Abstract
Although 30% of individuals diagnosed with CRC report a family history of the disease, only 5–6% carry germline mutations in genes associated with known hereditary cancer syndromes. The evaluation and management of families affected with CRC can be complicated by variability in disease phenotypes and limited sensitivity of genetic tests. In this review we examine what is currently known about familial CRC and what we have yet to learn, and explore how novel genomic approaches might be used to identify additional genetic and epigenetic factors implicated in heritable risk for cancer.
The average American’s lifetime risk for developing colorectal cancer (CRC) is estimated to be 5–6%. The implementation of routine screening for CRC among individuals age 50 and older has been associated with significant reductions in morbidity and mortality from the disease in the U.S.1 Family history of CRC remains a key factor in algorithms used to risk-stratify individuals for screening and surveillance. Approximately 30% of individuals with CRC report having one or more relatives also diagnosed with the disease. History of CRC in a first-degree relative has been associated with a two-fold increase in an individual’s risk; in the case of numerous affected relatives and/or diagnoses at young ages the risk for CRC is even higher.2 In the setting of specific hereditary cancer syndromes, lifetime risk of CRC may approach 70–90% in the absence of any medical or surgical interventions.3 Given the effectiveness of colonoscopy with polypectomy and surgical resection, identifying individuals who are high risk for CRC at pre-symptomatic stages provides the opportunity for cancer prevention.
Germline mutations in known cancer-causing genes have been implicated in up to 5–6% of all CRC cases. Making the diagnosis of a hereditary cancer syndrome has significant implications for the medical management of CRC patients and their families. Genetic testing can be useful for confirming the diagnosis and provides at-risk relatives the opportunity to pursue predictive testing. Lynch syndrome is the most common of the hereditary CRC syndromes and discovery of the genetic basis of the disease has resulted in the implementation of population-based screening for individuals diagnosed with CRC. Yet as awareness of familial CRC continues to grow and as more patients are referred for genetic evaluation, we are discovering that for many of these families with striking family histories a genetic cause cannot be identified.
Historically, our approaches to evaluating families with cancer have focused on searching for mutations in single genes associated with highly-penetrant disease phenotypes. This strategy has resulted in the identification of the genetic basis of a number of hereditary cancer syndromes (Table 1), but the majority of familial CRC cases are not associated with known germline mutations which suggests other mechanisms may be involved in pathogenesis. As more information related to the chromosomal instability, microsatellite instability, and serrated pathways of colorectal neoplasia becomes available, our understanding of the genetic and epigenetic events involved in carcinogenesis continues to evolve. The potential roles of low-penetrance loci, gene-gene interactions, epigenetic modification, environmental exposures, and/or a combination of these factors are being investigated. This review will summarize what we currently know and explores what we have yet to learn about familial CRC.
Table 1.
Clinical Features and Genes Associated with Hereditary Colorectal Cancer Syndromes
| Syndrome | Clinical Features | Gene (s) | Management* | Evidence for Recommendation |
|---|---|---|---|---|
| Lynch Syndrome |
MMR deficiency phenotype in tumors (MSI) Accelerated adenoma-carcinoma sequence CRC risk= 30–70% over lifetime Risk for extracolonic cancers |
MLH1 MSH2 MSH6 PMS2 Tacstd1/EpCAM Mutations detected in 70% |
Colonoscopy q 1–2 years starting at age 20–25y Consider Upper endoscopy 3–5 years, starting at age 30–35y Consider endometrial cancer screening vs prophylactic hysterectomy |
Cohort Studies10, 12 Expert Opinion14 |
| Familial Adenomatous Polyposis (FAP) | ||||
|
Classic Attenuated |
100s–1000s colorectal adenomas Risk for duodenal and ampullary adenoca Risk for desmoid tumors, thyroid CA CRC risk 90% without surgery 10–99 colorectal adenomas CRC risk is variable |
APC mutations detected in 90% MutYH (biallelic) APC, MutYH mutations detected in ~10% |
Colonoscopy q 1–2 years, starting at age 10–12y, colectomy for large polyp burden Upper endoscopy q 1–3 years Consider thyroid ultrasound Colonoscopy q 1–2 years, beginning at age 20–25y Upper endoscopy q 1–3 years |
Expert Opinion Expert Opinion13 Expert Opinion13 |
| Peutz Jeghers syndrome | 2 ≥Hamartomatous polyps in small bowel Mucocutaneous pigmentation (mouth/ lips, fingers) Cumulative cancer risks 80–90% (colorectal, breast, gastric, pancreatic) |
STK11 mutations detected in 50–70% | Upper endoscopy every 2–3 years starting in late teens Small bowel visualization (eg. capsule endoscopy, CT/MR enterography, small bowel follow through) every 1–3y starting at age 8–10y Colonoscopy every 2–3 years, starting in late teens Pancreas screening (MRCP or EUS) every 1–2 y, starting at age 25–30y. Mammogram and Breast MRI, yearly, starting at age 25y. Testicular exam/ultrasound yearly, starting at age 10y Transvaginal Ultrasound, yearly, starting at age 18y |
Expert Opinion13,27 |
| Juvenile Polyposis Syndrome | >3–5 juvenile polyps in GI tract Some associated with congenital heart disease, Hereditary hemorrhagic telangiectasia |
SMAD4 BMPR1A ENG Mutations detected in <50% |
Upper endoscopy q 1–3 years starting age 15y Colonoscopy q 1–3y starting age 15y |
Expert Opinion13 |
| Cowden Syndrome | Macrocephaly Increased risk for cancer (breast, thyroid, endometrial) Variable colorectal polyp phenotype (adenoma, hamartoma, sessile serrated, ganglioneuroma) CRC risk can be variable |
PTEN Mutations detected in 65–80% |
Colonoscopy q 3–5 years, beginning age 30–35y Mammogram and Breast MRI, yearly, starting at age 30–35y. Annual thyroid ultrasound starting by age 18y |
Expert Opinion13,30 |
CRC= colorectal cancer
MMR= Mismatch repair
MSI= Microsatellite Instability
Reference: *NCCN Clinical Practice Guidelines in Oncology Version 1.2013, nccn.org
Hereditary CRC Syndromes associated with Mutations in Known Genes
Lynch Syndrome
Lynch Syndrome is the most common of the hereditary CRC syndromes and has been implicated in 2–4% of CRC cases.4 It is characterized by a predisposition to develop colorectal, endometrial and selected other cancers, which often arise at young ages. Affected families frequently include multiple relatives with cancer and display autosomal dominant pattern of inheritance. Estimated lifetime risks for developing cancer range from 22% to 75% for CRC and 32% to 45%, for endometrial cancer,5–8 with risks for other cancers (including ovarian, gastric, small intestinal, urinary tract, brain, pancreatic, and sebaceous neoplasms of the skin) also increased. While the Amsterdam criteria (3 cases of CRC, involving 2 generations with one case diagnosed at age less than 50 years)9 were originally used for identifying affected families, the discovery of germline mutations in the DNA mismatch repair (MMR) genes hMLH1, hMSH2, hMSH6, and hPMS2 elucidated the genetic basis of the disease. More than 90% of Lynch-associated CRC tumors exhibit phenotypes of high DNA microsatellite instability (MSI-H) and loss of expression of MMR proteins MLH1, MSH2, MSH6 or PMS2 by immunohistochemistry (IHC). Although these tumors tend to develop at younger ages and feature accelerated neoplastic progression, early initiation of colonoscopy with frequent surveillance intervals is effective in reducing CRC incidence and mortality and has altered the natural history of the disease for many affected families.10–12 Consensus recommendations for CRC screening include intensive surveillance with colonoscopy every 1–2 years starting at age 20 to 25 years.13,14 Although evidence supporting the effectiveness of screening for extracolonic cancers is limited, upper GI endoscopy at age 3–5 year intervals starting at 30 to 35 years, and annual endometrial biopsy and transvaginal ultrasound for women starting at age 30 to 35 years may be considered14 (Table 1).
