Timely phosphorylation of SLD-2 by CDK is essential for proper replication initiation and cell proliferation in the germline of C. elegans.
Abstract
Cyclin-dependent kinase (CDK) plays a vital role in proliferation control across eukaryotes. Despite this, how CDK mediates cell cycle and developmental transitions in metazoa is poorly understood. In this paper, we identify orthologues of Sld2, a CDK target that is important for DNA replication in yeast, and characterize SLD-2 in the nematode worm Caenorhabditis elegans. We demonstrate that SLD-2 is required for replication initiation and the nuclear retention of a critical component of the replicative helicase CDC-45 in embryos. SLD-2 is a CDK target in vivo, and phosphorylation regulates the interaction with another replication factor, MUS-101. By mutation of the CDK sites in sld-2, we show that CDK phosphorylation of SLD-2 is essential in C. elegans. Finally, using a phosphomimicking sld-2 mutant, we demonstrate that timely CDK phosphorylation of SLD-2 is an important control mechanism to allow normal proliferation in the germline. These results determine an essential function of CDK in metazoa and identify a developmental role for regulated SLD-2 phosphorylation.
Introduction
The development of a multicellular organism requires the coordination of cell proliferation with differentiation. CDK plays a critical role in cell division across eukaryotes, but little is known about how CDK executes its essential functions in metazoa. As the cell cycle itself changes dramatically during development (Budirahardja and Gönczy, 2009), understanding cell proliferation in metazoa requires knowledge of both cell division control and the response of the cell cycle machinery to developmental cues.
One aspect of the cell cycle that changes during development is S-phase length (Nordman and Orr-Weaver, 2012), yet how DNA replication is regulated in vivo in metazoa is poorly understood. Perfect duplication of the eukaryotic genome is achieved by a two-step replication initiation mechanism that is regulated by transition through the cell cycle. The first step of replication initiation (licensing) results in the loading of the MCM2–7 helicase onto DNA in an inactive form called the prereplicative complex (pre-RC; Remus and Diffley, 2009). Licensing can only occur in late M/G1 phase of the cell cycle because pre-RC formation is inhibited outside of this phase of the cell cycle. In yeast, CDK is responsible for inhibiting pre-RC assembly by multiple mechanisms from late G1 phase to mitosis (Arias and Walter, 2007), whereas in metazoa, there are additional CDK-independent mechanisms for preventing licensing, for example, the inhibition of the pre-RC component Cdt1 both by Geminin binding and by ubiquitin-mediated proteolysis (Blow and Dutta, 2005; Kim and Kipreos, 2007a).
The second step in initiation occurs after the G1–S transition, when the APC/C is shut off and S-phase CDK activity accumulates. In the budding yeast Saccharomyces cerevisiae, the essential function of CDK for replication initiation is the phosphorylation of two proteins: Sld2 (Synthetic Lethal with Dpb11 2) and Sld3 (Tanaka et al., 2007; Zegerman and Diffley, 2007), which facilitates their subsequent interaction with the tandem BRCA1 C terminus (BRCT) repeat protein Dpb11 (Labib, 2010). This phosphodependent complex is important both for the recruitment of the leading strand DNA polymerase and for helicase activation through the recruitment of cofactors Cdc45 and GINS (Go-Ichi-Ni-San complex) to loaded MCM2–7 complexes to form the active CMG helicase complex (Muramatsu et al., 2010; Mueller et al., 2011). The dual role of CDK, both as an inhibitor of licensing and as an essential regulator of initiation, underscores the importance of this kinase in coordinating the fidelity of DNA synthesis.
Although much attention has focused on how licensing is regulated by multiple mechanisms in diverse organisms, how CDK controls replication initiation outside of fungi has remained unclear. One possible explanation for this is that surprisingly, although Sld2 and Sld3 are essential replication initiation factors in yeast, they are extremely poorly conserved. A metazoan orthologue of Sld3 (Treslin/Ticrr/C15orf42) has been identified (Fu and Walter, 2010; Kumagai et al., 2010; Sanchez-Pulido et al., 2010; Sansam et al., 2010; Pagliuca et al., 2011), and CDK phosphorylation of this protein has been shown to be important for S-phase progression in human cells (Boos et al., 2011; Kumagai et al., 2011). Despite this, CDK phosphorylation of Sld2, but not Sld3, is essential in the fission yeast Schizosaccharomyces pombe (Fukuura et al., 2011), and Sld3 orthologues are so far absent in Drosophila melanogaster (Sanchez-Pulido et al., 2010; Mueller et al., 2011). This suggests that Sld3 alone is not the single evolutionarily conserved CDK target required for replication initiation. Putative Sld2 orthologues have been identified in vertebrates and insects (Liu, 2010), and these have indeed been shown to be important for DNA replication (Sangrithi et al., 2005; Matsuno et al., 2006; Wu et al., 2008; Xu et al., 2009b; Thangavel et al., 2010), but there is little evidence to suggest that Sld2 is an important CDK target outside of yeast.
In this study, we identify Sld2 orthologues across eukaryotes and characterize SLD-2 function in the nematode worm Caenorhabditis elegans. We show that CDK-dependent phosphorylation of this key DNA replication target is vital for cell division in a metazoan, and we demonstrate a role for CDK-dependent regulation of SLD-2 in the developmental context of the C. elegans germline.
Results
Identification of Sld2 orthologues across eukaryotes
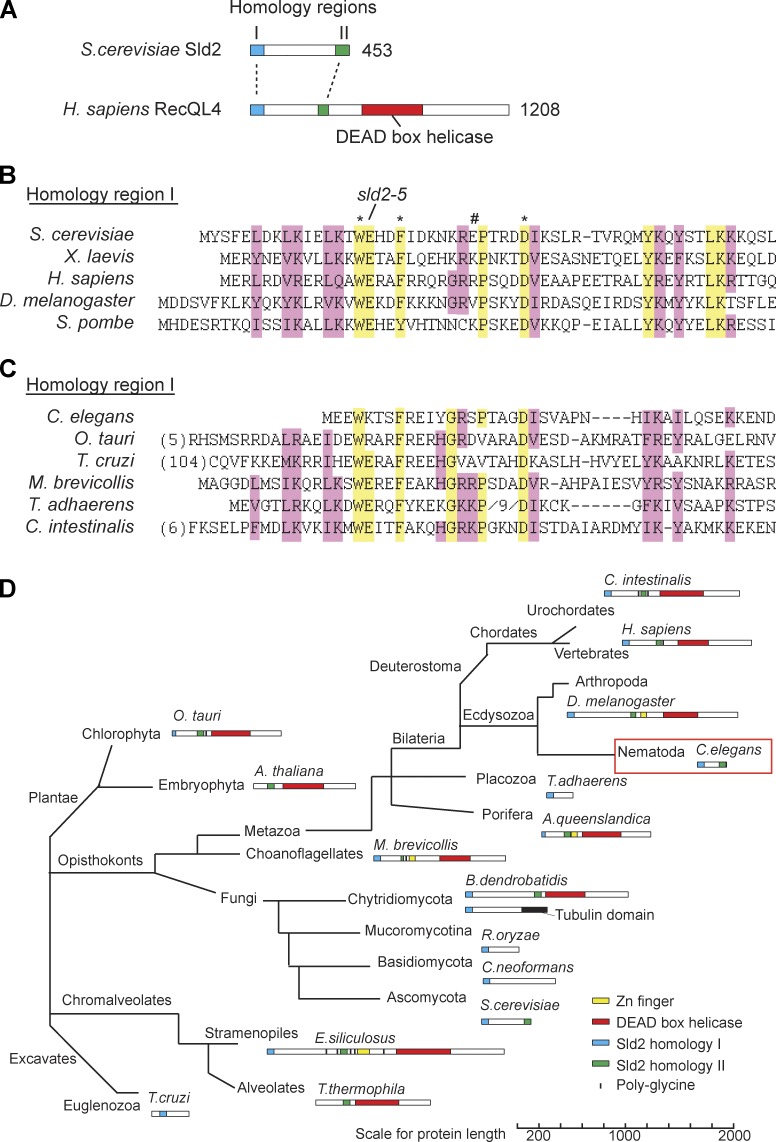
To understand the evolutionarily conserved functions of Sld2, we set out to identify orthologues of this protein across eukaryotes. To increase the sensitivity of this screen, we used HMMER (Finn et al., 2011), a program that determines homology via hidden Markov models (HMMs), rather than between individual proteins. To generate an HMM for Sld2, we first aligned yeast Sld2 with the putative Sld2 orthologues from vertebrates and flies (RecQ4/RecQL4). These RecQ4 proteins contain weak regions of homology to Sld2 at their N termini and possess an additional DEAD box helicase domain (Fig. 1 A; Liu, 2010). Alignment of characterized Sld2 and RecQ4 proteins identified two regions of significant homology (Matsuno et al., 2006; Marino et al., 2013), one at the very N terminus of the proteins (Fig. 1, A and B, region I) and the other at the very C terminus of yeast Sld2 (Fig. 1 A and Fig. S1 B, region II). To confirm the significance of the alignment of region I, we mutated residues in budding yeast Sld2 that are either conserved or not conserved in this alignment (Fig. 1 B, asterisks and number sign, respectively). Mutation of conserved residues caused synthetic lethality with a hypomorphic allele of the Sld2-binding protein Dpb11 (Fig. S1 A, dpb11-1), in accordance with a previously published mutant (sld2-5; Kamimura et al., 1998), whereas mutation of a nonconserved residue (Fig. 1 B, number sign) showed no synthetic lethality (Fig. S1 A). This mutational analysis confirms the biological significance of this alignment of homology region I.
Figure 1.
Bioinformatic screen for novel Sld2 orthologues. (A) Scale diagram of the two regions of homology (I [blue] and II [green]) between Sld2 from budding yeast and human RecQL4. (B) Alignment of homology region I between two yeast Sld2 proteins and three Sld2-RecQ4 proteins (see Table S1 for accession numbers). The first residue is the start methionine of these proteins. Residues with the greatest identity are highlighted in yellow, and regions of similarity are highlighted in pink. The position of the sld2-5 mutation and residues important (*) or unimportant (#) for Sld2 function in budding yeast are indicated above. (C) Alignment of Sld2 homology region I of some of the newly identified Sld2/RecQ4 orthologues as in B. The first residue is the start methionine, or the number of amino acids away from the first methionine is given in brackets. (D) Representation of Sld2/RecQ4 protein family as a phylogenetic tree. This tree is unrooted and not to scale. Multiple orthologues were found for each phylum (Table S1), but for each branch, only a single species is represented. H. sapiens, Homo sapiens; O. tauri, Ostreococcus tauri; M. brevicollis, Monosiga brevicollis; T. adhaerens, Trichoplax adhaerens; C. intestinalis, Ciona intestinalis; A. queenslandica, Amphimedon queenslandica; R. oryzae, Rhizopus oryzae; E. siliculosus, Ectocarpus siliculosus.
We used the alignment of region I to generate an HMM to screen for homologous Sld2 proteins across eukaryotes by HMMER. This computational screen identified several new Sld2 homologues (Fig. 1 C and Table S1). Notably, some of these newly identified Sld2 homologues appear to lack homology region II, for example, in the euglenozoan Trypanosoma cruzi and the fungus Cryptococcus neoformans. From additional BLAST (Basic Local Alignment Search Tool) searches with the novel Sld2-like proteins, we also found other Sld2 proteins that contain homology region II but appear to lack region I, for example, in the plant Arabidopsis thaliana and the alveolate Tetrahymena thermophila (Fig. S1 B and Table S1).
To verify that these new proteins that contain homology region I, II, or both are genuine Sld2 homologues, we performed both an HMM profile-to-profile comparison (HHpred; Sanchez-Pulido et al., 2010) and a reciprocal best-hit analysis (Fig. S1, C and D). Comparison of the HMMs between novel and characterized fungal Sld2 proteins verified that they are highly significantly similar, indicating that these novel proteins are Sld2/RecQL4 homologues (Fig. S1 C). In addition, a reciprocal best-hit analysis by BLAST showed that all the novel Sld2/RecQ4 proteins aligned in Fig. 1 C are the “best hit” with at least one of the characterized proteins aligned in Fig. 1 B (Fig. S1 D). Collectively, this analysis strongly suggests that the Sld2 and RecQ4 proteins identified from the HMMER screen are orthologous.
