In growing Dictyostelium discoideum, PIP3 is unnecessary for migration toward folate and actively inhibits chemotaxis and pseudopod formation by promoting macropinocytosis.
Abstract
In eukaryotic chemotaxis, the mechanisms connecting external signals to the motile apparatus remain unclear. The role of the lipid phosphatidylinositol 3,4,5-trisphosphate (PIP3) has been particularly controversial. PIP3 has many cellular roles, notably in growth control and macropinocytosis as well as cell motility. Here we show that PIP3 is not only unnecessary for Dictyostelium discoideum to migrate toward folate, but actively inhibits chemotaxis. We find that macropinosomes, but not pseudopods, in growing cells are dependent on PIP3. PIP3 patches in these cells show no directional bias, and overall only PIP3-free pseudopods orient up-gradient. The pseudopod driver suppressor of cAR mutations (SCAR)/WASP and verprolin homologue (WAVE) is not recruited to the center of PIP3 patches, just the edges, where it causes macropinosome formation. Wild-type cells, unlike the widely used axenic mutants, show little macropinocytosis and few large PIP3 patches, but migrate more efficiently toward folate. Tellingly, folate chemotaxis in axenic cells is rescued by knocking out phosphatidylinositide 3-kinases (PI 3-kinases). Thus PIP3 promotes macropinocytosis and interferes with pseudopod orientation during chemotaxis of growing cells.
Introduction
Eukaryotic cells sense chemoattractants using transmembrane receptors. When receptors detect attractants, they activate intracellular second messengers that relay information to the molecules that drive cell movement. One principal group of attractants, including cAMP in Dictyostelium discoideum and fMLP in mammalian neutrophils, act through on G proteins. Receptors with bound attractant cause G proteins to split from heterotrimers into active GTP-bound α subunits and βγ subunits, and these free subunits in turn activate further intracellular messengers. In cells such as neutrophils, with a vast range of receptors, this process simplifies the cell’s responses by limiting the number of active species that cause intracellular effects. In D. discoideum, one of the best organisms for studying chemotaxis (King and Insall, 2009; Cai and Devreotes, 2011), this simplification is even clearer: there are single genes for β and γ subunits (Wu et al., 1995; Zhang et al., 2001), and the βγ subunit is thought to mediate most responses. Thus, in theory all attractants (for example, cAMP and folate) should elicit fundamentally similar chemotaxis (Srinivasan et al., 2013). However folate chemotaxis is often difficult to measure, for reasons explored in this paper.
Cells can be steered using many mechanisms. In shallow gradients, the migration process is fundamentally random, and attractants can bias multiple features of the normal motility cycle, for example the direction of new protrusions after pseudopod splitting (Bosgraaf and Van Haastert, 2009), and the probability of old pseudopods being retracted (Andrew and Insall, 2007). However, the second messengers that are used are still not understood. In one model, phosphatidylinositol 3,4,5-trisphosphate (PIP3) is a “compass needle” (Bourne and Weiner, 2002); intracellular signal processing somehow resolves the ideal future direction of the cell, resulting in local generation of PIP3 by the p110 phosphatidylinositide 3-kinase (PI 3-kinase) family. Discrete patches of PIP3 appear at the leading edge of some cells during chemotaxis (Postma et al., 2004), which suggests that these represent the outcome of a nonlinear response, perhaps by discretizing spatial information to control protrusion (Cai and Devreotes, 2011). However, newer data have shown that PIP3 is not essential for chemotaxis in D. discoideum (Hoeller and Kay, 2007) or in neutrophils (Ferguson et al., 2007). This, among other data, has led to alternate models such as the pseudopod-centered model (Insall, 2010), in which multiple signaling processes act by biasing a normal, random pseudopod cycle. Despite this, the concept of a chemotactic compass (perhaps operating through an alternative second messenger) and a central role for PIP3 in chemotaxis are frequently cited in the literature.
More recently, PIP3 has been strongly associated with macropinocytosis (Posor et al., 2013), in which cells use actin-driven cups to endocytose large volumes of liquid. Macropinosomes are induced by growth factors such as PDGF in mammalian cells, and the small GTPase Ras, which directly activates PI 3-kinases, causes massive macropinocytosis when it is inappropriately activated (Commisso et al., 2013). Thus, macropinocytosis and PIP3 are associated.
Macropinocytosis also occurs in D. discoideum. In the wild, cells feed by phagocytosing bacteria. However, most laboratory work is done in axenic strains (“axenic” in this case means “able to grow in the absence of living prey”). These strains have been selected by growth in medium without bacteria, and are able to grow because of a constitutive high rate of macropinocytosis. D. discoideum macropinosomes are large structures (up to 5 µm) that efficiently take up liquid and soluble nutrients (Swanson, 2008). The mutations that allow axenic growth have been mapped to three separate loci, but their identity remains unknown (Clarke and Kayman, 1987). The use of axenic D. discoideum strains is so widespread that they are often, incorrectly, referred to as wild-type cells. In this work we show an unexpected conflict between chemotaxis and macropinocytosis that raises fundamental questions about the physiological role of PIP3.