The distinctive MMR deficient tumor phenotype, high cancer risk and effectiveness of surveillance make Lynch Syndrome an attractive target for population-based screening among individuals diagnosed with CRC. Systematic screening of tumors for MMR deficiency, using MSI and/or IHC, has emerged as a sensitive means to identify individuals who develop CRC as a result of heritable MMR mutations. Models suggest the strategy of screening all CRC cases for features of Lynch Syndrome is cost-effective, mainly as a result of benefits derived from implementation of early interventions that prevent cancers in at-risk family members.15,16 The use of risk assessment models which rely on personal and family cancer history to estimate an individual’s probability of carrying a MMR gene mutation (eg. PREMM1,2,6,17 and MMRPro18) has also been proposed as a cost effective means for screening all individuals for Lynch Syndrome, regardless of cancer status.19
Familial Adenomatous Polyposis (FAP)
Familial adenomatous polyposis (FAP) is the second most common of the inherited CRC syndromes following Lynch syndrome and accounts for approximately 1% of newly diagnosed CRC cases. In cases of “classic” polyposis, the phenotype of 100s to 1000s of adenomatous polyps in the colon makes FAP easily recognizable. In most cases, affected individuals develop colorectal adenomas by the second or third decade of life. Lifetime risk for CRC is estimated to exceed 90% for individuals who do not undergo surgical colectomy. Over half of individuals with FAP develop adenomas in the upper gastrointestinal tract and cancers of the duodenum/ampulla are the second leading cause of cancer death for FAP patients, following CRC. Risks for other cancers, including papillary thyroid cancer, adrenal carcinomas, and central nervous system tumors are also increased. Intra-abdominal desmoid tumors appear in some individuals with FAP and can be associated with significant morbidity and mortality.
In 90% of classic FAP cases, germline mutations in the adenomatous polyposis coli (APC) gene can be detected through clinical genetic testing. APC is a tumor suppressor gene involved in the WNT signaling pathway and somatic loss of function of APC is one of the first steps in the colorectal adenoma-carcinoma sequence. Individuals with germline mutations in APC develop multiple adenomas at very young ages as a result of inactivation of the remaining allele in colonic epithelial cells. Although FAP is associated with autosomal dominant inheritance, approximately 30% of affected individuals report no family history of the disease. While most of these represent de-novo APC mutations, biallelic mutations in MutYH20, a DNA base excision repair gene involved in the repair of oxidative damage, have also been identified in some patients with classic polyposis. Unlike FAP, MutYH-associated polyposis (MAP) is associated with an autosomal-recessive pattern of inheritance. Biallelic MutYH mutation carriers can exhibit a wide range of phenotypes;while some individuals have colonic and extracolonic manifestations indistinguishable from classic FAP, most cases are associated with attenuated pheonotypes exhibiting fewer than 100 adenomas.21,22 The Y179C and G396D mutations are the two most pathogenic altterations in MutYH in individuals of western European ancestry but other mutations are commonly reported among individuals of other races and ethnicities. Population based studies have identified monoallelic and biallelic MutYH mutations in 0.7% and 1.7% of unselected CRC cases, respectively.23 Biallelic gene mutation carriers have a 28-fold increased risk of developing CRC compared to the general population whereas the risk in monoallelic carriers is increased by less than 2-fold.24
Although clinical genetic testing for individuals affected with adenomatous polyposis has been available since the early 1990s, the sensitivity of testing has improved over time. Full sequencing of the APC gene, in conjunction with multiplex ligation dependent probe amplification (MLPA) detects mutations in 90% of individuals with classic polyposis. In the absence of an identifiable APC mutation, testing for the Y165C and G382D mutations in MutYH is indicated, with full gene sequencing of MutYH recommended for individuals who are found to have one of these two mutations or whose racial/ethnic ancestry is not western European. However, approximately 1 in 10 individuals with the classic FAP phenotype do not have identifiable mutations in APC or MutYH.22 While there are reports of somatic mosaicism for APC mutations,25 this likely explains only a small fraction of cases. Efforts to identify other genes implicated in cases of classic FAP without APC or MutYH mutations are underway.
Identification of mutations in APC or MutYH in families with adenomatous polyposis has significant implications for at-risk family members. If a mutation is identified in an affected individual, predictive genetic testing of other family members provides an opportunity to identify those which require intensive surveillance. Individuals who are confirmed carriers of APC mutations should begin annual colorectal surveillance at age 10–12 years, while individuals who test negative for the known mutation in the family can have screening according to population-based guidelines. While the severity of polyposis phenotypes among carriers of MutYH mutations can be variable, it is generally recommended that biallelic carriers undergo surveillance similar to APC mutation carriers, while monoallelic carriers can wait to begin surveillance according to moderate-risk guidelines for CRC.13
Hamartomatous Polyposis Syndromes
Hamartomatous polyposis, defined as greater than 3–5 hamartomatous polyps in the gastrointestinal tract, is implicated in less than 0.5% of all CRC cases. Although rare, the hamartomatous polyposis syndromes can be associated with increased risks for a variety of extraintestinal cancers and the diagnosis has significant implications for medical management. Consequently, the finding of one or more gastrointestinal hamartomas in the setting of a suspicious family history of cancer is considered an indication for genetic evaluation.
