Abstract
Free radicals are formed as a result of cellular processes and play a key role in predisposition to and development of numerous diseases and of premature aging. Recently, we reported the syntheses of a number of novel phenolic antioxidants for possible application in food industry. In the present study, analyses of the cellular processes and molecular gene expression effects of some of the novel antioxidants in normal human tissues and in cancer cells were undertaken. Results indicated that whereas the examined antioxidants showed no effects on morphology and gene expression of normal human oral and gingival epithelial tissues, they exerted a profound cell killing effect on breast cancer cells, including on chemotherapy-resistant breast cancer cells and on oral squamous carcinoma cells. Among the tested antioxidants, N-decyl-N-(3-methoxy-4-hydroxybenzyl)-3-(3,4-dihydroxyphenyl) propanamide and N-decyl-N-(3,5-dimethoxy-4-hydroxybenzyl)-3-(3,4-dihydroxyphenyl) propanamide were the most promising, with excellent potential for cancer treatment. Moreover, our gene expression databases can be used as a roadmap for future analysis of mechanisms of antioxidant action.
Keywords: free radicals, antioxidants, gene expression, human 3D tissues, breast cancer, oral cancer
Introduction
Free radicals and reactive oxygen species (ROS) are formed as a result of numerous cellular processes. Their roles in health and disease have been a subject of intensive research over the past decades.1-3 Free radicals and ROS cause a wide variety of effects in the cells and tissues, including DNA damage. They also mediate inflammation and immune response and alter cellular signaling processes. Furthermore, ROS cause potent induction of gene and protein expression in cells and tissues.4 As a result, it is currently well established that free radicals play key parts in predisposition to and development of numerous diseases, such as atherosclerosis, inflammatory joint disease, asthma, diabetes, senile dementia, Alzheimer disease, degenerative eye disease, and cancer.5,6 Moreover, they are also associated with premature aging.5,7,8
Free radicals and their roles in food have been extensively studied as well, especially in regards to oxidative and thermooxidative deterioration of polyunsaturated fatty acids.9 Although synthetic antioxidants, such as butylated hydroxy toluene (BHT), butylated hydroxy anisole (BHA), and tertiary-butylatedhydroquinone (TBHQ), have shown good efficiency, their use has been hampered due to their possible detrimental effect on human health. Generally, in addition to demonstrating high efficiency, an ideal antioxidant for food application must exhibit no or significantly low toxicity.
Phenolic compounds, such as phenolic acids, tocopherol, and flavonoids, are the most widely used natural antioxidants, principally because of their high radical scavenging activity and low toxicity. Additionally, they exhibit a spectrum of biological activity, including antiviral, anti-bacterial, anti-hypertension, anti-thrombosis, anti-fibrosis, and even anti-cancer properties.10 Therefore, the development of potent and safe phenolic antioxidants is always a worthwhile task. Recently, we reported the synthesis of 21 novel phenolic antioxidants using precursors naturally present in oil seeds.7,10
Establishing safety and effectiveness of novel compounds is a key aspect in antioxidant research. Even though there has been a long history of the use of antioxidants for food preservation and other purposes, there remains an important concern about their possible effects on human health and their overall significance for human health.11 Indeed, for some dietary phytochemicals, their antioxidant properties are less important for human health than the other effects that they may cause, including changes in gene expression and in cell signaling.11
In sum, there currently is a significant interest in the field of antioxidant research. There still exists an important concern about the possible effects of antioxidants on human health and their overall significance for human health. Therefore, establishing safety and effectiveness of novel compounds is a key aspect of antioxidant research.
Here, we aimed to analyze the cellular and molecular effects of selected novel antioxidants in normal human tissues and in cancer cells. We, for the first time, show that while being non-toxic for normal tissues (human buccal and gingival tissues), the examined antioxidants exerted a profound cell killing effects on oral and breast cancer cells, including chemotherapy-resistant breast cancer cells.
Results and Discussion
The key goal of this study was to determine if new lipid antioxidants exert any detrimental effects on human cells and tissues. In the present studies, we focused on four of the recently reported novel antioxidants: trolox trihydroxybenzoate (1a)12; trolox 3,5-dimethoxy-4-hydroxycinnamate (1b)13; and derivatives of dihydrocaffeic acid amide, (2a) and (2b)10 (Fig. 1). Based on the literature suggestions,11 we specifically focused our study on gene expression and signaling effects of the novel antioxidants.
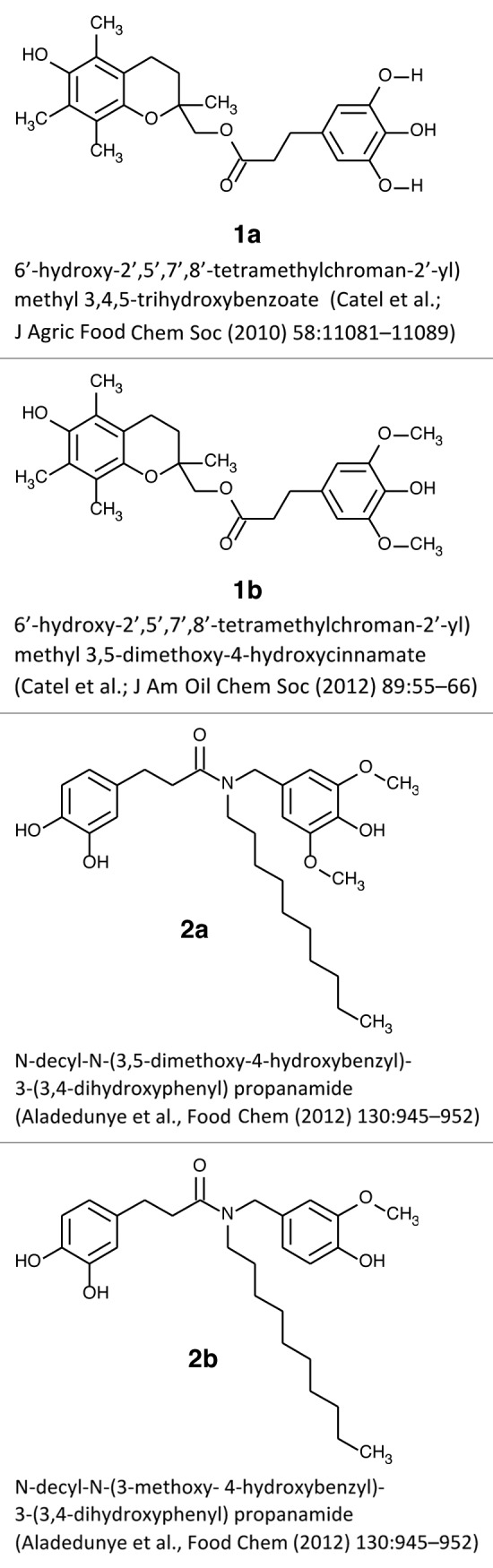
Figure 1. Studied antioxidant compounds.