Results and discussion
Axenic D. discoideum cells are defective in chemotaxis to folate
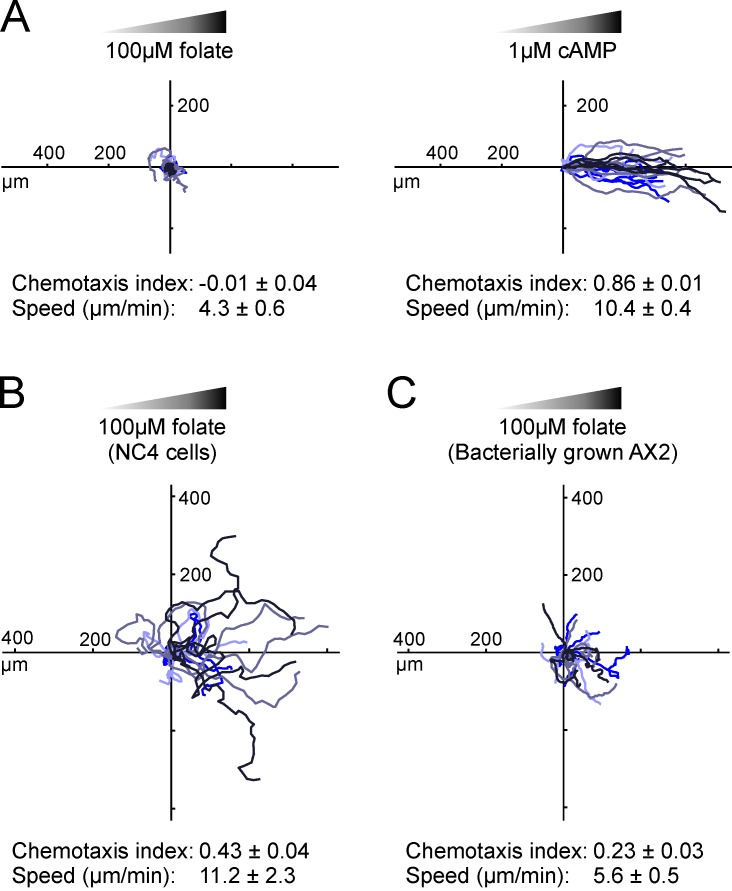
The main reason for studying chemotaxis in model organisms like D. discoideum is to find simple but generalizable results. It is therefore desirable to study multiple attractants to separate global from agonist-specific mechanisms. We have therefore studied chemotaxis toward folate. Compared with the well-studied cAMP system, folate uses different receptors and G protein α subunits (Srinivasan et al., 2013), which are expressed in growing cells that are not responsive to cAMP. However it has been difficult to measure folate chemotaxis with most assays (though under-agar and some micropipette assays succeed). To discover why, we exposed AX2 cells to folate gradients in a standard chemotaxis chamber (Muinonen-Martin et al., 2010). Under these conditions, starved cells chemotax efficiently toward cAMP. However, growing cells consistently failed to migrate up the folate gradient (Fig. 1 A). This was surprising, as folate is thought to be a potent attractant. To analyze the problem, we switched to using wild-type NC4, the parent strain of AX2, which does not have axenic mutations and must thus be grown on bacteria. In contrast to axenic cells, wild-type cells robustly migrated up the folate gradient (Fig. 1 B). AX2 cells grown on bacteria also show a little chemotaxis (Fig. 1 C), but still far less than NC4, which indicates that the loss of folate chemotaxis is caused by genetic differences between the wild type and the axenic AX2 strain.
Figure 1.
Folate chemotaxis is inefficient in axenic cells. Cells were grown under different conditions, then examined responding to linear attractant gradients in Insall chambers. Numbers are the means ± SEM of at least four independent experiments of at least 20 cells each. Tracks of cells from the same experiment have the same color. (A) Axenically cultivated AX2 cells responding to folate (left) and 4 h–starved cells responding to cAMP (right). (B) Bacterially grown NC4 cells (the parent of AX2) migrating toward folate. (C) Bacterially grown AX2 cells migrating toward folate.
Actin, pseudopod orientation, and PIP3
This defect in a strain that is widely used to study motility and chemotaxis is worrying, so we determined its cause. Chemoattractants steer cells by altering pseudopod behavior, so we measured pseudopod directions in axenic and nonaxenic cells. We also examined PIP3 because of its clear association with both macropinocytosis during axenic growth and chemotaxis. Cells were transfected with Lifeact-mRFP (Riedl et al., 2008) and PH-CRAC-GFP (Parent et al., 1998) to show actin pseudopods in red and PIP3 in green.
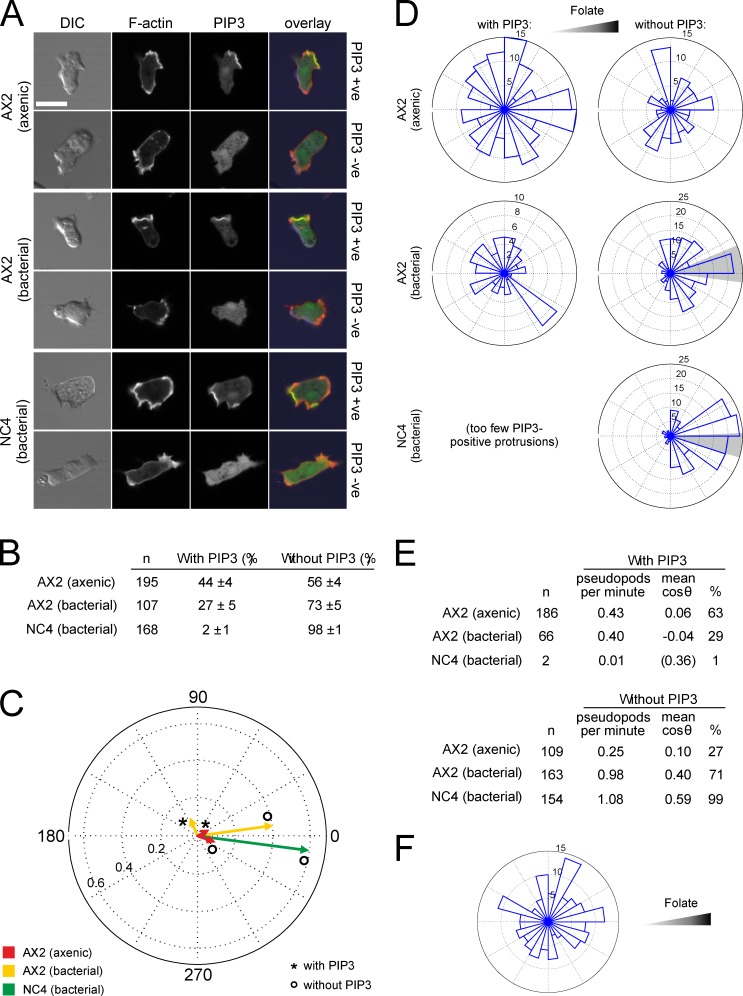
We first determined the fraction of F-actin–rich protrusions labeled with PIP3 during random migration (as observed by others, the PIP3-positive and -negative populations were essentially discrete, allowing an all-or-nothing score). In axenically cultivated AX2 cells, ∼50% of random protrusions accumulate PIP3 at the edge (Fig. 2, A and B). In AX2 grown on bacteria, PIP3-containing pseudopods are only about half as frequent, though these cells migrate more efficiently.
Figure 2.
PIP3–labeled protrusions do not orient up folate gradients. (A) AX2 and NC4 cells transfected with the F-actin reporter Lifeact-mRFP and the PIP3 reporter PH-CRAC-GFP were examined by confocal microscopy, and the presence or absence of a PIP3 patch in each cell was determined during random migration. Bar, 10 µm. (B) Numbers of pseudopods with and without PIP3 patches during random migration. Percentages are the means and standard deviations from three experiments. (C and D) Mean direction of protrusions during folate chemotaxis. (C) The displacements from the migration vectors of all pseudopods were measured, and the mean is shown in a circular plot. If all protrusions were extended in the same direction, a vector with length 1 would result. PIP3-containing pseudopods from NC4 were too rare to give a reliable direction. (D) Rose plots of the directions of all protrusions with and without PIP3. The shaded area indicates the 95% confidence interval of the mean angle. (E) Frequency and direction of pseudopods in C and D. (F) Rose plot of the direction of PIP3 patches that form macropinosomes during folate chemotaxis.