Phylogenetic analysis of these Sld2 orthologues shows that the Sld2-RecQ helicase fusion is ancestral because it is present in both animal and plant lineages (Fig. 1 D). Importantly however, there are several lineages in which the Sld2 homology domain is not fused to a helicase domain for example in the fungi, nematoda, placozoa, and euglenozoa (Fig. 1 D). In the absence of additional paralogues identified from our HMMER and subsequent BLAST searches, the most parsimonious explanation for the relationship between the Sld2 and RecQ4 proteins is that the helicase domain has separated from the Sld2 domain several times during evolution. This computational screen demonstrates that although Sld2 is rapidly evolving, Sld2 orthologues exist across eukaryotes.
C. elegans SLD-2 is essential for replication initiation
To address the evolutionarily conserved functions of Sld2, we set out to characterize Sld2 in a genetically tractable metazoan model organism. The bioinformatic screen identified the Sld2 orthologue in the nematode worm C. elegans (gene ID obtained from WormBase, T12F5.1) as a 249–amino acid protein that has no RecQ helicase domain (Fig. 1, C and D). Therefore, the characterization of this protein would allow the analysis of a metazoan Sld2 domain in isolation. From the homology of T12F5.1 to Sld2 and from the data presented in this study, we hereafter refer to T12F5.1 as C. elegans sld-2. We did not find an orthologue of the RecQ4 helicase domain in C. elegans (see Discussion and Fig. S5 A).
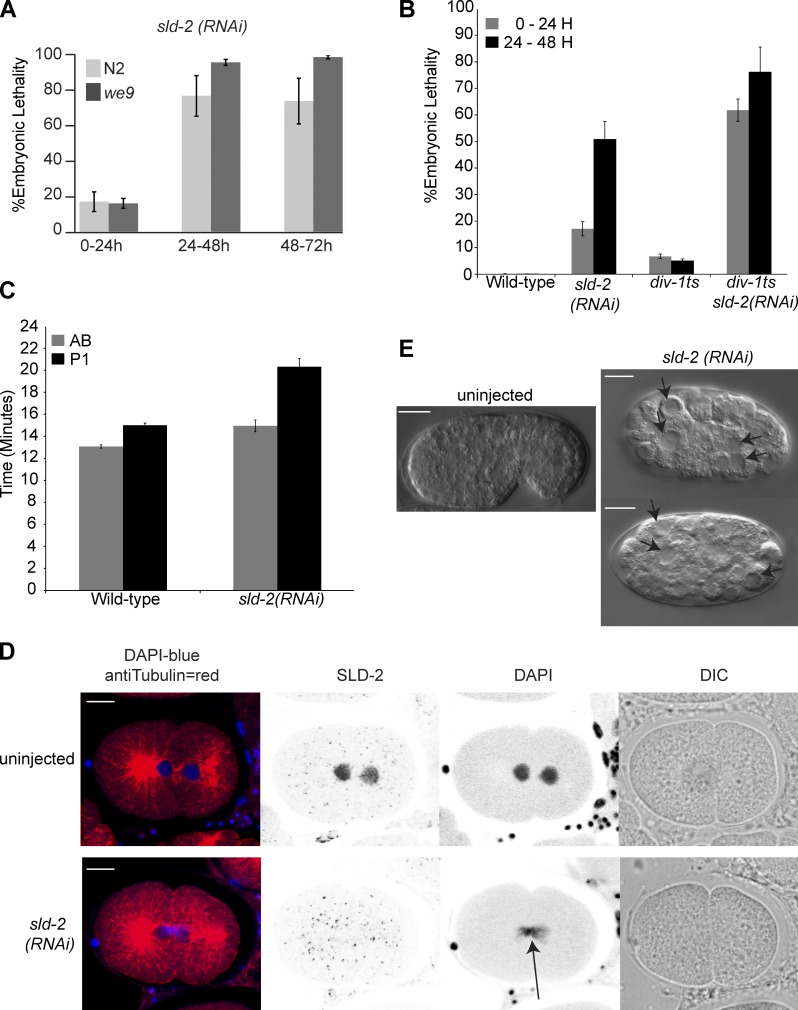
Knockdown of sld-2 by RNAi results in embryonic lethality, and in an RNAi-sensitive worm background (we9), this embryonic lethality approaches 100% after 24 h, confirming that sld-2 is an essential gene in C. elegans (Fig. 2 A). Consistent with a role for sld-2 in DNA replication, sld-2 RNAi results in synthetic lethality with a temperature-sensitive mutant in the B subunit of DNA polymerase α (div-1) at the semipermissive temperature (Fig. 2 B; Encalada et al., 2000). Replication defects in C. elegans embryos are characterized by a long delay in the division of the P1 blastomere after division of the AB blastomere in the second embryonic cell cycle (Encalada et al., 2000). In accordance with a role for sld-2 in DNA replication, RNAi of sld-2 produces a robust P1 division delay (Fig. 2 C). Further analysis of the embryonic phenotype after knockdown of sld-2 revealed that chromosomes are not effectively segregated before division (the “cut” phenotype; Fig. 2 D), and embryos arrest division with abnormally large and undifferentiated cells (Fig. 2 E). These defects are all characteristic of DNA replication mutants (Hirano et al., 1986; Encalada et al., 2000).
Figure 2.
C. elegans sld-2 is essential. (A) Mean embryonic lethality after sld-2(RNAi) by injection into young adult N2 or RNAi-sensitive we9 worms at 15°C (n = 11). Embryo lethality is scored as the number of unhatched eggs relative to the total number of F1 eggs. (B) Mean embryonic lethality for wild-type N2 and div-1(or148ts) worms with and without sld-2(RNAi) injection at 21.5°C, 0–24 and 24–48 h after egg laying. n = 20 except for sld-2(RNAi) in which n = 51. (C) The timing of cell division in early embryos, extruded from N2 worms with or without injection of sld-2 RNAi, was analyzed by time-lapse differential interference contrast (DIC) imaging. The lengths of time after P0 cell nuclear envelope breakdown until the onset of nuclear envelope breakdown in the AB blastomere or the P1 blastomere are shown. n = 9. (D) Images of the early C. elegans embryo from we9 worms with or without sld-2 (RNAi) by injection. SLD-2 was visualized by anti–SLD-2 immunofluorescence using affinity-purified Ab 5058. An arrow indicates DNA stuck in the plane of cytokinesis—the cut phenotype. (E) Differential interference contrast images showing C. elegans embryos of the same age, from mothers of the we9 strain either uninjected or injected with sld-2 (RNAi). Arrows indicate large undifferentiated cells. Error bars are SEMs. Bars, 10 µM.
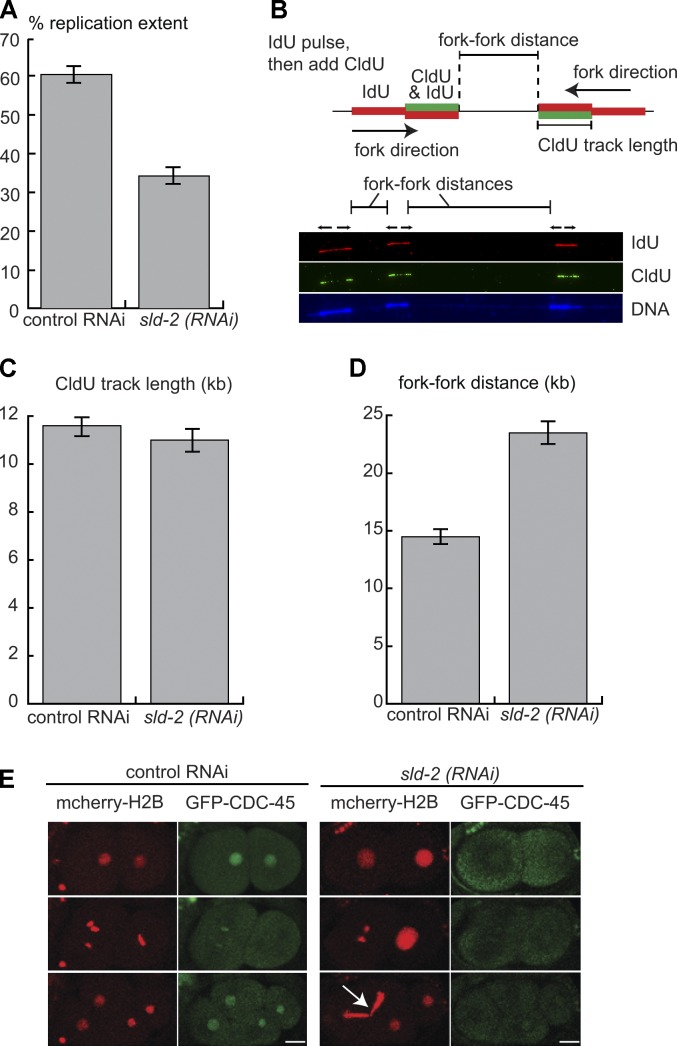
To directly assess the role of SLD-2 in DNA replication, we developed a method to analyze the replication dynamics of individual DNA molecules isolated from C. elegans embryos. For this, we pulse labeled permeabilized embryos with the nucleoside analogue 5-iodo-2′-deoxyuridine (IdU) and visualized replicated DNA by anti-IdU immunofluorescence after stretching the DNA onto slides. This single pulse labeling shows that sld-2 RNAi resulted in an ∼50% reduction in the amount of replicated DNA during the IdU pulse compared with wild type (Fig. 3 A). To control for possible effects of sld-2 RNAi on cell cycle progression, only DNA fibers that contained some IdU tracks (and hence from S-phase cells) were analyzed. To distinguish whether the reduction in DNA synthesis after sld-2 RNAi is caused by replication elongation or initiation defects, we double pulse labeled replicating DNA in embryos first with IdU and then with 5-chloro-2′-deoxyuridine (CldU; Fig. 3 B). Ongoing replication forks are labeled as tracks that contain IdU followed by CldU, and measurement of the length of CldU tracks allows the analysis of replication fork rates, whereas the gap between forks (fork–fork distances) is a measure of initiation frequencies (Fig. 3 B). From this analysis, we found that CldU track lengths are very similar in wild-type and sld-2 RNAi embryos (Fig. 3 C), which shows that replication elongation is not affected by loss of SLD-2. Importantly, however, the gap between replication forks is significantly increased in sld-2 RNAi embryos (Fig. 3 D). Therefore, the reduction in replication extent caused by knockdown of sld-2 (Fig. 3 A) is likely a result of a reduced frequency of origin firing, and from this, we conclude that C. elegans SLD-2 is required for DNA replication initiation.
Figure 3.
C. elegans sld-2 is required for replication initiation. (A) Replication extent of DNA fibers from control or sld-2 RNAi embryos from the we9 worm strain. Chitinase-treated embryos from synchronized adults were pulse labeled by 20-min incubation with IdU followed by DNA isolation and stretching onto silanized slides. Replicating DNA was visualized by anti-IdU immunofluorescence, and replication extent was calculated by dividing the summed lengths of IdU tracks on a single fiber by the total length of the fiber. n = 53. (B) Double pulse labeling of replicating embryonic cells allows the discrimination of replication initiation and elongation events. Chitinase-treated embryos are first pulse labeled with IdU (red) and then CldU (green). Tracks that are red followed by red/green are elongating forks, and the green track length provides a measure of fork rate. Gaps between two red/green tracks indicate interfork distances. (bottom) Example of a labeled C. elegans embryo DNA fiber. The DNA label cross reacts with the IdU staining, which makes the DNA labeling more intense over IdU tracks. (C) CldU track length in double pulse-labeled embryos isolated from control (n = 89) or sld-2 RNAi (n = 55) we9 worms after feeding. (D) Mean fork–fork distances from the experiment in C. For control RNAi, n = 116; for sld-2 RNAi by feeding, n = 98. (E) Visualization of the replicative helicase component CDC-45 in worm embryos extruded from mothers either injected with buffer (control) or with sld-2 (RNAi) 36 h after injection. The arrow indicates DNA stuck in the plane of cytokinesis—the cut phenotype. Error bars are SEMs. Bars, 10 µM.