Effects of antioxidants on normal tissues
First, we evaluated the toxicity of the phenolic antioxidants on normal human oral and gingival epithelial tissues, since they are the first to be exposed to food and its constituents, including antioxidants. We conducted Illumina microarray-based gene expression profiling to determine gene expression patterns upon exposure of EpiOral and EpiGingival tissues to selected antioxidants. The summary of the gene expression results is presented in Table 1.
Table 1. Summary of the gene expression in EpiOral and EpiGingival tissues upon treatment with novel antioxidant compounds 1a, 1b, 2a, and 2b.
| Antioxidant compounds | Gene expression fold change as compared with control | |||
|---|---|---|---|---|
| 1.5-fold cut-off | 2-fold cut-off | |||
| EpiOral | EpiGingival | EpiOral | EpiGingival | |
| 1a | 51 | 15 | 10 | 3 |
| 1b | 12 | 52 | 1 | 3 |
| 2a | 15 | 131 | 1 | 5 |
| 2b | 5 | 8 | 0 | 1 |
Effects in EpiOral model
Our analysis revealed that treatment with 1b led to altered expression of 12 genes when a 1.5-fold cut-off was applied. Yet, when a more conservative 2-fold cut-off was used, only 1 gene was deregulated (Table 1; Table S1). Treatment with 2a led to deregulated expression of 15 genes at the 1.5-fold cut-off level and only 3 genes at a 2-fold cut-off (Table 1; Table S2). Interestingly, treatment with 2b affected 5 genes when the 1.5-fold cut-off was applied, while none of these genes changed their expression more than 2 fold (Table 1; Table S3). Among all the studied compounds, changes induced by exposure to 1a were the most pronounced; overall, when 1.5 was used as a cut-off level, exposure to 1a influenced expression of 52 genes. Among those, expression of 10 genes was changed more than 2 fold (Table 1; Table S4). A direct relationship between their effects on gene expression and the number of oxygenated substituents, especially the hydroxy functional group, was evident; trolox derivative 1a has 4 OHs, whereas the much milder 1b has only 2 OHs. Although the dihydrocaffeic acid amides both have 3 OHs, the more active 2a possesses 2 additional methoxy substituents in comparison to the 1 methoxy group in 2b.
Even though 1a affected the expression of many genes, some of these gene expression changes may be viewed as protective. Among those, 1a suppressed the levels of the MYC oncogene, CDK5, and other genes involved in oncogenesis (Table S4). It also downregulated the MMP10 gene, which codes for a protein involved in cell–cell communication and metastasis.14 Interestingly, a recent study has shown that eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) inhibit gastric cancer cell migration by suppressing the expression of matrix metalloproteinase 10.14
Among the numerous genes altered by exposure of EpiOral tissues to antioxidants 1a, 1b, 2a, and 2b, only one gene was common in all four groups, ADRB2. This gene encodes a β-2-adrenergic receptor, which is a member of the G protein-coupled receptor super-family that is reported to be important in salivary gland function.15 Although the roles of ADRB2 in the function of oral epithelium are unclear,16 a recent study indicated that it might be involved in wound healing responses of oral epithelial cells.16
Four genes were common between the 1a and 2a groups; among those is the EGR1 gene. The protein encoded by this gene belongs to the EGR family of C2H2-type zinc-finger proteins. It is a nuclear protein and functions as a transcriptional regulator. It activates numerous genes, the products of which are required for differentiation and mitogenesis, such as PTEN, p53, and fibronectin.17 EGR1 is considered an important tumor suppressor.17 Therefore, the fact that EGR1 is upregulated by antioxidants in normal oral tissues may be viewed as a positive protective effect.
The DDIT4 gene encodes for protein regulation in development and DNA damage responses. Specifically, it inhibits cell growth by regulating the TOR signaling pathway upstream of the TSC1-TSC2 complex.18 Therefore, this gene has a protective, growth inhibitory effect. Another commonly changed gene is ADM. It encodes a vasodilator peptide, and its role in antioxidant response needs to be further established. The function of the ankyrin repeat domain 37 gene remains unknown.
Effects in EpiGingival tissue
Our analysis revealed that treatment of EpiGingival tissues with 1b led to altered expression of 52 genes when a 1.5-fold cut-off was applied. Yet, when a more conservative 2-fold cut-off was used, only 3 genes were deregulated (Table 1; Table S5). Treatment with 2a led to deregulated expression of 131 genes at the 1.5-fold cut-off level, and only 5 genes at a 2-fold cut-off level (Table 1; Table S6). Interestingly, treatment with 2b affected 8 genes when the 1.5-fold cut-off was applied, while only one of these genes changed its expression more than 2-fold (Table 1; Table S7). Treatment with 1a led to altered expression of 15 genes when the 1.5-fold cut-off level was used. Upon employment of a more conservative 2-fold cut-off, only 3 genes were deregulated (Table 1; Table S8).
Interestingly, in EpiGingival tissues among all four studied compounds, changes induced by exposure to 2a were the most pronounced. However, in both tissues, compound 2b induced the least amount of changes (Table 1).
Having observed changes in gene expression induced by antioxidants in normal oral tissues, we further proceeded to analyze the levels of several proteins involved in cell proliferation and damage responses (Fig. 2). We focused on the proliferating cells’ nuclear antigen (PCNA), phosphorylated histone H2AX (γH2AX), meiotic recombination 11 homolog A (MRE11), and p21/WAF1, a cyclin-dependent kinase inhibitor (p21).19-21
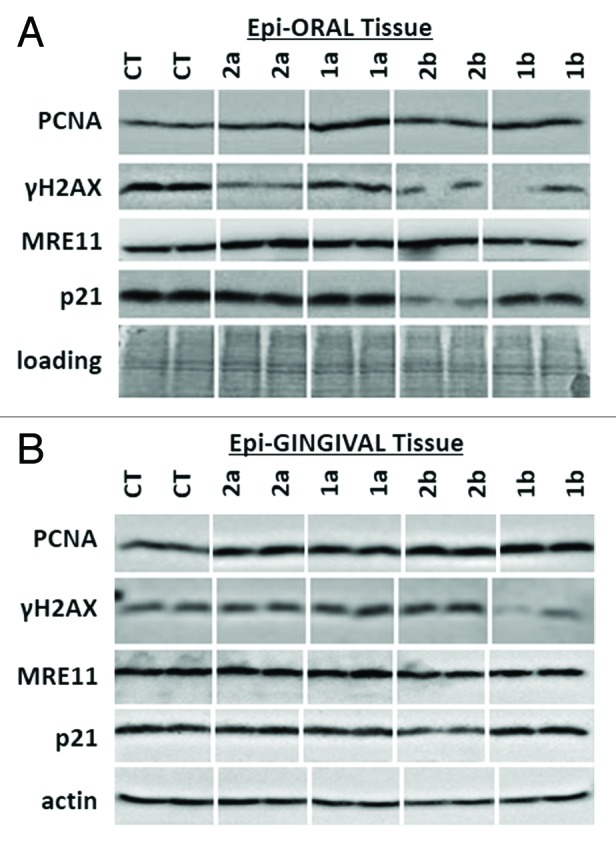
Figure 2. Expression of PCNA, γH2AX, MRE11 and p21 in EpiOral (A) and EpiGingival (B) tissues treated with 1a, 1b, 2a, and 2b antioxidants. CT, control tissues; 1a, 1b, 2a, 2b, tissues treated with antioxidants. Antioxidants: 1a, 6′-hydroxy-2′,5′,7′,8′-tetramethylchroman-2′-yl) methyl 3,4,5-trihydroxybenzoate; 1b, 6′-hydroxy-2′,5′,7′,8′-tetramethylchroman-2′-yl) methyl 3,5-dimethoxy-4-hydroxycinnamate; 2a, N-decyl-N-(3,5-dimethoxy-4-hydroxybenzyl)-3-(3,4-dihydroxyphenyl) propanamide; 2b, N-decyl-N-(3-methoxy-4-hydroxybenzyl)-3-(3,4-dihydroxyphenyl) propanamide.