Peutz-Jeghers Syndrome (PJS) is characterized by multiple intestinal hamartomatous polyps, mucocutaneous pigmentation, and a high lifetime risk of gastrointestinal, pancreatic, and breast cancers. PJS is quite rare with incidence estimated at 1 in 150,000. The clinical diagnosis requires two or more of the following features: (1) mucocutaneous pigmentation (eg. freckling in mouth/lips, fingers) (2) 2 or more Peutz-Jeghers type gastrointestinal hamartomas or (3) family history of PJS.26 Individuals with PJS can develop hamartomatous polyps throughout their gastrointestinal tract and often present with symptoms of abdominal pain, gastrointestinal bleeding with anemia, intestinal obstruction, or intussusception. Lifetime risk for developing any cancer by age 70 has been estimated at 85–90%, with gastrointestinal cancers (colon, small intestine, stomach, pancreas) and breast seen most commonly.26,27 Mutations in the serine threonine kinase 11 (STK-11 also known as LKB-1) tumor suppressor gene involved in the mTOR pathway have been found in approximately 50–70% of PJS patients. While genetic testing can be helpful in confirming the diagnosis, it is not informative in many individuals with a clinical diagnosis of PJS. To date no genes other than STK-11 have been associated with PJS. Patients with PJS and their at-risk relatives require frequent endoscopic surveillance for removal of polyps throughout the GI tract, as well as screening for extraintestinal cancers (Table 1). While techniques such as CT/MR enterography and capsule endoscopy have facilitated detection of small bowel polyps in patients with PJS, the evidence to support use of one imaging modality over others is limited.
Juvenile Polyposis Syndrome (JPS) is characterized by multiple (3–5 or more) juvenile polyps and increased risk for gastrointestinal cancers. Affected individuals often present in childhood with symptoms of anemia, bleeding, or abdominal pain. Juvenile polyps are most often found in the stomach or colon and less often in the small bowel. Certain congenital abnormalities, including cardiac valvular disease and/or atrial and ventricular septal defects, can be seen in some affected families. Individuals with JPS are at increased risk for gastric cancer and CRC, with lifetime risk approaching 40–50%.28
Mutations in the SMAD4 and BMPR1a genes are found in approximately 50% of individuals with a clinical diagnosis of JPS. These genes encode proteins involved in the transforming growth factor (TGF) beta signaling pathway. More recently mutations in ENG, also involved in the TGF-beta pathway, have been found in a small number of patients with JPS.29 Although clinical genetic testing can be useful for risk-stratifying relatives when a gene mutation is identified in the family, for many patients who meet clinical criteria for JPS genetic testing is clinically uninformative. Individuals with a personal or family history of juvenile polyposis should begin upper and lower endoscopy starting at age 15, with a goal of removal of all large polyps.
Cowden Syndrome, also known as Bannayan Riley Ruvalcaba Syndrome (BRRS) and PTEN-Hamartoma Tumor Syndrome (PHTS), has been associated with a broad range of clinical phenotypes. It is caused by mutations the phosphatase and tensin homolog (PTEN) gene which confers increased risk for cancers, most commonly breast, thyroid, and endometrial. Although Cowden Syndrome is often included among the colorectal hamartoma syndromes, there is significant variability in the colonic polyp phenotype. A retrospective review of findings of gastrointestinal endoscopy exams in 64 individuals with PTEN mutations reported heterogeneity in polyp number and histologic types (hamartomas, adenomas, serrated polyps, hyperplastic polyps, and ganglioneuromas); however the finding that 13% had been diagnosed with CRC at less than 50 years30 suggests early colonoscopic screening may be justified in these individuals.
Familial CRC without identifiable gene mutations
Familial Colorectal Cancer Type X (FCCX)
Most CRC cases with a familial component are referred for genetic evaluation because of the striking history of cancer affecting multiple family members, often at young ages. While the clinical phenotypes of some of these families resemble those of known hereditary syndromes, such as Lynch Syndrome or FAP, others appear to constitute distinct disease entities. Families with history of CRC that meet Amsterdam Criteria were originally referred to as hereditary “non-polyposis” colorectal cancer (HNPCC) to distinguish them from those with “polyposis” phenotypes. After the discovery of the role of MMR gene mutations in the pathogenesis of Lynch Syndrome, HNPCC families could be subdivided based on whether their CRC tumors had MMR deficient or MMR proficient phenotypes. Amsterdam criteria families with MMR proficient tumors have been found to differ from Lynch Syndrome families in a number of ways: 1) affected individuals tend to develop CRC at slightly older ages, 2) risks for extracolonic tumors do not seem to be increased, and 3) risk for CRC among relatives isincreased by only 2-fold.31 As a result, it appears these cases represent a disease entity distinct from Lynch syndrome, now referred to as Familial Colorectal Cancer Type X (FCCX).31
Approximately half of families that meet Amsterdam Criteria have Lynch Syndrome, and the remaining ones with MMR proficient CRC tumors without germline mutations in MMR genes are assumed to be FCCX. Although defining the genetic basis for FCCX has been a topic of intensive research, the cause for the increased risk of cancer in these families remains unknown.32 Genome Wide Association Studies (GWAS) have reported linkage to 4q, 8q, 12q, and 15q33 and a study of sibling pairs with microsatellite stable CRC found statistically significant linkage to 9q2234 but potential candidate genes have not been well-characterized. The difficulty in identifying genes implicated in FCCX has led some to suspect that it may not be a monogenic condition but rather a polygenic one resulting from the interaction of several low-penetrance gene variants.32 Another possibility is that epigenetic events which affect expression of oncogenes and tumor suppressor genes, rather than specific gene mutations themselves, may have a role in carcinogenesis. In comparing methylation profiles of CRC tumors, MMR proficient tumors from FCCX families had lower levels of global methylation at long interspersed nucleotide element-1 (LINE-1) when compared with sporadic CRC tumors and MMR deficient tumors associated with Lynch Syndrome.35,36 While epigenetic alterations have not generally been considered heritable, germline hypermethylation of MLH1 has been identified in a few families with presumed Lynch Syndrome without germline MLH1 mutations37,38 and some have proposed that hypomethylation may also be implicated in familial colorectal carcinogenesis.39,40
The management of families with FCCX remains a topic of debate. Since CRC risk appears lower than for Lynch Syndrome and risk for extracolonic cancers does not appear increased, recommendations based on expert opinion suggest initiating colonoscopy in at-risk individuals 5–10 years earlier than the youngest CRC in the family and repeating at least every 5 years.31
“Attenuated” Adenomatous Polyposis
Attenuated adenomatous polyposis is defined clinically as greater than 10 but less than 100 adenomatous colonic polyps. Because of the significant variability observed among the polyposis phenotypes , current guidelines recommend that genetic evaluation for APC and MutYH mutations be considered for individuals with 10 or more adenomas.3,13 However, the yield of clinical genetic testing among individuals with attenuated polyposis is significantly lower than in cases of classic polyposis, and the likelihood of finding a mutation in APC or MutYH depends on the number of adenomatous polyps. A review of genetic test results among individuals with classic and attenuated polyposis referred for clinical genetic testing found prevalence of mutations in APC and MutYH (biallelic) of 10% and 7% among individuals with 20–99 adenomas and 5% and 4% among those with 10–19 adenomas.22 While identification of mutations in APC and MutYH has implications for the management of family members, these data suggest that in the majority of cases of attenuated polyposis genetic testing is uninformative. The roles of other genetic or environmental factors in the pathogenesis of attenuated polyposis remain to be determined. The clinical management of these individuals focuses on removal of all adenomas, if possible, with surgical resection reserved for cases which cannot be managed endoscopically. The role of chemoprevention agents in the management of attenuated polyposis is being investigated.
