Abstract
Purpose of the review
Inherited bone marrow failure syndromes (IBMFS) are a diverse set of genetic disorders characterized by the inability of the bone marrow to produce sufficient circulating blood cells. The purpose of this review is to highlight novel findings in the last years and their impact into the understanding of IBMFS.
Recent Findings
Mutations in over 80 different genes have been associated with the development of bone marrow failure (BMF). The products of the genes mutated in IBMFS frequently participate in housekeeping pathways, which are important for cell growth and division rather than being specific for hematopoiesis. The common theme of these pathways, when disturbed, is the activation of p53, leading to cell cycle arrest, senescence, and cell death. With continued improvement in therapy for IBMFS, late complications, such as development of malignancies, are seen more frequently. This highlights the importance of understanding the affected pathways and their roles in cancer development.
Summary
The recent advancement of our understanding of IBMFS has come largely through the identification of the genetic lesions responsible for disease and the investigations of their pathways. Applied in clinical practice, these findings make it possible to unambiguously identify mutation carriers even before the development of BMF and exclude or confirm a suspected clinical diagnosis for many of the more common IBMFS. The further characterization of the pathways leading to IBMFS are likely to reveal novel targets for screening tests, prognostic biomarkers, and improved and specific therapeutics.
Keywords: Inherited bone marrow failure syndromes, Fanconi Anemia, Dyskeratosis Congenita, Diamond-Blackfan Anemia, Shwachman Diamond Syndrome, Congenital Neutropenia
Introduction
The inherited bone marrow failure syndromes (IBMFS) are a diverse set of genetic disorders characterized by insufficient blood cell production. Bone marrow failure (BMF) can affect all blood cell lineages causing clinical symptoms similar to aplastic anemia, or be restricted to one or two blood cell lineages with symptoms specific to the affected cell lineage. Early and correct disease recognition is important, as there are implications for management and for surveillance of long-term sequelae of the disease. Furthermore, as these conditions are often heritable, other family members and future generations may be affected, requiring insightful genetic counseling.
Current advances in the field of IBMFS have come largely through the rapid identification of new genes, that when mutated, cause or predispose to the development of BMF, and through the investigation of the pathways in which the genes’ products are involved. The discovery of new genes currently is driven by the sequencing of candidate genes in pathways identified to be impaired in IBMFS. In the future, development of high-throughput gene sequence technologies is likely to lead to a flood of new genes that are potentially associated with BMF. Understanding the pathways responsible for the development of BMF will be important for determining the clinical significance of newly identified genes, for the development of new screening strategies, and for the identification of potential therapeutic targets.
Excellent reviews in the general field of IBMFS have recently been published. [1–4] In this review we will refer to these reviews and focus on the most recent discoveries, how they advance our understanding in the disease pathophysiology, and their possible implication to clinical practice.
Recent developments in Fanconi Anemia (FA)
Fanconi anemia (FA, MIM #227650) is characterized by progressive bone-marrow failure, congenital abnormalities, and a pronounced susceptibility for malignancy including myelodysplastic syndrome (MDS), leukemia (especially acute myeloblastic leukemia, AML) and solid tumors (particularly carcinoma of the oropharynx and skin) [5]. Cells from FA patients exhibit a characteristic hypersensitivity to DNA interstrand cross-linking agents such as mitomycin C. Mutations in 15 different genes (FANC genes) (Table 1) have been identified, the products of which all participate in the sensing, responding, and resolving of DNA interstrand cross-links during replication (Figure 1) [6, 7]. With the exception of X-linked recessive FANCB, FA genes are inherited in an autosomal recessive pattern. [6, 8]
Table 1.
Genes associated with inherited bone marrow failure syndromes.
| Disease / Genes | Chromosome Locus |
Mode of Inheritance |
Frequency (% of pts) |
|---|---|---|---|
| Fanconi Anemia | |||
| FANCA | 16q24.3 | AR | 60 |
| FANCB | Xp22.31 | XLR | 2 |
| FANCC | 9q22.3 | AR | 14 |
| FANCD1/BRCA2 | 13q12.3 | AR | 3 |
| FANCD2 | 3p25.3 | AR | 3 |
| FANCE | 6p21.3 | AR | 3 |
| FANCF | 11p15 | AR | 2 |
| FANCG/XRCC9 | 9p13 | AR | 10 |
| FANCI | 15q25-26 | AR | 1 |
| FANCJ/BACH1/BRIP1 | 17q22.3 | AR | 2 |
| FANCL | 2p16.1 | AR | 0.2 |
| FANCM | 14q21.3 | AR | 0.2 |
| FANCN/PALB2 | 16p12.1 | AR | 0.7 |
| FANCO | 17q22 | AR | I case |
| FANCP | 16p13.3 | AR | I case |
| Dyskeratosis Congenita | |||
| DKC1 | Xq28 | XLR,DN | 30 |
| TINF2 | 14q11.2 | AD,DN | 15 |
| TERC | 3q26.3 | AD | 10 |
| TERT | 5p15.53 | AD, AR,DN | 5-10 |
| NOP10/NOLA3 | 15q14-q15 | AR | <1 |
| NHP2/NOLA2 | 5q35.5 | AR | <1 |
| TCAB1/WDR79/WRP53 | 17p13.1 | AR | 2 cases |
| Diamond-Blackfan Anemia | |||
| RPS19 | 19q13.2 | AD,DN | 25 |
| RPL5 | 1p22.1 | AD,DN | 7 |
| RPL11 | 1p36.1-p35 | AD,DN | 5 |
| RPL35a | 3q29-qter | AD,DN | 3 |
| RPS26 | 12q13.2 | AD,DN | 6 |
| RPS24 | 10q22-q23 | AD,DN | 2 |
| RPS17 | 15q25.2 | AD,DN | 1 |
| RPS7 | 2p25.3 | AD,DN | 1 |
| RPS10 | 6p21.31 | AD,DN | 3 |
| Shwachman-Diamond Syndrome | |||
| SBDS | 7q11 | AR | >90 |
| Severe Congenital Neutropenia | |||
| ELA-2/ELANE | 19p13.3 | AD,DN | 75 |
| GFI1 | 1p22 | AD | <1 |
| HAX1 | 1q21.3 | AR | 1 |
| WAS | Xp11.33-11.22 | XLR | <1 |
| G6PC3 | 17q21.31 | AR | <1 |
| CSF3R | 1p35-34.3 | AD,DN | <1 |
AR=autosomal recessive, XLR=X-linked recessive, DN=de novo, AD=autosomal dominant.
