Abstract
Carcinomas initiate and progress due to genetic and epigenetic alterations in epithelial cells. However, recently, these alterations have also been reported in stromal fibroblasts. The gain-of-function mutations in the PI3K p110 catalytic subunit (PIK3CA) have been identified in many cancers with a current global incidence of 26% (18–40%) in breast carcinomas. We analyzed the mutational frequency of PIK3CA of three hotspots (exons 1, 9, and 20) in 81 primary invasive breast cancers (BC) and 25 cultured breast cancer-associated fibroblast (CAF) samples by Sanger sequencing in Arab breast cancer patients. Associations between the incidence of any PIK3CA mutation and several clinicopathologic characteristics were assessed using chi-square tests for categorical or t test for continuous variables. Furthermore, survival curves were estimated using the Kaplan–Meier method with the log rank test to evaluate the significance of their differences. We identified a total of 21 PIK3CA missense mutations with a frequency of 25.9%. The majority of the mutations, 17 out of 21 (81%), were in exon 20 (p.His1047Arg, p.His1047Lys, p.Thr1025Ala, p.Gly1049Arg, p.Asp1056Asn) while the remainder, 4 out of 21 (19%) were in exon 9 (p.Glu545Lys). PIK3CA mutations were significantly associated with lower grade and hormone receptor positivity. Although there was a favorable trend in overall survival for patients whose tumor harbored PIK3CA mutations, the difference was not statistically significant (P = 0.10). However, we did not detect any somatic mutations in CAFs. Furthermore, we have shown a high prevalence (8.2-fold) of a silent variant (SNP, rs17849079) in the Arab breast cancer population compared with disease-free individuals.
Keywords: PI3K, PIK3CA, mutation, breast cancer, SNP, CAF
Introduction
Breast cancer is the most common cancer type among women worldwide including in the Arabian Peninsula.1,2 Although the highest incidence and mortalities occur in developed countries (i.e., United States and Europe), as the Western lifestyle is being adopted by increasing parts of the world, breast cancer cases continue to increase, including in the Middle East.3 Carcinomas are known to initiate and progress due to genetic and epigenetic changes such as loss of heterozygocity (LOH), copy number variations (CNV), somatic mutations, and methylation in epithelial cells.4-7 However, in recent years, genetic and epigenetic alterations have been also reported in stromal cells such as carcinoma-associated fibroblasts (CAFs) that may play a role in human carcinogenesis.8,9
The phosphoinositide 3-kinase (PI3K) signaling pathway regulates many important cellular processes, including proliferation and cell death.10,11 Gain-of-function mutations in the PI3K p110 α catalytic subunit (PIK3CA) have been frequently linked to human cancer development. PIK3CA is considered to be one of the “mountains” in the genomes of the human cancer landscape.12 Samuels et al. first reported the high prevalence of pathogenic mutations of PIK3CA in colon cancers while a lower rate was described in breast cancers, though due to the small number of breast tumor samples, the reported frequency was not reliable.13 A subsequent study expanding the sample size of primary breast tumors along with cell lines revealed a greater and true mutation frequency.14 Since these two initial publications, a high prevalence of PIK3CA mutations has been reported in many types of cancers and populations.15 The functional significance of these identified mutations has been studied in vitro revealing promising results for therapeutic interventions.16-18
The global frequency of PIK3CA somatic mutations in all major malignancies is about 10%, which makes PIK3CA one of the highest mutated oncogenes in human cancers.15 The great majority of PIK3CA mutations occur in three hotspot regions namely, exon 1 (p85 binding), exon 9 (helical), and exon 20 (kinase) domains.15 The current overall PIK3CA mutation frequency for breast cancers reported in the Catalogue of Somatic Mutations in Cancer database (COSMIC) ranges from 18% to 40%, depending on the studied population with an average of 26%.
As we are entering into the era of personalized medicine,19 cancer therapeutics that specifically target a tumor’s genetic and epigenetic alterations are also gaining ground. Although genomic alterations often give cancer cells a selective growth advantage during tumorigenesis,20,21 they could also serve as targets for anticancer agents that can selectively kill the tumor carrying DNA alterations with relatively low toxicity. In addition, most of the past and current genomic characterizations have been done in Western populations, and whether such studies apply to all ethnic and other diverse demographics remains an open question. Therefore, it is essential to identify population-specific genomic alterations in cancer genes and determine their frequencies in order to validate existing data as therapeutic targets or biomarkers for other populations.
In this study, we assessed PIK3CA somatic mutation frequency in 81 primary breast tumors and 25 cultured CAF cells in Arab breast cancer patients by direct DNA sequencing. Similar to other reports, we identified a PIK3CA mutation frequency of 25.9% in BC tissues, with the majority located in exon 20. However, no mutations were detected in CAF cells. Intriguingly, we have found a 8.2-fold higher prevalence of a silent variant (SNP, rs17849079) in Arab breast cancer patients as compared with disease-free normal individuals. Further implications of these results are discussed below.
Results
High frequency of PIK3CA missense mutations in Arab breast cancer patients
To evaluate the PIK3CA somatic mutation frequencies in Arab BC patients, we isolated genomic DNA from 81 primary BC tissues and 189 peripheral blood samples from breast cancer free individuals, and sequenced PIK3CA exons 1, 9, and 20. As previously reported, about 99% of the somatic mutations identified for this gene are located in these exons.15
We detected 21 missense mutations in 81 BC samples (25.9%). Seventeen of these mutations (81%) were in exon 20 (kinase domain) and the remaining 4 (19%) in exon 9 (helical domain). Furthermore, 62% (13 out of 21) of the mutations were identified in the most common hotspot mutation within exon 20, His1047Arg (Fig. 1A; Table 1). However, we did not detect any alterations in exon 1, and all the mutations identified in this breast cancer population have been previously reported (Fig. 1A; Table 1). In most cases, the mutations were either A > G or G > A purine-to-purine transition mutations except in one sample wherein a purine-to-pyrimidine (G > C) transversion mutation was detected (Fig. 1A; Table 1).