PCNA is a well-established marker of S-phase cells. Phosphorylated histone H2AX is a marker of DNA damage. Histone H2AX is rapidly phosphorylated at Ser139 upon induction of DNA strand breaks, and it can be effectively detected using specific antibodies.22,23 Analysis of histone H2AX phosphorylation is widely used to assess the extent of damage to cellular DNA.24 MRE 11, on the other hand, is a key DNA repair protein.25 P21 is one of the key tumor suppressors and plays numerous essential roles in DNA damage response, cell cycle arrest, apoptosis, and transcription.26
In EpiOral tissue, we noted that exposure to antioxidants did not lead to any significant changes in the levels of PCNA and MRE11 (Fig. 2A). The levels of γH2AX were reduced by 2b, 1b, and 1a compounds (Fig. 2A). This is an interesting observation, as γH2AX is usually induced upon DNA damage.27 Therefore, a decrease in γH2AX levels may be viewed as a positive, protective effect.
Interestingly, we noted that compound 1a strongly suppressed p21 in EpiOral tissues. Notwithstanding, gene expression data did not show any changes in the levels of p21 gene expression. Thus, its expression may be regulated posttranscriptionally. Indeed, small RNAs may effectively suppress the protein levels.28 In the future, it would be interesting to analyze the effects of the selected antioxidants on the small RNA levels and to correlate those with the levels of gene expression and protein levels.
Similar to observations in EpiOral tissues, we did not see any changes in the levels of MRE11 and p21 in EpiGingival tissues upon exposure to antioxidants (Fig. 2B). Levels of γH2AX were unaffected by 1a, 2a, and 2b compounds; however, exposure to antioxidant 1b led to a decrease in the levels of γH2AX, suggesting a protective effect similar to the one observed in EpiOral tissues.
Additionally, we have analyzed the level of cleaved caspase 3 as an established marker for apoptosis.29 None of the studied antioxidants led to caspase 3 cleavage. The apparent lack of caspase 3 cleavage indicates that novel antioxidants do not cause cell death in normal oral and gingival tissues (Fig. 3).
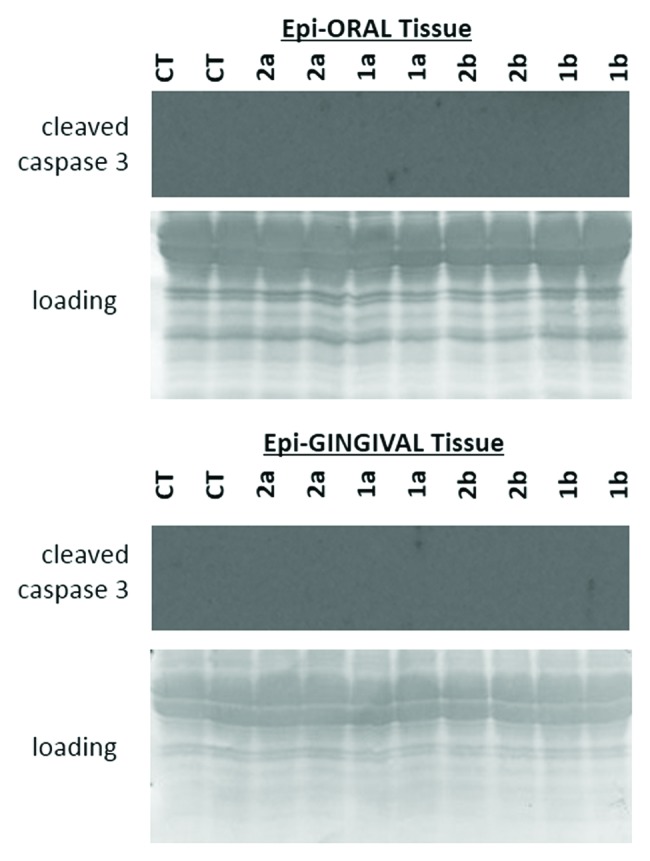
Figure 3. Levels of cleaved caspase 3 in EpiOral and EpiGingival tissues treated with 1a, 1b, 2a, and 2b antioxidants.CT, control tissues; 1a, 1b, 2a, 2b, tissues treated with antioxidants. Antioxidants: 1a, 6′-hydroxy-2′,5′,7′,8′-tetramethylchroman-2′-yl) methyl 3,4,5-trihydroxybenzoate; 1b, 6′-hydroxy-2′,5′,7′,8′-tetramethylchroman-2′-yl) methyl 3,5-dimethoxy-4-hydroxycinnamate; 2a, N-decyl-N-(3,5-dimethoxy-4-hydroxybenzyl)-3-(3,4-dihydroxyphenyl) propanamide; 2b, N-decyl-N-(3-methoxy-4-hydroxybenzyl)-3-(3,4-dihydroxyphenyl) propanamide.
Overall, based on the gene expression and protein expression levels, we suggest that the studied novel antioxidants are not toxic to the oral and gingival epithelial tissues, and some of the noticed changes may in fact be protective in nature. In the future, studies will be needed to conduct a further in-depth analysis of molecular, cellular, and histological effects of these antioxidants on a wide array of normal tissues as a function of time and concentration.