Serrated Polyposis
Initially described as hyperplastic polyposis, the condition now known as Serrated Polyposis is characterized by large and/or multiple serrated polyps with few, if any, adenomas. Sessile serrated polyps/adenomas and traditional serrated polyps are found in 2% of individuals41 and are believed to be the precursor lesions of serrated colorectal cancers which account for approximately 15–30 % of CRC tumors.42,43 Estimates for CRC risk associated with serrated polyposis range from 7–50% and vary with phenotype.44,45 Until recently, serrated polyps were categorized as hyperplastic polyps and were not believed to have malignant potential. However sessile serrated polyps/adenomas and traditional serrated polyps have histopathological characteristics which distinguish them from hyperplastic polyps and their association with an increased risk for CRC has led to the reclassification as premalignant lesions.45
The mechanism for carcinogenesis in the serrated pathway of colorectal neoplasia is presumed to be epigenetic hypermethylation of CpG islands resulting in silencing of tumor suppressor genes.45 These tumors are characterized by the phenotype of global hypermethylation at CpG islands (CIMP) and those with hypermethylation of the promoter for MLH1 are often MSI-H with loss of expression of MLH1 and PMS2 proteins. Unlike the MSI-H tumors which arise as a result of germline DNA MMR gene mutations in patients with Lynch Syndrome, the MSI-H tumors arising through the serrated pathway frequently have somatic mutations in the BRAF proto-oncogene and CIMP-high phenotypes. These serrated pathway CIMP-high tumors are more often found in the proximal colon and are more common in women and older individuals. Serrated Polyposis Syndrome (formerly referred to as Hyperplastic Polyposis Syndrome) has been defined as a distinct entity by the World Health Organization (WHO) on the basis of having any one of the following criteria:46 1) >5 serrated polyps proximal to the sigmoid colon, with at least 2 measuring >10mm; 2) any number of serrated polyps in the proximal colon in an individual who has a first-degree relative with serrated polyposis; or 3) >20 serrated polyps of any size, distributed throughout the colon. However, the use of terms hyperplastic polyposis and serrated polyposis interchangeably, along with revisions to the diagnostic criteria, have contributed to significant confusion in characterizing the Serrated Polyposis Syndrome. Furthermore, significant heterogeneity in clinical, endoscopic, and histologic features associated with these cases has raised concerns that these might not be part of a single syndrome, but may instead represent different disease entities associated with distinct epidemiologic and molecular characteristics.47
The genetic basis for serrated polyposis remains elusive. The prevalence of serrated polyps is higher in females, older individuals, and cigarette smokers which raises the question whether genetic or epigenetic factors are involved in pathogenesis. However, reports of familial cases of serrated polyposis and observations of increased CRC risk among first degree relatives of patients with serrated polyposis (standardized incidence ratio of approximately 5) suggest a possible hereditary component.48,49 Although biallelic MutYH mutations have been reported in some individuals meeting WHO criteria for serrated polyposis,50 clinical genetic testing in these patients has been low yield. Studies in individual families have reported linkage to loci on chromosomes 1p51 and 2q52; however no definitive candidate genes have been identified.
Based on expert consensus, the clinical management of patients with serrated polyposis is similar to that of attenuated polyposis, with the goal of removal of as many polyps as possible. In cases in which the polyp burden cannot be managed endoscopically, surgical resection may be considered.
Familial CRC “Not-otherwise specified”
Approximately 30% of CRC patients report having one or more relatives diagnosed with CRC, yet germline mutations in known cancer causing genes are implicated in only 5–6% of cases. A strong family history of cancer and/or CRC diagnosis at a young age are “red flags” which should prompt consideration of genetic testing. The National Comprehensive Cancer Network (NCCN) has proposed guidelines for identifying individuals at increased risk for CRC who may be candidates for further risk evaluation (Table 2). It is worth noting that these criteria are quite broad and could result in referral of large numbers of patients for genetic testing. For most individuals who do not meet clinical criteria for any of the hereditary CRC syndromes, testing for mutations in genes known to be associated with CRC risk will be uninformative. However, the sensitivity and specificity of clinical criteria for identifying mutation carriers are limited; similarly an uninformative result for one genetic test does not exclude the possibility that hereditary factors may be involved.
Table 2.