It has previously been shown in C. elegans that the conversion of pre-RCs into replisomes results in the nuclear enrichment of the CDC-45 protein, which is a component of the active form of the replicative helicase, the CMG complex (Sonneville et al., 2012). To address at which stage of replication initiation SLD-2 functions, we analyzed the nuclear accumulation of CDC-45 in worm embryos. In wild-type embryos, GFP-tagged CDC-45 is visible in the nucleus before division and disperses from chromatin during mitosis (Fig. 3 E, control). After sld-2 RNAi, however, nuclear retention of CDC-45 is drastically reduced, and CDC-45 fails to visibly colocalize with chromatin throughout cell division (Fig. 3 E). This abrogation of nuclear CDC-45 is similar to the effect observed after RNAi of pre-RC components (Sonneville et al., 2012) and shows that SLD-2 likely acts at an early step in replication initiation in worms, at a point before formation of the CMG helicase.
SLD-2 is CDK phosphorylated in vivo and interacts with MUS-101/Dpb11 in a phosphodependent manner
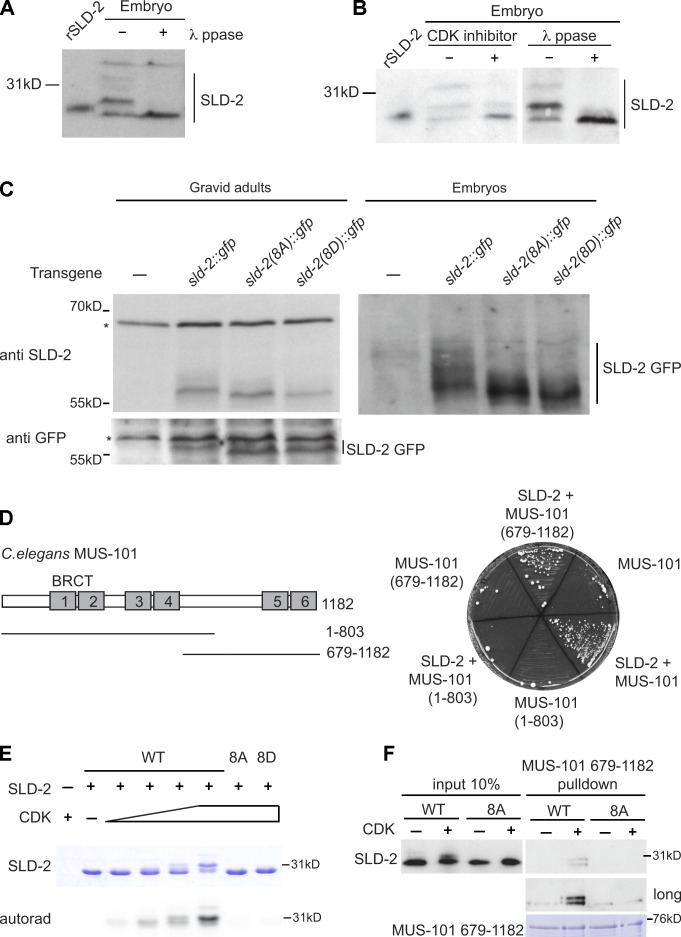
CDK phosphorylation of Sld2 is a critical step in DNA replication initiation in both fission yeast and budding yeast (Masumoto et al., 2002; Tanaka et al., 2007; Zegerman and Diffley, 2007; Fukuura et al., 2011), yet the importance of CDK regulation of metazoan Sld2/RecQ4 is not established. To test whether C. elegans SLD-2 is a CDK target in vivo, we analyzed endogenous SLD-2 protein in embryonic extracts. A Western blot for SLD-2 in embryonic extracts identified multiple bands that increase in mobility after the addition of phosphatase (Fig. 4 A). The specificity of these phosphorylated forms was confirmed with a second polyclonal antibody raised against SLD-2 (Fig. S2 A). This phosphorylation is at least partly CDK dependent because the addition of a CDK inhibitor to embryos rapidly reduces the amount of phosphorylated SLD-2 (Fig. 4 B and Fig. S2 B).
Figure 4.
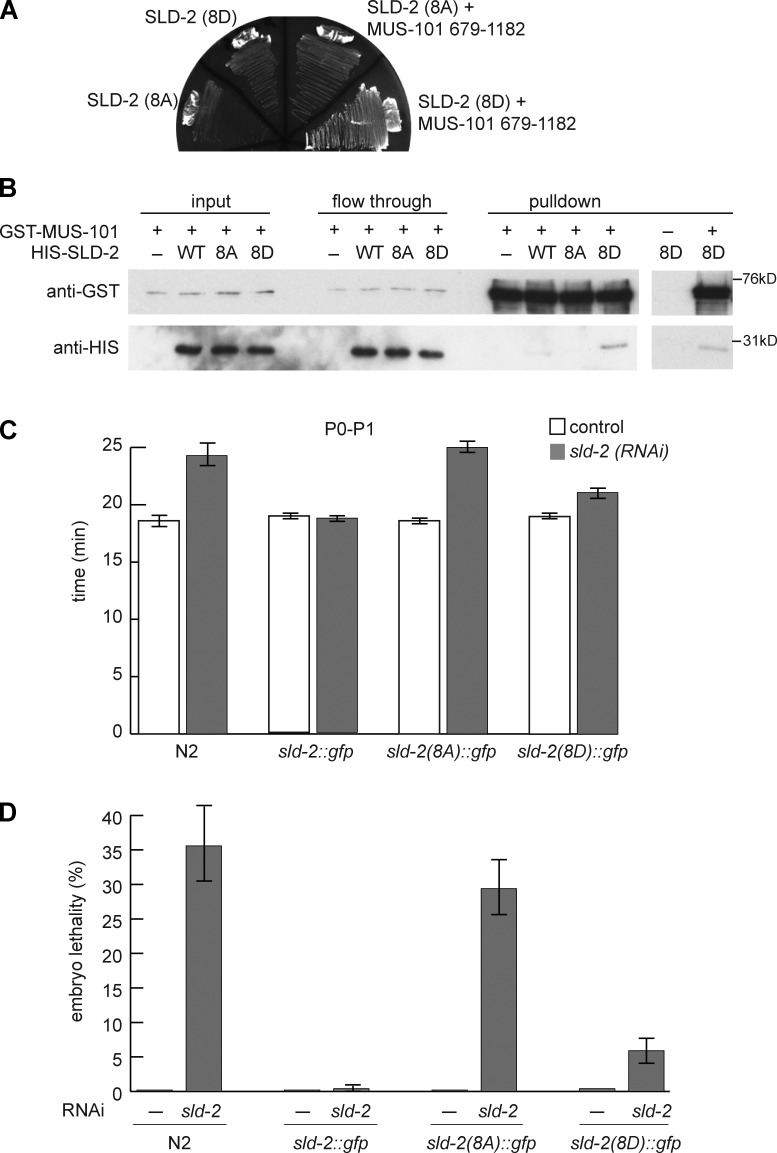
C. elegans SLD-2 is CDK phosphorylated in vivo and binds to MUS-101 in a phosphodependent manner. (A) Chitinase-treated embryos were treated with or without λ phosphatase (ppase) for 30 min at 25°C and analyzed by anti–SLD-2 (Ab 5058) Western blotting. Recombinant, E. coli–expressed SLD-2 (rSLD-2) has slightly lower mobility than endogenous, dephosphorylated SLD-2 because it is 6HIS tagged. (B) As in A, Chitinase-treated embryos were treated with or without CDK inhibitor III for 30 min at 25°C. A different exposure of the right-hand blot is also shown in Fig. S2 A. (C) Anti–SLD-2 (top) or anti-GFP Western (bottom) of SLD-2::GFP expressed from the indicated transgenes in gravid adult extracts (left) and embryo extracts (right). Asterisks mark nonspecific bands. The 8A or 8D sld-2 alleles correspond to mutants with all eight CDK consensus sites mutated to alanine or aspartic acid, respectively. (D, left) Diagram of C. elegans MUS-101 showing the position of the six BRCT repeats. (right) Selective growth medium after yeast two-hybrid analysis between MUS-101 (bait) and SLD-2 (prey). (E) Coomassie stain (top) and autoradiogram (bottom) of in vitro CDK phosphorylation of recombinant T7/6HIS-tagged SLD-2. (F) GST pull-down with GST–MUS-101 (679–1,182) and T7/6HIS SLD-2. (top) An anti-T7 Western of recombinant SLD-2. The 10% input and the short exposure are equal exposure times. (bottom) Coomassie stain of GST MUS-101 after the pull-down. WT, wild type.
C. elegans SLD-2 has eight CDK consensus sites (Fig. S5 C), and we set out to address whether SLD-2 is phosphorylated at these CDK sites in vivo. For this, we expressed sld-2 transgenes that are either wild type or have all eight CDK sites mutated to alanine (8A) or aspartic acid (8D). These transgenes have C-terminal GFP fusions and are expressed from the germline-specific mex-5 promoter integrated as a single copy at a Mos transposon site (Frøkjær-Jensen et al., 2008; Zeiser et al., 2011). Importantly although the SLD-2–GFP wild-type protein is heavily modified, particularly in embryos, removal of the eight CDK consensus sites, either in the 8A or the 8D mutant, greatly increases the mobility of SLD-2 (Fig. 4 C). From these experiments, we conclude that SLD-2 is CDK phosphorylated in vivo in C. elegans.
To understand the physiological importance of CDK phosphorylation of C. elegans SLD-2, we set out to determine whether this phosphorylation regulates the interaction of SLD-2 with other proteins. The essential function of CDK phosphorylation of Sld2 in yeast is to facilitate the interaction with the BRCT repeats of the replication initiation factor Dpb11 (Masumoto et al., 2002; Tanaka et al., 2007; Zegerman and Diffley, 2007). To test whether this phosphodependent interaction is conserved, we first performed a yeast two-hybrid assay between SLD-2 and the C. elegans orthologue of Dpb11 (MUS-101). This assay shows that SLD-2 indeed interacts with the C terminus of MUS-101 encompassing BRCT repeats 5 and 6 (Fig. 4 D). Although this yeast two-hybrid assay does not measure whether this interaction is phosphodependent, we note that C. elegans SLD-2 is modified in a cell cycle–dependent manner when expressed in yeast (Fig. S2 C). To address whether this interaction is direct and CDK dependent, we phosphorylated SLD-2 in vitro and analyzed interactions with MUS-101 by pull-down assay. SLD-2 is effectively phosphorylated by CDK in vitro, whereas a mutant lacking any CDK consensus sites (8A or 8D) is not (Fig. 4 E). Importantly, the pull-down assay shows that SLD-2 interacts directly with the C terminus of MUS-101 specifically after CDK phosphorylation (Fig. 4 F). The enrichment for the phosphorylated forms of SLD-2 in the MUS-101 pull-down and the abrogation of this interaction in the SLD-2 8A mutant (Fig. 4 F) indicate that this interaction is CDK phosphorylation dependent.
CDK phosphorylation of SLD-2 is essential in vivo
Having demonstrated that SLD-2 is targeted by CDK and that this phosphorylation event mediates the interaction with MUS-101, we next set out to address whether this CDK phosphorylation of SLD-2 is important in vivo. Although the SLD-2 8A mutant does not interact with MUS-101 either in vitro (Fig. 4 F) or by yeast two-hybrid analysis (Fig. 5 A), the SLD-2 8D mutant interacts with MUS-101 both in a yeast two-hybrid analysis (Fig. 5 A) and in a pull-down in the absence of CDK activity (Fig. 5 B). From this, we conclude that the 8D mutant is an effective phosphomimic for the interaction between SLD-2 and MUS-101. As a result, if CDK-dependent regulation of SLD-2 is essential in C. elegans, mutation of the CDK sites in sld-2 to alanine should not be compatible with viability, but mutation of these CDK sites to aspartic acid, which acts as a phosphomimetic (Fig. 5, A and B), should be viable. To test this, we analyzed the activity of transgenes expressing sld-2 wild type or the CDK mutant 8A and 8D versions of sld-2 (described in Fig. 4 C). These sld-2 transgenes are codon altered to be refractory to RNAi of endogenous sld-2, and we confirmed that these transgenes are similarly expressed and that their levels are indeed unaffected by RNAi of endogenous sld-2 (Fig. S3).