Effects of antioxidants on cancer cells
Having seen no toxic effects of antioxidants on normal oral and gingival cells, we proceeded to analyze their effects on tumor cells. In the present study, we used three oral squamous cell carcinoma lines—CAL-27, UMSCC1, and UMSCC47. CAL-27 is a tongue squamous cell carcinoma; UMSCC1 and UMSCC47 are oral cavity squamous carcinoma cell lines.30
We noted that in cell line CAL27, the levels of γH2AX were upregulated by 2b, 2b, and 1a compounds (Fig. 4). Among them, 2b caused the highest upregulation of γH2AX. Similarly, in the oral cavity cancer cell line UMSCC47, exposure to 2a, 2b, and 1a compounds resulted in a strong upregulation of γH2AX. Contrarily, in the UMSCC1 line, γH2AX phosphorylation was increased only upon exposure to compound 2a. The increased levels of γH2AX may be indicative of DNA damage; therefore future studies are needed to dissect the precise nature of DNA damage induced by novel antioxidants in cancer cells.
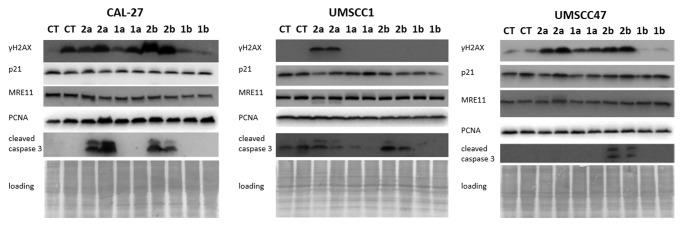
Figure 4. Expression of γH2AX, PCNA, MRE11, p21, and cleaved caspase 3 in CAL27, UMSCC1 and UMSCC47 oral squamous cell carcinoma lines treated with 1a, 1b, 2a, and 2b antioxidants. CT, control tissues; 1a, 1b, 2a, 2b, tissues treated with antioxidants. Antioxidants: 1a, 6′-hydroxy-2′,5′,7′,8′-tetramethylchroman-2′-yl) methyl 3,4,5-trihydroxybenzoate; 1b, 6′-hydroxy-2′,5′,7′,8′-tetramethylchroman-2′-yl) methyl 3,5-dimethoxy-4-hydroxycinnamate; 2a, N-decyl-N-(3,5-dimethoxy-4-hydroxybenzyl)-3-(3,4-dihydroxyphenyl) propanamide; 2b, N-decyl-N-(3-methoxy-4-hydroxybenzyl)-3-(3,4-dihydroxyphenyl) propanamide.
Our analysis also showed that exposure to antioxidants did not lead to any significant changes in the levels of p21 and MRE11 in all studied OSCC lines. The levels of PCNA were affected only in UMSCC1 upon exposure to 2b compound that caused a slight decrease in PCNA levels.
But most interestingly, we found that novel antioxidants 2a and 2b caused a profound increase in the levels of cleaved caspase 3 in CAL 27 and UMSCC47 cells. In UMSCC1 cells, caspase 3 cleavage was caused only by 2b compound. Caspase-3 is a critical executioner of apoptosis, and the induction of caspase 3 cleavage is a well-established indicator of apoptosis29 (Fig. 4).
To gain further insight into the apoptosis-inducing potential of 2a and 2b, we analyzed the level of apoptosis in OSCC cell lines by the Annexin V assay. Our analysis revealed that that both compounds caused a profound induction of apoptosis in the OSCC cell lines (Fig. 5). The profound apoptosis-inducing effects of novel antioxidants indicate their excellent potential as novel anti-cancer regimens.
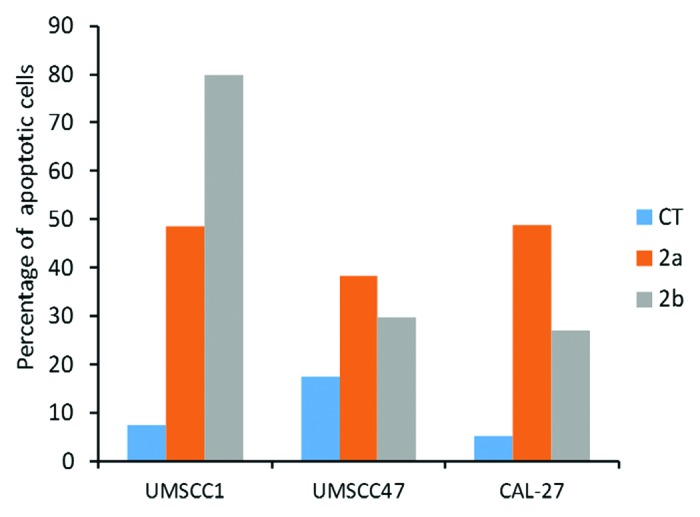
Figure 5. Induction of apoptosis by 1a, 1b, 2a, and 2b antioxidants in CAL27, UMSCC1 and UMSCC47 oral squamous cell carcinoma lines as analyzed by the Annexin V assay. CT, control tissues; 1a, 1b, 2a, 2b, tissues treated with antioxidants. Antioxidants: 1a, 6′-hydroxy-2′,5′,7′,8′-tetramethylchroman-2′-yl) methyl 3,4,5-trihydroxybenzoate; 1b, 6′-hydroxy-2′,5′,7′,8′-tetramethylchroman-2′-yl) methyl 3,5-dimethoxy-4-hydroxycinnamate; 2a, N-decyl-N-(3,5-dimethoxy-4-hydroxybenzyl)-3-(3,4-dihydroxyphenyl) propanamide; 2b, N-decyl-N-(3-methoxy-4-hydroxybenzyl)-3-(3,4-dihydroxyphenyl) propanamide.
To analyze the anticancer effects of the novel antioxidants further, we used human breast adenocarcinoma MCF-7 cells, an ER-positive, hormone-sensitive cell line that represents early stage human breast cancer. In parallel, we used MCF-7 variants resistant to common chemotherapy drugs, doxorubicin (MCF-7/DOX) and cis-dichlorodiammine platinum (II) or cisplatinum (MCF-7/CIS).31 The drug-resistant variants of the MCF-7 cell lines were previously established by stepwise selection after prolonged (>6 mo) treatment of the original MCF-7 cells with escalating concentrations of DOX or CIS at a range of 0.5 to 15 μg/mL in the medium. After 6 mo of culturing in the presence of drugs, the IC50 (inhibitory concentration to produce 50% cell death) values were 19.2 and 3.6 mg/L for the MCF-7/DOX and MCF-7/CIS cells, respectively.
At 70% confluency, the cells were treated with the studied antioxidant compounds at a concentration of 350 μg/mL for 2 h. Interestingly, we noted that all cells were dead after 30 min of such exposure. The experiment was reproduced a second time, and the same phenomenon of total cell death was observed.
Next, we reduced the concentration of antioxidants to 117 μg/mL. After 2 h of treatment, the cells were analyzed for survival and for gene expression. As with normal tissues, we used Illumina HumanHT-12 v4 Expression BeadChip arrays. At the same concentrations that did not cause any effects on normal cells, antioxidants led to significant alterations in gene expression in cancer lines.
