Activation of the TAK1 kinase drives RIPK3-dependent necrosis and inhibits apoptosis downstream of TNF-α stimulation.
Abstract
TNF activates three distinct intracellular signaling cascades leading to cell survival, caspase-8–mediated apoptosis, or receptor interacting protein kinase 3 (RIPK3)–dependent necrosis, also called necroptosis. Depending on the cellular context, one of these pathways is activated upon TNF challenge. When caspase-8 is activated, it drives the apoptosis cascade and blocks RIPK3-dependent necrosis. Here we report the biological event switching to activate necrosis over apoptosis. TAK1 kinase is normally transiently activated upon TNF stimulation. We found that prolonged and hyperactivation of TAK1 induced phosphorylation and activation of RIPK3, leading to necrosis without caspase activation. In addition, we also demonstrated that activation of RIPK1 and RIPK3 promoted TAK1 activation, suggesting a positive feedforward loop of RIPK1, RIPK3, and TAK1. Conversely, ablation of TAK1 caused caspase-dependent apoptosis, in which Ripk3 deletion did not block cell death either in vivo or in vitro. Our results reveal that TAK1 activation drives RIPK3-dependent necrosis and inhibits apoptosis. TAK1 acts as a switch between apoptosis and necrosis.
Introduction
On tumor necrosis factor-α (TNF-α) binding, TNF receptor 1 (TNFR1) triggers the intracellular assembly of the so-called TNFR complex I, which includes TNF receptor–associated death domain, receptor-interacting protein kinase 1 (RIPK1), cellular inhibitor of apoptosis proteins (cIAPs), and TNF receptor–associated factor 2 (TRAF2; Micheau and Tschopp, 2003). Within the complex, RIPK1 is polyubiquitinated by several ubiquitin ligases including cIAPs, which further recruits TGF-β–activated kinase 1 (TAK1) and IκB kinase (IKK), leading to the activation of nuclear factor-κB (NF-κB) and transactivation of cytoprotective genes such as cellular FLICE-like inhibitory protein (c-FLIP) to facilitate cell survival (Green et al., 2011). The molecular composition of the TNFR1 complex is subsequently changed and leads to the formation of protein complex II, the so-called cell death-inducing signaling complex (DISC; Micheau and Tschopp, 2003). In complex II, RIPK1, an adaptor molecule, Fas-associated death domain (FADD), and caspase-8 activate the pro-apoptotic caspase activation cascade (Vandenabeele et al., 2010). RIPK1 is de-ubiquitinated by de-ubiquitination enzymes such as CYLD concomitantly with the formation of complex II (Wang et al., 2008; O’Donnell et al., 2011). If caspases are inhibited or CYLD is hyperactivated, complex II cannot execute apoptosis but triggers phosphorylation and activation of RIPK1 and RIPK3 to initiate necrotic cell death (Hitomi et al., 2008; Vandenabeele et al., 2010; Yuan and Kroemer, 2010; Green et al., 2011; Oberst and Green, 2011; O’Donnell et al., 2011; Vandenabeele and Melino, 2012). Catalytic activity of RIPK1 is not required for complex I–induced pro-survival signaling (Degterev et al., 2005), whereas RIPK1 activation is required for RIPK3 activation and necrotic cell death (Degterev et al., 2008). In addition, when RIPK1 is activated by down-regulation of cIAP, RIPK1 induces not only necrosis but also caspase activation and apoptosis (Wang et al., 2008; Feoktistova et al., 2011; Tenev et al., 2011; Dondelinger et al., 2013). However, relatively little is known about the regulations by which RIPK1 activates RIPK3 and/or caspases.
TAK1 is a member of the mitogen-activated protein kinase kinase kinase (MAP3K) family that is activated by inflammatory cytokines such as IL-1, TNF, or Toll-like receptor ligands (Ninomiya-Tsuji et al., 1999; Hayden and Ghosh, 2008). TAK1 is known to be essential for prevention of TNF-induced cell death in both in vitro and in vivo settings (Omori et al., 2006; Kajino-Sakamoto et al., 2008; Inokuchi et al., 2010; Xiao et al., 2011; Morioka et al., 2012). Tak1-deficient cells are hypersensitive to TNF-induced cell death due to diminished pro-survival pathways including loss of NF-κB activation and reduced antioxidant enzymes, which results in activation of caspases (Omori et al., 2008; Morioka et al., 2009; Arslan and Scheidereit, 2011; Vanlangenakker et al., 2011; Lamothe et al., 2013). It has been recently reported that inhibition of RIPK1 blocks caspase activation and cell death in Tak1-deficienct cells (Arslan and Scheidereit, 2011; Vanlangenakker et al., 2011; Lamothe et al., 2013), suggesting that TNF-induced TAK1 activation inhibits RIPK1-induced caspase activation. TNF-induced activation of TAK1 is transient, peaking at 5–10 min, and quickly deactivated by type 2 protein phosphatases such as protein phosphatases 2A and 6 (PP2A and PP6; Kajino et al., 2006; Kim et al., 2008; Broglie et al., 2010), which is sufficient to activate the cytoprotective pathways. We have recently discovered that a binding partner of TAK1, TAK1-binding protein 2 (TAB2), is essential for TAK1 deactivation by recruiting PP6 to the TAK1 complex, and that ablation of TAB2 causes hyperactivation of TAK1 in fibroblasts (Broglie et al., 2010). Here we report that hyperactivation of TAK1 unexpectedly induces RIPK3-dependent cell death.