Expression of PIP3 markers in wild-type cells
It has previously not been practical to express ectopic genes in wild-type D. discoideum because standard selectable markers and promoters work poorly in bacterially grown cells. We constructed new vectors to allow expression of markers for PIP3 and F-actin in wild-type cells, and developed methods for transfection and selection using bacteria as food (see Materials and methods). We successfully cotransfected NC4 cells with Lifeact-mRFP and PH-CRAC-GFP to examine the distribution of F-actin and PIP3. To our surprise, however, PIP3-containing pseudopods were extremely rare in randomly moving wild-type cells (Fig. 2 B); 98% of pseudopods had no discernible PIP3. Thus, in growing wild-type D. discoideum, unlike in axenic cells, PIP3 appears not to be used at all in random migration, even though the cells migrate far more robustly.
PIP3 and folate chemotaxis
We next examined PIP3 during folate chemotaxis. Cells were exposed to a relatively steep folate gradient in a chemotaxis chamber, and the angle between each protrusion and the gradient was determined. As before, axenic AX2 cells did not show chemotaxis. The mean cosine of the angle between the protrusions and the chemoattractant gradient was close to zero, which indicates no alignment, regardless of whether PIP3 was present. Also, protrusions with PIP3 made a negligible contribution to net migration (Fig. 2, C, D, and E). Macropinosomes were not oriented by the folate gradient (Fig. 2 F).
As before, AX2 cells grown on bacteria chemotaxed weakly, and the proportion of pseudopods with PIP3 patches decreased substantially (from 63% to 29%; Fig. 2 E). Furthermore, PIP3-labeled protrusions did not orient up the gradient (Fig. 2 E). In contrast, the protrusions that showed no discernible PIP3 oriented up the folate gradient with a mean cosine of 0.40. Thus, unexpectedly, absence of PIP3 patches correlated with improved chemotaxis.
During chemotaxis, pseudopods in vegetative wild-type cells were still almost never labeled with PIP3 (Fig. 2 E). In a folate gradient, the protrusions of wild-type cells showed high directionality (mean cosine 0.59) despite the lack of PIP3 at the front (Fig. 2, C and D). Thus, in all conditions we observed, only PIP3-negative pseudopods are directed by folate.
PIP3 and macropinocytosis in wild-type and axenic cells
The negative correlation between the number of PIP3 patches and the chemotaxis efficiency was unexpected, as PIP3 signaling is thought to be fundamentally involved in directional sensing. However, PIP3 is involved in several other physiological processes, in particular macropinocytosis (Dormann et al., 2004; Hoeller et al., 2013). We speculated that this could negatively impact chemotaxis efficiency, and we therefore examined macropinocytosis and its regulation by PIP3 in more detail.
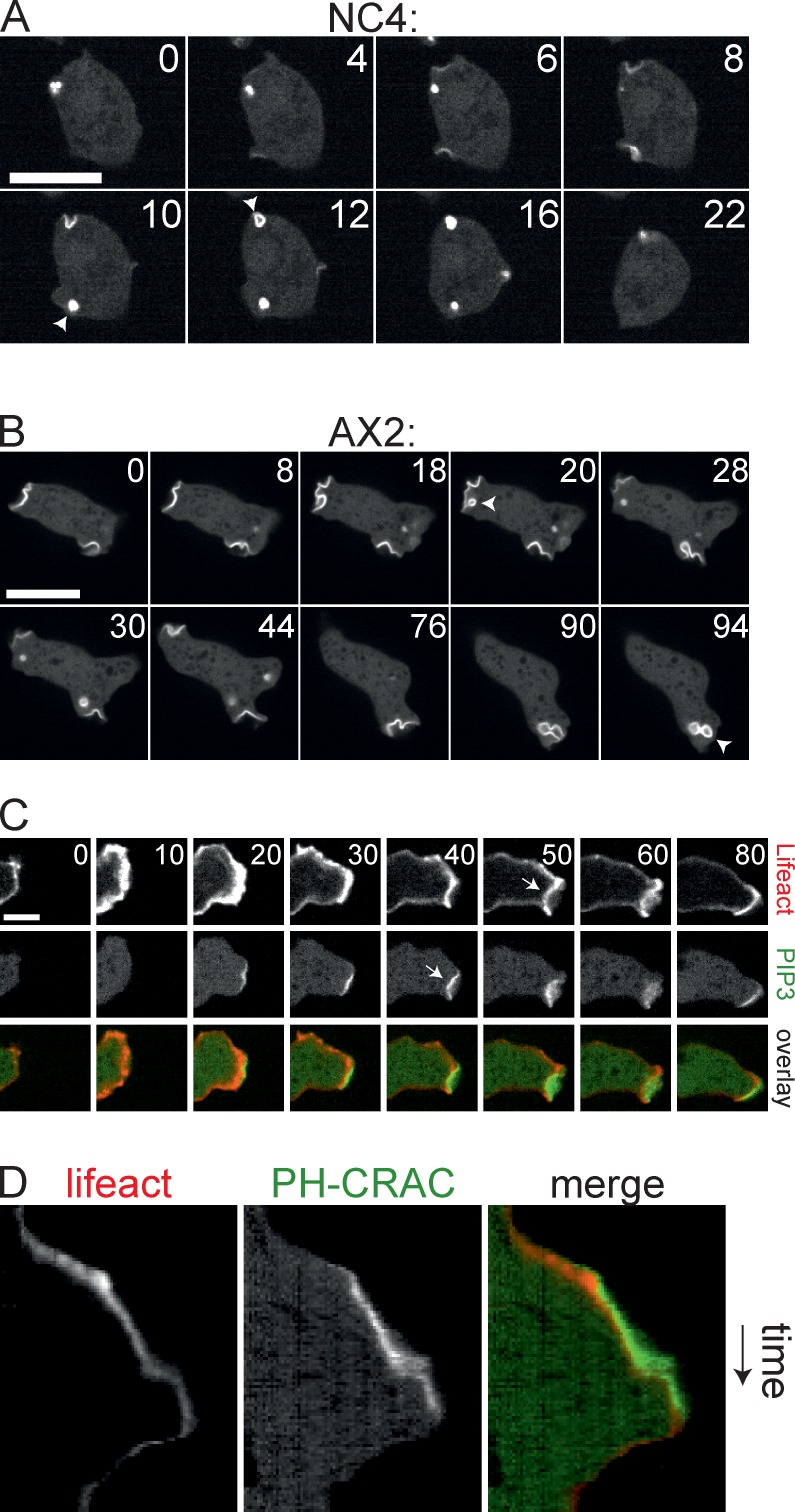
In wild-type cells, macropinocytosis could readily be visualized with the PIP3 marker PH-CRAC-GFP (Fig. 3 A), despite the substantially lower rate of fluid uptake. The PIP3 is strictly confined to the nascent vesicle; levels increase during invagination and reach a maximum as the vesicle closes. The entire process is very fast, with a mean time of 8 s between appearance of PIP3 and budding of the vesicle (n = 27, SD = 4 s). Macropinosomes in wild-type cells are small and do not contribute to cell movement, and are readily distinguishable from the large, convex, and spiky F-actin pseudopods.