Exposure of MCF-7 to 1b led to changes in expression of 28 genes. Application of 2b to MCF-7 cells altered the expression of 845 genes. Interestingly, 1a and 2a were so toxic to the cells that, even though we were able to extract the RNA from the cells, the RNA did not yield any good cDNA libraries. This is indicative of apoptotic-related processes and further confirms a very profound and rapid death of MCF-7, DOX and CIS cells induced by antioxidants.
Compounds 1a and 1b affected gene expression in DOX and CIS cells as well. Indeed, exposure of DOX cells to 1b resulted in altered expression of 50 genes, and exposure to 1a led to altered expression of 7 genes. Three genes were common in both groups (Table S9). In CIS cells, 1a induced expression changes of 56 genes, and 1b induced changes of 24 genes. Three genes were common between 1a- and 1b-exposed CIS cells (Table S10). Interestingly, compounds 2a and 2b were extremely toxic to DOX and CIS cells.
Overall, we noted that 2b affected expression of MCF-7 cells, but killed DOX and CIS cells. Therefore, among all the studied compounds, 2b appeared to be the most interesting and promising one; it did not affect normal tissues, but it causes profound deregulation of gene expression in breast adenocarcinoma tissues and effectively eradicated drug resistant cells.
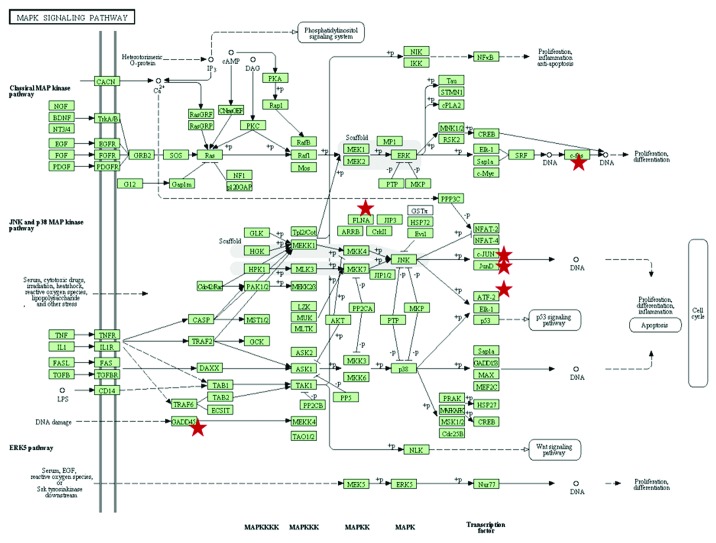
Treatment with 2b led to altered expression of 845 genes, thus affecting numerous cellular pathways (Table S11). It caused upregulation of the MAPK signaling pathway (Fig. 6). Additionally, it downregulated key metabolic pathways, such as oxidative phosphorylation, ribosome, aminoacyl-tRNA biosynthesis, citrate cycle (TCA cycle), proteasome, spliceosome, fatty acid elongation in mitochondria, glutathione metabolism, and the pyruvate metabolism pathways (Table 2). These pathways are critically important for general cellular wellbeing, and their downregulation may lead to cell death. Indeed, altered functioning of these key networks may underlie the mechanisms of antioxidant-induced killing of MCF-7 cells.
Figure 6. Schematic representation of the MAPK pathways that is upregulated in MCF-7 cells upon treatment with compound 2b (N-decyl-N-[3-methoxy-4-hydroxybenzyl]-3-[3,4-dihydroxyphenyl] propanamide). DAVID Bioinformatic resource (http://david.abcc.ncifcrf.gov/), the KEGG Pathways.42 The KEGG (Kyoto Encyclopedia of Genes and Genomes) MAPK pathway map can be found online at http://www.genome.jp/kegg/. Stars denote genes which exhibited altered expression upon antioxidant treatment.
Table 2. List of pathways differentially regulated in MCF-7 cells treated with 2b compound.
| Upregulated | ||
|---|---|---|
| Term | P value | Altered genes |
| hsa05322:Systemic lupus erythematosus | P < 0.0001 | HIST2H2AA3, HIST1H2BC, HIST1H2BD, HIST1H2BE, HIST1H2BF, HIST1H2BG, HIST1H2AE, HIST1H2BH, H2AFJ, HIST2H3C, HIST2H3D, HIST2H2AB, HIST2H2BE, HIST2H2AC, HIST1H2BJ, HIST1H4E, HIST1H2AH, HIST1H3D, HIST1H3F, HIST1H3G, HIST1H3H, HIST1H4H |
| hsa00900:Terpenoid backbone biosynthesis | P < 0.0001 | MVD, HMGCS1, FDPS, IDI1 |
| hsa04010:MAPK signaling pathway | P < 0.0001 | FOS, ATF4, JUN, JUND, HSPA6, HSPA1A, HSPA1B, GADD45B, FLNA, DDIT3, ATF2 |
| Downregulated | ||
| Term | P value | Altered genes |
| hsa05012:Parkinson disease | P < 0.0001 | ATP5D, NDUFB5, UQCRC1, COX7A2, NDUFA8, LOC729317, NDUFA9, NDUFB9, COX8A, COX7A2L, COX5A, COX5B, PARK7, VDAC1, LOC402175, SDHD, COX6A1, UBB |
| hsa00190:Oxidative phosphorylation | P < 0.0001 | ATP5D, NDUFB5, UQCRC1, COX7A2, NDUFA8, NDUFA9, COX10, NDUFB9, COX8A, COX7A2L, COX5A, COX5B, ATP6V1A, ATP6V0E2, LOC402175, SDHD, COX6A1 |
| hsa05010:Alzheimer disease | P < 0.0001 | ATP5D, NDUFB5, COX7A2, UQCRC1, NDUFA8, APH1A, NDUFA9, NDUFB9, COX8A, COX7A2L, COX5A, COX5B, ATP2A2, LOC402175, SDHD, PPP3CB, COX6A1, CHP, CALM1 |
| hsa05016:Huntington disease | 6.21E-04 | ATP5D, NDUFB5, UQCRC1, COX7A2, NDUFA8, LOC729317, NDUFA9, NDUFB9, COX8A, COX7A2L, COX5A, COX5B, DCTN2, VDAC1, LOC402175, SDHD, CREB3L2, COX6A1, AP2M1 |
| hsa03010:Ribosome | 0.001104 | MRPL13, RPS3A, RPL34, RPL8, RPL15, RPL27, RPS13, FAU, RPL24, RPL7A, RPS11, RPS3 |
| hsa00970:Aminoacyl-tRNA biosynthesis | 0.001563 | WARS, YARS, DARS, EPRS, IARS2, DARS2, KARS, EARS2 |
| hsa00020:Citrate cycle (TCA cycle) | 0.009441 | SUCLG2, SUCLG1, SDHD, IDH1, PDHB, MDH1 |
| hsa03050:Proteasome | 0.0143 | PSMB5, PSMB7, PSMD14, PSMB6, PSMA6, PSMB2, PSMD2 |
| hsa04142:Lysosome | 0.027671 | LAMP1, LAMP2, AP4E1, AP1G1, PSAP, IGF2R, HEXB, PPT1, SCARB2, GBA, AP3B1 |
| hsa03040:Spliceosome | 0.042939 | LOC100129585, EFTUD2, ZMAT2, SF3B5, RBMX, CTNNBL1, PRPF19, SFRS7, PLRG1, SFRS9, SNRPC, LOC100131735, PUF60 |
| hsa00062:Fatty acid elongation in mitochondria | 0.043188 | ACAA2, PPT1, HADHB |
| hsa04260:Cardiac muscle contraction | 0.048592 | COX7A2, UQCRC1, ATP2A2, COX8A, COX6A1, COX7A2L, COX5A, COX5B |
| hsa00860:Porphyrin and chlorophyll metabolism | 0.050819 | BLVRA, COX10, FTHL3, EPRS, EARS2 |
| hsa00480:Glutathione metabolism | 0.061389 | MGST3, GPX4, RRM1, PGD, LOC642590, IDH1, LOC646347, SMS |
| hsa05120:Epithelial cell signaling in Helicobacter pylori infection | 0.069935 | ATP6V1A, TJP1, ATP6V0E2, MAPK13, RAC1, NFKB1, PTPN11 |