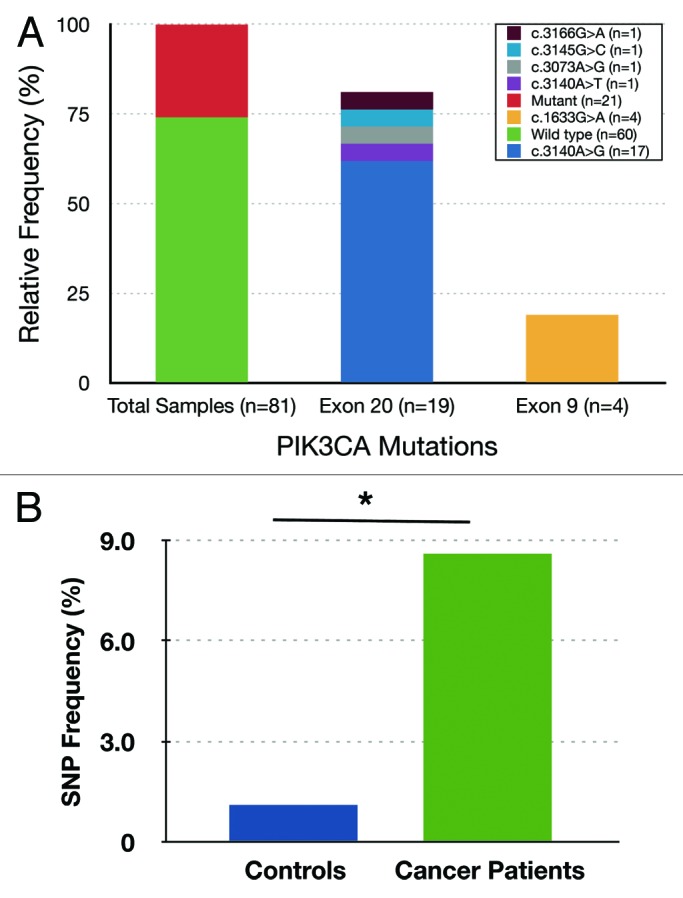
Figure 1. (A) PIK3CA mutation status in invasive epithelial tumors of Saudi Breast Cancer patients. The x-axis indicates relative frequency and y-axis shows mutation and change in amino acid. The total 21 missense mutations out of 81 total primary tumor samples (25.9%), 17 in exon 20 (81.0%), and 4 in exon 9 (19.0%). (B) The frequency of C3075T SNP (rs17849079) in disease-free control samples and in breast cancer patient population in Saudi Arabia. The frequency of SNP in breast cancer patients (7/81 or 8.6%) is significantly higher than those with disease-free controls (2/189 or 1.1%). *P value = 0.0032.
Table 1. Summary of PIK3CA mutational status and their clinicopathological features in BC samples.
| Tumor # | Age | Grade | ER | PR | HER2 | Epithelium mutation Nucleotide: codon (exon) |
|---|---|---|---|---|---|---|
| 1 | 36 | II | + | - | 1+ | WT |
| 2 | 48 | II | - | - | 3+ | WT |
| 3 | 72 | III | - | - | 1+ | WT |
| 4 | 32 | II | + | + | 1+ | c.3140A > G: p.His1047Arg (20) |
| 5 | 44 | II | + | - | 1+ | WT |
| 6 | 34 | III | - | - | 3+ | WT |
| 7 | 62 | III | + | + | 3+ | WT |
| 8 | 67 | III | - | - | 3+ | WT |
| 9 | 47 | III | + | + | 1+ | c.3145C > G: p.Gly1049Arg (20) |
| 10 | 68 | III | + | + | 1+ | WT |
| 11 | 40 | III | + | - | 3+ | WT |
| 12 | 32 | III | - | - | 3+ | WT |
| 13 | 57 | II | + | + | 1+ | WT |
| 14 | 31 | III | - | - | 1+ | WT |
| 15 | 34 | II | - | - | 1+ | WT |
| 16 | 27 | II | + | + | 3+ | c.3140A > G: p.His1047Arg (20) |
| 17 | 42 | II | - | - | 1+ | WT |
| 18 | 54 | III | + | - | 1+ | WT |
| 19 | 66 | III | + | + | 3+ | WT |
| 20 | 55 | III | + | + | 3+ | c.3075C > T: SNP (20) |
| 21 | 61 | II | + | + | 1+ | WT |
| 22 | 50 | II | + | + | 1+ | WT |
| 23 | 40 | I | + | - | 1+ | c.3140A > G: p.His1047Arg (20) |
| 24 | 60 | II | + | + | 1+ | c.3140A > G: p.His1047Arg (20) |
| 25 | 57 | III | + | + | 1+ | WT |
| 26 | 42 | III | - | - | 3+ | c.3075C > T: SNP (20) c.3140A > G: p.His1047Arg (20) |
| 27 | 42 | III | - | - | 3+ | WT |
| 28 | 54 | III | + | - | 1+ | c.3140A > G: p.His1047Arg (20) |
| 29 | 38 | III | - | - | 3+ | WT |
| 30 | 32 | II | + | - | 1+ | WT |
| 31 | 51 | II | + | + | 3+ | c.3073A > G: p.Thr1025Ala (20) |
| 32 | 56 | III | + | + | 3+ | c.3075C > T: SNP (20) |
| 33 | 58 | II | + | + | 1+ | c.3140A > G: p.His1047Arg (20) |
| 34 | 61 | II | + | + | 1+ | WT |
| 35 | 45 | I | + | + | 1+ | c.3075C > T: SNP (20) |
| 36 | 40 | II | - | - | 1+ | WT |
| 37 | 39 | III | - | - | 3+ | WT |
| 38 | 36 | II | + | + | 3+ | WT |
| 39 | 55 | III | - | - | 1+ | WT |
| 40 | 51 | II | + | - | 3+ | c.3140A > G: p.His1047Arg (20) |
| 41 | 40 | III | - | - | 3+ | c.3140A > G: p.His1047Arg (20) |
| 42 | 41 | III | + | + | 1+ | WT |
| 43 | 46 | III | - | - | 1+ | WT |
| 44 | 50 | II | + | + | 1+ | WT |
| 45 | 35 | III | - | - | 1+ | WT |
| 46 | 51 | II | + | - | 1+ | WT |
| 47 | 44 | III | - | - | 3+ | WT |
| 48 | 37 | III | - | - | 1+ | WT |
| 49 | 45 | II | + | + | 1+ | WT |
| 50 | 34 | III | + | + | 1+ | WT |
| 51 | 70 | I | + | + | 1+ | WT |
| 52 | 47 | II | - | - | 3+ | WT |
| 53 | 65 | I | + | + | 1+ | c.3140A > G: p.His1047Arg (20) |