NCCN Criteria for Further Risk Evaluation for High Risk Syndromes Associated with CRC*
|
NCCN Clinical Practice Guidelines in Oncology Version 1.2013, nccn.org
To date, the search for causes for familial CRC has focused on identifying mutations in highly penetrant genes. As was the case for Lynch Syndrome, linkage analysis in affected families has continued to be instrumental in identifying loci associated with cancer risk. Linkage analysis conducted in sibling-pairs affected with CRC has identified chromosomal regions of interest, among them 9q22.34 Similarly, large population-based GWAS involving thousands of CRC cases and controls have identified additional potential loci including 8q23, 8q24, 9p24, 11q23,18q21 among others.53 However, these appear to be associated with relatively small effect sizes with relative risks ranging from 1.1 to 1.26 and resequencing has failed to identify common coding sequence variants. Furthermore, most of these single nucleotide polymorphisms (SNPs) appear to be located in regions of non-coding DNA, making it less likely that they are closely linked with genes associated with high risk for cancer. Consequently, many experts believe that most (if not all) of the highly penetrant cancer genes have already been discovered54 and that a large part of the variability in CRC risk results from the additive effects of combining common, less penetrant risk alleles and/or epigenetic regulation of gene expression. Polymorphisms in several genes, including TGF-beta receptor 1, methylenetetrahydrofolate reductase (MTHFR), N-acetyl transferase 1 and 2 (NAT1 and NAT2), and glutathione-S transferase Mu (GSTM1) have been implicated in modest increase in cancer risk through gene-environment interactions and/or modification of expression of other cancer-associated genes.55
Even though the etiology of most familial CRC remains unclear, novel genomic technologies such as next generation sequencing (NGS) are making it possible to exhaustively examine the whole genome and epigenome of individual patients, families, and large multinational cohorts of CRC cases. In the search for factors which influence CRC risk, investigators are considering a number of potential mechanisms for causality, including traditional autosomal dominant inheritance, autosomal recessive inheritance, intermediate/low penetrance susceptibility alleles, as well as the possibility of complex gene-gene and gene-environment interactions. Molecular characterization of CRC tumors using whole genome sequencing, methylation and gene expression analyses has provided a detailed outline of the different molecular pathways involved in carcinogenesis.56 Epigenetic alterations (including hypermethylation as well as hypomethylation) have been associated with risk for CRC and there is growing appreciation for the role of post-transcriptional gene regulation by microRNAs in pathogenesis of colorectal neoplasms.54 The expectation is that identification of mechanisms involved in carcinogenesis will facilitate discovery of factors affecting cancer risk as well as therapeutic targets for cancer treatment and prevention.
Clinical Approach to Familial CRC: Past, Present and Future
Historically, making the diagnosis of a hereditary cancer syndrome has depended on clinicians to recognize specific clinical criteria and the process of discovery of the genetic basis for familial cancers has been painstaking and time-consuming. In the case of Lynch Syndrome, the timeline from describing the clinical features of affected families, to identifying the genes implicated in the pathogenesis of the cancers, to implementing clinical algorithms for population based screening has spanned more than 30 years. However NGS technologies offer opportunities to analyze the entire genome and epigenome rapidly and relatively inexpensively. As the $1,000 genome comes closer to becoming reality, we can expect that the timeframe for discovery of additional genomic factors implicated in familial CRC will be greatly accelerated.
Growing awareness of the association of family history with cancer risk, along with direct-to-consumer marketing of genetic tests, has fueled patients’ interest in clinical genetic testing. As the experience with genetic testing for mutations in BRCA1 and BRCA2 associated with hereditary breast ovarian cancer syndrome has shown, we may expect that growing numbers of patients with and without a personal diagnosis of CRC will seek genetic testing to guide medical decision-making about surveillance and perhaps even chemoprevention. The integration of molecular diagnostic techniques into clinical laboratories will continue to make genetic testing more widely available. Although the high cost of genetic testing (ranging from $300 to $2,000 for sequencing of individual genes) has been a major barrier, use of NGS technologies makes it possible to sequence multiple genes simultaneously at lower cost. A number of clinical laboratories now offer multiplex genetic tests which include several pre-selected highly penetrant and moderately penetrant genes in cancer-specific panels. Although requesting mutation analysis of 14 or more genes associated with 8 different hereditary syndromes through one genetic panel test may seem fairly straightforward, the interpretation of these results can be complicated, particularly with regard to determining the clinical significance of test results which identify one or more mutations in genes with low or moderate penetrance and/or genetic variants of uncertain pathogenicity.59 The work to compile and analyze the data needed to re-classify genetic variants and quantify the magnitude of cancer risks will continue to require multidisciplinary collaborations between clinicians, geneticists, molecular biologists, and statisticians.
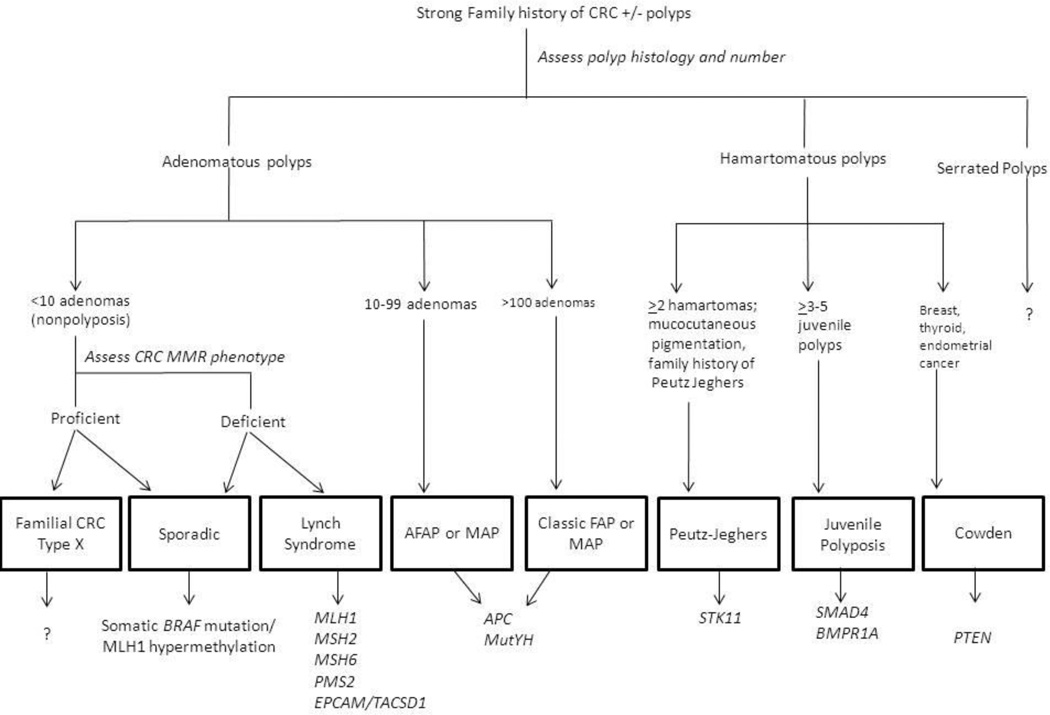
Consensus statements from several professional societies have recommended that genetic testing for hereditary cancer syndromes be performed in conjunction with pre and post-test counseling by providers with expertise in genetic testing whenever possible;3,59–61 however increasing demand for testing and limited availability of clinical “genetics experts” make this model increasingly impractical. In this era of personalized medicine, oncologists, gastroenterologists, and primary care doctors will be expected to possess a working knowledge of the diagnostic evaluation and management of hereditary cancer syndromes. Yet even as clinical genetic tests become more widely available, it is important to recognize that ordering a genetic test is only one small part of cancer risk assessment. The clinical approach to patients with presumed familial CRC should still begin with a comprehensive assessment of patients’ personal and family history. As cancer predisposition syndromes associated with risk for CRC frequently include other cancers, it is important to elicit information about all cancer diagnoses in first and second degree relatives. Clinical presentations of hereditary CRC syndromes can vary and the differential diagnosis may be broad. Reviewing pathology and endoscopy reports to ascertain size, number, and histology of colorectal polyps and categorizing the number of adenomas (<10, 10–100, and 100s–1000s) can be useful for classifying cases into non-polyposis, attenuated polyposis, and classic polyposis phenotypes, respectively (figure 1.) Routine screening of CRC tumors for MMR deficiency with mechanisms for ensuring those at risk for Lynch Syndrome undergo genetic evaluation is becoming the expected standard of care.