| hsa05014:Amyotrophic lateral sclerosis (ALS) | 0.075133 | DERL1, MAPK13, RAC1, PPP3CB, BCL2L1, CHP |
| hsa04722:Neurotrophin signaling pathway | 0.083546 | MAP2K1, MAPK13, RAC1, YWHAB, YWHAQ, RHOA, NFKB1, CRK, CALM1, PTPN11 |
| hsa00620:Pyruvate metabolism | 0.090327 | ME1, LDHA, GLO1, PDHB, MDH1 |
| hsa04114:Oocyte meiosis | 0.098472 | PPP1CA, MAP2K1, YWHAB, YWHAQ, PPP3CB, PPP2R5E, CHP, PPP1CC, CALM1 |
Future Perspectives
Based on the data presented herein, compounds 2a and 2b seem to be the most promising ones. They spare the normal cells, but effectively kill both oral cavity and breast cancer cells. Moreover, compound 2b is especially active in killing multi-drug resistant breast cancer cells. The latter finding is of crucial importance, as multi-drug resistance is a major clinical problem.31 It usually leads to tumor recurrence and a poor clinical outcome. In the future, it would be important to conduct a detailed analysis of the potential of compound 2b for killing multi-drug resistant cells and tumors of other tissue types, including colon, lung, melanoma, prostate cancer.
Of special interest and importance would be applicability of compounds 2a and 2b to treat oral squamous cell carcinoma (OSCC). Since compounds 2a and 2b are non-toxic against normal oral and gingival tissues and effectively induced DNA damage and apoptosis in OSCC cell lines, in the future, these compounds should be further investigated for their potential in oral cancer treatment. OSCC accounts for approximately 300 000 new cases worldwide, including 30 000 in North America, annually. This debilitating tumor is very difficult to treat and has a dismal prognosis.32,33 Despite advances in diagnostic, surgical, and chemo-radiation techniques, only 40–50% of OSCC patients survive beyond 5 years—in fact, survival rates in OSCC have remained unchanged for the last five decades. Even patients surviving OSCC often endure chronic and debilitating consequences of treatment. These novel antioxidants may hold potential for cancer treatment.
Furthermore, it would be interesting to check the protective effects of novel antioxidants. Sometimes, an application of a given substance may lead to a decrease in the rates of carcinogen-induced cancer. Would the same apply for pre-treatment of cells with novel antioxidant compounds? This question remains to be answered. Additionally, animal studies are clearly needed to analyze whole organism and tissue-specific effects of novel antioxidants.
Materials and Methods
Antioxidants
The syntheses of the novel compounds have previously been reported: 1a,12 1b,13 2a, and 2b10 (Fig. 1).
Tissues and treatment
We used EpiOralTM and EpiGingivalTM tissues (Mattek Inc.). EpiOralTM tissues are 3-dimensional, highly differentiated, metabolically and mitotically active tissues. They are produced from normal human cells and represent buccal (inner cheek) cell phenotypes. EpiOral tissues are multilayered with organized basal layer and multiple non-cornified layers, harbor in vivo-like lipid profiles, produce human β defensins, and overall are known to be ideal for irritation, toxicity, and oral pathology studies.
The EpiGingival tissue is also multilayered except that the apical layers are cornified, similar to in vivo gingival tissue. Both of these tissue models have been used to study effects of DNA damaging agents34 as well as toxic effects of ethanol and the mouth rinse Listerine.35
EpiOral and EpiGingival tissues were cultured as recommended by the manufacturer (Mattek Inc.). Tissues were treated with 117 μg/mL of aforementioned antioxidants in the culture media for 2 h.
Cell lines and treatment
To analyze potential anti-cancer effects of novel antioxidants, we used three oral squamous cell carcinoma lines—CAL-27, UMSCC1, and UMSCC47. CAL-27 is a tongue squamous cell carcinoma; UMSCC1 and UMSCC47 are oral cavity squamous carcinoma cell lines.30,36,37 These lines were a kind gift of Professor Joseph Dort. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM)/High Glucose (Hyclone) with 100 nmol/L nonessential amino acids (Gibcon) and 2 mmol/L l-glutamine (Hyclone). All culture media were supplemented with 10% fetal bovine serum (Gibco), as recommended.
Additionally, we used human breast adenocarcinoma MCF-7 cells, an ER-positive, hormone-sensitive cell line that represents early stage human breast cancer. In parallel to MCF-7, we used MCF-7 variants resistant to common chemotherapy drugs—doxorubicin (MCF-7/DOX) and cis-dichlorodiammine platinum(II) or cisplatimun (MCF-7/CIS).31 The cells were cultured as recommended using Dulbecco IS-OV (Sigma) containing 10% newborn calf serum (HyClone) and 40 μg/mL gentamicin at 37 °C in a 5% CO2 atmosphere.31 The drug-resistant variants of MCF-7 cell lines were previously established by stepwise selection after prolonged (>6 mo) treatment of original MCF-7 cells with escalating concentrations of DOX or CIS at a range of 0.5 to 15 μg/mL in the medium. After 6 mo of culturing in the presence of drugs, the IC50 (inhibitory concentration to produce 50% cell death) values were 19.2 and 3.6 mg/L for the MCF-7/DOX and MCF-7/CIS cells, respectively.