Figure 5.
C. elegans SLD-2 is an essential CDK target. (A) Selective growth medium of yeast two-hybrid analysis between MUS-101 (679–1,182; bait) and SLD-2 mutants (prey). (B) GST pull-down with GST–MUS-101 (679–1,182) and T7/6HIS SLD-2. The GST–MUS-101 lane (−) is with glutathione–Sepharose beads alone. (C) The length of time from P0 nuclear envelope breakdown until division of the P1 blastomere in N2 and transgenic worms either uninjected or after sld-2 RNAi injection. These data are the means of five separate experiments. For N2 and sld-2 wild type, n = 7; for 8A, n = 11; and for 8D, n = 14. (D) Percentage of embryonic lethality at 25°C of uninjected (n = 12–15) and sld-2 RNAi-injected adult worms (n = 40) from 2–24 h. These data are the means of three experiments. Error bars are SEMs. WT, wild type.
We first tested whether these sld-2, sld-2(8A), or sld-2(8D) transgenes can rescue the replication defect after knockdown of endogenous sld-2. For this, we analyzed the delay in the division time of the P1 cell in the early embryo, which is an indicator of defects in DNA replication (Fig. 2 C). As expected, RNAi of sld-2 results in a delay in P1 division, and this delay is fully rescued by expression of a wild-type sld-2::gfp transgene (Fig. 5 C). Importantly, the sld-2(8A) mutant does not rescue the P1 division delay, whereas the sld-2(8D) mutant largely rescues this defect (Fig. 5 C). This experiment suggests that CDK phosphorylation of SLD-2 is required for its function in replication.
To test whether CDK phosphorylation of SLD-2 is essential for viability, we measured the embryonic lethality in these strains after sld-2 RNAi. As expected, the wild-type sld-2 transgene fully rescues the lethality of sld-2 RNAi (Fig. 5 D). The sld-2(8A) mutant does not rescue lethality, suggesting that CDK phosphorylation of SLD-2 is essential (Fig. 5 D). In contrast, the sld-2(8D) mutant that bypasses the requirement for CDK for the interaction of SLD-2 with MUS-101 (Fig. 5, A and B), nearly fully rescues the lethality of the sld-2 RNAi (Fig. 5 D). From these results, we conclude that CDK phosphorylation of SLD-2 is essential for viability in C. elegans.
CDK regulates SLD-2 localization in the germline
In this paper, we have demonstrated the physiological importance of CDK phosphorylation of SLD-2 for embryonic viability. To investigate SLD-2 regulation in a developmental context, we analyzed the C. elegans germline, which contains both proliferating and differentiating nuclei. The distal end of the germline consists of ∼200 mitotically dividing germ nuclei, and immediately adjacent to this mitotic zone, germline nuclei begin differentiation by transitioning into meiosis. The transition zone is distinguished from the mitotic zone by crescent-shaped DAPI or histone-stained nuclei (Jeong et al., 2011).
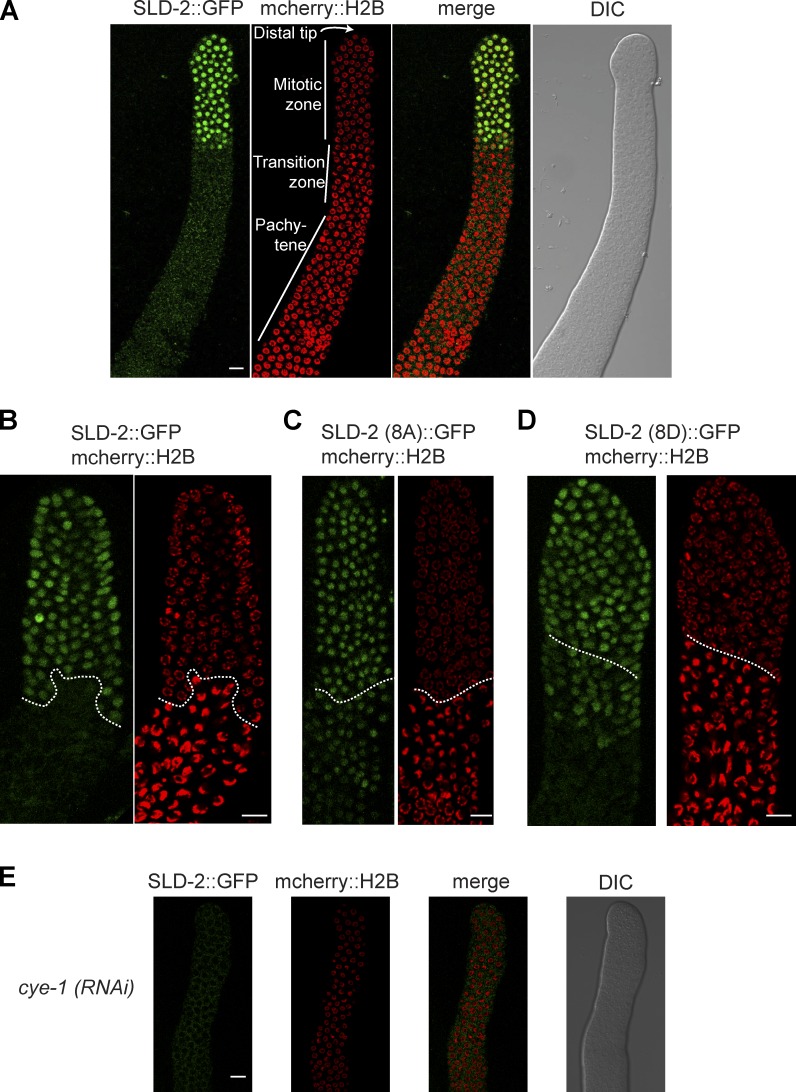
Visualization of SLD-2::GFP revealed localization to the majority of nuclei in the mitotic zone of the germline (Fig. 6 A), regardless of cell cycle stage. Similarly, we observed in early embryonic cell cycles that SLD-2 is nuclear from as soon as the nuclear membrane forms after mitosis until nuclear envelope breakdown in the next mitosis (unpublished data), suggesting that nuclear localization of SLD-2 is not regulated during the mitotic cell cycle. Interestingly, however, SLD-2::GFP localization ends abruptly at the meiotic transition zone, as determined by the virtual absence of GFP signal in the crescent-shaped meiotic nuclei (Fig. 6 A). Although absent from the meiotic region of the germline, SLD-2::GFP reappears in developing oocyte nuclei in the loop region of the gonad and in mature oocytes (Fig. S4 A). It is likely that this pattern of SLD-2 protein in the germline is generated posttranslationally because the sld-2::gfp transgenes analyzed here are expressed from the mex-5 promoter with a tbb-2 3′UTR, which drives transcription/translation throughout the germline (Fig. S4 C, H2B control; Merritt et al., 2008; Zeiser et al., 2011).
Figure 6.
CDK regulates SLD-2 localization in the germline. (A) Confocal images of the distal region of the C. elegans gonad extruded from an sld-2::gfp, mcherry::histone h2b adult worm. This sld-2::gfp transgene is integrated as a single copy at a Mos transposon site and is expressed from the mex-5 promoter with a tbb-2 3′UTR. (B–D) As in A. The dotted line delineates the start of the transition zone as determined by the appearance of crescent-shaped nuclear histone signal. (E) As in A, but after cye-1 RNAi by feeding at 25°C. DIC, differential interference contrast. Bars, 10 µM.
To address whether the sharp transition in SLD-2 abundance at the mitotic–meiotic boundary is regulated by CDK phosphorylation, we analyzed the localization of the SLD-2 mutants that cannot be phosphorylated by CDK. SLD-2 (wild type)::GFP protein is undetectable in the transition zone nuclei in 82% of gonads analyzed, with 18% showing very faint expression in a small number of transition zone nuclei and 0% showing any expression in the pachytene nuclei (Fig. 6 B, n = 22). In contrast, the SLD-2 (8A)::GFP protein is present in the nuclei of the transition zone in 85% of gonads (Fig. 6 C, n = 20), and 45% show SLD-2 (8A) nuclear localization in the pachytene nuclei (not depicted). This indicates that the CDK consensus sites of SLD-2 are important for restricting the expression of SLD-2 in the meiotic region of the germline.
Previously in this paper, we showed that the SLD-2 8D mutant is an effective phosphomimetic for the interaction with MUS-101 (Fig. 5). Despite this, we found that SLD-2 (8D)::GFP is present in the transition zone nuclei in 90% of analyzed gonads and in the pachytene nuclei of 60% of gonads (Fig. 6 D, n = 20). This pattern of SLD-2 localization is similar to the 8A mutant not to the wild-type protein. From this, we conclude that the 8D mutant is not able to confer regulation of SLD-2 localization at the transition zone border and that this localization is likely to be independent of the interaction with MUS-101. The persistence of the SLD-2 CDK site mutant proteins in the meiotic region of the germline suggests that CDK phosphorylation contributes to the restricted meiotic localization of SLD-2.
Cyclin E/Cdk2 is required for proliferation of the C. elegans germline, and Cyclin E/CYE-1 has a restricted localization in the germline that is similar to that of SLD-2, with high expression in the mitotic zone and oocytes and absence from the transition zone and pachytene region (Brodigan et al., 2003). RNAi of cye-1 results in abnormal enlarged distal germline nuclei that arrest proliferation before S phase (Fox et al., 2011; Jeong et al., 2011). Given the similar localization of CYE-1 and SLD-2, we tested whether the SLD-2 localization pattern was CYE-1 dependent. Strikingly, we found that SLD-2::GFP is undetectable in the distal regions of cye-1(RNAi) germlines (Fig. 6 E). RNAi of cye-1 did not affect the localization of an H2B transgene with the same regulatory sequences (Fig. S4 C), confirming that this regulation is specific to SLD-2. The loss of SLD-2 signal in the distal tip region of the germline after cye-1 RNAi (Fig. 6 E) is unlikely caused by direct phosphorylation because the SLD-2 (8A) mutant that cannot be CDK phosphorylated is still present in this region (Fig. 6 C). Therefore, although distal germline SLD-2 expression requires CYE-1, the loss of SLD-2 signal may be because cye-1 (RNAi) causes cell cycle exit.
To test this further, we examined SLD-2 expression in gld-1 mutant germlines, which display hyperproliferation and ectopic CYE-1 expression (Biedermann et al., 2009). Consistent with a requirement for proliferation for SLD-2 expression, we observed that gld-1(RNAi) animals have widespread expression of SLD-2::GFP in the germline (Fig. S4 B). Together, these data indicate that the SLD-2 protein is regulated by both proliferative signals and by a CDK-dependent mechanism specifically at the meiotic transition zone in the C. elegans germline.
Regulated CDK phosphorylation of SLD-2 is important for proliferation
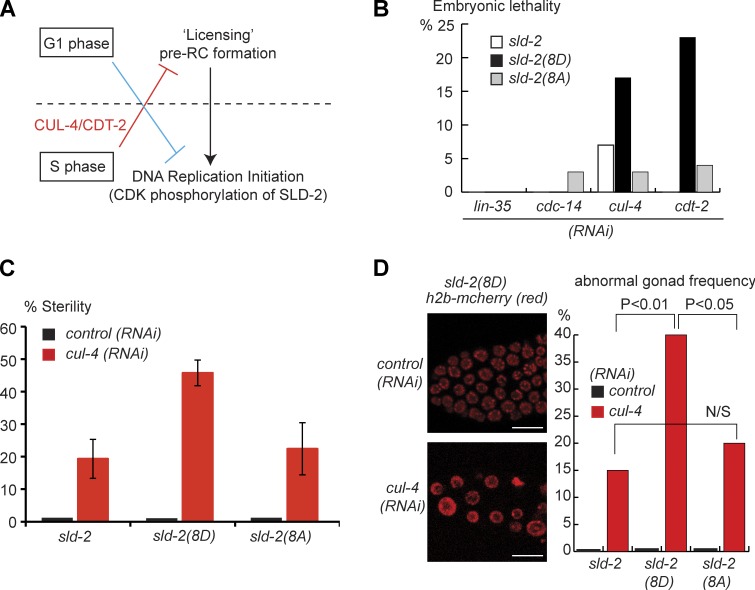
DNA synthesis can only occur once per cell division because the two steps of replication initiation are separated into different parts of the cell cycle (Fig. 7 A). We have demonstrated that one of the critical steps in the initiation of replication in C. elegans is the CDK phosphorylation of SLD-2. Importantly, the sld-2(8D) mutant can constitutively mimic the essential requirement for CDK phosphorylation of SLD-2 (Fig. 5). Therefore, the sld-2(8D) mutant can, at least in part, bypass one aspect of the cell cycle regulation of the replication initiation mechanism (Fig. 7 A, blue line).