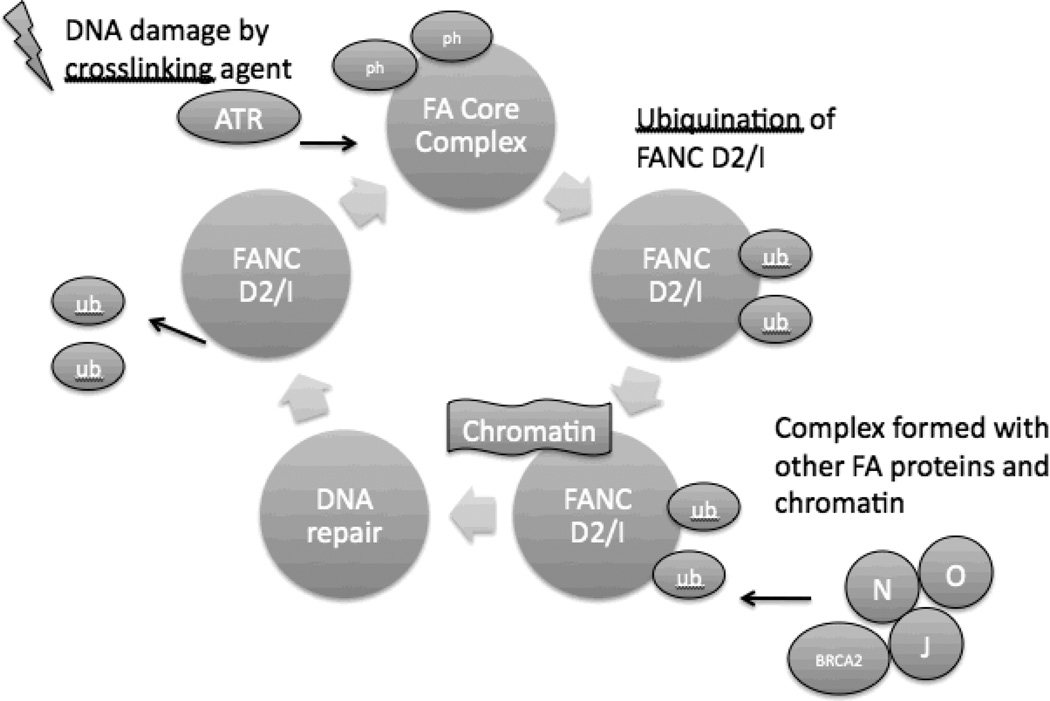
Figure 1. DNA repair and FANC proteins.
After DNA damage, the Ataxia Telangiectasia and RAD3-related kinase (ATR) activates the FA core complex (consisting of FANC A/B/C/E/F/G/L/M) by phosphorylation (ph). The core complex then acts as a ubiquitin ligase and monoubiquinates (ub) FANCD2 and FANCI. This monoubiquinated complex then is targeted to chromatin where it forms a complex with additional FA proteins and DNA repair proteins. DNA repair occurs through a variety of mechanisms. FANC D2/I are then deubiquinated, allowing their release from chromatin. Mutations in any of the FANC proteins will lead to defective DNA repair.
FANCO and FANCP – new disease-causing members in the FA pathway
Vaz et al [9]** identified biallelic mutations in RAD51C, a member of the RAD51-like family that is essential for RAD51-mediated homologous recombination (HR) in a family with “FA-like” phenotypes. The two affected siblings carrying biallelic RAD51C mutations died at a very young age, with multiple congenital abnormalities. Their lymphocytes showed an increased sensitivity to DNA cross-linking agents, characteristic for genes of the FA pathway. The only surviving patient exhibited many characteristic FA phenotypes, including increased sensitivity to DNA cross-linking agents and pronounced cell cycle arrest at G2/M which were rescued by exogenous RAD51C cDNA expression, suggesting that RAD51C is an FA gene (FANCO). At the time of diagnosis none of the affected individuals showed signs of BMF. However the onset of BMF may vary in FA patients.[9]**
SLX4 is the most recent member of the FANC protein family. SLX4 forms a complex with multiple other nucleases that all are required to prevent sensitivity to DNA cross-linking agents. Biallelic SLX4 mutations were identified in two individuals with typical clinical features of Fanconi anemia and the cellular defects were rescued by wildtype SLX4, defining SLX4 as a new FANC protein, FANCP. [10]**
Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia
The FA pathway promotes HR mediated repair and inhibits the error prone repair by non-homologous end joining (NHEJ) of DNA interstrand crosslinks. Thus FA cells have an increased use of the NHEJ repair mechanism, which in part accounts for cell toxicity in FA. Interestingly, recent reports have shown that the inhibition of the NHEJ pathway in cell lines derived from patients with FA can reduce the toxicity of interstrand crosslinks inducing drugs, thereby identifying an exciting and new drug target for the possible treatment of FA.[11]*
Progression MDS and Leukemia in patients with FA
Patients with FA have an increased risk of myelodysplastic syndrome (MDS) and acute leukemia, typically AML. Recent studies of the FA patient cohort of the National Cancer Institute determined a cumulative incidence of 40% for MDS and of 10% for AML in patients with FA by the age of 50 years[12]*.
Recent studies of disease progression have found that MDS and AML in FA are associated with a specific pattern of acquired genomic abnormalities[13–15]. Transformation to MDS in FA was found to be associated with a pattern of recurrent chromosomal abnormalities, such as monosomy 7 (−7), deletion of the long arm of chromosome 7 (7q-), gain of chromosome 3q (+3q), and gain of chromosome 1q (+1q). Cryptic chromosome 21 abnormalities, including the RUNX1 gene mutations, were seen in about 20% of FA patients with MDS. Chromosome 7 and 3q abnormalities had a poor prognostic value, whereas an isolated +1q was observed before the development of dysplasia and was not predictive for the development of AML[13]**.