Results
Tab2 deficiency causes necrotic cell death
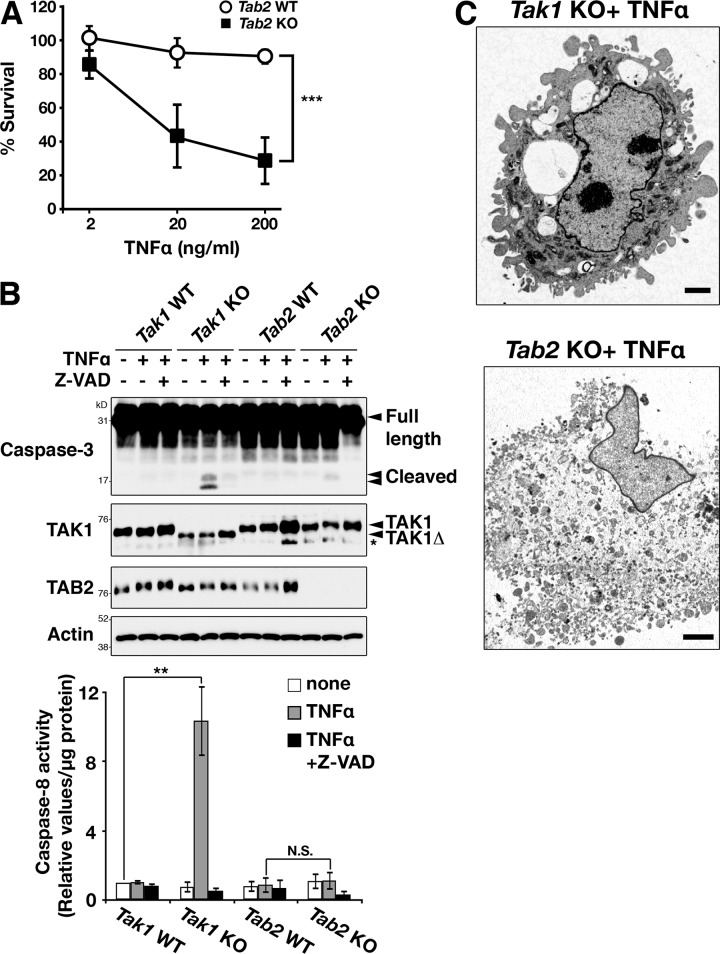
Tab2-deficient fibroblasts show enhanced and prolonged TAK1 activation after TNF stimulation (Fig. S1 A; Broglie et al., 2010). Because TAK1 has a protective role to inhibit TNF-induced cell death, we initially anticipated that Tab2 deletion would be protective in TNF-induced cell death. However, to our surprise, we found that Tab2-deficient fibroblasts were also sensitive to TNF-induced cell death (Fig. 1 A). To understand the mechanism of TNF-induced cell death in Tab2-deficient fibroblasts, we compared cell death in Tab2-deficient fibroblasts with that in Tak1-deficient fibroblasts. We found that TNF-induced cell death in Tab2-deficient fibroblasts exhibited characteristics different from those by Tak1 deficiency. TNF stimulation up-regulated activity of caspase-3 and caspase-8 in Tak1-deficient fibroblasts within 6 h after stimulation (Fig. 1 B). In contrast, Tab2-deficient fibroblasts did not show a pronounced increase in caspase activation (Fig. 1 B), even though they were already morphologically disrupted at 6 h after TNF stimulation (Fig. S1 B). These results suggest that TNF causes caspase-independent cell death in Tab2-deficient fibroblasts. We examined morphology of TNF-treated Tak1-deficient and Tab2-deficient fibroblasts by transmission electron microscopy in order to understand the type of cell death they undergo. Tak1-deficient fibroblasts exhibited the characteristic morphological features associated with apoptosis such as plasma membrane blebbing, whereas Tab2-deficient fibroblasts exhibited the features associated with necrosis such as plasma membrane rupture, translucent cytoplasm, and organelle swelling (Fig. 1 C and Fig. S1 C). These results suggest that Tak1 deficiency causes TNF-induced apoptosis, whereas Tab2 deficiency causes necrotic cell death.
Figure 1.
Tab2-deficient fibroblasts are sensitive to TNF-induced cell death. (A) Tab2 wild-type (Tab2 WT) and -deficient (Tab2 KO) fibroblasts were seeded on 24-well plates and treated with 2, 20, or 200 ng/ml of TNF for 24 h. Cells attached on the plates were determined by the crystal violet assay. Values of unstimulated fibroblasts were set at 100%. The x axis is a log scale (five independent experiments; mean ± SD; ***, P = 0.00002). (B) Tak1 wild-type (Tak1 WT), -deficient (Tak1 KO), Tab2 WT, and Tab2 KO fibroblasts were pretreated with vehicle (DMSO) or pan-caspase inhibitor, Z-VAD(OMe)FMK (Z-VAD; 20 µM) for 1 h and stimulated with TNF (20 ng/ml for Tak1 fibroblasts or 200 ng/ml for Tab2 fibroblasts) for 6 h. Caspase-3 was analyzed by immunoblotting. Immunoblots of TAK1, TAB2, and β-actin are shown as controls. Tak1 KO fibroblasts express a truncated (kinase-negative) form of TAK1 (TAK1Δ). Asterisk indicates a nonspecific band. Caspase-8 activity in cellular extracts from samples treated with the same procedure was measured. Data are shown as caspase-8 activity relative to that in unstimulated Tak1 WT samples (three independent experiments; mean ± SD; one-way ANOVA; **, P < 0.01; N.S., not significant, P = 0.0013 and P = 0.59 from the left). (C) Tak1 KO and Tab2 KO fibroblasts were exposed to TNF (20 ng/ml for Tak1 KO and 200 ng/ml for Tab2 KO) for 6 h and samples were analyzed using a transmission electron microscope. Bars, 2 µm.