Figure 7.
Constitutive CDK phosphorylation SLD-2 disrupts germline integrity. (A) Schematic diagram showing the two steps in eukaryotic replication initiation. Licensing can only occur in G1 phase and is inhibited by multiple mechanisms outside of G1 (red line). Conversely, replication from licensed origins cannot occur in G1 phase (blue line) because initiation requires the accumulation of CDK activity in S phase. (B) Analysis of synthetic lethality between four factors involved in cell cycle regulation and the sld-2 transgenes. L1 worms fed with the indicated RNAi were incubated at 20°C until 24 and 36 h after they reached the adult stage. For each of these time points, a subset of adult worms were isolated and allowed to lay eggs (F1) for 1 h at 20°C. Adult F1 worms were then singled (n = 32) and allowed to lay eggs (F2) for 15 h. The embryonic lethality refers to the percentage of F2 embryos that did not hatch. Note that endogenous sld-2 is still present in this experiment, and thus, any effects of the transgenes are dominant. (C) As in B, except singled adult F1 worms were scored for those that laid no eggs (percentage of sterile). These data are the means of three experiments (n = >18 for each experiment), and error bars are SEMs. (D) L4 sld-2(8D)::GFP/h2b::mcherry worms were fed with control or cul-4 RNAi as in B. Adult worms were isolated and allowed to lay eggs (F1 progeny) for 1 h at 20°C. (left) Image of control or abnormal gonads from F1 worms. Bars, 10 µM. (right) The frequency of abnormal gonads in F1 worms (n = 50) under each condition. An abnormal gonad refers to germlines with enlarged nuclei in the distal tip region, as in the image on the left. P-values from a t test are represented between columns.
To investigate the importance of timely phosphorylation of SLD-2, we tested whether the sld-2(8D) mutant showed genetic interactions with factors that are required for cell cycle regulation. Knockdown of either cdc-14 or lin-35 (Rb), which are required to regulate entry into the cell cycle in C. elegans (Boxem and van den Heuvel, 2002; Saito et al., 2004), showed virtually no synthetic lethality in strains harboring the sld-2 mutant transgenes (Fig. 7 B). We next tested two factors that have been shown to be important for coordinating replication licensing in C. elegans cul-4 and cdt-2 (Zhong et al., 2003; Kim et al., 2008). CUL-4 and CDT-2 form part of a cullin RING ubiquitin ligase that regulates replication licensing by the degradation of several factors, including CDT-1, outside of G1 phase (Fig. 7 A, red; Kim et al., 2008). We found that RNAi of either cul-4 or cdt-2 resulted in synthetic lethality with the phosphomimetic sld-2(8D) mutant but not with sld-2(8A) (Fig. 7 B). Importantly, endogenous SLD-2 is still present in this experiment, and the dominance of the sld-2(8D) mutant in this genetic interaction with cul-4/cdt-2 likely reflects the fact that the aspartate residues act as a gain-of-function mutation to constitutively bypass the CDK phosphorylation of SLD-2. This synthetic lethality between the sld-2(8D) mutant and deregulated licensing suggests that the proper timing of CDK phosphorylation of SLD-2 during the cell cycle is important for viability (Fig. 7 A, blue line).
Previous studies have shown that RNAi of cul-4 or cdt-2 result in sterility (Simmer et al., 2003; Ceron et al., 2007; Kim and Kipreos, 2007b), suggestive of failures in germline development. Because the sld-2(8D) mutant enhances embryonic lethality when replication licensing is deregulated (Fig. 7 B), we tested whether timely CDK phosphorylation of SLD-2 is also important to prevent sterility. Under conditions at which RNAi of cul-4 induces a low level of sterility, we observed an enhancement in sterility in the sld-2(8D) but not the sld-2(8A) mutant background (Fig. 7 C). This suggests that both CUL-4 and timely CDK phosphorylation of SLD-2 are important regulators of germline development. Consistent with this, we observed that the combination of the sld-2(8D) mutant with cul-4 RNAi resulted in a synergistic increase in worms with abnormal gonads (Fig. 7 D). The nuclei in the proliferative zone of these abnormal gonads are enlarged and are fewer in number relative to wild-type gonads (Fig. 7 D). This germline phenotype is similar to that observed after ionizing radiation exposure (Craig et al., 2012) and possibly reflects arrest in the cell cycle as a result of the accumulation of DNA damage arising from rereplication of the genome. Together, the genetic interactions of the sld-2(8D) mutant suggest that timely CDK-dependent regulation of replication initiation is important for proliferation fidelity in metazoa.
Discussion
Evolution of Sld2/RecQ4
From a bioinformatic screen, we have identified Sld2 orthologues across eukaryotes and characterized sld-2 function in the nematode worm C. elegans. The functional and phenotypic similarities of worm sld-2 to yeast Sld2 confirm that this bioinformatic screen was effective in identifying genuine Sld2 orthologues. Reciprocal best-hit analysis shows that the Sld2 and RecQ4 proteins are very likely orthologous, and phylogenetic analysis suggests that although the Sld2-RecQ helicase protein is ancestral, these domains have separated multiple times in evolution (Fig. 1 D). For example, in the fungal lineage, the chytridiomycete Batrachochytrium dendrobatidis contains two Sld2-like proteins, one with a RecQ helicase domain and the other with a tubulin-like domain (Fig. 1 D and Table S1). The presence of an Sld2-RecQ4 protein in this organism suggests that the separation of the Sld2 domain from the helicase domain occurred after the divergence of the chytrids in the fungal lineage.
Although the helicase domain is essential in flies (Capp et al., 2009; Xu et al., 2009b; Crevel et al., 2012), the N-terminal Sld2 homology region alone of RecQL4 is sufficient for viability in chicken cells (Abe et al., 2011) and mice (Mann et al., 2005) and can largely complement the loss of replication initiation activity of RecQ4-depleted frog egg extracts (Matsuno et al., 2006). These studies show that the N-terminal Sld2-like domain can be functionally separated from the helicase domain in several organisms, which may explain the frequency of separation of these domains during evolution (Fig. 1 D).
C. elegans SLD-2 is a 249–amino acid protein that lacks a helicase domain. Interestingly, although we find orthologues of the other members of the RecQ helicase family in C. elegans, we do not find any orthologue of the RecQ4 helicase in this organism (Fig. S5 A). This suggests that during the evolution of the nematoda, the helicase domain of RecQ4 was lost, whereas the Sld2 domain was conserved. This does not exclude the possibility that the RecQ4 helicase domain may play roles in replication as well as repair in other organisms (Capp et al., 2010; Liu, 2010) and that such functions may be fulfilled by the other RecQ helicases in C. elegans.
Conservation of the essential functions of CDK in replication initiation
In yeast, phosphorylation of Sld2 and Sld3 and their subsequent interaction with Dpb11 are the minimal roles of CDK in replication initiation (Tanaka et al., 2007; Zegerman and Diffley, 2007). Recent work has shown that CDK phosphorylation of Sld3 (Treslin/Ticrr/C15orf42) is important for S-phase progression in human cells (Boos et al., 2011; Kumagai et al., 2011). Despite this, Sld3 phosphorylation is not essential in the fission yeast S. pombe (Fukuura et al., 2011), and Sld3 orthologues have not yet been identified in D. melanogaster (Sanchez-Pulido et al., 2010). Sld2, on the other hand, is an essential CDK target in both budding and fission yeast (Masumoto et al., 2002; Noguchi et al., 2002; Fukuura et al., 2011) and orthologues exist across eukaryotes (Fig. 1 D).
One argument that has been put forward to suggest that Sld2/RecQ4 is not an important CDK target in metazoa is that the essential CDK site in budding and fission yeast Sld2 (T84 and T111, respectively; Fig. S5 B; Tak et al., 2006; Fukuura et al., 2011) is not conserved in RecQ4 (Boos et al., 2011). Here, we show that C. elegans SLD-2 is CDK phosphorylated in vivo (Fig. 4), and this phosphorylation is essential for viability (Fig. 5). Importantly, none of the eight CDK sites in C. elegans SLD-2 show homology to the essential CDK site in yeast Sld2 (Fig. S5, B and C). We note that C. elegans SLD-2 interacts with the C-terminal region of MUS-101 encompassing BRCT repeats 5/6, whereas budding yeast Sld2 binds to a different region of Dpb11-BRCT repeats 3/4 (Tak et al., 2006). This may reflect the fact that CDK sites can evolve rapidly, as has been demonstrated for pre-RC components (Moses et al., 2007), resulting in a coevolution in the interaction region with Dpb11/MUS-101. Interestingly, as in worms, human RecQL4 has been shown to bind to the C-terminal BRCT repeats of the Dpb11 orthologue TopBP1 (Ohlenschläger et al., 2012). Because Xenopus laevis/human RecQ4/RecQL4 are CDK phosphorylated in extracts and in cells, respectively (Matsuno et al., 2006; Xu et al., 2009a), it is likely that Sld2/RecQ4 is an important CDK target in vivo across eukaryotes.
In yeast, the phospho-Sld2–Dpb11 interaction is important to bring the active helicase component GINS and the leading strand polymerase to replication origins (Muramatsu et al., 2010). Here, we demonstrate a similarly important phosphodependent interaction between SLD-2 and MUS-101. Although the sld-2(8A) mutant that cannot bind to MUS-101 is inviable, whereas the sld-2(8D) mutant that restores binding to MUS-101 is viable (Fig. 5), we cannot rule out that other CDK-dependent interactions involving SLD-2 are also important in C. elegans. The exact role of SLD-2 in the replication initiation reaction is not currently clear, but the failure of embryos after sld-2 RNAi to retain the helicase component CDC-45 in the nucleus (Fig. 3 E) strongly suggests that SLD-2 acts at an early step in replication initiation, before establishment of the CMG helicase. This is consistent with work in human cells demonstrating a requirement for RecQL4 for CMG assembly (Im et al., 2009), providing further evidence that SLD-2 and RecQ4 proteins are indeed functional orthologues.
Control of SLD-2 localization in the germline
Although CDK is critical for cell cycle regulation, we understand very little about how the targets of CDK contribute to developmental changes in cell proliferation (Budirahardja and Gönczy, 2009). To our knowledge, this characterization of SLD-2 is the first description of an essential CDK target required for replication initiation in an animal. SLD-2 localization is strictly limited to the proliferative nuclei of the C. elegans germline (Fig. 6). This regulation of SLD-2 is at least in part CDK dependent because the 8A and 8D mutants show abnormal localization in the meiotic region of the gonad. Recent work has shown that Cyclin E directly phosphorylates the translational repressor GLD-1 to form a negative feedback loop that is vital for the balance of germline proliferation and differentiation (Jeong et al., 2011). Although the physiological importance of the CDK-dependent restriction of SLD-2 in the mitotic zone of the germline is not currently clear, this regulation may represent a control mechanism that together with repression of GLD-1 is important to balance mitotic cycles with meiotic entry. Despite this, we do not detect significant differences in the number of nuclei in the proliferative zone of sld-2(wt), sld-2(8A), or sld-2(8D) gonads (unpublished data). From this, we hypothesize that other replication factors may also be targeted by CDK to facilitate meiotic entry.