Recent Developments in Dyskeratosis Congenita (DC)
Dyskeratosis congenita (DC, MIM 305000, 127550, 224230) classically presents with skin pigmentation changes, leukoplakia, and nail dystrophy. BMF develops in about 80% of patients by the age of 20 years old. Patients with DC have increased risk of MDS, AML and solid tumors with a cumulative incidence of 30%, 10% and 20–30%, respectively[12]*. There are mutations found in 7 genes, which are all involved in telomere maintenance (Table 1) [16].
Telomeres are at the ends of linear chromosomes. In vertebrates, they are composed of many tandem repeats of a hexanucleotide—TTAGGG—and associated proteins, collectively termed the shelterin complex [17]. Telomeres shorten with cell replication. The telomerase enzyme, a ribonucleoprotein complex, is responsible for the elongation of telomeres by adding telomeric repeats to the end of telomeres in early progenitor cells. Six of the DC genes encode proteins that are part of the telomerase complex, whereas the seventh, TINF2, encodes the TIN2 protein, which is part of the shelterin complex (Figure 2). No mutations were identified in the 5 other proteins of the shelterin complex in patients with DC [18]. The clinical symptoms caused by mutations in DC genes vary with the gene, the mutation, and the age of onset of disease. [19, 20]* [21]
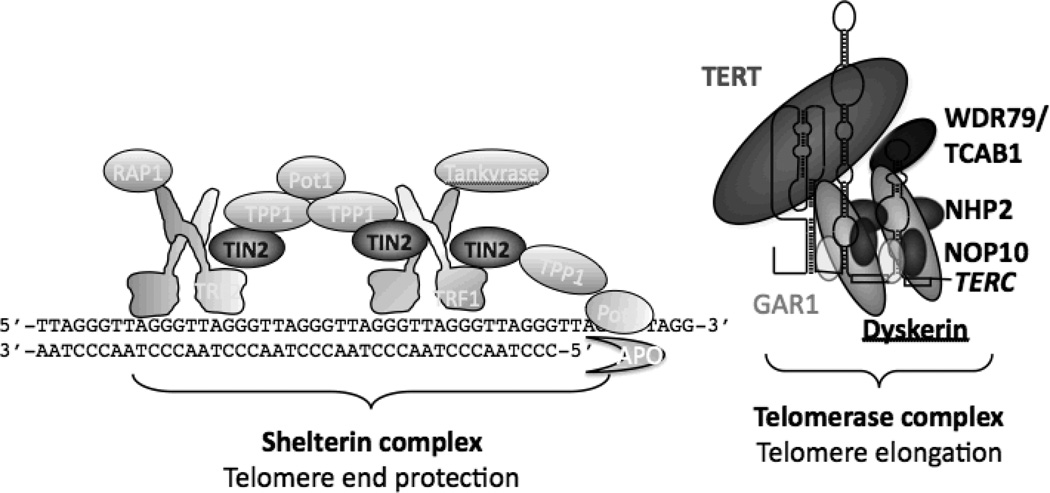
Figure 2. Telomere biology.
Telomeres are at the ends of linear chromosomes and are composed of many tandem repeats of a hexanucleotide and associated proteins, called the shelterin complex. As telomeres shorten with cell replication, the telomerase complex is responsible for the elongation of telomeres by adding telomeric repeats to the ends of telomeres. Six of the DC gene products are a part of the telomerase complex (Dyskerin, TERT, TERC, NHP2, NOP10, and WDR79/TCAB1). The seventh (TIN2) is part of the shelterin protein complex.
Telomere length measurement in peripheral blood mononuclear cells or lymphocytes is a sensitive screening test for patients presenting with BMF. The presence of short telomeres in peripheral blood lymphocytes is suggestive for DC to be the cause of BMF but not necessarily specific as only the demonstration of a pathogenic DC gene mutation can confirm the diagnosis. [22–24].
TCAB1, a new disease-causing member in the DC pathway
WDR79/TCAB1 (WRAP53) binds the telomerase RNA TERC and is required for the trafficking of the telomerase complex to the Cajal bodies, which is essential for telomerase function[25, 26]. Mutations in the TERC/TCAB1 binding domain (CAB domain) or the loss of TCAB1 results in the mislocalization of the telomerase RNA TERC and telomere shortening. Biallelic missense mutations in these genes have been identified in 2 patients with the classic presentation of DC, including BMF and shortened telomeres. The heterozygous parents and siblings demonstrated no clinical manifestations and had a telomere length within the normal distribution.
Clericuzio-type Poikiloderma with Neutropenia - a disease with clinical similarities but normal telomeres
Neutropenia and poikiloderma (Clericuzio-type Poikiloderma with Neutropenia, CPN, MIM # 604173) is an autosomal recessive disease with clinical features that share similarities to those of classic DC, including hyperpigmenation of the skin, nail changes, marrow failure with a propensity to develop MDS and AML[16]. Recently, biallelic mutations in the C16orf57 gene had been identified in patients with CPN. However, in contrast to all other forms of DC, telomere lengths are normal in peripheral blood cells of these individuals. As the exact function of the C16orf57 protein remains unknown, it remains to be determined whether the gene product shares or overlaps with pathways of other DC genes.