Cells were seeded at a density of 0.5 × 106 viable cells per 100-mm plate, and the medium was changed every other day for 6 d. At 70% confluency, the cells were treated with the studied antioxidant compounds at concentrations 350 μg/mL for 2 h. Cell survival and morphology was evaluated under the microscope after 2 h of treatment.
RNA extraction and gene expression
Upon treatment, cells and tissues were flash-frozen in liquid nitrogen for storage and further analysis. Then, cells and tissues were homogenized in TRIzol® Reagent (Invitrogen). RNA isolation was performed according to manufacturer’s instructions (Invitrogen). The RNA was quantified and the quality was found to be of a high quality using a Bioanalyser 2100 (Agilent). RNA labeling and microarray hybridization were performed by the Lethbridge Epigenetics Laboratory Illumina Facility. HumanHT-12 v4 Expression BeadChip whole-genome expression arrays (Illumina) were used in this study. Each array on the HumanHT-12 v4 Expression BeadChip targets more than 47 000 probes derived from the National Center for Biotechnology Information Reference Sequence (NCBI) RefSeq Release 38 and other sources (Illumina). Three biological replicates were used per each experimental group. In brief, each RNA sample was amplified using the Ambion Illumina RNA amplification kit with biotin UTP (Enzo) labeling. The Ambion Illumina RNA amplification kit uses T7 oligo(dT) primer to generate single stranded cDNA followed by a second strand synthesis to generate double-stranded cDNA, which is then column purified. In vitro transcription was conducted to synthesize biotin-labeled cRNA using T7 RNA polymerase. The cRNA was column purified and checked for size and yield. cRNA was hybridized using standard Illumina protocols with streptavidin-Cy3 (Amersham). Arrays were scanned on an Illumina Beadstation and analyzed using BeadStudio (Illumina) as recommended.38 Normalization, clustering, and significance analysis were done by the Lethbridge Epigenetics Laboratory Illumina Facility as previously described.38
Protein extraction and western blotting
Protein extraction and western blotting was conducted as described.39 Membranes were incubated with antibodies against MRE11 and PCNA (1:1000; Santa Cruz Biotechnology), cleaved caspase, γH2AX, p21 (1:1000; Cell Signaling). Antibody binding was revealed by incubation with horseradish peroxidase-conjugated secondary antibodies and the ECL Plus TM immunoblotting detection system (GE Healthcare). Chemiluminescence was detected by Biomax MR film (Eastman Kodak) and scanned, and by the FluoChemHD2 system (ProteinSimple). Protein loading was visualized by Coomassie brilliant blue R250 staining (BioRad, Mississauga).40
The Annexin V assay
For apoptosis analysis, we used the Annexin V-FITC Apoptosis Detection Kit I (BD Biosciences) according to the manufacturer’s protocol as described earlier. Cells were grown on 75 cm2 cell culture flasks and treated with 2a and 2b compounds as previously described (see section on tissues and Treatments). The analysis was performed 2 h after treatment. In brief, cells were harvested, washed with PBS, resuspended in 1× binding buffer, stained with Annexin V and propidium iodide (PI) for 15 min at 25 °C in the dark and analyzed by flow cytometry using the BD FACS Canto flow cytometer. The results were represented as the percentage of gated Annexin V positive cells.41
Supplementary Material
Acknowledgments
We are grateful to Jody Filkowski for running the Illumina array expression, to Dr Vasyl Chekhun for the gift of MCF-7/DOX and MCF-7/cisDDP cells, and to Dr Joseph Dort for the gift of OSCC cell lines. Research in the Kovalchuk laboratory was supported by the Alberta Cancer Foundation, CIHR, and NSERC. Research in the Przybylski laboratory was supported by the Alberta Funding Consortium and the Alberta Value Added Corporation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cbt/article/25935
References
- 1.Rosanna DP, Salvatore C. Reactive oxygen species, inflammation, and lung diseases. Curr Pharm Des. 2012;18:3889–900. doi: 10.2174/138161212802083716. [DOI] [PubMed] [Google Scholar]
- 2.Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Dröge W. Free radicals in the physiological control of cell function. Physiol Rev. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 4.Ray PD, Huang BW, Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal. 2012;24:981–90. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Florence TM. The role of free radicals in disease. Aust N Z J Ophthalmol. 1995;23:3–7. doi: 10.1111/j.1442-9071.1995.tb01638.x. [DOI] [PubMed] [Google Scholar]
- 6.Moreira PI, Santos MS, Oliveira CR, Shenk JC, Nunomura A, Smith MA, Zhu X, Perry G. Alzheimer disease and the role of free radicals in the pathogenesis of the disease. CNS Neurol Disord Drug Targets. 2008;7:3–10. doi: 10.2174/187152708783885156. [DOI] [PubMed] [Google Scholar]
- 7.Catel Y, Aladedunye F, Przybylski R. Synthesis, Radical Scavenging Activity, Protection during Storage, and Frying by Novel Antioxidants. J Agric Food Chem. 2010 doi: 10.1021/jf102287h. [DOI] [PubMed] [Google Scholar]
- 8.Flora SJ. Role of free radicals and antioxidants in health and disease. Cell Mol Biol (Noisy-le-grand) 2007;53:1–2. [PubMed] [Google Scholar]
- 9.German JB. Food processing and lipid oxidation. Adv Exp Med Biol. 1999;459:23–50. doi: 10.1007/978-1-4615-4853-9_3. [DOI] [PubMed] [Google Scholar]
- 10.Aladedunye F, Catel Y, Przybylski R. Novel caffeic acid amide antioxidants: Synthesis, radical scavenging activity and performance under storage and frying conditions. Food Chem. 2012;130:945–52. doi: 10.1016/j.foodchem.2011.08.021. [DOI] [PubMed] [Google Scholar]
- 11.Gordon MH. Significance of dietary antioxidants for health. Int J Mol Sci. 2012;13:173–9. doi: 10.3390/ijms13010173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Catel Y, Aladedunye F, Przybylski R. Synthesis, Radical Scavenging Activity, Protection during Storage, and Frying by Novel Antioxidants. J Agric Food Chem. 2010;58:11081–9. doi: 10.1021/jf102287h. [DOI] [PubMed] [Google Scholar]
- 13.Catel Y, Aladedunye F, Przybylski R. Radical Scavenging Activity and Performance of Novel Phenolic Antioxidants in Oils During Storage and Frying. J Am Oil Chem Soc. 2012;89:55–66. doi: 10.1007/s11746-011-1889-6. [DOI] [Google Scholar]