In addition to direct CDK-dependent regulation of SLD-2 in the germline, we also provide evidence for other levels of regulation of SLD-2 localization. RNAi of cye-1 results in loss of SLD-2::GFP signal from the distal germline nuclei (Fig. 6 E). This loss of SLD-2::GFP signal is unlikely to be directly CDK dependent because it is not mirrored by the SLD-2 8A mutant that cannot be phosphorylated by CDK (Fig. 6 C). It may be that the lower SLD-2 abundance in the distal tip region of cye-1(RNAi) animals reflects the fact that these nuclei are no longer proliferative (Brodigan et al., 2003). A reduction in the levels of RecQ4 in nondividing cells has indeed been observed in the wing and eye imaginal discs in D. melanogaster (Wu et al., 2008). The multiple levels of regulation of SLD-2 demonstrated here suggest that the C. elegans germline will be a good model to study mechanisms that control DNA replication initiation during development.
Regulated CDK phosphorylation of SLD-2 is important during germline development
Although much attention has been given to how eukaryotes regulate replication licensing in different cell cycle stages, in diverse organisms, very little is known about the importance of restricting the replication initiation activity of CDK to S phase of the cell cycle. To our knowledge, the gain-of-function sld-2(8D) allele, which bypasses the essential requirement for CDK phosphorylation of SLD-2, is the first tool to directly address the importance of this regulation in metazoa. With this allele, we show that deregulation of CDK phosphorylation of SLD-2 results in embryonic lethality and sterility when replication licensing is compromised. We hypothesize from this genetic interaction that by bypassing at least some of the requirement for CDK activation at the G1–S transition, the sld-2(8D) allele may reduce the time window to inhibit licensing before replication initiation begins (Fig. 7 A), leading to a greater risk of rereplication and genome instability.
The maintenance of genome integrity is not only important in the germline, but the accumulation of replication errors is also likely to be an early step in tumorigenesis (Blow and Gillespie, 2008). Significantly, mutations in RecQ4, the human orthologue of SLD-2, are the cause of several human disorders, including Rothmund–Thomson syndrome, which leads to a cancer predisposition (Liu, 2010). Investigating the functions of SLD-2/RecQ4 and understanding how replication fidelity is maintained in diverse cell types in a whole animal will therefore be important to understand how cancers develop in humans.
Materials and methods
Bioinformatics
Multiple sequence alignments were performed using ClustalX (Thompson et al., 1994) and manually refined. HMM were generated from the multiple sequence alignments using HMMER 3.0 (Eddy, 2009) and were used to search against the NCBI proteomes with the associated tools. HMM profiles were compared using HHPred as previously described (Söding et al., 2005; Sanchez-Pulido et al., 2010). The orthologue network was generated using a reciprocal best hit with BLAST as described in the legend of Fig. S1.
Strains
The C. elegans Bristol strain N2 was used as wild-type strain. The div-1(ts) strain used was EU548 div-1(or148ts) (B. Bowerman, University of Oregon, Eugene, OR). For immunofluorescence and nucleoside analogue labeling of embryos, the RNAi supersensitive strain JA1320 (we9) was used. Transgenic strains were generated via Mos1-mediated single copy transgene insertion transformation (Frøkjær-Jensen et al., 2008). Transgenes were inserted into the Mos1 insertion strain EG4322: ttTi5605 II; unc-119(ed9) III. The cdc-45::gfp strain (TG1754) was a gift of A. Gartner and J. Blow (University of Dundee, Dundee, Scotland, UK; Sonneville et al., 2012).
Mutations and transgenes used in this study are listed as follows: N2 wild type, JA1320 (we9), JA1564 (weSi35 [Pmex-5::sld-2(wt)::egfp/tbb-2 3′UTR; cb-unc-119(+)] II; unc-119(ed9) III), JA1563 (weSi34 [Pmex-5::sld-2(8D)::egfp/tbb-2 3’UTR; cb-unc-119(+)] II; unc-119(ed9) III), JA1566 (weSi37 [Pmex-5::sld-2(8A)::egfp::tbb-2 3′UTR, cb-unc-119(+)] II; unc-119(ed9) III), JA1570 (weSi35 [Pmex-5::sld-2(wt)::egfp/tbb-2 3′UTR; cb-unc-119(+)] II; weSi14 [Pmex-5::mCherry::his-58/tbb-2 3′UTR, cb-unc-119(+)] IV; unc-119(ed9) III), JA1589 (weSi34 [Pmex-5::sld-2(8D)::egfp/tbb-2 3′UTR; cb-unc-119(+)] II; weSi14 [Pmex-5::mCherry::his-58/tbb-2 3′UTR, cb-unc-119(+)] IV; unc-119(ed9) III), JA1590 (weSi37 [Pmex-5::sld-2(8A)::egfp::tbb-2 3′UTR, cb-unc-119(+)] II; weSi14 [Pmex-5::mCherry::his-58/tbb-2 3′UTR, cb-unc-119(+)] IV; unc-119(ed9) III), JA1522 (weSi6 [Pmex-5::his-58::egfp/tbb-2 3′UTR; cb-unc-119(+)] II; unc-119(ed9) III), EU548 div-1(or148ts) (B. Bowerman), and TG1754 (gtIs65 [pie-1p::GFP(lap)::cdc-45 + unc-119(+)]; ltIs37 [pie-1p::mCherry::his-58 + unc-119(+)]; unc-119(ed9) III; Sonneville et al., 2012). Transgenes were inserted into the Mos1 insertion strain EG4322 (ttTi5605 II; unc-119(ed9) III) using the method of Frøkjær-Jensen et al. (2008). Note that egfp is the GFP-F64LS65T variant.
DNA fiber analysis
To allow large amounts of DNA to be recovered for fiber analysis, we used the JA1320 (we9) C. elegans strain that has increased sensitivity to RNAi by feeding. In RNAi control experiments, bacteria contained only the empty vector L4440. RNAi or control was fed to L3 larvae at 15°C for 60 h. Then, adult worms were collected by filtration on a 41-µm nylon filter. Embryos were collected from adult worms by bleaching. To allow incorporation of nucleoside analogues, the embryos were treated with 0.2 U/ml chitinase (C6137; Sigma-Aldrich) for 25 min at RT. Double pulse labeling of embryonic replication intermediates was performed with a first pulse of 50 µM IdU (Sigma-Aldrich) for 10 min directly followed by a second pulse of 500 µM CldU (Sigma-Aldrich) for 10 min. Embryos were then fixed in 4% PFA (Sigma-Aldrich) for 15 min at RT.
Processing of the DNA samples.
Fixed embryos were included in an agarose plug (1% final low melting point agarose; UltraPure Low Melting Point Agarose; Invitrogen) to prevent DNA breakage during the processing of the sample. Agarose plugs were incubated in 0.5 M EDTA, 1% sarkosyl (N-lauroylsarcosine sodium salt; Sigma-Aldrich), and 1 mg/ml Proteinase K (Roche) for 12 h. This incubation was repeated once. Agarose plugs were equilibrated for 3 h (3 × 1 h) in 50 mM MES (Sigma-Aldrich) and 1 mM EDTA. Finally, the plugs were melted at 68°C for 20 min in 400 µl of 50-mM MES and 1-mM EDTA. The resulting solution was transferred to 42°C for 5 min before incubation with 2 U β-agarase I (New England Biolabs, Inc.) for 12 h at 42°C.
Stretching of DNA fibers.
DNA fibers were manually stretched on silanized cover slides (Labit et al., 2008; Demczuk and Norio, 2009). Slides were fixed for 10 min in methanol/acetic acid (3:1) and air dried.
Immunolabeling.
The slides were treated with 2.5 M HCl for 1 h and dehydrated in ethanol 70, 90, and 100%, 1 min each. Slides were washed four times in PBS and 0.1% Tween 20 for 20 min and blocked in PBS, 0.1% Tween 20, and 3% BSA for 1 h at 37°C. The slides were then incubated at 37°C with the following antibodies, rinsed four times in PBS and 0.1% Tween 20 for 20 min between each incubation: (a) 30 min in 1:750 rat anti-BrdU (detect CldU only; OBT0030G; AbD Serotec). (b) 20 min in 1:50 Alexa Fluor 488–conjugated chicken anti–rat antibody (A-21470; Molecular Probes). (c) 20 min in 1:50 Alexa Fluor 488–conjugated goat anti–chicken antibody (A-11039; Molecular Probes). (d) 1 h in 1:5 mouse anti-BrdU (detect both IdU and CldU; 347580; BD). (e) 20 min in 1:50 Alexa Fluor 594–conjugated rabbit anti–mouse antibody (A-11062; Molecular Probes). (f) 20 min in 1:50 Alexa Fluor 594–conjugated donkey anti–rabbit antibody (A-21207; Molecular Probes). (g) 30 min in 1:1,000 mouse anti-DNA antibody (MAB3034; EMD Millipore). (h) 20 min in 1:50 Alexa Fluor 350–conjugated rabbit anti–mouse antibody (A21062; Molecular Probes). (i) 20 min in 1:50 Alexa Fluor 350–conjugated donkey anti–rabbit antibody (A10039; Molecular Probes). Finally, the slides were rinsed four times in PBS for 20 min, air dried quickly, mounted in 50% glycerol in PBS, and sealed. Slides were stored at 4°C until observation. Microscopy was performed using a microscope (DeltaVision; Applied Precision).
Analysis of replication stretches.
Using ImageJ (National Institutes of Health), we estimated the size of the background signal, and according to the distribution of these parameters, we then defined a threshold defining the minimal size of a replication stretch.
Preparation of adult worm and embryonic lysates
Worms were collected at the adult stage, filtered, washed in M9, and resuspended in 4× Laemmli buffer. For embryonic lysates, adult worms were bleached, washed in M9 buffer, and resuspended in 4× Laemmli buffer. Samples were sonicated for 15 min (15 × 30–s pulse followed by 30-s pause in a sonication device [Bioruptor; Diagenode]) and incubated at 95°C for 5 min. This operation was repeated once. Finally, samples were centrifugated at 14,000 g for 5 min before SDS-PAGE analysis. For Western blots of endogenous SLD-2, we raised two rabbit polyclonal antibodies against full-length SLD-2–6HIS (Ab 5057 and Ab 5058). These antibodies were affinity purified against GST–SLD-2 and were shown to be specific in Western blots of worm extracts both by analysis of GFP-tagged SLD-2 and by the disappearance of SLD-2 bands after RNAi.
Yeast two-hybrid analysis and expression of C. elegans SLD-2 and MUS-101 in budding yeast
MUS-101 full length, 1–803 aa, and 679–1,183 aa were fused to the LexA protein in pBTM116 that was coexpressed with C. elegans SLD-2 fused to the GAL4 activation domain in pACT2 in the yeast strain L40. Interactions were assayed using the two reporter systems: (LexAop)4-HIS3 and (LexAop)8-LacZ.
C. elegans MUS-101 and SLD-2 were expressed in budding yeast from the GAL1–10 galactose inducible promoter according to the following scheme. Cell cultures exponentially growing in YP (yeast peptone)-raffinose at 30°C were synchronized in G1 with 5 µg/ml α-factor. When >95% cells were arrested in G1 phase, 2% galactose was added to the cell cultures. After 30 min of induction, cells were washed and released in YP-galactose medium without α-factor to allow entry into S phase with high levels of the overexpressed proteins.
In vitro CDK phosphorylation and GST pull-downs
SLD-2 was cloned into pET21a to generate an N-terminal T7 tag and a C-terminal 6HIS tag. Protein was expressed in BL21 codon+ cells after induction with 1 mM IPTG for 4 h at 37°C. T7-SLD2-HIS protein was purified on a HisTrap HP column (GE Healthcare) in buffer A (40 mM sodium phosphate buffer, pH 7.1, 1% Triton X-100, 0.2% Tween 20, 0.05% NP-40, 10% glycerol, 300 mM NaCl, 20 mM imidazole, pH 8, 1 mM benzamidine HCl, 0.1 mM PMSF, 1 mg/ml leupeptin, and 1 mg/ml pepstatin A) and eluted in buffer A + 400 mM imidazole. Protein fractions were subsequently dialyzed against buffer A.