Recent developments in Diamond Blackfan Anemia
Diamond–Blackfan anemia (DBA, MIM 105650) is an inherited form of pure red cell aplasia. DBA was the first human disease that was directly linked to a defect in ribosome biogenesis, with the discovery of the ribosomal protein (RP) S19 mutation. Despite this discovery, it took several years before DBA was recognized as a disease of impaired ribosome biogenesis (Figure 3). There was an initial disbelief that a disorder that primarily affects erythropoiesis may be caused by a defect in a housekeeping pathway essential for every dividing cell. Now, mutations in 9 proteins of the large and small ribosomal subunits have been identified as accounting for about half of DBA cases (Table 1 and DBA mutation Database http://www.dbagenes.unito.it). Haploinsufficiency in RPs is thought to be responsible for disease. [27]
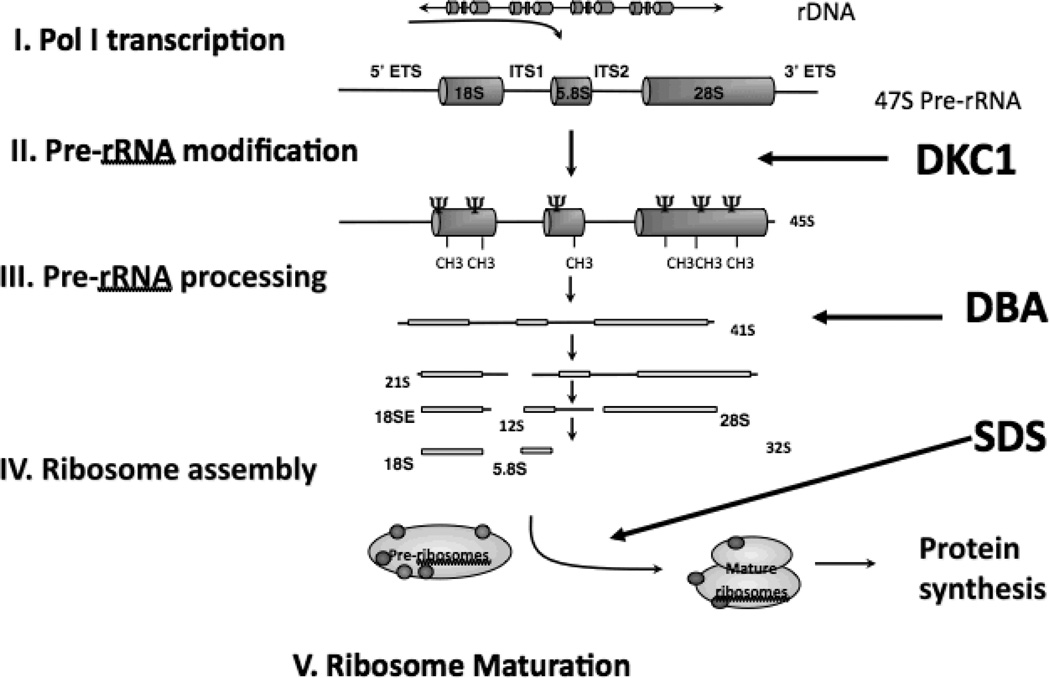
Figure 3. Eukaryotic ribosome biogenesis.
This process requires the coordinated synthesis of 4 ribosomal rRNAs (18S, 28S, 5.8S, 5S), several additional core ribosomal proteins and other associated proteins. Several diseases are implicated along the process of ribosome biogenesis, including Diamond-Blackfan Anemia (DBA), Shwachman Diamond Syndrome SDS), and Dyskeratosis Congenita (DC) secondary to DKC1 mutations.
RP S7, S15, S26, S27A, L11, L5, and L36, - new disease causing genes in the DBA pathway
Mutations in RP S19, S24, S17, and L35A have been identified in approximately one-third of patients with DBA. RP L5 and L11 were found to be mutated in an additional 11.4% of patients[28]. A large-scale screen of additional 45 RP genes in a large DBA cohort revealed pathogenic mutations in S7, S15, S27A, S10, S26, and L36. [29–31]. Together, mutations in 9 of these RPs account for about 53% of patients with DBA (Table 1). This screen did not include the looking for intragenic or large gene deletions, which may account for possibly another 20–30% of patients. Interestingly, mutations have been found only in some RPs and some are more frequently mutated in patients with DBA than others. A possible explanation is that there is a delicate balance in the synthesis of functional RPs and that haploinsuffciency of specific RPs is compatible with life and leads to the DBA specific red cell aplasia.
RPs are important in the processing of the ribosomal RNA (rRNA). Defects in RPs often lead to the accumulation normal and abnormal rRNA precursors. Quantitation of rRNA precursors in immortalized lymphoblastoid cell lines has been suggested as a screening test for DBA. In addition, it is thought that quantitation of rRNA precursors may indicate the RP that is affected. However, the changes in the accumulation of the rRNA precursors are very subtle and are shared between RPs. Therefore, results would have to be carefully interpreted.
Genotype-phenotype correlations
While the phenotype of DBA is highly variable, recent studies suggested some genotype-phenotype correlations. Mutations in L5 were associated with a higher frequency of physical abnormalities, including cleft lip and/or palate, whereas mutations in L11 had more isolated thumb abnormalities compared with patients with mutations in S19.[28, 32]
p53 activation as a result of ribosome dysfunction
There is experimental evidence through a variety of animal and cellular disease models that impairment of ribosome biosynthesis causes the activation of p53. The p53 protein and RNA interacts with a several RPs on different levels. An area of major regulation occurs through MDM2 binding to L11 and L5 [33].
Haploinsufficiency of S19 caused selective activation of the p53 pathway in erythroid progenitor cells compared with cells from other hematopoietic lineages[34]* suggesting that terminal erythroid differentiation is particularly sensitive to reduced expression levels of RPs. Decreased expression of p53 was shown to rescue the red cell phenotype. Unfortunately the lack of p53 is also associated with leukemia transformation in patients with 5q-, a form of MDS that is associated with red cell aplasia due to the deletion of S14. [35]
Dexamethasone and lenalidomide have distinct functional effects on erythropoiesis
The efficacy of lenalidomide in 5q- MDS has raised the possibility that lenalidomide might have therapeutic utility in DBA. While dexamethasone increases selectively the number of burst-forming units-erythroid (BFU-E), lenalidomide was found to specifically increase colony-forming units-erythroid (CFU-E). Use of the drugs in combination demonstrated that their effects are not overlapping.[36]* However the long-term effects of lenalidomide, particularly an increased risk of developing MDS/AML is a concern, as DBA with current treatment has a low incidence of transformation to MDS/AML.