| 54 | 36 | III | - | - | 3+ | WT |
| 55 | 39 | I | + | + | 3+ | c.1633G > A: p.Glu545Lys (9) c.3075C > T: SNP (20) |
| 56 | 60 | II | + | + | 1+ | WT |
| 57 | 63 | III | + | + | 1+ | c.1633G > A: p.Glu545Lys (9) |
| 58 | 42 | I | + | + | 1+ | WT |
| 59 | 52 | II | + | + | 3+ | c.3075C > T: SNP (20) c.3140A > G: p.His1047Arg (20) |
| 60 | 49 | I | + | + | 1+ | WT |
| 61 | 60 | I | + | + | 1+ | c.3166G > A: p.Asp1056Asn (20) |
| 62 | 48 | II | + | - | 1+ | WT |
| 63 | 56 | II | + | - | 3+ | WT |
| 64 | 53 | II | + | + | 1+ | WT |
| 65 | 28 | DCIS | NA | NA | 1+ | c.3140A > T: p.His1047Lys (20) |
| 66 | 40 | II | - | - | 3+ | c.1633G > A: p.Glu545Lys (9) |
| 67 | 45 | III | - | - | 3+ | WT |
| 68 | 44 | NA | NA | NA | NA | c.1633G > A: p.Glu545Lys (9) |
| 69 | 45 | MET | - | - | 1+ | c.3075C > T: SNP (20) |
| 70 | 22 | II | + | + | 1+ | WT |
| 71 | 37 | III | - | - | 1+ | WT |
| 72 | 45 | III | - | - | 3+ | c.3140A > G: p.His1047Arg (20) |
| 73 | 46 | II | - | - | 1+ | WT |
| 74 | 49 | III | + | + | 3+ | WT |
| 75 | 46 | III | + | - | 3+ | WT |
| 76 | 51 | III | - | - | 3+ | WT |
| 77 | 53 | I | + | - | 1+ | c.3140A > G: p.His1047Arg (20) |
| 78 | 48 | III | + | + | 1+ | WT |
| 79 | 39 | II | + | + | 3+ | WT |
| 80 | 40 | II | - | - | 3+ | WT |
| 81 | 36 | II | - | - | 3+ | WT |
Mutations are shown as nucleotide change followed by amino acid change and the number of exon in parenthesis. Abbreviations: ER, estrogen receptor; PR, progesterone receptor; Her2, human epidermal growth factor receptor 2; WT, wild type; NA, information not available; MET, metastatic; DCIS, ductal carcinoma in situ.
High prevalence of the PIK3CA C3075T polymorphism (SNP) in Arab breast cancer patients
Next, we studied the prevalence of the rs17849079 SNP in Arab BC patients. We have found a high prevalence (7 out of 81, 8.6%) of this silent polymorphism in the analyzed 81 breast cancer tissues (Fig. 1B). To determine the overall frequency of this SNP in the Saudi population, we sequenced the same region using genomic DNA isolated from peripheral blood of 189 cancer-free individuals. Interestingly, the frequency of this SNP was 1.05% in healthy normal individuals compared with 8.6% in our cancer patients (Fig. 1B), which accounts for 8.2-fold enrichment in women with breast cancer. The frequency of this SNP in individuals with breast cancer was significantly higher than that observed in the controls (the Fisher exact test P = 0.0032). The estimated odds ratio of the rs17849079 SNP in affected individuals relative to disease-free individuals was OR = 8.85 (95% CI 1.8–43.6) (Fig. 1B).
It is noteworthy that 3 out of 7 (43%) breast tumors with this SNP also harbored a PIK3CA somatic mutation (Table 1).
Absence of PIK3CA mutations in CAFs
The presence of somatic mutations in CAFs remains controversial. Therefore, we decided to investigate the possible presence of PIK3CA mutations in 25 CAFs isolated from 25 BC tissues.
Importantly, we did not detect PIK3CA mutations in any of the analyzed CAF cells (Table 2). Additionally, in 3 tissues, tumors that harbored mutated PIK3CA, did not display mutant PIK3CA in the associated CAFs (Table 2).