Figure 1.
Approach to Patients with Familial CRC
Legend:
MMR: mismatch repair
AFAP: attenuated familial adenomatous polyposis
MAP: MutYH associated polyposis
FAP: familial adenomatous polyposis
The ultimate goal of screening for familial CRC is to identify high risk individuals early enough to change the natural history of the disease. In the two decades since genetic testing for FAP and Lynch Syndrome became clinically available, we have seen how pre-symptomatic risk assessment and implementation of specialized endoscopic screening and/or surgery has resulted in dramatic improvements in clinical outcomes for many of these families. However current strategies to identify individuals at risk for hereditary CRC focus primarily on evaluating patients who already have a cancer diagnosis. Cost-effectiveness models have demonstrated that a substantial portion of the benefit of genetic testing is derived from preventing cancers among at-risk family members.16 Ensuring that information about the diagnosis of a hereditary cancer syndrome reaches other family members remains a clinical challenge. How results of genetic testing are interpreted by patients and their physicians and how this information influences health behaviors, clinical management, and outcomes will continue to be a focus of implementation research.
Although current algorithms for CRC risk stratification rely primarily upon an individual’s age, family history and personal history of colorectal neoplasia; we know that the sensitivity and specificity of clinical criteria for identifying individuals with hereditary cancer syndromes is limited. The Bethesda Guidelines identify nearly 20% of CRC patients as potentially high risk for Lynch Syndrome57 and risk assessment tools suggest as many as 15–20% of individuals referred for screening colonoscopy may meet criteria for genetic evaluation.58 As awareness of the role of genetics in cancer increases, we can expect that more people without cancer diagnoses will seek genetic testing. As we learn more about the effects of lower penetrance susceptibility genes and gene-environment interactions on risk for CRC, approaches to risk assessment which integrate both family history and genomic data may have a bigger role in clinical care.
At present we are able to identify a genetic cause in only a minority of familial CRC cases. The expectation is that knowledge of the genomic factors implicated in familial, as well as sporadic, CRC will improve our ability to risk stratify individuals, making it possible to tailor screening and surveillance recommendations on the basis of individual patients’ personal history, family history and genomic risk profile. Discovery of novel heritable factors associated with risk for CRC will not only enhance our understanding of the mechanisms of disease but will also guide strategic approaches to cancer prevention and therapeutics.
Acknowledgments
Funding support: NIH/NCI K07CA120448-5 (E. Stoffel), NIH/NCI K07 CA151769 (F. Kastrinos) Louis V. Gerstner, Jr Scholars Program (F. Kastrinos).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: None
References
- 1.Levin B, Lieberman DA, McFarland B, et al. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. Gastroenterology. 2008;134:1570–1595. doi: 10.1053/j.gastro.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Fuchs CS, Giovannucci EL, Colditz GA, et al. A prospective study of family history and the risk of colorectal cancer. N Engl J Med. 1994;331:1669–1674. doi: 10.1056/NEJM199412223312501. [DOI] [PubMed] [Google Scholar]
- 3.American Gastroenterological Association medical position statement: hereditary colorectal cancer and genetic testing. Gastroenterology. 2001;121:195–197. doi: 10.1053/gast.2001.25580. [DOI] [PubMed] [Google Scholar]
- 4.Hampel H, Frankel WL, Martin E, et al. Feasibility of screening for Lynch syndrome among patients with colorectal cancer. J Clin Oncol. 2008;26:5783–5788. doi: 10.1200/JCO.2008.17.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quehenberger F, Vasen HF, van Houwelingen HC. Risk of colorectal and endometrial cancer for carriers of mutations of the hMLH1 and hMSH2 gene: correction for ascertainment. J Med Genet. 2005;42:491–496. doi: 10.1136/jmg.2004.024299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jenkins MA, Baglietto L, Dowty JG, et al. Cancer risks for mismatch repair gene mutation carriers: a population-based early onset case-family study. Clin Gastroenterol Hepatol. 2006;4:489–498. doi: 10.1016/j.cgh.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Stoffel E, Mukherjee B, Raymond VM, et al. Calculation of risk of colorectal and endometrial cancer among patients with Lynch syndrome. Gastroenterology. 2009;137:1621–1627. doi: 10.1053/j.gastro.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aarnio M, Sankila R, Pukkala E, et al. Cancer risk in mutation carriers of DNA-mismatch-repair genes. Int J Cancer. 1999;81:214–218. doi: 10.1002/(sici)1097-0215(19990412)81:2<214::aid-ijc8>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 9.Vasen HF, Mecklin JP, Khan PM, et al. The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC) Dis Colon Rectum. 1991;34:424–425. doi: 10.1007/BF02053699. [DOI] [PubMed] [Google Scholar]
- 10.Jarvinen HJ, Aarnio M, Mustonen H, et al. Controlled 15-year trial on screening for colorectal cancer in families with hereditary nonpolyposis colorectal cancer. Gastroenterology. 2000;118:829–834. doi: 10.1016/s0016-5085(00)70168-5. [DOI] [PubMed] [Google Scholar]