Recent developments in Shwachman Diamond Syndrome
Shwachman–Diamond syndrome (SDS, MIM 260400) is an autosomal recessive multisystem disorder characterized by bone-marrow failure, pancreatic insufficiency, and skeletal abnormalities. Biallelic SBDS gene mutations inherited in an autosomal recessive fashion is causative of the disease (Table 1). In over 90% of patients diagnosed with SDS, the most common mutations are 258+2T>C, followed by 183−184TA>CT. Compound heterozygote mutations at 183−184TA>CT and 258+2T>C account for approximately 81% of patients with SDS. Patients with SDS have an increased propensity for MDS and leukemia of about 30–40% by the age of 30 years old. [37]
Multifunctional role for the SBDS protein
The SBDS protein is localized throughout the cell and shuttles in and out of the nucleolus, the major cellular site of ribosome biosynthesis. Mammalian SBDS binds to the 60S large ribosomal subunit. While involvement of SBDS in ribosome biogenesis had long been suspected, other possible functions of the SBDS protein, including chemotaxis and mitotic spindle destabilization, have been postulated. Recent data using slime mold as a disease model showed that SBDS-depleted cells had a defect in ribosome subunit joining.[38] A recent study also showed that SBDS evicted eIF6 from interaction with the 60S subunit. eIF6 is a translation initiation factor that binds to the 60S subunit in the nucleolus and inhibits subunit joining. Defective ribosomal subunit joining was also demonstrated in lymphoblasts from SDS patients.[38]* This work supports the idea that SDS is a ribosomopathy[29].
Progression to clonal hematopoiesis, MDS, and Leukemia in patients with SDS
Abnormalities of chromosome 7 and 20q deletions are the most frequent chromosomal changes in patients with SDS. If these chromosomal abnormalities are isolated findings without dysplastic changes in the marrow, they may be transient and not necessarily indicative for progression to advanced MDS or AML.
Isochromosome i(7)is the most frequent karyotypic anomaly in patients with SDS. It causes a duplication of the SBDS locus and usually occurs on the allele with some residual SBDS function. Interestingly, this chromosomal abnormality is not found in AML cells arising in SBDS patients suggesting that this karyotypic abnormality does not contribute to leukemia transformation. [39]
Recent developments in Congenital Neutropenia
Congenital neutropenia is genetically and phenotypically heterogeneous. To date several genetic causes associated with neutropenia have been identified (Table 1 and for review, see [40, 41]). There is currently no consensus on the classification of congenital neutropenias. Patients with severe congenial neutropenia (SCN) typically present with recurrent life-threatening infections in the first few moths of life and have neutrophil count of less than 500/ul for at least 3 months. SCN often shows a characteristic a maturation arrest of myeloid precursors at the promyelocyte stage of differentiation. Heterozygous ELANE mutations are the most frequent cause of SCN and are found in approximately 30% of patients with SCN.
Despite the progress in gene discovery an all-encompassing mechanism causing SCN has not yet emerged. Pathways of the unfolded protein response, endoplasmic reticulum stress [42], lysosomal trafficking, mitochondrial dysfunction, and defective glycosylation[43] have all been implicated in the apoptosis of the maturing myeloid cell.
Interestingly many, but not all, forms of SCN are associated with an increased risk transformation to MDS and AML (30% by the age of 40 years in SCN patients with ELANE mutations). The development of MDS/AML in SCN patients with ELANE, HAX1, and activating WASP mutations is often preceded by clonal expansion of cells harboring nonsense mutations in the CSF3R gene, which results in the expression of truncated CSF3R proteins. These mutations in SCN patients are highly predictive of leukemic transformation [44]. This suggests a common early common pathway of leukemogenesis that is unique for patients with SCN. Chromosome 7 abnormalities, RUNX1, RAS, and KIT gene mutations have also been described in MDS/AML of patients with ELANE mediated SCN [45].
Ongoing controversy of G-CSF treatment and leukemic transformation
Patients with severe congenital neutropenia present early in infancy with severe infections. With the introduction of GCSF treatment has significantly improved survival and morbidly for patients with congenital neutropenias, however, there is ongoing concern that the long-term use of G-CSF is associated with the development of MDS/AML. Transformation to MDS/AML is highest for patients with a poor G-CSF response requiring higher doses of G-CSF. Whether this is directly caused by G-CSF or by the underlying condition remains controversial.
Clinical Impact
What are the insights gained by the recent discoveries and how do they translate to clinical practice? The increasing number of new genes associated with BMF indicates that genetic causes or susceptibly to develop BMF is probably more common than previously thought and that mutations in BMF genes may account for a far broader clinical spectrum than originally anticipated, both in the pediatric and adult patient population. The increasing numbers of genes associated with BMF make genetic testing for a large number of genes prohibitively expensive and impractical at the moment. However in the future, with the development of parallel testing of multiple BMF genes in a single sample using next-generation sequencing, comprehensive genetic testing will be available. The interpretation of comprehensive genetic testing as well as of screening tests however also adds to the complexity of making a clinical diagnosis.
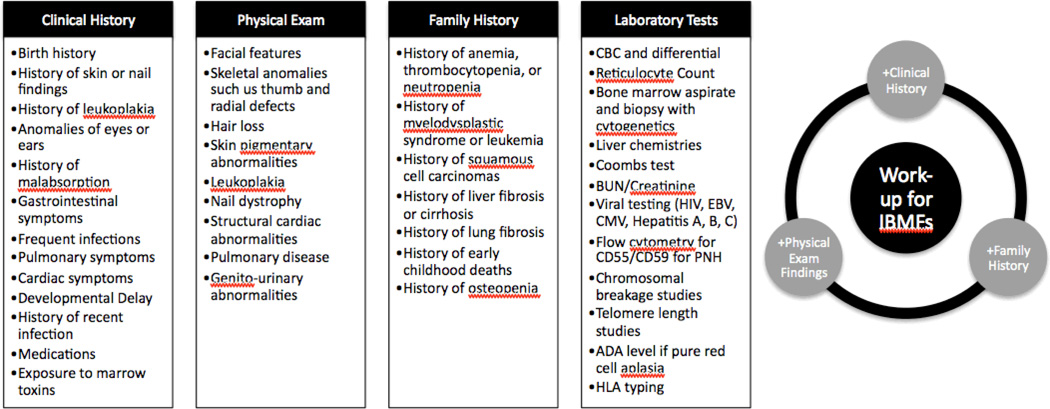
Thus, interpretation should be performed by a specialist or preferably a team of specialists familiar with the clinical presentation of the patient. Family and personal history, physical examination, and laboratory findings may raise the suspicion of an IBMFS. Screening tests, in particular for genetic predisposition that would drastically alter the therapeutic decisions should be performed not only in children but also in the adult patient with BMF. Chromosomal breakage studies or telomere length measurements, for example, should be performed in all patients with BMF for whom HSCT is considered a possible therapy. Figure 4 highlights some of the findings in clinical history, physical exam, family history and suggested laboratory tests that may indicate an IBMFS.