Table 2. Summary of PIK3CA mutational status and their clinicopathological features in CAF and their associated BC samples.
| Sample # | Age | Grade | ER | PR | HER2 | Fibroblast mutation (Exon) | Tumor mutation (Exon) |
|---|---|---|---|---|---|---|---|
| 1 | 55 | II | + | - | 3+ | WT | NA |
| 2 | 40 | II | + | + | 1+ | WT | NA |
| 3 | 54 | III | + | - | 3+ | WT | NA |
| 4 | 40 | III | NA | NA | NA | WT | NA |
| 5 | 47 | II | + | + | 1+ | WT | NA |
| 6 | 35 | III | - | - | 1+ | WT | NA |
| 7 | 54 | II | + | - | 1+ | WT | NA |
| 8 | 47 | II | + | + | 3+ | WT | NA |
| 9 | 32 | II | + | + | 1+ | WT | c.3140A > G: p.His1047Arg (20) |
| 10 | 52 | II | + | - | 1+ | WT | WT |
| 11 | 60 | III | - | - | 3+ | WT | WT |
| 12 | 60 | II | + | + | 1+ | WT | NA |
| 13 | 52 | II | - | - | 3+ | WT | NA |
| 14 | 40 | III | + | - | 3+ | WT | WT |
| 15 | 57 | II | + | + | 1+ | WT | WT |
| 16 | 63 | III | + | - | 3+ | WT | NA |
| 17 | 54 | III | - | - | 3+ | WT | WT |
| 18 | 42 | II | + | + | 3+ | WT c.3075C > T: SNP (20) |
c.3140A > G: p.His1047Arg (20) c.3075C > T: SNP (20) |
| 19 | 54 | II | + | - | 1+ | WT | NA |
| 20 | 64 | II | + | + | 3+ | WT | NA |
| 21 | 60 | I | + | + | 1+ | WT | NA |
| 22 | 43 | DCIS | NA | NA | NA | WT | NA |
| 23 | 68 | DCIS | NA | NA | NA | WT | NA |
| 24 | 40 | II | - | - | 3+ | WT | c.1633G > A: p.Glu545Lys (9) |
| 25 | 33 | III | - | - | 1+ | WT | NA |
Mutations are shown as nucleotide change followed by amino acid change and the number of exon in parenthesis. Abbreviations: ER, estrogen receptor; PR, progesterone receptor; Her2, human epidermal growth factor receptor 2; WT, wild type; NA, information not available; DCIS, ductal carcinoma in situ.
Associations of PIK3CA mutations with tumor clinicopathological features and patient survival
Next, we sought to determine the association of PIK3CA mutations with tumor clinicopathologic features. Features significantly associated with PIK3CA mutations included lower grade (P = 0.02) and estrogen receptor (ER) positivity (P = 0.03), while there was a marginal association that was not statistically significant with progesterone receptor (PR) (P = 0.07). However, PIK3CA mutations did not correlate with age nor HER2 status (Table 3).
Table 3. The associations between PIK3CA somatic mutation status and BC samples’ clinicopathologic features.
| Characteristic | Overall cohort | Wild type (n = 56) | SNP (n = 7) | Any PIK3CA mutation (n = 21) | P* value |
|---|---|---|---|---|---|
| Age (SD) | 46.8 (10.8) | 46.4 (11.1) | 47.7 (6.5) | 47.2 (11.0) | 0.79 |
| Grade | |||||
| I | 9 | 3 | 2 | 5 | 0.02 |
| II | 34 | 25 | 2 | 9 | |
| III | 35 | 28 | 2 | 5 | |
| Unknown | 3 | 2 | |||
| ER | |||||
| Negative | 29 | 25 | 1 | 3 | 0.03 |
| Positive | 50 | 31 | 6 | 16 | |
| Unknown | 2 | 2 | |||
| PR | |||||
| Negative | 42 | 34 | 1 | 7 | 0.07 |
| Positive | 37 | 22 | 6 | 12 | |
| Unknown | 2 | 2 | |||
| HER2 | |||||
| Negative | 46 | 34 | 2 | 10 | 0.59 |
| Positive | 33 | 22 | 5 | 9 | |
| Unknown | 2 |
Statistical tests are based on available data; samples with SNP-only and unknown information are not included in p-value calculations.. *χ2 test (or the Fisher exact test when an expected cell count was less than 5) used for binary/categorical variables; t test used for continuous variables. Abbreviations: ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2 status; SD, standard deviation.
The presence of the exon 20 hotspot mutation, His1047Arg, was significantly associated with low-grade tumors (P = 0.03) and ER positivity (P = 0.02) (Table 4). However, the Glu545Lys (exon 9) mutation was not statistically associated with any of the clinicopathologic features (Table 4).
Table 4. The associations between PIK3CA mutation type and clinicopathological variables.
| Mutation type | Mutation type | |||||
|---|---|---|---|---|---|---|
| Characteristic | Wild type | Exon 9 mutation | P*-value | Wild type | Exon 20 mutation | P*-value |
| Age (SD) | 46.4(11.1) | 46.5 (13.6) | 0.99 | 46.4 (11.1) | 47.4 (11.4) | 0.77 |
| Grade | ||||||
| I | 3 | 1 | 0.27 | 3 | 4 | 0.03 |
| II | 25 | 1 | 25 | 8 | ||
| III | 28 | 1 | 28 | 4 | ||
| Unknown | 1 | 1 | ||||
| ER | ||||||
| Negative | 25 | 1 | 1 | 25 | 2 | 0.02 |
| Positive | 31 | 2 | 31 | 14 | ||
| Unknown | 1 | 1 | ||||
| PR | ||||||
| Negative | 34 | 1 | 0.56 | 34 | 6 | 0.09 |
| Positive | 22 | 2 | 22 | 10 | ||
| Unknown | 1 | |||||
| HER2 | ||||||
| Negative | 34 | 1 | 0.56 | 34 | 9 | 0.75 |
| Positive | 22 | 2 | 22 | 7 | ||
| Unknown | 1 | 1 | ||||
Statistical tests are based on available data; samples with SNP-only and unknown information are not included in p-value calculations. *χ2 test (or the Fisher exact test when an expected cell count was less than 5) used for binary/categorical variables; t test used for continuous variables. Abbreviations: ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2 status; SD, standard deviation.