Figure 3.
PIP3 patches organize macropinosomes. (A and B) Confocal series showing turnover of PIP3 patches in NC4 and axenically grown AX2 cells expressing the PIP3 marker PH-CRAC-GFP. Arrowheads mark budding. Bar, 10 µm. (C) Confocal series showing protrusions in an axenically cultivated AX2 cell expressing PH-CRAC-GFP and the F-actin marker Lifeact-mRFP. Arrows indicate negative curvature. Bar, 5 µm. This corresponds with Video 1. (D) Kymograph of a line drawn through the protrusion in C.
Unlike wild-type cells, axenic AX2 cells generate macropinosomes from preassembled PIP3 patches. The PI 3-kinases responsible for this process have recently been analyzed in depth (Hoeller et al., 2013); the patches are presumably the structures organized by PIKs 1 and 2. The diameter of the patches is a substantial fraction of the size of the cell (Fig. 3 B). Progression of negative curvature to drive vesicle formation frequently stalls, causing patches to remain planar or slightly inwardly curved for prolonged periods of time. Moreover, PIP3 patches often disappear without any vesicle formation, which indicates that macropinocytosis was aborted.
The PIP3 patches that are involved in axenic macropinocytosis are a similar size to pseudopods and recruit large amounts of F-actin. Nascent macropinosomes are therefore difficult to discriminate from pseudopods, especially if they abort before vesicle formation. This is further complicated because pseudopods and macropinosomes are able to interconvert. Fig. 3 C and Video 1 show an example. The protrusion is initially free of PIP3 and resembles a normal pseudopod. At 20 s, a PIP3 patch appears at the leading edge, and shortly after, negative curvature is induced at the site of the patch. The image plainly resembles a forming macropinocytic cup. However, it does not fully close and aborts after 80 s. A kymograph of this event (Fig. 3 D) shows that the forward movement of the leading edge reduces after the appearance of PIP3.
Suppressor of cAR mutations (SCAR) and F-actin organization in macropinosomes
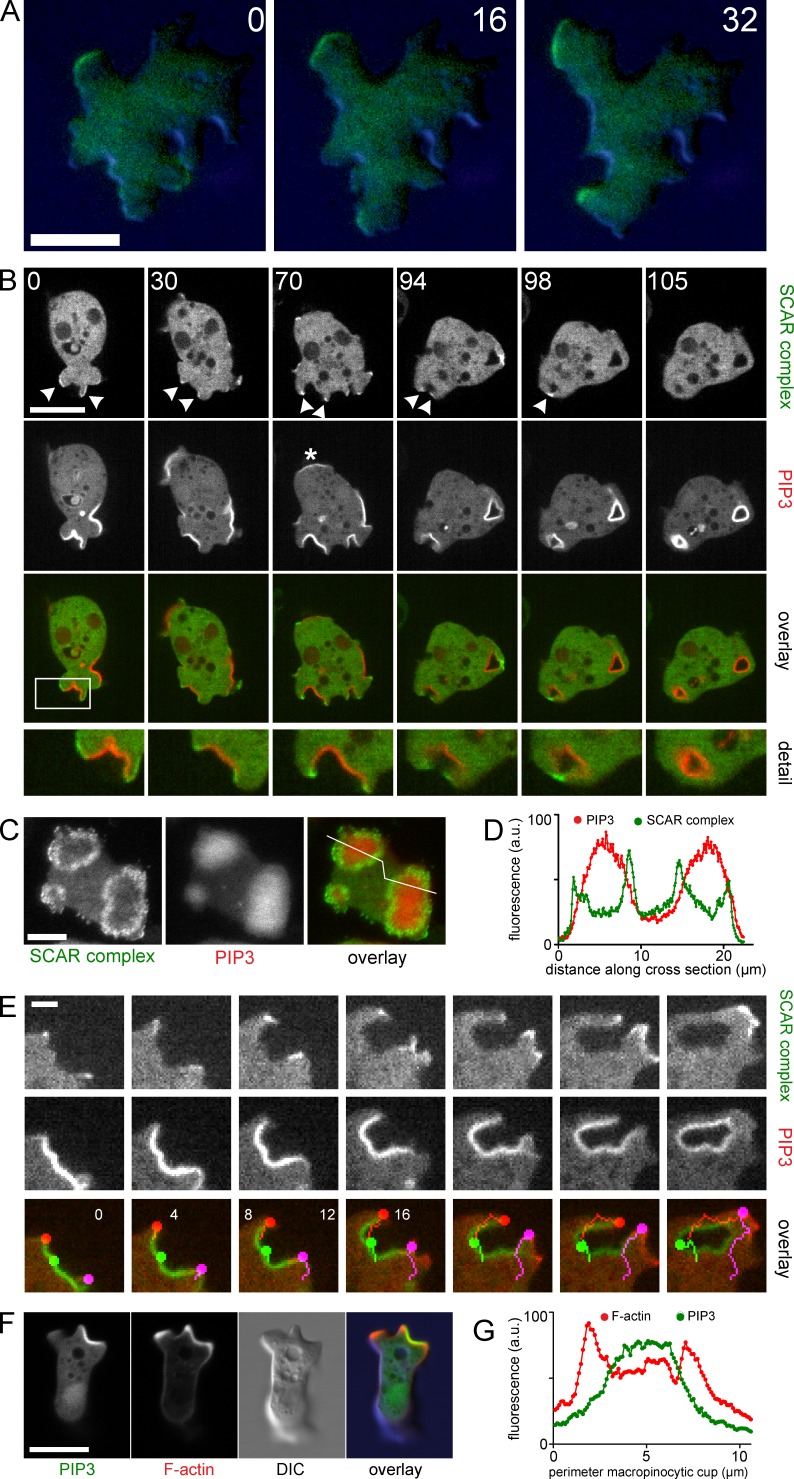
The ambiguous character of these actin structures in axenic cells complicates analysis. To more accurately distinguish the potential role of PIP3 in pseudopods and macropinosomes, we examined the behavior of the SCAR/WASP and verprolin homologue (WAVE) complex, which is the definitive marker for pseudopods. It remains associated with the leading edge throughout the progression of a pseudopod and disappears from the leading edge when the pseudopod stops advancing (Fig. 4 A; Hahne et al., 2001; Veltman et al., 2012). Interestingly, high concentrations of PIP3 do not colocalize with the SCAR complex, which is recruited to a small margin surrounding each patch (Fig. 4, B–D; and Video 2). This distribution is not only visible in the PIP3 patches that develop to form macropinosomes, but also in ones that remain planar and that cannot readily be classified as either a pseudopod or a nascent macropinosome (Video 1 and Fig. 4 B, asterisk). We were unable to find any event where a PIP3 patch was followed by recruitment of SCAR to the leading edge in the fashion that is typical for pseudopods.