We previously reported that the pan-caspase inhibitor Z-VAD(OMe)-FMK (Z-VAD) could block cell death in Tak1-deficient keratinocytes at 2–4 h after TNF stimulation (Omori et al., 2008). To examine whether caspase is responsible for the cell death in Tak1-deficient dermal fibroblasts, we treated cells with Z-VAD. Z-VAD partially rescued cell death at an early time point (6 h) after TNF stimulation in Tak1-deficient cells (Fig. S1 B), which is consistent with the study in keratinocytes. However, we found that Z-VAD did not rescue the cell death in Tak1-deficient cells after 24 h (Fig. S1 D). Caspase inhibition might activate TNF-induced necrosis in Tak1-deficient cells, which is consistent with previous studies showing that Z-VAD does not block but rather induces TNF-induced necrotic cell death (Han et al., 2011).
TNF-induced cell death in Tab2-deficient fibroblasts requires RIPK1 activity
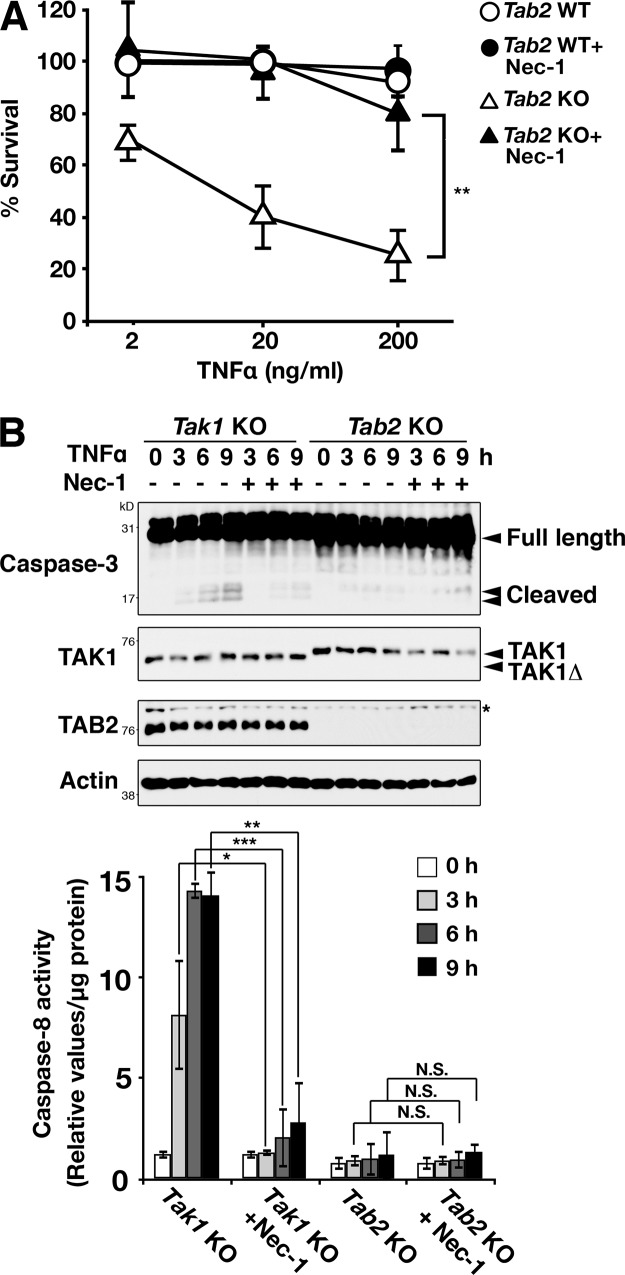
RIPK1 catalytic activity is generally implicated in necrotic cell death in TNF signaling pathways (Vandenabeele et al., 2010; Yuan and Kroemer, 2010). Necrotic morphology seen in TNF-treated Tab2-deficient fibroblasts prompted us to investigate the involvement of RIPK1. Treatment of Tab2-deficient fibroblasts with a specific inhibitor of RIPK1 kinase, necrostatin-1 (Nec-1), completely blocked TNF-induced cell death, indicating that Tab2 deficiency causes RIPK1-dependent cell death in response to TNF (Fig. 2 A). Although Tak1-deficient cells showed apoptotic morphology in response to TNF, Nec-1 also blocked TNF-induced cell death in Tak1-deficient fibroblasts (Fig. S2 A), consistent with previous studies (Arslan and Scheidereit, 2011; Vanlangenakker et al., 2011; Lamothe et al., 2013). This rescue effect may be caused by an inhibitory effect of Nec-1 on apoptosis, as RIPK1 catalytic activity was previously shown to be also required for induction of apoptosis (Vanlangenakker et al., 2012). Consistently, Nec-1 inhibited TNF-dependent caspase-3 and caspase-8 activation in Tak1-deficient cells, whereas Tab2 deficiency did not induce TNF-induced caspase activation (Fig. 2 B). Tab2-deficient fibroblasts undergo RIPK1-dependent cell death without engaging caspase activation, whereas Tak1-deficient fibroblasts die with RIPK1-dependent caspase activation.