Moreover, we assessed the association between the presence of PIK3CA gain-of-function mutations and overall survival of patients. Figure 2 shows an overall survival advantage for patients with tumors harboring PIK3CA mutations as compared with those tumors that do not contain such alterations. However, the difference between the two groups was not statistically significant (P = 0.10).
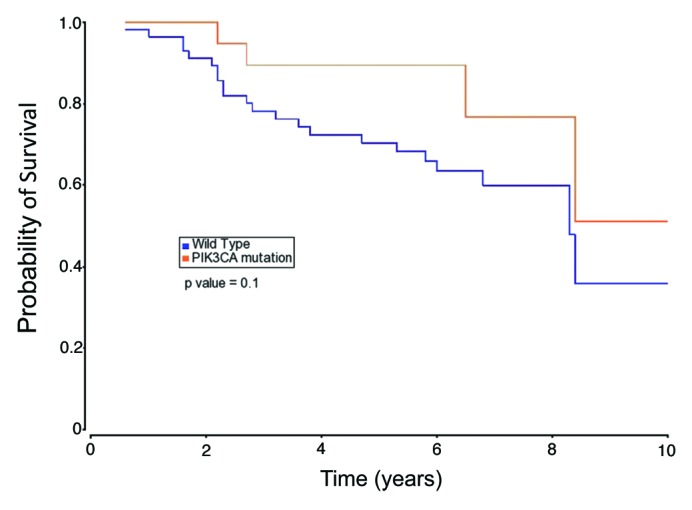
Figure 2. The Kaplan–Meier curves illustrating overall survival based on tumor PIK3CA mutational status.
Discussion
In the era of personalized medicine, we sought to determine the frequency and nature of PIK3CA mutations in Arab BC patients. The obtained results are summarized in the following points: first, the overall frequency of PIK3CA missense mutations in the Arab breast cancer population is 25.9% with a great majority of mutations being in exon 20 (Fig. 1A); second, the silent polymorphism (rs17849079) in the exon 20 coding region is 8.2-fold enriched in the BC population as compared with disease-free individuals (Fig. 1B); third, CAF cells do not harbor PIK3CA somatic missense mutations in the mutation hotspot regions of this gene (Table 2). Lastly, PIK3CA mutations have a slight but not significant effect in patient’s overall survival (Fig. 2). A larger cohort may be needed to clarify this important issue.
The frequency of PIK3CA mutations in the Arab breast cancer population (25.9%) is similar to the global average22 and all the identified mutations have been previously reported. The exon 20 His1047Arg alteration comprises 61.9% (13 out of 21) of total PIK3CA mutations, which represents a significant percentage of human breast cancers, warranting the development of targeted therapies against this particular PIK3CA mutation.
The functional significance of the His1047Arg and Glu545Lys mutations have been analyzed in both in vitro and in vivo model systems and reported to be gain-of-function mutations and pathogenic. The initial in vitro model systems created by Samuels et al. in 2005 and Gustin et al. in 2009 confirmed the pathogenicity of these two mutations16,17 Gustin et al. knocked in two PIK3CA hotspot mutations, p.Glu545Lys (helicase domain) and p.His1047Arg (kinase domain), in the non-cancerous mammary epithelial cell line MCF-10A by somatic cell gene targeting strategies. The authors have shown that both knocked in mutants had significantly lower cell proliferation when treated with lithium chloride as compared with their isogenic parental counterparts and importantly, unexpectedly activated signaling pathways that were not predicted based upon the known pathways of PI3 kinase activation.17
In a more recent study, Di Nicalantonio et al. reported that patients’ tumor cells with PIK3CA mutations were sensitive to everolimus, an inhibitor of the mammalian target of rapamycin (mTOR). These authors also knocked in Glu545Lys and His1047Arg mutations into two immortalized human breast epithelial cell lines, hTERT-HME1 and MCF-10A. Similar to primary tumors, both cell lines with either His1047Arg or Glu545Lys knocked in mutations were more sensitive to everolimus as compared with their respective isogenic parental cell lines.23,24
As the PI3K/AKT/mTOR pathway is frequently activated in human cancers, there have been intense efforts in using inhibitors (i.e., everolimus, wortmannin, LY294002, PX-866) that specifically target alterations in this pathway, some of which are currently in phase II and III clinical trials.25-28
There is currently no consensus in the literature on the role of PIK3CA mutations in breast cancer patient survival. This could be due to the lack of standard treatments for breast cancer, different samples size, different patient populations or combinations thereof. The disease-free or overall survival for patients whose tumors harbor PIK3CA mutations have been reported to be either favorable,29-31 unfavorable,32,33 or not statistically significant34-36 as compared with patients with tumors containing wild-type PIK3CA. Further studies are needed to provide clarity to this important issue.
The PIK3CA mutation status has been recently reported for colorectal and ovarian cancers in the Middle Eastern population37,38 with frequencies of 12.4% and 3.9%, respectively. We present here the first report on the frequency and nature of PIK3CA mutations related to breast cancer in the Arab population.