Figure 4.
SCAR, actin, and PIP3 in pseudopods and macropinosomes. (A) TIRF and DIC image of HSPC300-GFP during progression of a normal pseudopod of axenic AX3 cells. Cells were overlayed with 0.4% agarose to ensure contact with the glass surface. (B) Confocal series of an axenically grown AX2 cell expressing PH-CRAC-RFP and the SCAR complex marker HSPC300-GFP. Arrowheads show regions marked by HSPC300-GFP. Asterisk indicates a PIP3 patch that does not form a macropinosome. The boxed regions are shown in detail (2.2× magnification) in the bottom row. This corresponds with Video 2. (C) TIRF image of an axenically cultivated AX2 cell expressing PH-CRAC-RFP and HSPC300-GFP. (D) Profile plot of the line indicated in C. (E) Confocal image series of an axenic AX2 cell expressing PH-CRAC-RFP and HSPC300-GFP during macropinocytosis. Colored dots indicate the edges (red, purple) and center (green) of the PIP3 patch. (F) Confocal image of an axenic AX2 cell expressing Lifeact-mRFP and PH-CRAC-GFP with a pseudopod (top left) and a macropinosome (top right). (G) Profile plot of a line drawn along the perimeter of the developing macropinosome in F. All indicated times are in seconds. Bars: (A, B, C, and F) 10 µm; (E) 2 µm.
A close-up view demonstrates the complexity of the macropinocytic cup. The region labeled with PIP3 remains static, with little movement perpendicular to the membrane (Fig. 4 E, green dots). SCAR remains associated with the rim of the cup throughout the event. The SCAR-containing region moves outward and drives closure of the cup (Fig. 4 E, other dots). This outward movement at the edge of the PIP3 patch can generate cell movement, though it is retarded by the slow movement at the center of the PIP3 patch. It is also clear that the edge of the patch is a major site of actin filament nucleation (Fig. 4 F). The Lifeact-mRFP signal overlies the PIP3, though it is strongest at the extremities (Fig. 4 G), and there is a clear difference between the edges and the center; the SCAR-labeled edges are rough and their thickness is variable, like pseudopods, whereas the central F-actin is thinner and smooth.
Collectively, these data indicate that PIP3 marks membrane regions that are destined for invagination during axenic growth. In axenic cells, this results in a complex pattern of inward and outward movement of the membrane, which combines with F-actin to mediate both macropinocytosis and a subset of cell movement.
Inverse correlation between macropinocytosis and chemotaxis
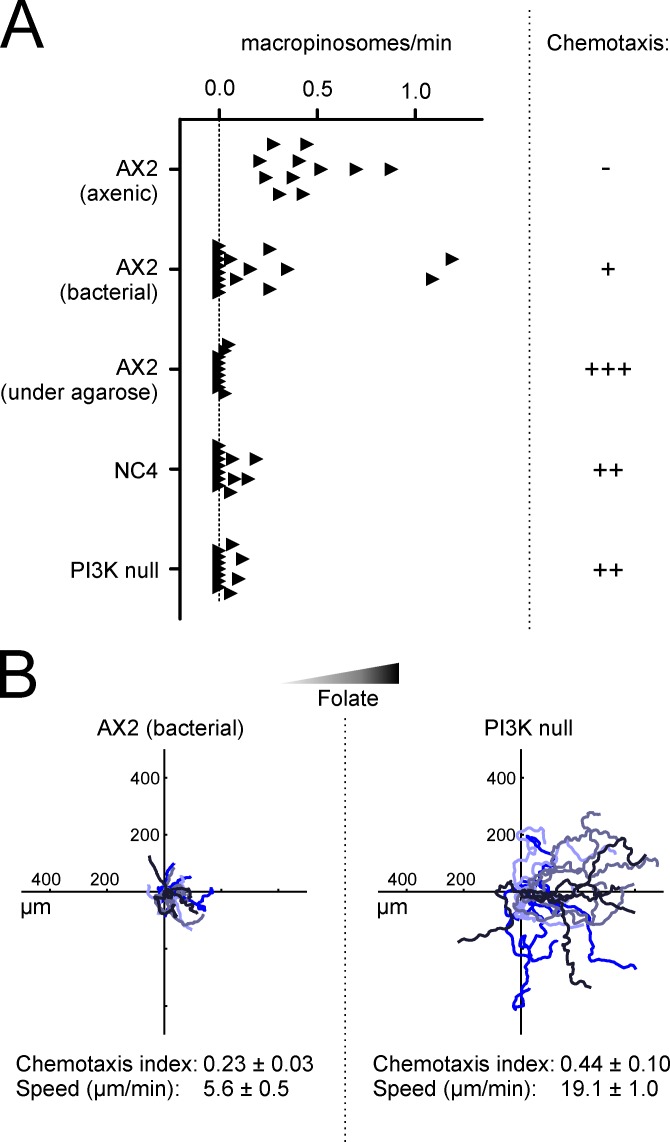
We assessed the relationship between macropinocytosis and chemotaxis by altering the macropinocytic rate. Axenically grown AX2 cells form FITC-labeled vesicles at a rate of 0.44/min (Fig. 5 A). However, AX2 cells that have been cultivated on bacteria have a diminished rate of macropinocytosis. Macropinosomes in NC4 cells are even rarer, with a mean rate of 0.05/min. These changes correlate with the efficiency of chemotaxis toward folate (Fig. 5 A).