Figure 4.
Findings in the clinical history, physical exam, and family history that can provide clues to IBMFs. Suggested laboratory testing is also listed.
Key: CBC=complete blood count, BUN=blood urea nitrogen, HIV=human immunodeficiency virus, EBV=Epstein Barr virus, CMV=cytomegalovirus, PNH=paroxysmal nocturnal hemoglobinuria, ADA= red cell adenosine deaminase activity, HLA=human leukocyte antigen
Conclusion
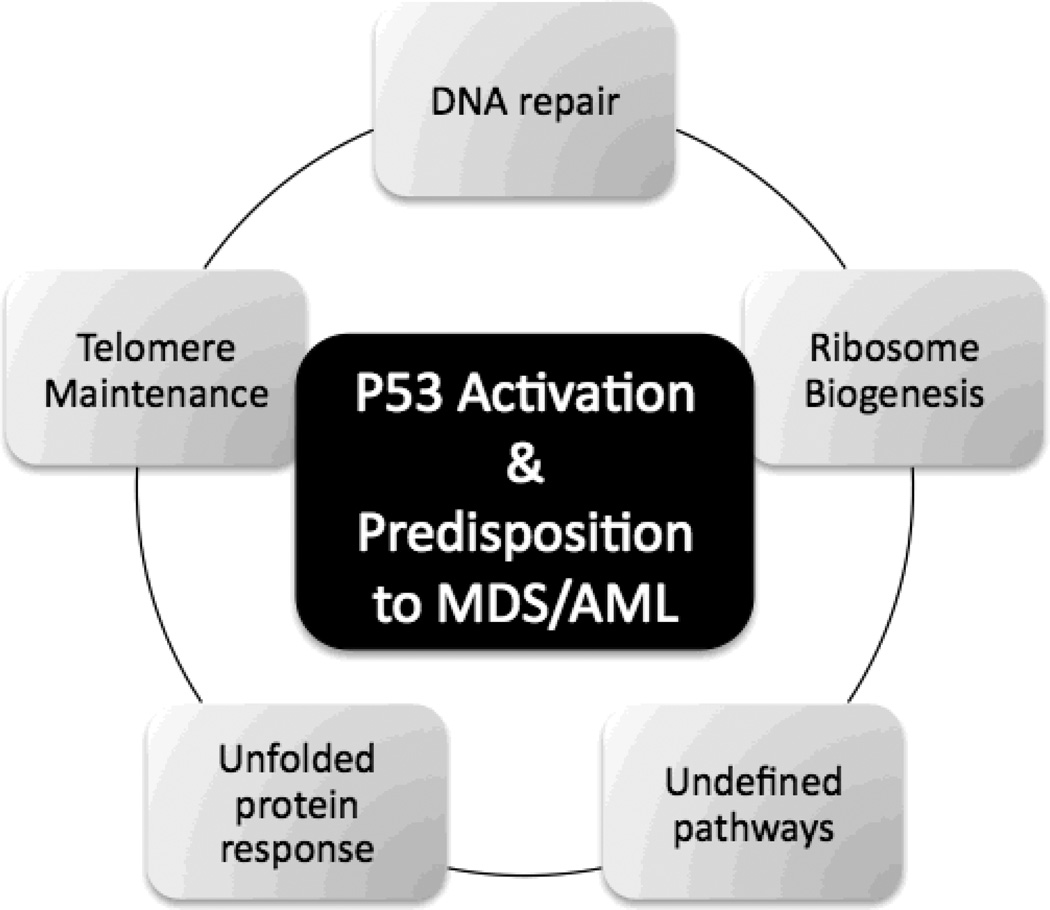
Understanding of the pathophysiology of IBMFS has exploded with the identification of genes. Many of these genes encode proteins involved in cellular housekeeping pathways that, when disturbed, cause cell senescence and apoptosis as well as predispose to malignant transformation (Figure 5). The understanding of the disease causing pathways will allow us to cope with the large number of genes likely to be discovered with the new sequence technologies. It will enable us to develop the appropriate screening tools, discover new prognostic markers, and develop more specific drugs.
Figure 5. Biologic Pathways Involved in IBMFs.
The pathways affected in the major forms of IBMFS are house-keeping pathways important for the growth and cell division of all cells: Defective DNA repair pathways is responsible for Fanconi Anemia, defective telomere maintenance causes Dyskeratosis Congenita, defective ribosomal processing is seen in Diamond-Blackfan Anemia, and is also thought to have implications in Shwachmann Diamond Syndrome and Dyskeratosis Congenita. Unfolded protein response is involved in some forms of Severe Congenital Neutropenia and is thought to play a role in apoptosis of granulocytic precursors. Additional pathways remain to be determined. The pathways involved in the IBMFs all lead to the activation of p53 causing cell cycle arrest, cell senescence or cell death, which are all features of bone marrow failure. Interestingly what these pathways also share is a predisposition to myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML), but the pathways(s) of leukemic transformations remain to be determined.
Key: MDS=myelodysplastic syndrome, AML=Acute myeloid leukemia
Key Phrases.
IBMFS are a diverse set of genetic disorders characterized by the inability of the bone marrow to produce sufficient blood cells.
Many of the pathways discovered in IBMFS involve cellular housekeeping pathways, that when disturbed, lead to cellular apoptosis and can predispose to malignant transformation.