In addition, we have shown the presence of 8.2-fold enrichment of the PIK3CA rs17849079 SNP among the Arab breast cancer patient population as compared with disease free controls (Fig. 1B). This silent polymorphism has been reported for the first time in retinoblastomas with no obvious functional consequence.39 This SNP has also been previously classified as a silent somatic mutation in 19 samples of various cancer types worldwide.22 Interestingly, the microRNA (miRNA), hsa-miR-4324 (accession #MI0015854), binds to the PIK3CA mRNA sequence at exon 20, which harbors this SNP.40,41 This miRNA seems to bind only the wild-type allele but not the variant sequence containing this SNP. The functional significance of this could be that miRNA regulation of PIK3CA expression is affected by the presence of this genetic variation.
Finally, we have also shown the absence of PIK3CA mutations in stromal fibroblasts. There have been numerous conflicting reports regarding genetic alterations in tumor stroma including cultured CAFs.42 The fact that CAFs maintain an activated-phenotype even after many passages in culture, has been thought to be due to somatic alterations in their genomes.43 The initial studies supporting wide spread somatic mutations in CAFs44,45 have been later challenged by others.46,47 Interestingly, in our study, three tumor samples that contained PIK3CA mutations were not identified in their corresponding CAFs. Our results support recent studies that CAFs are not likely to harbor somatic alterations in commonly mutated oncogenes.
In conclusion, the prevalence of PIK3CA mutation in Arab breast cancers is similar to the global average and the presence of these mutations may favor patient survival. On the other hand there were no PIK3CA somatic mutations in breast CAFs that were analyzed. Furthermore, we report a high prevalence of the SNP rs17849079 among Arab breast cancer patients compared with controls, which suggests its potential use as a breast cancer susceptibility and early diagnosis molecular marker for this population.
Materials and Methods
Breast cancer tumor and blood sample collection
In this retrospective study, 81 invasive breast cancer samples were obtained from patients that underwent surgery at King Faisal Special Hospital and Research Center (KFSH&RC) in Riyadh, Saudi Arabia. All patients gave written informed consent to participate in this study, which was approved by KFSH&RC Institutional Research Ethics Committee (project #2100017). Tumor samples were either provided as frozen tissue blocks or paraffin embedded slides and were coded and kept anonymous. We also collected 189 peripheral blood samples from cancer-free individuals to be used as controls.
Generation of primary fibroblasts in tissue culture
Twenty-five CAF cells from 25 separate breast tumors were generated as previously described.48 Isolated CAFs were cultured in a 5% CO2 incubator at 37 °C in Medium 199 and Ham’s F12 mixed 1:1 and supplemented with 10% FCS and 1% Pen/Strep.
Genomic DNA isolation
Genomic DNA (gDNA) was isolated from cultured CAFs, frozen tumor sections and peripheral blood lymphocytes using the Qiagen Blood and Tissue Kit whereas paraffin embedded slides containing tumor cells were subjected to the QIAamp DNA FFPE Tissue Kit following the manufacturer’s instructions.
Immunohistochemical analysis of tumor samples
Immunohistochemistry analysis for estrogen receptor (ER), progesterone receptor (PR), and Her2/neu were performed in the KFSH&RC diagnostic laboratory following optimized protocols. Slides were developed using an automated slide machine (BenchMark XT). Briefly, sections of formalin-fixed paraffin-embedded tumor samples were stained for ER using the mouse monoclonal antibody, ER 6F11 (Novacastra) with 1:30 dilutions, PR using the mouse monoclonal antibody, PGR 312 (Novacstra) with 1:100 dilutions, and HER2 using the rabbit monoclonal antibody, 4B5 (Ventana) and DAB Kit (Ventana). Stains were scored for HER2 as 1+ as negative and 3+ as positive. The normal tissue surrounding tumor was used as a negative control.
PCR amplification and DNA sequencing
PIK3CA hotspot exons (exons 1, 9, and 20) were PCR amplified in 81 BC and 25 CAF samples with specific primers for each exon including exon-intron junctions (Table S1). The PIK3CA exon 20 was also PCR amplified and sequenced in 189 normal blood samples and three tumor matching normal tissue to identify whether the silent polymorphism, rs17849079, was somatic or germline in origin.
About 20 to 100 ng of gDNA was added to the PCR reaction mix depending on the amount available for each sample. High fidelity Taq DNA Polymerase (Invitrogen, 11304-011) was used to reduce PCR artifacts during the amplification process.
PCR amplicons were sequenced by direct sequencing (Sanger sequencing) in the KFSH&RC Sequencing Core Facility. Samples with mutations were re-sequenced at least twice by repeating PCR reactions to rule out any amplification related artifacts as well as sequencing errors.
Statistical analysis
Associations between the mutation status and clinicopathologic features were assessed using the χ2 test for categorical variables or t test for continuous variables. The Fisher exact test was used when expected cell counts were less than 5 using the Monte Carlo method as implemented in SAS. The frequency of SNPs in patients was compared with that of controls without the disease. The risk of the disease was estimated by the odds ratio (OR) and 95% confidence intervals (CIs). Survival probabilities were estimated using the Kaplan–Meier method and log-rank tests were used to compare survival curves between the various subgroups of patients. All statistical tests were two-tailed. A P value of < 0.05 was considered significant. Statistical analyses were performed with SAS version 9.2 (SAS Institute Inc.) and the MATLAB software packages (Mathworks).
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Authors’ Contributions
BK designed and performed the experiments. DC conducted statistical analysis; NK provided disease free blood samples and reagents; HG provided cultured CAF samples; AAQ and FH extracted DNA from tumor samples; MT provided some of the tumor samples and conducted the pathological analyses; TAT collected patient clinical data; BK, BHP, and AA wrote the manuscript.
Acknowledgments
This project was funded by King Faisal Specialist and Research Centre (KFSH&RC). This work was performed under the RAC proposal #2100017 of KFSH&RC. We also thank M. Mohammed Rajab and Abdelmoneim Eldali (KFSH&RC) for assistance with the sequencing of our samples and data analysis.