Figure 5.
PIP3-induced macropinocytosis is incompatible with chemotaxis. (A) Negative correlation of macropinocytosis and chemotaxis. The rate of macropinosome formation under different conditions was measured using FITC-dextran and fluorescence microscopy. Each data point represents a single cell that was imaged for at least 5 min. Data were taken from at least two independent experiments. Chemotaxis is represented qualitatively to avoid quantitative comparisons made under noncomparable conditions. (B) Deletion of PI 3-kinases from axenically growing cells restores chemotaxis to folate. (B, right) Insall chamber chemotaxis of axenic AX2 cells quintuply deleted for all p110 PI 3-kinases. (B, left) Chemotaxis of axenic wild-type cells (see data from Fig. 1 C for comparison).
Folate chemotaxis is robust under a sheet of agarose (for example Laevsky and Knecht, 2001; and Blagg et al., 2003). We examined whether the difference between under-agar and chamber assays was connected with changes in macropinocytosis. Surprisingly, macropinocytosis in vegetative axenic cells is almost completely blocked under agar, with the number of macropinosomes dropping below those in nonaxenic cells (Fig. 5 A). It is not clear why physical constraint causes such a large suppression of macropinocytosis, but it is reminiscent of the induction of autophagy by compression (King et al., 2011). However, the effect on chemotaxis is clear: axenic cells become chemotactic when macropinocytosis is blocked under agarose despite being nonchemotactic when unrestricted.
Loss of PI 3-kinase blocks macropinocytosis and rescues migration and folate chemotaxis
The inverse correlation between macropinocytosis and chemotaxis raises an interesting conflict. Despite the belief that PIP3 is fundamental to chemotaxis, we have found that PIP3-driven macropinocytosis is not consistent with chemotaxis to folate. We therefore examined a strain lacking all five PI 3-kinases (Hoeller and Kay, 2007). These cells have no PIP3 patches, and their axenic growth is slow due to a defect in macropinocytosis (Hoeller et al., 2013). In our hands, the rate of macropinocytosis of PI 3-kinase–null cells was only 0.05/min, similar to that of nonaxenic cells. However, chemotaxis toward folate was greatly improved (Fig. 5 A). Bacterially grown AX2 migrated toward folate with a weak chemotaxis index of 0.23 and a speed of 5.6 µm/min; knocking out all the PI 3-kinases restored this to 0.44 and 19.1 µm/min, respectively (Fig. 5 B). Similarly, 10 µM LY294002, enough to inhibit PI 3-kinase without blocking the target of rapamycin (TOR) complex, significantly diminished macropinocytosis and increased cell speed (Fig. S2). Thus, it appears that PIP3 is not only inessential for folate chemotaxis in D. discoideum, but actively opposes it.
Axenic mutations, macropinocytosis, and chemotaxis
Our data clearly show that PIP3 patches are not used at all by cells chemotaxing to one of the two principal attractants used by D. discoideum cells. Thus, at the very least, PIP3 is not a universal player in chemotaxis. This is not a surprise, as a great deal of data has accumulated over the past six years showing that PI 3-kinases are not required for chemotaxis. What was a surprise, however, was that PIP3 actually opposes folate chemotaxis. Folate and cAMP mostly use the same signaling components, in particular the single G protein βγ subunit, which is thought to mediate most of the responses downstream of chemoattractant receptors. The key difference is therefore not likely to be in signaling pathways, but rather the developmental stages of the cells. While cells are growing, axenic mutations lead to massive, PIP3-driven macropinocytosis, which is clearly inconsistent with chemotaxis. Starved axenic cells cease macropinocytosis and thus regain chemotaxis at the time they become sensitive to cAMP.
The identity of the axenic mutations is not yet known, but it will be fascinating to unravel their influence on chemotaxis at all stages. This may fundamentally alter our understanding of chemotaxis in D. discoideum and metazoan cells.
Materials and methods
Cell strains and cultivation
D. discoideum AX2 cells were used as an axenic strain and NC4 cells were used as nonaxenic, wild-type cells. The PI3 kinase quintuple knockout strain HM1200 (Hoeller and Kay, 2007) was obtained from the Dicty Stock Center. For axenic growth, cells were cultivated on Petri dishes under HL5 medium (ForMedium). Nonaxenic cultivation was done on Petri dishes under SorMC buffer supplemented with live Klebsiella aerogenes at an OD600 of 2. SorMC buffer consists of 15 mM KH2PO4, 2 mM Na2HPO4, 50 µM MgCl2, and 50 µM CaCl2, pH 6.0. To reduce autofluorescence of axenically cultivated cells in preparation for fluorescence microscopy, cells were incubated overnight in LoFlo medium (ForMedium). For microscopy, cells were put on a 3.5-cm acid-washed glass-bottom MatTek dish.
DNA constructs and transfection
In brief, the act6 promoter driving the resistance marker of the modular pDM expression vector set (Veltman et al., 2009) was replaced by the coaA promoter (bp −293 to bp −1 relative to the start codon of coaA). The PIP3-binding PH domain of cytoplasmic regulator of adenylyl cyclase (CRAC; residue 1–126 of the protein encoded by dagA) was used as a PIP3 sensor. Full-length HSPC300 was used as a marker for the SCAR complex, and Lifeact (Riedl et al., 2008) was used as a marker for filamentous actin. Gene fragments were cloned into the BglII–SpeI site of the expression vector. For coexpression of two genes the shuttle vector was used as described previously (Veltman et al., 2009). avGFP S65T and mCherry were used for fluorescence labeling.
Transfection of nonaxenic cells was performed as follows: 5 × 106 cells were harvested from the plate, washed once in SorMC buffer, and resuspended in 400 µl SorMC buffer. Cells were mixed with 5 µl miniprep DNA (∼0.5–1 µg total) and put on ice. Cells were then electroporated at 500 V using the high-voltage setting of an ECM 399 Exponential Decay Wave Electroporation System (Harvard Apparatus) and immediately transferred to a Petri dish with SorMC buffer and live K. aerogenes at an OD600 of 2. Selection marker was added after 5 h (10 µg/ml G418 or 100 µg/ml hygromycin).