Figure 2.
TNF-induced cell death in Tab2-deficient cells is rescued by inhibition of RIPK1. (A) Tab2 WT and Tab2 KO fibroblasts were pretreated with either vehicle (DMSO) or Nec-1 (30 µM) for 1 h, and then treated with 2, 20, or 200 ng/ml of TNF for 24 h. Cells attached on the plates were determined by the crystal violet assay. Values of unstimulated fibroblasts were set at 100%. The x axis is a log scale (three independent experiments; mean ± SD; **, P = 0.0066). (B) Tak1 KO and Tab2 KO fibroblasts were pretreated with vehicle (DMSO) or Nec-1 (30 µM) for 1 h and stimulated with TNF (20 ng/ml for Tak1 fibroblasts or 200 ng/ml for Tab2 fibroblasts) for 0, 3, 6, and 9 h. Caspase-3 was analyzed by immunoblotting. Immunoblots of TAK1, TAB2, and β-actin are shown as controls. Asterisk indicates a nonspecific band. Caspase-8 activity in cellular extracts from samples treated with the same procedure was measured. Data are shown as caspase-8 activity relative to that in unstimulated Tak1 KO samples (three independent experiments; mean ± SD; *, P < 0.05; **, P < 0.01; ***, P < 0.001; N.S., not significant; P = 0.012, P = 0.00016, P = 0.0012, P = 0.97, P = 0.99, and P = 0.33 from the left).
Tab2-deficient fibroblasts die with RIPK3-dependent necrosis
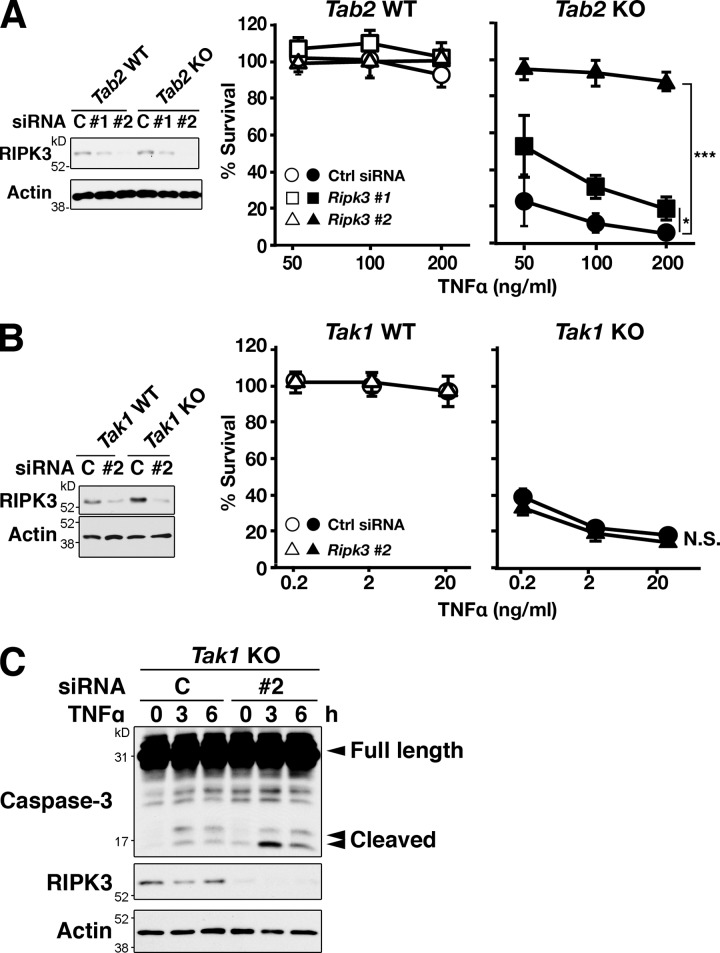
RIPK3 has been shown to interact with RIPK1 and induce necrotic cell death (Vandenabeele et al., 2010; Yuan and Kroemer, 2010; Green et al., 2011; Oberst and Green, 2011; Vandenabeele and Melino, 2012). Thus, we examined whether RIPK3 is required for TNF-induced cell death in Tab2-deficient fibroblasts. Ripk3 knockdown effectively blocked TNF-induced cell death in Tab2-deficient fibroblasts (Fig. 3 A). To confirm this result, we generated Tab2 and Ripk3 double-deficient mice using a ubiquitously expressing inducible Cre transgene system, Rosa26-CreERT (Badea et al., 2003). Dermal fibroblasts were isolated from Rosa26-CreERT Tab2flox/flox Ripk3−/− and control mice, and Tab2 deletion was induced in vitro by a CreERT activator, 4-hydroxytamoxifen. Tab2 and Ripk3 double-deficient fibroblasts were found to be resistant to TNF-induced cell death (Fig. S2 B). Thus, TNF-induced cell death in Tab2-deficient fibroblasts is RIPK3 dependent. In contrast, ablation of Ripk3 did not block TNF-induced cell death in Tak1-deficient fibroblasts (Fig. 3 B), and caspase activation in Tak1-deficient fibroblasts was not reduced by Ripk3 knockdown (Fig. 3 C). These results suggest that TNF induces RIPK1–RIPK3-dependent cell death in Tab2-deficient fibroblasts, whereas Tak1 deficiency engages RIPK3-independent cell death.