Glossary
Abbreviations:
- PIK3CA
phosphoinositide 3-kinase p110 alpha catalytic subunit
- BC
primary invasive breast cancer
- CAF
cancer associated fibroblasts
- ER
estrogen receptor
- PR
progesterone receptor
- HER2
human epidermal growth factor receptor 2
- SNP
single nucleotide polymorphism
- mTOR
mammalian target of rapamycin
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/cbt/articles/25945
Footnotes
Previously published online: www.landesbioscience.com/journals/cbt/article/25945
References
- 1.Garcia M, Jemal A, Ward E, Center MM, Hao Y. Global Cancer Facts and Figures 2007. American Cancer Society 2007. [Google Scholar]
- 2.Al-Eid HS. Cancer Incidence Report: Saudi Arabia. National Cancer Registry 2004. [Google Scholar]
- 3.Porter P. “Westernizing” women’s risks? Breast cancer in lower-income countries. N Engl J Med. 2008;358:213–6. doi: 10.1056/NEJMp0708307. [DOI] [PubMed] [Google Scholar]
- 4.Deramaudt T, Rustgi AK. Mutant KRAS in the initiation of pancreatic cancer. Biochim Biophys Acta. 2005;1756:97–101. doi: 10.1016/j.bbcan.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67. doi: 10.1016/0092-8674(90)90186-I. [DOI] [PubMed] [Google Scholar]
- 6.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–59. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 7.Warnecke PM, Bestor TH. Cytosine methylation and human cancer. Curr Opin Oncol. 2000;12:68–73. doi: 10.1097/00001622-200001000-00012. [DOI] [PubMed] [Google Scholar]
- 8.Orimo A, Weinberg RA. Stromal fibroblasts in cancer: a novel tumor-promoting cell type. Cell Cycle. 2006;5:1597–601. doi: 10.4161/cc.5.15.3112. [DOI] [PubMed] [Google Scholar]
- 9.Eng C, Leone G, Orloff MS, Ostrowski MC. Genomic alterations in tumor stroma. Cancer Res. 2009;69:6759–64. doi: 10.1158/0008-5472.CAN-09-0985. [DOI] [PubMed] [Google Scholar]
- 10.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–7. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 11.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–19. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 12.Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 13.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554–554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 14.Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S, Konishi H, Karakas B, Blair BG, Lin C, et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004;3:772–5. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- 15.Karakas B, Bachman KE, Park BH. Mutation of the PIK3CA oncogene in human cancers. Br J Cancer. 2006;94:455–9. doi: 10.1038/sj.bjc.6602970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samuels Y, Diaz LA, Jr., Schmidt-Kittler O, Cummins JM, Delong L, Cheong I, Rago C, Huso DL, Lengauer C, Kinzler KW, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–73. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 17.Gustin JP, Karakas B, Weiss MB, Abukhdeir AM, Lauring J, Garay JP, Cosgrove D, Tamaki A, Konishi H, Konishi Y, et al. Knockin of mutant PIK3CA activates multiple oncogenic pathways. Proc Natl Acad Sci U S A. 2009;106:2835–40. doi: 10.1073/pnas.0813351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–62. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 19.Allison M. Is personalized medicine finally arriving? Nat Biotechnol. 2008;26:509–17. doi: 10.1038/nbt0508-509. [DOI] [PubMed] [Google Scholar]
- 20.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 22. http://www.sanger.ac.uk/genetics/CGP/cosmic/
- 23.Di Nicolantonio F, Arena S, Tabernero J, Grosso S, Molinari F, Macarulla T, Russo M, Cancelliere C, Zecchin D, Mazzucchelli L, et al. Deregulation of the PI3K and KRAS signaling pathways in human cancer cells determines their response to everolimus. J Clin Invest. 2010;120:2858–66. doi: 10.1172/JCI37539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohseni M, Park BH. PIK3CA and KRAS mutations predict for response to everolimus therapy: now that’s RAD001. J Clin Invest. 2010;120:2655–8. doi: 10.1172/JCI44026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–44. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Courtney KD, Corcoran RB, Engelman JA. The PI3K pathway as drug target in human cancer. J Clin Oncol. 2010;28:1075–83. doi: 10.1200/JCO.2009.25.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maira S-M, Finan P, Garcia-Echeverria C. From the bench to the bed side: PI3K pathway inhibitors in clinical development. Curr Top Microbiol Immunol. 2010;347:209–39. doi: 10.1007/82_2010_60. [DOI] [PubMed] [Google Scholar]
- 28.Chavez-MacGregor M, Gonzalez-Angulo AM. Everolimus in the treatment of hormone receptor-positive breast cancer. Expert Opin Investig Drugs. 2012;21:1835–43. doi: 10.1517/13543784.2012.726218. [DOI] [PubMed] [Google Scholar]
- 29.Kalinsky K, Jacks LM, Heguy A, Patil S, Drobnjak M, Bhanot UK, Hedvat CV, Traina TA, Solit D, Gerald W, et al. PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res. 2009;15:5049–59. doi: 10.1158/1078-0432.CCR-09-0632. [DOI] [PubMed] [Google Scholar]