Microscopy and image analysis
All microscopy was performed at room temperature. Confocal laser scanning microscopy was performed on a confocal microscope (A1R; Nikon) using a 1.4 NA Plan-Apochromat 60× oil immersion objective lens and the manufacturer’s NIS Elements acquisition software (Nikon). Total internal reflection fluorescence (TIRF) microscopy was performed on an inverted microscope (Eclipse TE2000-U; Nikon) that was fitted with a custom TIRF condenser and a 1.45 NA 100× Plan-Apochromat TIRF objective lens. Red and green fluorescent signals were separated using a beam splitter and projected onto an EM charge-coupled device camera (Evolve512; Photometrics). MetaMorph software was used to control the camera, shutters, and light sources. Spinning disk confocal microscopy was performed on an Andor system using an Eclipse Ti body (Nikon) fitted with a Plan-Apochromat VC 100× oil immersion objective lens (NA 1.4). Images were captured on a Neo 5.5 sCMOS camera using the manufacturer’s iQ software (both from Andor Technology).
Fluorescence images were analyzed using the standard tools available in ImageJ. Images were not processed other than brightness/contrast adjustments and bleach correction for mRFP in Video 2. The Angle tool was used to determine the angle between a protrusion and the gradient. Graphs of the collected angle data were generated in MATLAB (MathWorks). Statistical analysis was done using the Circular Statistics Toolbox (Berens, 2009).
Chemotaxis assays
Chemotaxis assays were performed on an inverted microscope (Eclipse TE2000) using a 10× phase contrast objective lens (NA 0.3). The microscope was controlled using Micromanager (Edelstein et al., 2010). Insall chambers (Muinonen-Martin et al., 2010) were used to expose cells to a chemoattractant gradient of 0–100 µM folate across a bridge that was 1 mm wide. Typically, a recording of 60 min was captured, starting immediately after the assembly of the chemotaxis chamber. The position of at least 20 randomly selected cells in each recording was determined every minute using the ImageJ plugin mTrackJ (Meijering et al., 2012). The chemotaxis index of the population was calculated for each time point using a custom-written Excel (Microsoft) spreadsheet that uses the following formula:
where n is the number of cells analyzed, cosθ(t)i is the cosine of the angle between the displacement vector of cell i and the direction of the gradient at time point t, and a(t)i is the length of the displacement vector of cell i at time point t. The chemotaxis index of the population is then plotted over time and a 40-min window is selected where the chemotaxis is optimal. Typically, this would exclude the start of the recording, where the gradient has not yet stabilized, and sometimes the end of the recording, when the chamber dried out and the gradient collapsed. The mean chemotaxis index of the population was then calculated as follows:
where CIt is the mean CI of the cells at time point t, at is the mean displacement of the cells at time point t, and nt is the number of analyzed cells at time point t. The chemotaxis indices shown are the means of the population means of at least three experiments ± the standard error.
Quantification of the rate of macropinocytosis
Cells were seeded onto an acid-washed glass-bottom dish. The medium was supplemented with 800 µg/ml FITC-dextran. For the under-agarose condition, cells were overlaid with a 1-mm thick slab of 0.8% agarose in LoFlo medium + 800 µg/ml FITC-dextran. Cells were allowed to equilibrate for 20 min. A 15-min recording was made, simultaneously capturing the fluorescent FITC signal and the corresponding differential interference contrast (DIC) image on a line-scanning confocal microscope. Frames were taken every 2.5 s and across three different z planes spanning a total of 4 µm. Individual uptake events of fluorescent dextran were then identified by eye using both the data from the fluorescence channel and the DIC channel (see Fig. S1).
Macropinosome and speed experiment and quantification
Cells expressing Lifeact-mRFP were harvested in growth phase and centrifuged for 5 min at 400 g, then resuspended in LoFlo medium at a titer between 0.5 × 106 and 106 cells/ml, and 0.5 ml was added to a glass-bottom dish (Bioptechs, Inc.). Cells were incubated for 3 h to allow growth medium to be exported and then imaged every 20 s for 40 min on a confocal microscope (A1R; Nikon) as described in the Microscopy and image analysis section. LY294002 was added to a final concentration of 10.8 μM and imaging was continued.
Macropinosome quantification was done by counting the number of internalized macropinosomes (circular and Lifeact-mRFP labeled) formed in 30 min in each cell (n = 31). Cells were also tracked for speed, before and after LY294002 addition using the ImageJ and the MTrackJ plugin.
Online supplemental material
Fig. S1 shows the detailed process for determination of the rates of macropinocytosis. Fig. S2 shows that moderate levels of LY294002, an inhibitor of PI 3-kinases and PIP3 accumulation, cause axenically growing cells to migrate faster and macropinocytose more slowly. Video 1 shows a PIP3 patch inducing a negative (i.e., concave) curvature at the leading edge, unlike a normal pseudopod that shows positive/convex curvature. Video 2 shows that the SCAR complex, the principal driver of pseudopod extension, is only recruited to the edges, not the center, of PIP3 patches. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201309081/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201309081.dv.
Supplementary Material
Acknowledgments
We are grateful to Dr. G. Kalna for help with statistics and data presentation and Prof. L. Machesky for criticism of the manuscript. We are grateful to all the members of Beatson Advanced Imaging Resource (BAIR) for assistance with microscopy.