- 14.Wu MH, Tsai YT, Hua KT, Chang KC, Kuo ML, Lin MT. Eicosapentaenoic acid and docosahexaenoic acid inhibit macrophage-induced gastric cancer cell migration by attenuating the expression of matrix metalloproteinase 10. J Nutr Biochem. 2012;23:1434–9. doi: 10.1016/j.jnutbio.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 15.Proctor GB, Carpenter GH. Regulation of salivary gland function by autonomic nerves. Auton Neurosci. 2007;133:3–18. doi: 10.1016/j.autneu.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 16.Steenhuis P, Huntley RE, Gurenko Z, Yin L, Dale BA, Fazel N, Isseroff RR. Adrenergic signaling in human oral keratinocytes and wound repair. J Dent Res. 2011;90:186–92. doi: 10.1177/0022034510388034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baron V, Adamson ED, Calogero A, Ragona G, Mercola D. The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGFbeta1, PTEN, p53, and fibronectin. Cancer Gene Ther. 2006;13:115–24. doi: 10.1038/sj.cgt.7700896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellisen LW. Growth control under stress: mTOR regulation through the REDD1-TSC pathway. Cell Cycle. 2005;4:1500–2. doi: 10.4161/cc.4.11.2139. [DOI] [PubMed] [Google Scholar]
- 19.Stivala LA, Cazzalini O, Prosperi E. The cyclin-dependent kinase inhibitor p21CDKN1A as a target of anti-cancer drugs. Curr Cancer Drug Targets. 2012;12:85–96. doi: 10.2174/156800912799095126. [DOI] [PubMed] [Google Scholar]
- 20.Langerak P, Russell P. Regulatory networks integrating cell cycle control with DNA damage checkpoints and double-strand break repair. Philos Trans R Soc Lond B Biol Sci. 2011;366:3562–71. doi: 10.1098/rstb.2011.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mavers M, Balomenos D, Perlman H. The cyclin dependent kinase inhibitor p21((WAF1/C1P1)) Arthritis Rheum. 2008;58:S933–933. doi: 10.1002/art.33311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–68. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 23.Ivashkevich A, Redon CE, Nakamura AJ, Martin RF, Martin OA. Use of the γ-H2AX assay to monitor DNA damage and repair in translational cancer research. Cancer Lett. 2012;327:123–33. doi: 10.1016/j.canlet.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dickey JS, Redon CE, Nakamura AJ, Baird BJ, Sedelnikova OA, Bonner WM. H2AX: functional roles and potential applications. Chromosoma. 2009;118:683–92. doi: 10.1007/s00412-009-0234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85:509–20. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 26.Cazzalini O, Scovassi AI, Savio M, Stivala LA, Prosperi E. Multiple roles of the cell cycle inhibitor p21(CDKN1A) in the DNA damage response. Mutat Res. 2010;704:12–20. doi: 10.1016/j.mrrev.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 27.Sedelnikova OA, Pilch DR, Redon C, Bonner WM. Histone H2AX in DNA damage and repair. Cancer Biol Ther. 2003;2:233–5. doi: 10.4161/cbt.2.3.373. [DOI] [PubMed] [Google Scholar]
- 28.Iorio MV, Croce CM. microRNA involvement in human cancer. Carcinogenesis. 2012;33:1126–33. doi: 10.1093/carcin/bgs140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fesik SW. Promoting apoptosis as a strategy for cancer drug discovery. Nat Rev Cancer. 2005;5:876–85. doi: 10.1038/nrc1736. [DOI] [PubMed] [Google Scholar]
- 30.Lin CJ, Grandis JR, Carey TE, Gollin SM, Whiteside TL, Koch WM, Ferris RL, Lai SY. Head and neck squamous cell carcinoma cell lines: established models and rationale for selection. Head Neck. 2007;29:163–88. doi: 10.1002/hed.20478. [DOI] [PubMed] [Google Scholar]
- 31.Chekhun VF, Lukyanova NY, Kovalchuk O, Tryndyak VP, Pogribny IP. Epigenetic profiling of multidrug-resistant human MCF-7 breast adenocarcinoma cells reveals novel hyper- and hypomethylated targets. Mol Cancer Ther. 2007;6:1089–98. doi: 10.1158/1535-7163.MCT-06-0663. [DOI] [PubMed] [Google Scholar]
- 32.Ramqvist T, Dalianis T. An epidemic of oropharyngeal squamous cell carcinoma (OSCC) due to human papillomavirus (HPV) infection and aspects of treatment and prevention. Anticancer Res. 2011;31:1515–9. [PubMed] [Google Scholar]
- 33.Poh CF, Durham JS, Brasher PM, Anderson DW, Berean KW, MacAulay CE, Lee JJ, Rosin MP. Canadian Optically-guided approach for Oral Lesions Surgical (COOLS) trial: study protocol for a randomized controlled trial. BMC Cancer. 2011;11:462. doi: 10.1186/1471-2407-11-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitchell D, Paniker L, Godar D. Nucleotide excision repair is reduced in oral epithelial tissues compared with skin. Photochem Photobiol. 2012;88:1027–32. doi: 10.1111/j.1751-1097.2012.01163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koschier F, Kostrubsky V, Toole C, Gallo MA. In vitro effects of ethanol and mouthrinse on permeability in an oral buccal mucosal tissue construct. Food Chem Toxicol. 2011;49:2524–9. doi: 10.1016/j.fct.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 36.Harris T, Jimenez L, Kawachi N, Fan JB, Chen J, Belbin T, Ramnauth A, Loudig O, Keller CE, Smith R, et al. Low-level expression of miR-375 correlates with poor outcome and metastasis while altering the invasive properties of head and neck squamous cell carcinomas. Am J Pathol. 2012;180:917–28. doi: 10.1016/j.ajpath.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, Huang H, Wang C, Liu X, Hu F, Liu M. MicroRNA-375 sensitizes tumour necrosis factor-alpha (TNF-α)-induced apoptosis in head and neck squamous cell carcinoma in vitro. Int J Oral Maxillofac Surg. 2013;42:949–55. doi: 10.1016/j.ijom.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 38.Ding LH, Xie Y, Park S, Xiao G, Story MD. Enhanced identification and biological validation of differential gene expression via Illumina whole-genome expression arrays through the use of the model-based background correction methodology. Nucleic Acids Res. 2008;36:e58. doi: 10.1093/nar/gkn234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kovalchuk O, Zemp FJ, Filkowski JN, Altamirano AM, Dickey JS, Jenkins-Baker G, Marino SA, Brenner DJ, Bonner WM, Sedelnikova OA. microRNAome changes in bystander three-dimensional human tissue models suggest priming of apoptotic pathways. Carcinogenesis. 2010;31:1882–8. doi: 10.1093/carcin/bgq119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simpson RJ. Rapid coomassie blue staining of protein gels. Cold Spring Harb Protoc. 2010;2010:t5413. doi: 10.1101/pdb.prot5413. [DOI] [PubMed] [Google Scholar]
- 41.Koopman G, Reutelingsperger CP, Kuijten GA, Keehnen RM, Pals ST, van Oers MH. Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood. 1994;84:1415–20. [PubMed] [Google Scholar]
- 42.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.