- 30.Barbareschi M, Buttitta F, Felicioni L, Cotrupi S, Barassi F, Del Grammastro M, Ferro A, Dalla Palma P, Galligioni E, Marchetti A. Different prognostic roles of mutations in the helical and kinase domains of the PIK3CA gene in breast carcinomas. Clin Cancer Res. 2007;13:6064–9. doi: 10.1158/1078-0432.CCR-07-0266. [DOI] [PubMed] [Google Scholar]
- 31.Maruyama N, Miyoshi Y, Taguchi T, Tamaki Y, Monden M, Noguchi S. Clinicopathologic analysis of breast cancers with PIK3CA mutations in Japanese women. Clin Cancer Res. 2007;13:408–14. doi: 10.1158/1078-0432.CCR-06-0267. [DOI] [PubMed] [Google Scholar]
- 32.Li SY, Rong M, Grieu F, Iacopetta B. PIK3CA mutations in breast cancer are associated with poor outcome. Breast Cancer Res Treat. 2006;96:91–5. doi: 10.1007/s10549-005-9048-0. [DOI] [PubMed] [Google Scholar]
- 33.Lerma E, Catasus L, Gallardo A, Peiro G, Alonso C, Aranda I, Barnadas A, Prat J. Exon 20 PIK3CA mutations decreases survival in aggressive (HER-2 positive) breast carcinomas. Virchows Arch. 2008;453:133–9. doi: 10.1007/s00428-008-0643-4. [DOI] [PubMed] [Google Scholar]
- 34.Loi S, Haibe-Kains B, Majjaj S, Lallemand F, Durbecq V, Larsimont D, Gonzalez-Angulo AM, Pusztai L, Symmans WF, Bardelli A, et al. PIK3CA mutations associated with gene signature of low mTORC1 signaling and better outcomes in estrogen receptor-positive breast cancer. Proc Natl Acad Sci U S A. 2010;107:10208–13. doi: 10.1073/pnas.0907011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michelucci A, Di Cristofano C, Lami A, Collecchi P, Caligo A, Decarli N, Leopizzi M, Aretini P, Bertacca G, Porta RP, et al. PIK3CA in breast carcinoma: a mutational analysis of sporadic and hereditary cases. Diagn Mol Pathol. 2009;18:200–5. doi: 10.1097/PDM.0b013e31818e5fa4. [DOI] [PubMed] [Google Scholar]
- 36.Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, Yu JS, Malmström PO, Mansukhani M, Enoksson J, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–9. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 37.Abubaker J, Bavi P, Al-Harbi S, Ibrahim M, Siraj AK, Al-Sanea N, Abduljabbar A, Ashari LH, Alhomoud S, Al-Dayel F, et al. Clinicopathological analysis of colorectal cancers with PIK3CA mutations in Middle Eastern population. Oncogene. 2008;27:3539–45. doi: 10.1038/sj.onc.1211013. [DOI] [PubMed] [Google Scholar]
- 38.Abubaker J, Bavi P, Al-Haqawi W, Jehan Z, Munkarah A, Uddin S, Al-Kuraya KS. PIK3CA alterations in Middle Eastern ovarian cancers. Mol Cancer. 2009;8:51. doi: 10.1186/1476-4598-8-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen Y, Merhavi-Shoham E, Avraham-Lubin BCR, Savetsky M, Frenkel S, Pe’er J, Goldenberg-Cohen N. PI3K/Akt pathway mutations in retinoblastoma. Invest Ophthalmol Vis Sci. 2009;50:5054–6. doi: 10.1167/iovs.09-3617. [DOI] [PubMed] [Google Scholar]
- 40.Goff LA, Davila J, Swerdel MR, Moore JC, Cohen RI, Wu H, Sun YE, Hart RP. Ago2 immunoprecipitation identifies predicted microRNAs in human embryonic stem cells and neural precursors. PLoS One. 2009;4:e7192. doi: 10.1371/journal.pone.0007192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.miRBase. http://www.mirbase.org/search.shtml
- 42.Campbell I, Qiu W, Haviv I. Genetic changes in tumour microenvironments. J Pathol. 2011;223:450–8. doi: 10.1002/path.2842. [DOI] [PubMed] [Google Scholar]
- 43.Orimo A, Weinberg RA. Stromal fibroblasts in cancer: a novel tumor-promoting cell type. Cell Cycle. 2006;5:1597–601. doi: 10.4161/cc.5.15.3112. [DOI] [PubMed] [Google Scholar]
- 44.Kurose K, Gilley K, Matsumoto S, Watson PH, Zhou X-P, Eng C. Frequent somatic mutations in PTEN and TP53 are mutually exclusive in the stroma of breast carcinomas. Nat Genet. 2002;32:355–7. doi: 10.1038/ng1013. [DOI] [PubMed] [Google Scholar]
- 45.Patocs A, Zhang L, Xu Y, Weber F, Caldes T, Mutter GL, Platzer P, Eng C. Breast-cancer stromal cells with TP53 mutations and nodal metastases. N Engl J Med. 2007;357:2543–51. doi: 10.1056/NEJMoa071825. [DOI] [PubMed] [Google Scholar]
- 46.Qiu W, Hu M, Sridhar A, Opeskin K, Fox S, Shipitsin M, Trivett M, Thompson ER, Ramakrishna M, Gorringe KL, et al. No evidence of clonal somatic genetic alterations in cancer-associated fibroblasts from human breast and ovarian carcinomas. Nat Genet. 2008;40:650–5. doi: 10.1038/ng.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hosein AN, Wu M, Arcand SL, Lavallée S, Hébert J, Tonin PN, Basik M. Breast carcinoma-associated fibroblasts rarely contain p53 mutations or chromosomal aberrations. Cancer Res. 2010;70:5770–7. doi: 10.1158/0008-5472.CAN-10-0673. [DOI] [PubMed] [Google Scholar]
- 48.Hawsawi NM, Ghebeh H, Hendrayani S-F, Tulbah A, Al-Eid M, Al-Tweigeri T, Ajarim D, Alaiya A, Dermime S, Aboussekhra A. Breast carcinoma-associated fibroblasts and their counterparts display neoplastic-specific changes. Cancer Res. 2008;68:2717–25. doi: 10.1158/0008-5472.CAN-08-0192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