We thank Cancer Research UK for core funding to R.H. Insall.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CRAC
- cytoplasmic regulator of adenylyl cyclase
- DIC
- differential interference contrast
- PI 3-kinase
- phosphatidylinositide 3-kinase
- PIP3
- phosphatidylinositol 3,4,5-trisphosphate
- SCAR
- suppressor of cAR mutations
- TIRF
- total internal reflection fluorescence
References
- Andrew N., Insall R.H. 2007. Chemotaxis in shallow gradients is mediated independently of PtdIns 3-kinase by biased choices between random protrusions. Nat. Cell Biol. 9:193–200 10.1038/ncb1536 [DOI] [PubMed] [Google Scholar]
- Berens Philipp. 2009. CircStat: A MATLAB Toolbox for Circular Statistics. Journal of Statistical Significance. 31:1–21 http://www.jstatsoft.org/v31/i10 (accessed January 30, 2014) [Google Scholar]
- Blagg S.L., Stewart M., Sambles C., Insall R.H. 2003. PIR121 regulates pseudopod dynamics and SCAR activity in Dictyostelium. Curr. Biol. 13:1480–1487 10.1016/S0960-9822(03)00580-3 [DOI] [PubMed] [Google Scholar]
- Bosgraaf L., Van Haastert P.J. 2009. Navigation of chemotactic cells by parallel signaling to pseudopod persistence and orientation. PLoS ONE. 4:e6842 10.1371/journal.pone.0006842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne H.R., Weiner O. 2002. Cell Polarity: A chemical compass. Nature. 419:21 10.1038/419021a [DOI] [PubMed] [Google Scholar]
- Cai H., Devreotes P.N. 2011. Moving in the right direction: how eukaryotic cells migrate along chemical gradients. Semin. Cell Dev. Biol. 22:834–841 10.1016/j.semcdb.2011.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M., Kayman S.C. 1987. The axenic mutations and endocytosis in Dictyostelium. Methods Cell Biol. 28:157–176 10.1016/S0091-679X(08)61642-8 [DOI] [PubMed] [Google Scholar]
- Commisso C., Davidson S.M., Soydaner-Azeloglu R.G., Parker S.J., Kamphorst J.J., Hackett S., Grabocka E., Nofal M., Drebin J.A., Thompson C.B., et al. 2013. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 497:633–637 10.1038/nature12138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D., Weijer G., Dowler S., Weijer C.J. 2004. In vivo analysis of 3-phosphoinositide dynamics during Dictyostelium phagocytosis and chemotaxis. J. Cell Sci. 117:6497–6509 10.1242/jcs.01579 [DOI] [PubMed] [Google Scholar]
- Edelstein A., Amodaj N., Hoover K., Vale R., Stuurman N. 2010. Computer control of microscopes using µManager. Curr. Protoc. Mol. Biol. Chapter 14:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson G.J., Milne L., Kulkarni S., Sasaki T., Walker S., Andrews S., Crabbe T., Finan P., Jones G., Jackson S., et al. 2007. PI(3)Kγ has an important context-dependent role in neutrophil chemokinesis. Nat. Cell Biol. 9:86–91 10.1038/ncb1517 [DOI] [PubMed] [Google Scholar]
- Hahne P., Sechi A., Benesch S., Small J.V. 2001. Scar/WAVE is localised at the tips of protruding lamellipodia in living cells. FEBS Lett. 492:215–220 10.1016/S0014-5793(01)02239-6 [DOI] [PubMed] [Google Scholar]
- Hoeller O., Kay R.R. 2007. Chemotaxis in the absence of PIP3 gradients. Curr. Biol. 17:813–817 10.1016/j.cub.2007.04.004 [DOI] [PubMed] [Google Scholar]
- Hoeller O., Bolourani P., Clark J., Stephens L.R., Hawkins P.T., Weiner O.D., Weeks G., Kay R.R. 2013. Two distinct functions for PI3-kinases in macropinocytosis. J. Cell Sci. 126:4296–4307 10.1242/jcs.134015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insall R.H. 2010. Understanding eukaryotic chemotaxis: a pseudopod-centred view. Nat. Rev. Mol. Cell Biol. 11:453–458 10.1038/nrm2905 [DOI] [PubMed] [Google Scholar]
- King J.S., Insall R.H. 2009. Chemotaxis: finding the way forward with Dictyostelium. Trends Cell Biol. 19:523–530 10.1016/j.tcb.2009.07.004 [DOI] [PubMed] [Google Scholar]
- King J.S., Veltman D.M., Insall R.H. 2011. The induction of autophagy by mechanical stress. Autophagy. 7:1490–1499 10.4161/auto.7.12.17924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laevsky G., Knecht D.A. 2001. Under-agarose folate chemotaxis of Dictyostelium discoideum amoebae in permissive and mechanically inhibited conditions. Biotechniques. 31:1140–1142: 1144: 1146–1149 [DOI] [PubMed] [Google Scholar]
- Meijering E., Dzyubachyk O., Smal I. 2012. Methods for cell and particle tracking. Methods Enzymol. 504:183–200 10.1016/B978-0-12-391857-4.00009-4 [DOI] [PubMed] [Google Scholar]
- Muinonen-Martin A.J., Veltman D.M., Kalna G., Insall R.H. 2010. An improved chamber for direct visualisation of chemotaxis. PLoS ONE. 5:e15309 10.1371/journal.pone.0015309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent C.A., Blacklock B.J., Froehlich W.M., Murphy D.B., Devreotes P.N. 1998. G protein signaling events are activated at the leading edge of chemotactic cells. Cell. 95:81–91 10.1016/S0092-8674(00)81784-5 [DOI] [PubMed] [Google Scholar]
- Posor Y., Eichhorn-Gruenig M., Puchkov D., Schöneberg J., Ullrich A., Lampe A., Müller R., Zarbakhsh S., Gulluni F., Hirsch E., et al. 2013. Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature. 499:233–237 10.1038/nature12360 [DOI] [PubMed] [Google Scholar]
- Postma M., Roelofs J., Goedhart J., Loovers H.M., Visser A.J., Van Haastert P.J. 2004. Sensitization of Dictyostelium chemotaxis by phosphoinositide-3-kinase-mediated self-organizing signalling patches. J. Cell Sci. 117:2925–2935 10.1242/jcs.01143 [DOI] [PubMed] [Google Scholar]
- Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z., et al. 2008. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 5:605–607 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan K., Wright G.A., Hames N., Housman M., Roberts A., Aufderheide K.J., Janetopoulos C. 2013. Delineating the core regulatory elements crucial for directed cell migration by examining folic-acid-mediated responses. J. Cell Sci. 126:221–233 10.1242/jcs.113415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson J.A. 2008. Shaping cups into phagosomes and macropinosomes. Nat. Rev. Mol. Cell Biol. 9:639–649 10.1038/nrm2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman D.M., Akar G., Bosgraaf L., Van Haastert P.J. 2009. A new set of small, extrachromosomal expression vectors for Dictyostelium discoideum. Plasmid. 61:110–118 10.1016/j.plasmid.2008.11.003 [DOI] [PubMed] [Google Scholar]
- Veltman D.M., King J.S., Machesky L.M., Insall R.H. 2012. SCAR knockouts in Dictyostelium: WASP assumes SCAR’s position and upstream regulators in pseudopods. J. Cell Biol. 198:501–508 10.1083/jcb.201205058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Valkema R., Van Haastert P.J., Devreotes P.N. 1995. The G protein beta subunit is essential for multiple responses to chemoattractants in Dictyostelium. J. Cell Biol. 129:1667–1675 10.1083/jcb.129.6.1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Long Y., Devreotes P.N. 2001. Gγ in Dictyostelium: Its Role in Localization of Gβγ to the Membrane is Required for Chemotaxis in Shallow Gradients. Mol. Biol. Cell. 12:3204–3213 10.1091/mbc.12.10.3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.