.1371/journal.pone.0003839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.DeVeale B, van der Kooy D, Babak T. Critical evaluation of imprinted gene expression by RNA-Seq: a new perspective. PLoS Genet. 2012;8:e1002600. doi: 10.1371/journal.pgen.1002600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–5. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 74.Hark AT, et al. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–9. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 75.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harris RA, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 2010;28:1097–105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li M, et al. An atlas of DNA methylomes in porcine adipose and muscle tissues. Nat Commun. 2012;3:850. doi: 10.1038/ncomms1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hager R, Cheverud JM, Wolf JB. Relative contribution of additive, dominance and imprinting effects to phenotypic variation in body size and growth between divergent selection lines of mice. Evolution. 2009;63:1118–1128. doi: 10.1111/j.1558-5646.2009.00638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Falconer DS, Mackay TFC. Introduction to quantitative genetics. 4. Longman; Essex: 1996. [Google Scholar]
- 80.Varrault A, et al. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Developmental Cell. 2006;11:711–722. doi: 10.1016/j.devcel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 81.Wolf JB, Cheverud JM. A framework for detecting and characterizing genetic backgroun-dependent imprinting effects. Mammalian Genome. 2009;20:681–698. doi: 10.1007/s00335-009-9209-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li S, et al. Bayesian mapping of genome-wide epistatic imprinted loci for quantitative traits. Theoretical and Applied Genetics. 2012;124:1561–1571. doi: 10.1007/s00122-012-1810-1. [DOI] [PubMed] [Google Scholar]
- 83.Reilly KM, et al. An imprinted locus epistatically influences Nstr1 and Nstr2 to control resistance to nerve sheath tumors in a neuorofibromatosis type 1 mouse model. Cancer Research. 2006;66:62–68. doi: 10.1158/0008-5472.CAN-05-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cattanach BM, Beechey CV, Peters J. Interactions between imprinting effects: summary and review. Cytogenetics and Genome Research. 2006;113:17–23. doi: 10.1159/000090810. [DOI] [PubMed] [Google Scholar]