Figure 3.
Ripk3 knockdown rescues TNF-induced cell death in Tab2-deficient but not Tak1-deficient fibroblasts. (A) Tab2 WT and Tab2 KO fibroblasts were transfected with Ripk3 siRNA #1 and #2 and stimulated with 50, 100, or 200 ng/ml of TNF at 72 h after transfection. The protein levels of RIPK3 were determined by immunoblotting. Cell viability was determined at 24 h after TNF stimulation by the crystal violet assay. Values of unstimulated fibroblasts were set at 100%. The x axis is a log scale (three independent experiments; mean ± SD; ***, P < 0.001; P = 0.026 and P = 0.000024 from the bottom). (B) Tak1 WT and Tak1 KO fibroblasts were transfected with Ripk3 siRNA #2, and stimulated with 0.2, 2, or 20 ng/ml of TNF at 72 h after transfection. Cell viability was determined at 24 h after TNF stimulation by the crystal violet assay. Values of unstimulated fibroblasts were set at 100%. The x axis is a log scale (three independent experiments; mean ± SD; N.S., not significant; P = 0.99). (C) Tak1 KO fibroblasts were transfected with Ripk3 siRNA #2, and stimulated with 20 ng/ml TNF at 72 h after transfection. Caspase-3 was analyzed by immunoblotting. Immunoblots of RIPK3 and β-actin are shown as controls.
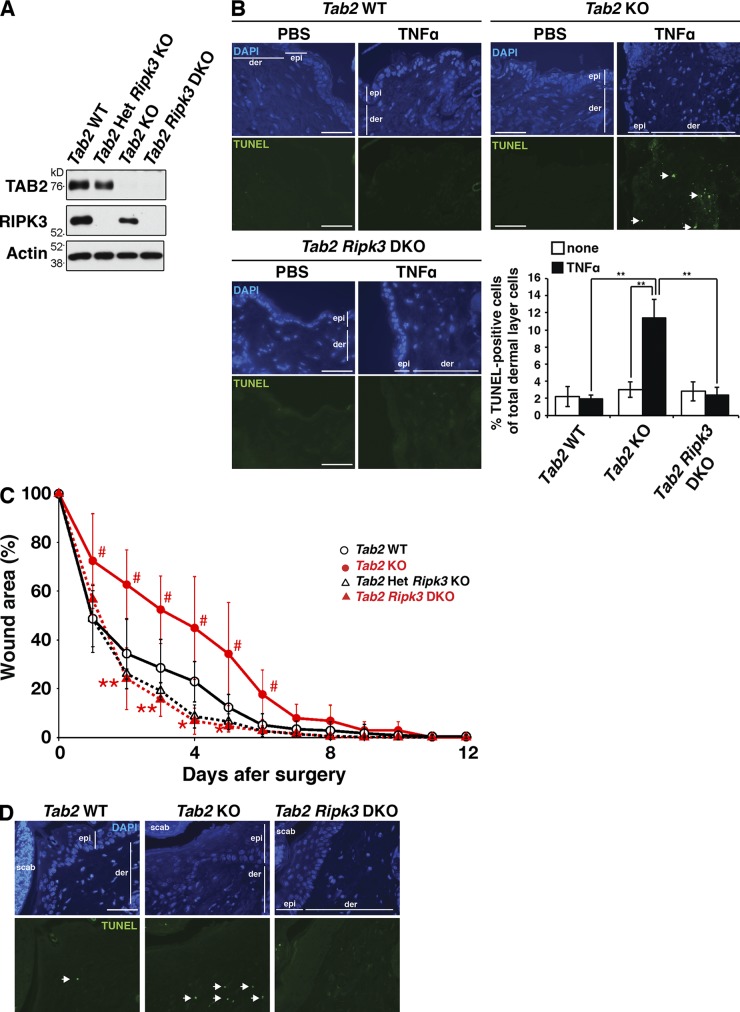
To investigate the role of TAB2 in vivo, we used the inducible Tab2 deletion (Rosa26-CreERT Tab2flox/flox) mice and generated Tab2-deficient mice by treatment of a CreERT activator, tamoxifen. We confirmed that TAB2 protein was diminished by the gene deletion (Fig. 4 A). Because our fibroblasts were isolated from the dermal layer of the skin, we intradermally injected TNF to examine whether our results in culture cells can be reproduced in the in vivo skin (Fig. 4 B). We found that Tab2-deficient mice exhibited extensive cell death in the dermal fibroblasts in vivo, which was rescued by Ripk3 deletion (Fig. 4 B). This demonstrates that TNF induces RIPK3-dependent cell death in Tab2-deficient mice in vivo. To further examine the relationship between TAB2 and RIPK3 in vivo, we performed a wound-healing assay. The wound-healing process is characterized by regeneration of the cellular and extracellular components of the skin. This process involves inflammation, proliferation, and remodeling (Werner and Grose, 2003). The proliferative phase of healing includes the re-epithelialization of the superficial surface layer and the reconstitution of the underlying dermis by dermal fibroblasts. Upon cutaneous injury, TNF is known to be secreted by immune cells (Werner and Grose, 2003). Thus, we hypothesized that Tab2 deficiency in dermal fibroblasts would induce cell death upon skin injury, which would delay the wound-healing process. Indeed, Tab2-deficient mice showed slower wound healing compared with wild-type mice, and the delay in Tab2-deficient mice was rescued by Ripk3 deletion (Fig. 4 C). TUNEL-positive dermal fibroblasts were increased in the injured Tab2-deficient skin, which was also reduced by Ripk3 deletion (Fig. 4 D). Thus, TAB2 participates in skin wound healing in vivo by preventing RIPK3-dependent cell death.