For in vitro phosphorylation, cyclin A/CDK2 purified from Escherichia coli (Zegerman and Diffley, 2007) was used to phosphorylate 34 pmol SLD-2 for 30 min at 37°C in CDK buffer (10 mM sodium phosphate buffer, pH 7.1, 150 mM NaCl, 10 mM MgCl2, 1 mM ATP, and 1 mCi γ-[32P]ATP) to a total volume of 20 µl.
For GST pull-downs, MUS-101 (679–1,182) was cloned into pET2lb to generate an N-terminal 6HIS and a C-terminal GST tag. Protein was expressed in BL21 codon+ cells after induction with 1 mM IPTG for 4 h at 37°C. HIS-MUS-101-GST protein was purified on a HisTrap HP column in buffer A as previously stated. Protein fractions were dialyzed into GST binding buffer (40 mM Tris, pH 7.5, 1% Triton X-100, 0.2% Tween 20, 0.05% NP-40, 10% glycerol, 150 mM NaCl, 1 mM benzamidine HCl, 0.1 mM PMSF, 1 µg/ml leupeptin, and 1 µg/ml pepstatin A) and incubated with glutathione–Sepharose 4B (GE Healthcare) for 4 h at 4°C. Sepharose-bound protein was washed into pull-down buffer (10 mM Tris, pH 7.5, 10% glycerol, 0.1% NP-40, 1 mM EDTA, and 150 mM NaCl). For pull-downs, 0.4 µg CDK phosphorylated or unphosphorylated protein was incubated with 0.8 µg glutathione-Sepharose–bound HIS-Mus-101-GST in 200 µl of pull-down buffer (made up of 350 mM NaCl with 50 ng/ml BSA added). Pull-downs were incubated at RT for 30 min, and bound protein was washed with 2× pull-down buffer and 350 mM NaCl and pull-down buffer and 150 mM NaCl. SLD-2 protein was visualized with anti-T7 Western (Ab18611; Abcam), 1 in 500 overnight incubation in 5% milk and TBST (0.1% Tween). GST–MUS-101 was visualized by Coomassie stain.
For the GST pull-down between GST–MUS-101 and the SLD-2 8D mutant, E. coli lysates prepared in the following buffer (10 mM, Tris, pH 7.5, 1 mM EDTA, 0.1% NP-40, 10% glycerol, 300 mM NaCl, 1 mM benzamidine HCl, 0.1 mM PMSF, 1 mg/ml leupeptin, and 1 mg/ml pepstatin A) were premixed to a final volume of 250 µl before the addition of 30 µl glutathione–Sepharose beads, prewashed in lysis buffer. Lysate and beads were incubated for 1 h at 4°C before washing three times in 1 ml lysis buffer.
In vivo CDK phosphorylation of C. elegans SLD-2
N2 worms were grown at 20°C until they reach the adult stage. Adult worms were collected by filtration on 41-µm nylon filter. Embryos were collected from adult worms by bleaching. Embryos were then treated with 0.2 U/ml chitinase for 25 min at RT. Chitinase-treated embryos were then treated with λ phosphatase (P0753; New England Biolabs, Inc.) or 10 µM Cdk 1/2 inhibitor III (217714; EMD Millipore) as described in the figure legends. Reactions were stopped by the addition of 4× Laemmli buffer. Samples were processed for SDS-PAGE analysis as described in the previous paragraph.
RNAi injection and AB–P1 delay measurement
Before RNAi injection, JA1563, JA1564, JA1566, and N2 were synchronized and grown at 25°C to the young adult stage. Worms were injected with sld-2 double-stranded RNA, corresponding to the entire sld-2 ORF, and subsequently incubated at 25°C. For AB–P1 division measurements, worms were cut open in egg buffer to release the eggs and placed on 0.5% egg buffer and 3% agar pads. Embryo videos were made 20–31 h after injection at 20°C. Videos were taken with an upright light microscope (DMRE; Leica) and a digital camera (ORCA-ER C4742-80-12AG; Hamamatsu Photonics) using Openlab software (PerkinElmer). Videos consist of stacks of 12 levels taken every 10 s for durations that covered the P0 and P1 nuclear envelope breakdown.
Genetic interaction between sld-2 and cul-4/cdt-2
A small-scale screen for genetic interaction between sld-2 and factors involved in the G1–S transition was performed (see Fig. 7 B for details). L1 larvae were seeded on RNAi plates and incubated at 20°C until they reached the adult stage. Adult P0 worms were placed on RNAi plates and were allowed to lay eggs for 1 h at 20°C. Embryonic lethality was scored by dividing the number of unhatched F2 eggs by the number of hatched L2 larvae.
Germline defects were detected by seeding L4 larvae on RNAi plates and incubating at 20°C until 24 and 36 h after they reached the adult stage. For each of these time points, P0 adult worms were isolated and allowed to lay eggs for 1 h at 20°C. From these plates, L1 stage worms (n = 50) were isolated onto fresh plates. The total brood sizes of these F1 worms were counted. F1 worms laying no eggs were scored as sterile.
SLD-2 Immunofluorescence
After freeze cracking embryos as in Andrews and Ahringer (2007), fixation was performed for 10 s in ice-cold methanol and 20 min at RT in 4% PFA (4% PFA from a freshly opened vial in PBS, 80 mM Hepes, 1.6 mM MgSO4, 0.8 mM EGTA; Hepes is added at a pH of 6.9, and the pH of the final solution should reach values around 7–7.5). Samples were washed five times in PBST (PBS and 0.2% Tween 20), and blocked in PBSTB (PBST and 0.1% BSA) for 30 min at RT before overnight incubation with primary antibody at 4°C. Slides were washed 3× for 10 min with PBST, incubated with secondary antibody for 1 h at 37°C, and washed twice with PBST and once with PBS before mounting with Mowiol. Primary antibodies were SLD-2 (Ab 5058; this study) and tubulin (rat monoclonal MAB1864; EMD Millipore). Secondary antibodies labeled with Alexa Fluor dyes were obtained from Molecular Probes. Primary and secondary antibodies were diluted in PBSTB before administration to the samples.
RNAi of cye-1 and gld-1
RNAi of cye-1 (C37A2.4) and gld-1 (T23G11.3) was performed by feeding L1 or L3 worms as indicated in the legend of Fig. S4 and Fig. 6 at 25°C for 3.5 and 2.5 d, respectively, using RNAi clones from Fraser et al. (2000) and Kamath et al. (2003).
Microscope image acquisition
Embryos/gonads were imaged at RT using a 40×, 1.3 NA and 63×, 1.4 NA objectives (Carl Zeiss) on an microscope (Axioplan 2; Carl Zeiss) fitted with a confocal detection system (LSM 510 Meta; Carl Zeiss). Images were acquired using the LSM 510 Meta software (Carl Zeiss). Live worms were immobilized in 2 mM tetramisole (Sigma-Aldrich). Composite images were obtained using ImageJ software.
DNA fibers were imaged using a 60×, 1.42 NA objective on an wide-field microscope (IX81; Olympus) fitted with a camera (CoolSNAP HQ2; Photometrics). Images were acquired using the DeltaVision software and further processed using ImageJ.
Online supplemental material
Fig. S1 demonstrates the significance of the Sld2 alignment in Fig. 1 B and the subsequent bioinformatic screen for Sld2 orthologues. Fig. S2 shows that two separate rabbit polyclonal antibodies identify phosphorylated forms of SLD-2 in worm extracts and that this protein is modified in a cell cycle–dependent manner when expressed in budding yeast. Fig. S3 shows the expression of codon altered sld-2::gfp transgenes relative to endogenous SLD-2 and shows that RNAi of endogenous sld-2 does not affect the expression of these transgenes. Fig. S4 shows the expression of SLD-2::GFP in the distal tip and loop region of the germline and shows the expression of SLD-2::GFP after gld-1 and cye-1 (RNAi), with a control showing that the effect of CYE-1 on SLD-2 localization is independent of the regulatory elements of the transgene. Fig. S5 shows the conservation of RecQ helicase family members in C. elegans and an alignment of the CDK sites in yeast and worm Sld2 proteins. Table S1 provides the accession numbers of the Sld2/RecQ4 orthologues identified in the bioinformatic screen for Sld2 homologues. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201310083/DC1.
Supplementary Material
Acknowledgments
We are grateful to T. Down and S. Maslau for bioinformatics discussions. The div-1(or148ts) worm strain was a kind gift from Bruce Bowerman, and the cdc-45::gfp strain (TG1754) was a kind gift from Julian Blow and Anton Gartner. We thank Jon Pines for critical reading of the manuscript and Judith Kimble for suggestions.