Continued identification of new genes and pathways involved in IBMFS is likely allow for development of new screening tools, discovery of new prognostic markers, and development of new therapeutics.
Acknowledgements
We thank Amanda McCarthy and Mariela JMorales Jimenez MD study coordinators for their assistance in our Studies on Bone Marrow Failure at CHOP and HUP UPENN and Philip J Mason PhD for his input and interesting discussions.
The work has been supported by the Buck Family Endowed Chair in Hematology and NCI NHI 2R01 CA105312 to MB and the NIH 2KL2RR024132-06 to SP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shimamura A, Alter BP. Pathophysiology and management of inherited bone marrow failure syndromes. Blood Rev. 2010;24:101–122. doi: 10.1016/j.blre.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dokal I, Vulliamy T. Inherited bone marrow failure syndromes. Haematologica. 2010;95:1236–1240. doi: 10.3324/haematol.2010.025619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klein C. Genetic defects in severe congenital neutropenia: emerging insights into life and death of human neutrophil granulocytes. Annu Rev Immunol. 2011;29:399–413. doi: 10.1146/annurev-immunol-030409-101259. [DOI] [PubMed] [Google Scholar]
- 4.Orkin SH, Nathan DG. Nathan and Oski's hematology of infancy and childhood. Philadelphia: Saunders/Elsevier; 2009. [Google Scholar]
- 5.Green AM, Kupfer GM. Fanconi anemia. Hematol Oncol Clin North Am. 2009;23:193–214. doi: 10.1016/j.hoc.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seif AE. Pediatric leukemia predisposition syndromes: clues to understanding leukemogenesis. Cancer Genet. 2011;204:227–244. doi: 10.1016/j.cancergen.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 7.de Winter JP, Joenje H. The genetic and molecular basis of Fanconi anemia. Mutat Res. 2009;668:11–19. doi: 10.1016/j.mrfmmm.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 8.D'Andrea AD. Susceptibility pathways in Fanconi's anemia and breast cancer. N Engl J Med. 2010;362:1909–1919. doi: 10.1056/NEJMra0809889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vaz F, Hanenberg H, Schuster B, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 2010;42:406–409. doi: 10.1038/ng.570. This paper highlights how a homozygous missense mutation was discovered in the RAD51C gene in a family with characteristic Fanconi Anemia findings. RAD51C is involved in homologous recombination mediated DNA repair.
- 10. Kim Y, Lach FP, Desetty R, et al. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet. 2011;43:142–146. doi: 10.1038/ng.750. This paper highlights how the depletion of SLX4, a protein that interacts with multiple nucleases, resulted in increased sensitivity of cells to crosslinking agents. This finding was then linked to two individuals with clinical stigmata of Fanconi Anemia, where testing of these individuals' cells showed biallelic mutations in SLX4.
- 11. Adamo A, Collis SJ, Adelman CA, et al. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell. 2010;39:25–35. doi: 10.1016/j.molcel.2010.06.026. This article highlights the importance of FANCD2 in preventing DNA repair by non-homologous end joining, which is an error-prone repair pathway.
- 12. Alter BP, Giri N, Savage SA, et al. Malignancies and survival patterns in the National Cancer Institute inherited bone marrow failure syndromes cohort study. Br J Haematol. 2010;150:179–188. doi: 10.1111/j.1365-2141.2010.08212.x. This paper describes a cohort of patients with IBMFS that were followed for the development of BMF, MDS, AML, and solid tumors.
- 13. Quentin S, Cuccuini W, Ceccaldi R, et al. Myelodysplasia and leukemia of Fanconi anemia are associated with a specific pattern of genomic abnormalities that includes cryptic RUNX1/AML1 lesions. Blood. 2011;117:e161–e170. doi: 10.1182/blood-2010-09-308726. This article highlights the importance of acquired genetic changes in the bone marrow of patients with Fanconi Anemia, and illustrates that specific chromosomal abnormalities may be linked to the development of MDS/AML. This has significant implications for prognostic management of Fanconi Anemia patients.
- 14.Mehta PA, Harris RE, Davies SM, et al. Numerical chromosomal changes and risk of development of myelodysplastic syndrome--acute myeloid leukemia in patients with Fanconi anemia. Cancer Genet Cytogenet. 2010;203:180–186. doi: 10.1016/j.cancergencyto.2010.07.127. [DOI] [PubMed] [Google Scholar]
- 15.Cioc AM, Wagner JE, MacMillan ML, et al. Diagnosis of myelodysplastic syndrome among a cohort of 119 patients with fanconi anemia: morphologic and cytogenetic characteristics. Am J Clin Pathol. 2010;133:92–100. doi: 10.1309/AJCP7W9VMJENZOVG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dokal I, Vulliamy T, Mason P, Bessler M. Clinical utility gene card for: Dyskeratosis congenita. Eur J Hum Genet. 2011 doi: 10.1038/ejhg.2011.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calado RT, Young NS. Telomere diseases. N Engl J Med. 2009;361:2353–2365. doi: 10.1056/NEJMra0903373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savage SA, Giri N, Jessop L, et al. Sequence analysis of the shelterin telomere protection complex genes in dyskeratosis congenita. J Med Genet. 2011;48:285–288. doi: 10.1136/jmg.2010.082727. [DOI] [PubMed] [Google Scholar]
- 19.Bessler M, Wilson DB, Mason PJ. Dyskeratosis congenita. FEBS Lett. 2010;584:3831–3838. doi: 10.1016/j.febslet.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vulliamy TJ, Kirwan MJ, Beswick R, et al. Differences in disease severity but similar telomere lengths in genetic subgroups of patients with telomerase and shelterin mutations. PLoS One. 2011;6:e24383. doi: 10.1371/journal.pone.0024383. This paper reviewed genetic subtypes of dyskeratosis congenita by looking at telomere length, telomerase activity, and clinical presentation among 194 cases.