Figure 4.
Ripk3 deletion rescues cell death in the Tab2-deficient skin in vivo. (A) Rosa26-CreERT Tab2flox/+ Ripk3−/−, Rosa26-CreERT Tab2flox/flox, and Rosa26-CreERT Tab2flox/flox Ripk3−/− mice were treated with tamoxifen at 50 mg/kg for three consecutive days to generate Tab2 Het Ripk3 KO, Tab2 KO, and Tab2 Ripk3 DKO, respectively. Tab2flox/flox mice were treated with the same dose of tamoxifen and studied in parallel as Tab2 WT. 2 wks after the injection, protein extracts from spleen were isolated to confirm the deletion of Tab2 gene by immunoblotting. RIPK3 and β-actin are shown as controls. (B) 1 µg TNF was intradermally injected in Tab2 WT (n = 3), Tab2 KO (n = 3), and Tab2 Ripk3 DKO mice (n = 3). After 6 h, the skin was isolated and TUNEL staining was conducted on the sections. The arrows indicate TUNEL-positive dermal fibroblasts. epi, epidermis; der, dermis. Bars, 40 µm. The percentages of TUNEL-positive cells in the dermal fibroblasts are shown (mean ± SD; **, P < 0.01; P = 0.0018, P = 0.0035, and P = 0.0027 from the left). (C and D) Skin wound surgery was conducted on Tab2 WT (n = 6), Tab2 KO (n = 5), Tab2 Het Ripk3 KO (n = 4), and Tab2 Ripk3 DKO (n = 5) mice. Wound area was expressed as a percentage of initial wound size. Statistical significance between Tab2 WT and Tab2 KO or between Tab2 KO and Tab2 Ripk3 DKO is indicated by # or *, respectively (mean ± SD; ## and **, P < 0.01; # and *, P < 0.05; #, P = 0.034; #, P = 0.011; #, P = 0.014; #, P = 0.045; #, P = 0.038; #, P = 0.027; **, P = 0.0042; **, P = 0.0022; **, P = 0.01; *, P = 0.030; and *, P = 0.028 from the left). 4 d after skin surgery, the skin was isolated from the mice and TUNEL staining was conducted on the sections (D). Arrows indicate TUNEL-positive cells. epi, epidermis; der, dermis; scab, a scab formed over the wound. Bars, 40 µm.
We next examined the effect of Ripk3 deletion on TNF-induced cell death in a Tak1-deficient tissue in vivo. Because whole-body Tak1-deficient mice die within several days after gene deletion (Takaesu et al., 2012), we used mice having hepatocyte-specific deletion of Tak1, which is known to develop TNF-dependent liver damage by 1–2 months of age (Bettermann et al., 2010; Inokuchi et al., 2010). We generated mice with liver parenchymal cell-specific deletion of Tak1 (TAK1LPC-KO) in either a Ripk3−/− or a Tnfr1−/− background to determine the contribution of RIPK3 to TNF-mediated liver damage. Tak1 deficiency caused pronounced liver injury involving TUNEL-positive cells, and highly increased alanine aminotransferase (ALT) levels in serum and the activity of caspase-3 and caspase-8 in liver extracts (Fig. S3, A–C), which is consistent with the earlier studies (Bettermann et al., 2010; Inokuchi et al., 2010). Whereas ablation of TNF signaling diminished injury in Tak1-deficient livers, Ripk3 deficiency did not restore them (Fig. S3, A–C). Thus, liver damage in Tak1-deficient mice is TNF dependent but RIPK3 independent. To further analyze the role of RIPK3 in Tak1-deficient cell death in vivo, we used the LPS-induced acute hepatotoxicity injury model, as LPS injection induces TNF-dependent hepatocyte toxicity (Leist et al., 1995). Although TAK1LPC-KO mice spontaneously develop severe liver injury by 1–2 months of age, older TAK1LPC-KO mice have milder liver damage for unknown reasons (Bettermann et al., 2010; Inokuchi et al., 2010). In the acute toxicity assay, we used relatively aged (4–7 months old) mice to avoid having a high basal level of liver damage without LPS stimulation. LPS highly elevated hepatocyte death and damage in Tak1-deficient mice compared with control mice, and the liver damage was not rescued by Ripk3 deficiency (Fig. S3, D and E). These results demonstrate that RIPK3 has little contribution to cell death in Tak1-deficient cells in vivo as well as in vitro.