- 11.Mecklin JP, Aarnio M, Laara E, et al. Development of colorectal tumors in colonoscopic surveillance in lynch syndrome. Gastroenterology. 2007;133:1093–1098. doi: 10.1053/j.gastro.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 12.Vasen HF, Abdirahman M, Brohet R, et al. One to 2-year surveillance intervals reduce risk of colorectal cancer in families with Lynch syndrome. Gastroenterology. 2010;138:2300–2306. doi: 10.1053/j.gastro.2010.02.053. [DOI] [PubMed] [Google Scholar]
- 13.(NCCN) NCCN: Colorectal Cancer Screening. NCCN Clinical Practice Guidelines in Oncology nccn.org. 2012 [Google Scholar]
- 14.Lindor NM, Petersen GM, Hadley DW, et al. Recommendations for the care of individuals with an inherited predisposition to Lynch syndrome: a systematic review. Jama. 2006;296:1507–1517. doi: 10.1001/jama.296.12.1507. [DOI] [PubMed] [Google Scholar]
- 15.Moreira L, Balaguer F, Lindor N, et al. Identification of Lynch syndrome among patients with colorectal cancer. Jama. 2012;308:1555–1565. doi: 10.1001/jama.2012.13088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ladabaum U, Wang G, Terdiman J, et al. Strategies to identify the Lynch syndrome among patients with colorectal cancer: a cost-effectiveness analysis. Ann Intern Med. 2011;155:69–79. doi: 10.7326/0003-4819-155-2-201107190-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kastrinos F, Steyerberg EW, Mercado R, et al. The PREMM(1,2,6) model predicts risk of MLH1, MSH2, and MSH6 germline mutations based on cancer history. Gastroenterology. 2011;140:73–81. doi: 10.1053/j.gastro.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen S, Wang W, Lee S, et al. Prediction of germline mutations and cancer risk in the Lynch syndrome. Jama. 2006;296:1479–1487. doi: 10.1001/jama.296.12.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dinh TA, Rosner BI, Atwood JC, et al. Health benefits and cost-effectiveness of primary genetic screening for Lynch syndrome in the general population. Cancer Prev Res (Phila) 2011;4:9–22. doi: 10.1158/1940-6207.CAPR-10-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sieber OM, Lipton L, Crabtree M, et al. Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. N Engl J Med. 2003;348:791–799. doi: 10.1056/NEJMoa025283. [DOI] [PubMed] [Google Scholar]
- 21.Jo WS, Bandipalliam P, Shannon KM, et al. Correlation of polyp number and family history of colon cancer with germline MYH mutations. Clin Gastroenterol Hepatol. 2005;3:1022–1028. doi: 10.1016/s1542-3565(05)00411-8. [DOI] [PubMed] [Google Scholar]
- 22.Grover S, Kastrinos F, Steyerberg EW, et al. Prevalence and phenotypes of APC and MUTYH mutations in patients with multiple colorectal adenomas. Jama. 2012;308:485–492. doi: 10.1001/jama.2012.8780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balaguer F, Castellvi-Bel S, Castells A, et al. Identification of MYH mutation carriers in colorectal cancer: a multicenter, case-control, population-based study. Clin Gastroenterol Hepatol. 2007;5:379–387. doi: 10.1016/j.cgh.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 24.Theodoratou E, Campbell H, Tenesa A, et al. A large-scale meta-analysis to refine colorectal cancer risk estimates associated with MUTYH variants. Br J Cancer. 2010;103:1875–1884. doi: 10.1038/sj.bjc.6605966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aretz S, Stienen D, Friedrichs N, et al. Somatic APC mosaicism: a frequent cause of familial adenomatous polyposis (FAP) Hum Mutat. 2007;28:985–992. doi: 10.1002/humu.20549. [DOI] [PubMed] [Google Scholar]
- 26.van Lier MG, Wagner A, Mathus-Vliegen EM, et al. High cancer risk in Peutz-Jeghers syndrome: a systematic review and surveillance recommendations. Am J Gastroenterol. 2010;105:1258–1264. doi: 10.1038/ajg.2009.725. author reply 1265. [DOI] [PubMed] [Google Scholar]
- 27.Giardiello FM, Trimbath JD. Peutz-Jeghers syndrome and management recommendations. Clin Gastroenterol Hepatol. 2006;4:408–415. doi: 10.1016/j.cgh.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 28.Howe JR, Mitros FA, Summers RW. The risk of gastrointestinal carcinoma in familial juvenile polyposis. Ann Surg Oncol. 1998;5:751–756. doi: 10.1007/BF02303487. [DOI] [PubMed] [Google Scholar]
- 29.Sweet K, Willis J, Zhou XP, et al. Molecular classification of patients with unexplained hamartomatous and hyperplastic polyposis. Jama. 2005;294:2465–2473. doi: 10.1001/jama.294.19.2465. [DOI] [PubMed] [Google Scholar]
- 30.Heald B, Mester J, Rybicki L, et al. Frequent gastrointestinal polyps and colorectal adenocarcinomas in a prospective series of PTEN mutation carriers. Gastroenterology. 2010;139:1927–1933. doi: 10.1053/j.gastro.2010.06.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindor NM, Rabe K, Petersen GM, et al. Lower cancer incidence in Amsterdam-I criteria families without mismatch repair deficiency: familial colorectal cancer type X. Jama. 2005;293:1979–1985. doi: 10.1001/jama.293.16.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindor NM. Familial colorectal cancer type X: the other half of hereditary nonpolyposis colon cancer syndrome. Surg Oncol Clin N Am. 2009;18:637–645. doi: 10.1016/j.soc.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cicek MS, Cunningham JM, Fridley BL, et al. Colorectal cancer linkage on chromosomes 4q21, 8q13, 12q24, and 15q22. PLoS One. 2012;7:e38175. doi: 10.1371/journal.pone.0038175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gray-McGuire C, Guda K, Adrianto I, et al. Confirmation of linkage to and localization of familial colon cancer risk haplotype on chromosome 9q22. Cancer Res. 2010;70:5409–5418. doi: 10.1158/0008-5472.CAN-10-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pavicic W, Joensuu EI, Nieminen T, et al. LINE-1 hypomethylation in familial and sporadic cancer. J Mol Med (Berl) 2012;90:827–835. doi: 10.1007/s00109-011-0854-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goel A, Xicola RM, Nguyen TP, et al. Aberrant DNA methylation in hereditary nonpolyposis colorectal cancer without mismatch repair deficiency. Gastroenterology. 2010;138:1854–1862. doi: 10.1053/j.gastro.2010.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hitchins MP, Wong JJ, Suthers G, et al. Inheritance of a cancer-associated MLH1 germ-line epimutation. N Engl J Med. 2007;356:697–705. doi: 10.1056/NEJMoa064522. [DOI] [PubMed] [Google Scholar]