- 21.Savage SA, Alter BP. Dyskeratosis congenita. Hematol Oncol Clin North Am. 2009;23:215–231. doi: 10.1016/j.hoc.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vulliamy TJ, Marrone A, Knight SW, et al. Mutations in dyskeratosis congenita: their impact on telomere length and the diversity of clinical presentation. Blood. 2006;107:2680–2685. doi: 10.1182/blood-2005-07-2622. [DOI] [PubMed] [Google Scholar]
- 23.Du HY, Pumbo E, Ivanovich J, et al. TERC and TERT gene mutations in patients with bone marrow failure and the significance of telomere length measurements. Blood. 2009;113:309–316. doi: 10.1182/blood-2008-07-166421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alter BP, Baerlocher GM, Savage SA, et al. Very short telomere length by flow fluorescence in situ hybridization identifies patients with dyskeratosis congenita. Blood. 2007;110:1439–1447. doi: 10.1182/blood-2007-02-075598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tycowski KT, Shu MD, Kukoyi A, Steitz JA. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell. 2009;34:47–57. doi: 10.1016/j.molcel.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venteicher AS, Artandi SE. TCAB1: driving telomerase to Cajal bodies. Cell Cycle. 2009;8:1329–1331. doi: 10.4161/cc.8.9.8288. [DOI] [PubMed] [Google Scholar]
- 27.Vlachos A, Dahl N, Dianzani I, Lipton JM. Clinical utility gene card for: Diamond Blackfan anemia. Eur J Hum Genet. 2011;19 doi: 10.1038/ejhg.2010.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gazda HT, Sheen MR, Vlachos A, et al. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am J Hum Genet. 2008;83:769–780. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boria I, Garelli E, Gazda HT, et al. The ribosomal basis of Diamond-Blackfan Anemia: mutation and database update. Hum Mutat. 2010;31:1269–1279. doi: 10.1002/humu.21383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doherty L, Sheen MR, Vlachos A, et al. Ribosomal protein genes RPS10 and RPS26 are commonly mutated in Diamond-Blackfan anemia. Am J Hum Genet. 2010;86:222–228. doi: 10.1016/j.ajhg.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quarello P, Garelli E, Carando A, et al. Diamond-Blackfan anemia: genotype-phenotype correlations in Italian patients with RPL5 and RPL11 mutations. Haematologica. 2010;95:206–213. doi: 10.3324/haematol.2009.011783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boultwood J, Pellagatti A, Wainscoat JS. Haploinsufficiency of ribosomal proteins and p53 activation in anemia: Diamond-Blackfan anemia and the 5q- syndrome. Adv Enzyme Regul. 2011 doi: 10.1016/j.advenzreg.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 34. Dutt S, Narla A, Lin K, et al. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood. 2011;117:2567–2576. doi: 10.1182/blood-2010-07-295238. This article specifically describes the accumulation of p53 in erythroid progenitor cells after knockdown of ribosomal proteins S19 and S14. Indeed, inhibition of p53 rescued the erythroid defect.
- 35.Jadersten M, Saft L, Smith A, et al. TP53 mutations in low-risk myelodysplastic syndromes with del(5q) predict disease progression. J Clin Oncol. 2011;29:1971–1979. doi: 10.1200/JCO.2010.31.8576. [DOI] [PubMed] [Google Scholar]
- 36. Narla A, Dutt S, McAuley JR, et al. Dexamethasone and lenalidomide have distinct functional effects on erythropoiesis. Blood. 2011 doi: 10.1182/blood-2010-11-318543. Corticosteroids and Lenolidamide are used in patients with DBA and MDS, respectively. This article found that dexamethasone and lenolidomide promoted different stages of erythropoiesis and implies a potentially new combination of therapy for these patients.
- 37.Donadieu J, Leblanc T, Bader Meunier B, et al. Analysis of risk factors for myelodysplasias, leukemias and death from infection among patients with congenital neutropenia. Experience of the French Severe Chronic Neutropenia Study Group. Haematologica. 2005;90:45–53. [PubMed] [Google Scholar]
- 38. Wong CC, Traynor D, Basse N, et al. Defective ribosome assembly in Shwachman-Diamond syndrome. Blood. 2011 doi: 10.1182/blood-2011-06-353938. In a slime mold model, SDS mutants showed impairment in ribosomal subunit joining, supporting the idea that SDS is a ribosomopathy.
- 39.Minelli A, Maserati E, Nicolis E, et al. The isochromosome i(7)(q10) carrying c.258+2t>c mutation of the SBDS gene does not promote development of myeloid malignancies in patients with Shwachman syndrome. Leukemia. 2009;23:708–711. doi: 10.1038/leu.2008.369. [DOI] [PubMed] [Google Scholar]
- 40.Boztug K, Appaswamy G, Ashikov A, et al. A syndrome with congenital neutropenia and mutations in G6PC3. N Engl J Med. 2009;360:32–43. doi: 10.1056/NEJMoa0805051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Donadieu J, Fenneteau O, Beaupain B, et al. Congenital neutropenia: diagnosis, molecular bases and patient management. Orphanet J Rare Dis. 2011;6:26. doi: 10.1186/1750-1172-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grenda DS, Murakami M, Ghatak J, et al. Mutations of the ELA2 gene found in patients with severe congenital neutropenia induce the unfolded protein response and cellular apoptosis. Blood. 2007;110:4179–4187. doi: 10.1182/blood-2006-11-057299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayee B, Antonopoulos A, Murphy EJ, et al. G6PC3 mutations are associated with a major defect of glycosylation: a novel mechanism for neutrophil dysfunction. Glycobiology. 2011;21:914–924. doi: 10.1093/glycob/cwr023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Germeshausen M, Ballmaier M, Welte K. Incidence of CSF3R mutations in severe congenital neutropenia and relevance for leukemogenesis: Results of a long-term survey. Blood. 2007;109:93–99. doi: 10.1182/blood-2006-02-004275. [DOI] [PubMed] [Google Scholar]
- 45.Link DC, Kunter G, Kasai Y, et al. Distinct patterns of mutations occurring in de novo AML versus AML arising in the setting of severe congenital neutropenia. Blood. 2007;110:1648–1655. doi: 10.1182/blood-2007-03-081216. [DOI] [PMC free article] [PubMed] [Google Scholar]