Enforced activation of TAK1 induces RIPK3-dependent cell death
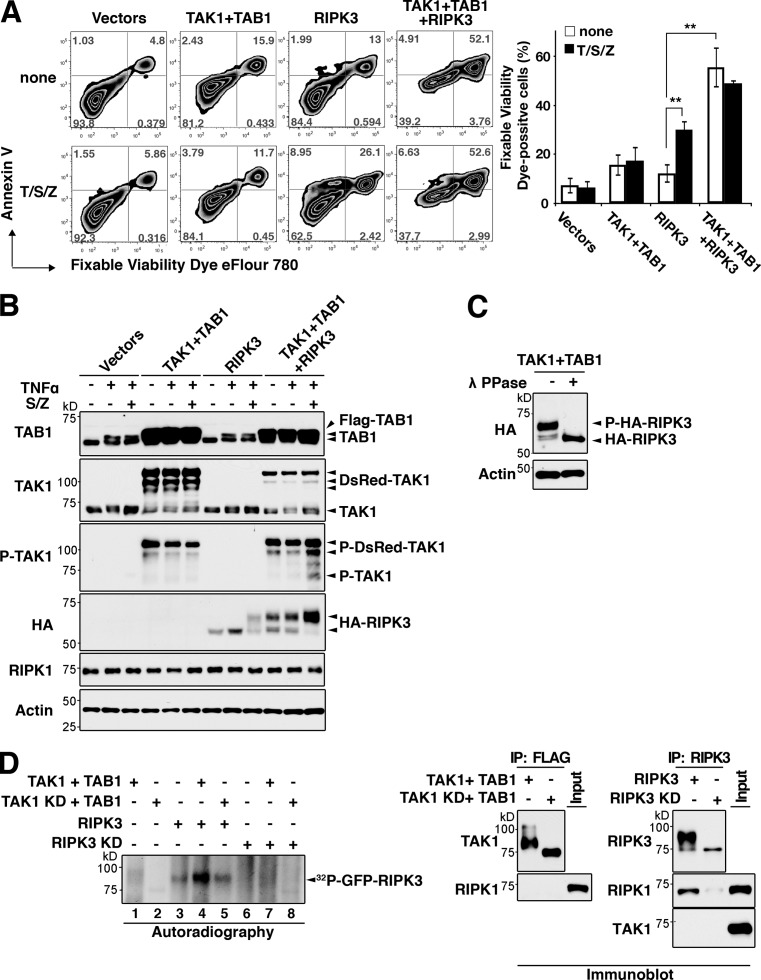
In an effort to determine the mechanism by which Tab2 deficiency activates TNF-induced RIPK3-dependent cell death, we focused on hyperactivation and sustained activation of TAK1 and asked whether it is the cause of RIPK3-dependent cell death. We used HeLa cells because they are highly transfectable and known to be deficient in RIPK3 expression (He et al., 2009). We activated TAK1 using TAK1-binding protein 1 (TAB1), which is a constitutively associated TAK1 binding partner, and co-overexpression of TAK1 and TAB1 highly activates TAK1 (Kishimoto et al., 2000). TAK1 and TAB1 were overexpressed in the presence and absence of RIPK3 in HeLa cells, which were analyzed for cell death (Fig. 5). Activation of TAK1 or expression of RIPK3 alone did not profoundly induce cell death but RIPK3 moderately increased annexin V– and cell permeability dye double-positive necrotic cell death in the presence of treatment with TNF together with an inhibitor of cIAP, Smac mimetic (Wang et al., 2008), and Z-VAD (T/S/Z) treatment (Fig. 5 A), which is consistent with the earlier study (He et al., 2009). However, when TAK1 and TAB1 were coexpressed with RIPK3, cell death was dramatically enhanced (Fig. 5 A). We also reproduced this result in mouse dermal fibroblasts. Overexpression of TAK1 and TAB1 or RIPK3 alone only marginally increased necrotic cell death in dermal fibroblasts, but overexpression of TAK1, TAB1, and RIPK3 together highly enhanced cell death (Fig. S4). Overexpression of TAK1 and TAB1 produces an active form of TAK1 and we found that it induced a slower migrating additional RIPK3 band (Fig. 5 B). The retardation of RIPK3 migration was abolished by lambda phosphatase treatment (Fig. 5 C), suggesting that RIPK3 is phosphorylated by TAK1. We then examined whether TAK1 activates RIPK3 by the in vitro kinase assay (Fig. 5 D). An active and kinase-dead TAK1, RIPK3, and kinase-dead RIPK3 were prepared from HEK293 cells separately overexpressing these proteins, and proteins were mixed as indicated and subjected to in vitro kinase assay. Kinase activity was assessed by its autophosphorylation of RIPK3. RIPK3 was basally active (lane 3), which is consistent with an earlier study (Cho et al., 2009), and the activity was further increased by TAK1 (lane 4). The kinase-dead RIPK3 was not detectably phosphorylated by TAK1 (lane 7). Thus, phosphorylation of wild-type RIPK3 (lanes 3–5) is likely to be predominantly mediated by RIPK3 autophosphorylation. We note that endogenous RIPK1 was co-precipitated with RIPK3 (Fig. 5 D, far right panels). This raises the possibility that co-precipitated RIPK1 in the RIPK3 preparation may be involved in activity of RIPK3 in these in vitro assays. However, endogenous RIPK1 was not coprecipitated with TAK1 (Fig. 5 D, second from right panels), indicating that adding TAK1 preparation to the kinase assay mixture did not increase total amount of RIPK1. Thus, the increase of RIPK3 activity (lane 4) over basal RIPK3 activity (lane 3) is mediated by TAK1 but not due to increase amount of RIPK1. These data demonstrate that an active TAK1 activates RIPK3, leading to necrosis.
Figure 5.