P. Zegerman and V. Gaggioli are funded by the Association for International Cancer Research 10-0908. E. Zeiser, D. Rivers, and J. Ahringer were supported by a Wellcome Trust Senior Research Fellowship to J. Ahringer (054523). All authors acknowledge the core support provided by Cancer Research UK (C6946/A14492) and the Wellcome Trust (092096).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- BRCT
- BRCA1 C terminus
- CldU
- 5-chloro-2′-deoxyuridine
- HMM
- hidden Markov model
- IdU
- 5-iodo-2′-deoxyuridine
- pre-RC
- prereplicative complex
References
- Abe T., Yoshimura A., Hosono Y., Tada S., Seki M., Enomoto T. 2011. The N-terminal region of RECQL4 lacking the helicase domain is both essential and sufficient for the viability of vertebrate cells. Role of the N-terminal region of RECQL4 in cells. Biochim. Biophys. Acta. 1813:473–479 10.1016/j.bbamcr.2011.01.001 [DOI] [PubMed] [Google Scholar]
- Andrews R., Ahringer J. 2007. Asymmetry of early endosome distribution in C. elegans embryos. PLoS ONE. 2:e493 10.1371/journal.pone.0000493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias E.E., Walter J.C. 2007. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 21:497–518 10.1101/gad.1508907 [DOI] [PubMed] [Google Scholar]
- Biedermann B., Wright J., Senften M., Kalchhauser I., Sarathy G., Lee M.H., Ciosk R. 2009. Translational repression of cyclin E prevents precocious mitosis and embryonic gene activation during C. elegans meiosis. Dev. Cell. 17:355–364 10.1016/j.devcel.2009.08.003 [DOI] [PubMed] [Google Scholar]
- Blow J.J., Dutta A. 2005. Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 6:476–486 10.1038/nrm1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow J.J., Gillespie P.J. 2008. Replication licensing and cancer—a fatal entanglement? Nat. Rev. Cancer. 8:799–806 10.1038/nrc2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boos D., Sanchez-Pulido L., Rappas M., Pearl L.H., Oliver A.W., Ponting C.P., Diffley J.F. 2011. Regulation of DNA replication through Sld3-Dpb11 interaction is conserved from yeast to humans. Curr. Biol. 21:1152–1157 10.1016/j.cub.2011.05.057 [DOI] [PubMed] [Google Scholar]
- Boxem M., van den Heuvel S. 2002. C. elegans class B synthetic multivulva genes act in G(1) regulation. Curr. Biol. 12:906–911 10.1016/S0960-9822(02)00844-8 [DOI] [PubMed] [Google Scholar]
- Brodigan T.M., Liu J., Park M., Kipreos E.T., Krause M. 2003. Cyclin E expression during development in Caenorhabditis elegans. Dev. Biol. 254:102–115 10.1016/S0012-1606(02)00032-5 [DOI] [PubMed] [Google Scholar]
- Budirahardja Y., Gönczy P. 2009. Coupling the cell cycle to development. Development. 136:2861–2872 10.1242/dev.021931 [DOI] [PubMed] [Google Scholar]
- Capp C., Wu J., Hsieh T.S. 2009. Drosophila RecQ4 has a 3′-5′ DNA helicase activity that is essential for viability. J. Biol. Chem. 284:30845–30852 10.1074/jbc.M109.008052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capp C., Wu J., Hsieh T.S. 2010. RecQ4: the second replicative helicase? Crit. Rev. Biochem. Mol. Biol. 45:233–242 10.3109/10409231003786086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceron J., Rual J.F., Chandra A., Dupuy D., Vidal M., van den Heuvel S. 2007. Large-scale RNAi screens identify novel genes that interact with the C. elegans retinoblastoma pathway as well as splicing-related components with synMuv B activity. BMC Dev. Biol. 7:30 10.1186/1471-213X-7-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig A.L., Moser S.C., Bailly A.P., Gartner A. 2012. Methods for studying the DNA damage response in the Caenorhabditis elegans germ line. Methods Cell Biol. 107:321–352 10.1016/B978-0-12-394620-1.00011-4 [DOI] [PubMed] [Google Scholar]
- Crevel G., Vo N., Crevel I., Hamid S., Hoa L., Miyata S., Cotterill S. 2012. Drosophila RecQ4 is directly involved in both DNA replication and the response to UV damage in S2 cells. PLoS ONE. 7:e49505 10.1371/journal.pone.0049505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demczuk A., Norio P. 2009. Determining the replication dynamics of specific gene loci by single-molecule analysis of replicated DNA. Methods Mol. Biol. 521:633–671 10.1007/978-1-60327-815-7_35 [DOI] [PubMed] [Google Scholar]
- Eddy S.R. 2009. A new generation of homology search tools based on probabilistic inference. Genome Inform. 23:205–211 10.1142/9781848165632_0019 [DOI] [PubMed] [Google Scholar]
- Encalada S.E., Martin P.R., Phillips J.B., Lyczak R., Hamill D.R., Swan K.A., Bowerman B. 2000. DNA replication defects delay cell division and disrupt cell polarity in early Caenorhabditis elegans embryos. Dev. Biol. 228:225–238 10.1006/dbio.2000.9965 [DOI] [PubMed] [Google Scholar]
- Finn R.D., Clements J., Eddy S.R. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39(Suppl. 2):W29–W37 10.1093/nar/gkr367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox P.M., Vought V.E., Hanazawa M., Lee M.H., Maine E.M., Schedl T. 2011. Cyclin E and CDK-2 regulate proliferative cell fate and cell cycle progression in the C. elegans germline. Development. 138:2223–2234 10.1242/dev.059535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser A.G., Kamath R.S., Zipperlen P., Martinez-Campos M., Sohrmann M., Ahringer J. 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 408:325–330 10.1038/35042517 [DOI] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M.W., Hopkins C.E., Newman B.J., Thummel J.M., Olesen S.P., Grunnet M., Jorgensen E.M. 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40:1375–1383 10.1038/ng.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.V., Walter J.C. 2010. DNA replication: metazoan Sld3 steps forward. Curr. Biol. 20:R515–R517 10.1016/j.cub.2010.05.033 [DOI] [PubMed] [Google Scholar]
- Fukuura M., Nagao K., Obuse C., Takahashi T.S., Nakagawa T., Masukata H. 2011. CDK promotes interactions of Sld3 and Drc1 with Cut5 for initiation of DNA replication in fission yeast. Mol. Biol. Cell. 22:2620–2633 10.1091/mbc.E10-12-0995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Funahashi S., Uemura T., Yanagida M. 1986. Isolation and characterization of Schizosaccharomyces pombe cutmutants that block nuclear division but not cytokinesis. EMBO J. 5:2973–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im J.S., Ki S.H., Farina A., Jung D.S., Hurwitz J., Lee J.K. 2009. Assembly of the Cdc45-Mcm2-7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc. Natl. Acad. Sci. USA. 106:15628–15632 10.1073/pnas.0908039106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J., Verheyden J.M., Kimble J. 2011. Cyclin E and Cdk2 control GLD-1, the mitosis/meiosis decision, and germline stem cells in Caenorhabditis elegans. PLoS Genet. 7:e1001348 10.1371/journal.pgen.1001348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R.S., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 421:231–237 10.1038/nature01278 [DOI] [PubMed] [Google Scholar]
- Kamimura Y., Masumoto H., Sugino A., Araki H. 1998. Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol. Cell. Biol. 18:6102–6109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Kipreos E.T. 2007a. Cdt1 degradation to prevent DNA re-replication: conserved and non-conserved pathways. Cell Div. 2:18 10.1186/1747-1028-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Kipreos E.T. 2007b. The Caenorhabditis elegans replication licensing factor CDT-1 is targeted for degradation by the CUL-4/DDB-1 complex. Mol. Cell. Biol. 27:1394–1406 10.1128/MCB.00736-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Starostina N.G., Kipreos E.T. 2008. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev. 22:2507–2519 10.1101/gad.1703708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A., Shevchenko A., Shevchenko A., Dunphy W.G. 2010. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell. 140:349–359 10.1016/j.cell.2009.12.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A., Shevchenko A., Shevchenko A., Dunphy W.G. 2011. Direct regulation of Treslin by cyclin-dependent kinase is essential for the onset of DNA replication. J. Cell Biol. 193:995–1007 10.1083/jcb.201102003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K. 2010. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 24:1208–1219 10.1101/gad.1933010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labit H., Goldar A., Guilbaud G., Douarche C., Hyrien O., Marheineke K. 2008. A simple and optimized method of producing silanized surfaces for FISH and replication mapping on combed DNA fibers. Biotechniques. 45:649–658 10.2144/000113002 [DOI] [PubMed] [Google Scholar]
- Liu Y. 2010. Rothmund-Thomson syndrome helicase, RECQ4: on the crossroad between DNA replication and repair. DNA Repair (Amst.). 9:325–330 10.1016/j.dnarep.2010.01.006 [DOI] [PubMed] [Google Scholar]
- Mann M.B., Hodges C.A., Barnes E., Vogel H., Hassold T.J., Luo G. 2005. Defective sister-chromatid cohesion, aneuploidy and cancer predisposition in a mouse model of type II Rothmund-Thomson syndrome. Hum. Mol. Genet. 14:813–825 10.1093/hmg/ddi075 [DOI] [PubMed] [Google Scholar]
- Marino F., Vindigni A., Onesti S. 2013. Bioinformatic analysis of RecQ4 helicases reveals the presence of a RQC domain and a Zn knuckle. Biophys. Chem. 177-178:34–39 10.1016/j.bpc.2013.02.009 [DOI] [PubMed] [Google Scholar]
- Masumoto H., Muramatsu S., Kamimura Y., Araki H. 2002. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature. 415:651–655 10.1038/nature713 [DOI] [PubMed] [Google Scholar]
- Matsuno K., Kumano M., Kubota Y., Hashimoto Y., Takisawa H. 2006. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase alpha in the initiation of DNA replication. Mol. Cell. Biol. 26:4843–4852 10.1128/MCB.02267-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C., Rasoloson D., Ko D., Seydoux G. 2008. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr. Biol. 18:1476–1482 10.1016/j.cub.2008.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses A.M., Liku M.E., Li J.J., Durbin R. 2007. Regulatory evolution in proteins by turnover and lineage-specific changes of cyclin-dependent kinase consensus sites. Proc. Natl. Acad. Sci. USA. 104:17713–17718 10.1073/pnas.0700997104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller A.C., Keaton M.A., Dutta A. 2011. DNA replication: mammalian Treslin-TopBP1 interaction mirrors yeast Sld3-Dpb11. Curr. Biol. 21:R638–R640 10.1016/j.cub.2011.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu S., Hirai K., Tak Y.S., Kamimura Y., Araki H. 2010. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol ε, and GINS in budding yeast. Genes Dev. 24:602–612 10.1101/gad.1883410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E., Shanahan P., Noguchi C., Russell P. 2002. CDK phosphorylation of Drc1 regulates DNA replication in fission yeast. Curr. Biol. 12:599–605 10.1016/S0960-9822(02)00739-X [DOI] [PubMed] [Google Scholar]
- Nordman J., Orr-Weaver T.L. 2012. Regulation of DNA replication during development. Development. 139:455–464 10.1242/dev.061838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlenschläger O., Kuhnert A., Schneider A., Haumann S., Bellstedt P., Keller H., Saluz H.P., Hortschansky P., Hänel F., Grosse F., et al. 2012. The N-terminus of the human RecQL4 helicase is a homeodomain-like DNA interaction motif. Nucleic Acids Res. 40:8309–8324 10.1093/nar/gks591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliuca F.W., Collins M.O., Lichawska A., Zegerman P., Choudhary J.S., Pines J. 2011. Quantitative proteomics reveals the basis for the biochemical specificity of the cell-cycle machinery. Mol. Cell. 43:406–417 10.1016/j.molcel.2011.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D., Diffley J.F. 2009. Eukaryotic DNA replication control: lock and load, then fire. Curr. Opin. Cell Biol. 21:771–777 10.1016/j.ceb.2009.08.002 [DOI] [PubMed] [Google Scholar]
- Saito R.M., Perreault A., Peach B., Satterlee J.S., van den Heuvel S. 2004. The CDC-14 phosphatase controls developmental cell-cycle arrest in C. elegans. Nat. Cell Biol. 6:777–783 10.1038/ncb1154 [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L., Diffley J.F., Ponting C.P. 2010. Homology explains the functional similarities of Treslin/Ticrr and Sld3. Curr. Biol. 20:R509–R510 10.1016/j.cub.2010.05.021 [DOI] [PubMed] [Google Scholar]
- Sangrithi M.N., Bernal J.A., Madine M., Philpott A., Lee J., Dunphy W.G., Venkitaraman A.R. 2005. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell. 121:887–898 10.1016/j.cell.2005.05.015 [DOI] [PubMed] [Google Scholar]
- Sansam C.L., Cruz N.M., Danielian P.S., Amsterdam A., Lau M.L., Hopkins N., Lees J.A. 2010. A vertebrate gene, ticrr, is an essential checkpoint and replication regulator. Genes Dev. 24:183–194 10.1101/gad.1860310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F., Moorman C., van der Linden A.M., Kuijk E., van den Berghe P.V., Kamath R.S., Fraser A.G., Ahringer J., Plasterk R.H. 2003. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 1:e12 10.1371/journal.pbio.0000012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding J., Biegert A., Lupas A.N. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33(Suppl. 2):W244–W248 10.1093/nar/gki408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneville R., Querenet M., Craig A., Gartner A., Blow J.J. 2012. The dynamics of replication licensing in live Caenorhabditis elegans embryos. J. Cell Biol. 196:233–246 10.1083/jcb.201110080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak Y.S., Tanaka Y., Endo S., Kamimura Y., Araki H. 2006. A CDK-catalysed regulatory phosphorylation for formation of the DNA replication complex Sld2-Dpb11. EMBO J. 25:1987–1996 10.1038/sj.emboj.7601075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Umemori T., Hirai K., Muramatsu S., Kamimura Y., Araki H. 2007. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 445:328–332 10.1038/nature05465 [DOI] [PubMed] [Google Scholar]
- Thangavel S., Mendoza-Maldonado R., Tissino E., Sidorova J.M., Yin J., Wang W., Monnat R.J., Jr, Falaschi A., Vindigni A. 2010. Human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation. Mol. Cell. Biol. 30:1382–1396 10.1128/MCB.01290-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Capp C., Feng L., Hsieh T.S. 2008. Drosophila homologue of the Rothmund-Thomson syndrome gene: essential function in DNA replication during development. Dev. Biol. 323:130–142 10.1016/j.ydbio.2008.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Rochette P.J., Feyissa E.A., Su T.V., Liu Y. 2009a. MCM10 mediates RECQ4 association with MCM2-7 helicase complex during DNA replication. EMBO J. 28:3005–3014 10.1038/emboj.2009.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Lei Z., Huang H., Dui W., Liang X., Ma J., Jiao R. 2009b. dRecQ4 is required for DNA synthesis and essential for cell proliferation in Drosophila. PLoS ONE. 4:e6107 10.1371/journal.pone.0006107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P., Diffley J.F. 2007. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 445:281–285 10.1038/nature05432 [DOI] [PubMed] [Google Scholar]
- Zeiser E., Frøkjær-Jensen C., Jorgensen E., Ahringer J. 2011. MosSCI and gateway compatible plasmid toolkit for constitutive and inducible expression of transgenes in the C. elegans germline. PLoS ONE. 6:e20082 10.1371/journal.pone.0020082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W., Feng H., Santiago F.E., Kipreos E.T. 2003. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature. 423:885–889 10.1038/nature01747 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.