- 38.Suter CM, Martin DI, Ward RL. Germline epimutation of MLH1 in individuals with multiple cancers. Nat Genet. 2004;36:497–501. doi: 10.1038/ng1342. [DOI] [PubMed] [Google Scholar]
- 39.Ogino S, Nishihara R, Lochhead P, et al. Prospective study of family history and colorectal cancer risk by tumor LINE-1 methylation level. J Natl Cancer Inst. 2013;105:130–140. doi: 10.1093/jnci/djs482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antelo M, Balaguer F, Shia J, et al. A high degree of LINE-1 hypomethylation is a unique feature of early-onset colorectal cancer. PLoS One. 2012;7:e45357. doi: 10.1371/journal.pone.0045357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lash RH, Genta RM, Schuler CM. Sessile serrated adenomas: prevalence of dysplasia and carcinoma in 2139 patients. J Clin Pathol. 2010;63:681–686. doi: 10.1136/jcp.2010.075507. [DOI] [PubMed] [Google Scholar]
- 42.Leggett B, Whitehall V. Role of the serrated pathway in colorectal cancer pathogenesis. Gastroenterology. 2010;138:2088–2100. doi: 10.1053/j.gastro.2009.12.066. [DOI] [PubMed] [Google Scholar]
- 43.Snover DC. Update on the serrated pathway to colorectal carcinoma. Hum Pathol. 2011;42:1–10. doi: 10.1016/j.humpath.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 44.Boparai KS, Mathus-Vliegen EM, Koornstra JJ, et al. Increased colorectal cancer risk during follow-up in patients with hyperplastic polyposis syndrome: a multicentre cohort study. Gut. 2010;59:1094–1100. doi: 10.1136/gut.2009.185884. [DOI] [PubMed] [Google Scholar]
- 45.Rex DK, Ahnen DJ, Baron JA, et al. Serrated lesions of the colorectum: review and recommendations from an expert panel. Am J Gastroenterol. 2012;107:1315–1329. doi: 10.1038/ajg.2012.161. quiz 1314, 1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Snover DC, Ahnen D, Burt R, et al. Serrated polyps of the colon and rectum and serrated polyposis. In: Bosman FTCF, Hruban R, et al., editors. WHO Classification of Tumours of the Digestive System. ed 4. Lyon, France: IARC; 2010. pp. 160–165. [Google Scholar]
- 47.Kalady MF, Jarrar A, Leach B, et al. Defining phenotypes and cancer risk in hyperplastic polyposis syndrome. Dis Colon Rectum. 2011;54:164–170. doi: 10.1007/DCR.0b013e3181fd4c15. [DOI] [PubMed] [Google Scholar]
- 48.Win AK, Walters RJ, Buchanan DD, et al. Cancer risks for relatives of patients with serrated polyposis. Am J Gastroenterol. 2012;107:770–778. doi: 10.1038/ajg.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boparai KS, Reitsma JB, Lemmens V, et al. Increased colorectal cancer risk in first-degree relatives of patients with hyperplastic polyposis syndrome. Gut. 2010;59:1222–1225. doi: 10.1136/gut.2009.200741. [DOI] [PubMed] [Google Scholar]
- 50.Boparai KS, Dekker E, Van Eeden S, et al. Hyperplastic polyps and sessile serrated adenomas as a phenotypic expression of MYH-associated polyposis. Gastroenterology. 2008;135:2014–2018. doi: 10.1053/j.gastro.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 51.Rashid A, Houlihan PS, Booker S, et al. Phenotypic and molecular characteristics of hyperplastic polyposis. Gastroenterology. 2000;119:323–332. doi: 10.1053/gast.2000.9361. [DOI] [PubMed] [Google Scholar]
- 52.Roberts A, Nancarrow D, Clendenning M, et al. Linkage to chromosome 2q32.2-q33.3 in familial serrated neoplasia (Jass syndrome) Fam Cancer. 2011;10:245–254. doi: 10.1007/s10689-010-9408-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tenesa A, Dunlop MG. New insights into the aetiology of colorectal cancer from genome-wide association studies. Nat Rev Genet. 2009;10:353–358. doi: 10.1038/nrg2574. [DOI] [PubMed] [Google Scholar]
- 54.Goel A, Boland CR. Recent insights into the pathogenesis of colorectal cancer. Curr Opin Gastroenterol. 2010;26:47–52. doi: 10.1097/MOG.0b013e328332b850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jasperson KW, Tuohy TM, Neklason DW, et al. Hereditary and familial colon cancer. Gastroenterology. 2010;138:2044–2058. doi: 10.1053/j.gastro.2010.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kastrinos F, Allen JI, Stockwell DH, et al. Development and validation of a colon cancer risk assessment tool for patients undergoing colonoscopy. Am J Gastroenterol. 2009;104:1508–1518. doi: 10.1038/ajg.2009.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Domchek SM, Bradbury A, Garber JE, et al. Multiplex Genetic Testing for Cancer Susceptibility: Out on the High Wire Without a Net? J Clin Oncol. 2013 doi: 10.1200/JCO.2012.46.9403. [DOI] [PubMed] [Google Scholar]
- 60.Robson ME, Storm CD, Weitzel J, et al. American Society of Clinical Oncology policy statement update: genetic and genomic testing for cancer susceptibility. J Clin Oncol. 2010;28:893–901. doi: 10.1200/JCO.2009.27.0660. [DOI] [PubMed] [Google Scholar]
- 61.Genetic testing for colon cancer: joint statement of the American College of Medical Genetics and American Society of Human Genetics. Joint Test and Technology Transfer Committee Working Group. Genet Med. 2000;2:362–366. doi: 10.1097/00125817-200011000-00012. [DOI] [PMC free article] [PubMed] [Google Scholar]