Hyperactivation of TAK1 promotes RIPK3-dependent cell death. (A) HeLa cells were transfected with 1 µg of Flag-TAB1 (TAB1), DsRedMT7-TAK1 (TAK1), HA-RIPK3 (RIPK3), or their control vectors for the total amount of 3 µg. At 48 h after transfection, cells were stimulated with 200 ng/ml TNF, 1 µM Smac mimetic, and 20 µM Z-VAD (T/S/Z) for 6 h. DsRed-positive transfected cells were gated, and cell death was analyzed by annexin V and fixable viability dye eFlour 780 staining. Percentages of fixable viability dye–positive cells in the transfected cells are shown (three independent experiments; mean ± SD; **, P < 0.01; P = 0.0043 and P = 0.0011 from the left). (B) HeLa cells were transfected as shown in A and treated with 200 ng/ml TNF alone or together with 1 µM Smac mimetic and 20 µM Z-VAD (S/Z). Activity of TAK1 was monitored by immunoblotting with anti-phospho-TAK1. Expression levels of TAB1, TAK1, RIPK1, and RIPK3 were also analyzed. β-Actin is shown as a control. (C) HeLa cells were transfected with expression vectors for RIPK3 with TAB1 and TAK1. Cell lysates were incubated with lambda protein phosphatase. Mobility shift of RIPK3 was observed by immunoblotting. β-Actin is shown as a control. (D) The Flag-tagged TAK1 wild-type (TAK1) or kinase-dead (TAK1 KD) TAK1 together with T7-tagged TAB1 (TAB1) and GFP-tagged wild-type (RIPK3) or kinase-dead (D161N; RIPK3 KD) RIPK3 were separately expressed in HEK293 cells. TAK1–TAB1 complex and RIPK3 were immunoprecipitated with anti-Flag or anti-RIPK3 antibody, respectively. TAK1–TAB1 complex was released from antibody beads, activated by incubation with ATP, and then mixed with RIPK3 or RIPK3 KD. RIPK3 kinase activity was assessed by in vitro kinase assay. Left panel, autoradiograph; right panels, immunoblot analysis of the immunoprecipitates with anti-TAK1 and anti-RIPK3 antibodies. Co-precipitated endogenous TAK1 and RIPK1 in the protein preparations were also determined by immunoblotting (right panels). Because HEK293 cells do not express RIPK3, endogenous RIPK3 was not examined.
TAK1 mediates TNF-induced cell death in Tab2-deficient cells
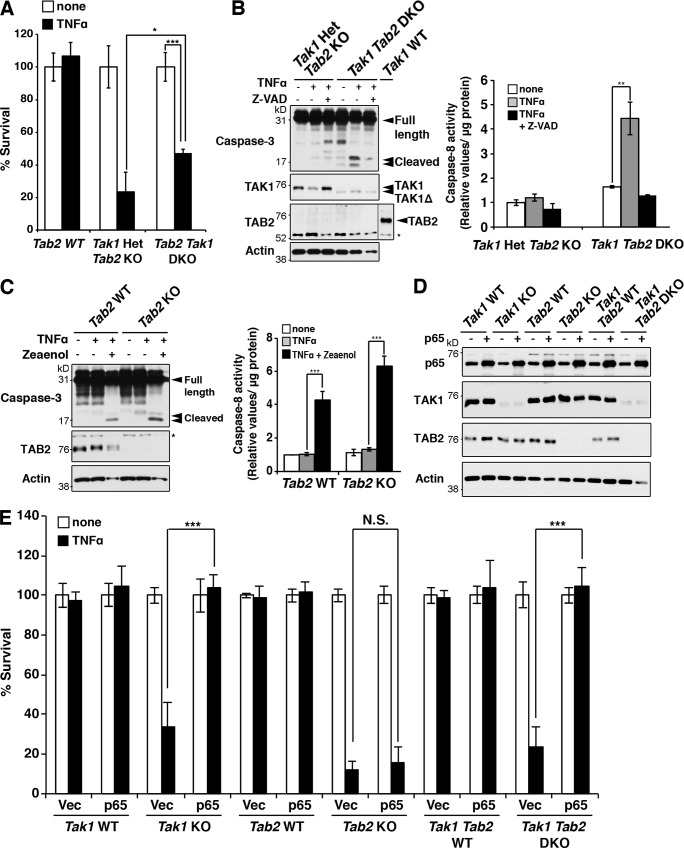
The results shown in Fig. S1 demonstrate that Tab2 deficiency induces hyperactivation of TAK1, which might be the cause of TNF-induced RIPK3-dependent cell death in Tab2-deficient fibroblasts. If Tab2 deficiency mediates TNF-induced cell death through enhanced TAK1 activity, inhibition of TAK1 should rescue the TNF-dependent cell death in Tab2-deficient cells. To address this, we generated inducible Tab2 and Tak1 double-deficient mice (Rosa26-CreERT Tab2flox/flox Tak1flox/flox), and dermal fibroblasts were isolated and used for the experiments. However, we found that Tak1 and Tab2 double-deficient fibroblasts still died (Fig. 6 A) and caspase activity was increased in response to TNF (Fig. 6 B), resembling Tak1 single-deficient fibroblasts. We confirmed these results by using a selective TAK1 inhibitor, 5Z-7-Oxozeaenol (Zeaenol; Fig. 6 C). Thus, inhibition of TAK1 activates apoptosis and does not rescue TNF-induced cell death in Tab2-deficient fibroblasts. To overcome this difficulty, we attempted to block TNF-induced apoptosis. TNF-induced NF-κB activation is known to be impaired in Tak1-deficient cells (Sato et al., 2005; Omori et al., 2008), and NF-κB is an inducer of anti-apoptotic proteins such as c-FLIP (Pasparakis, 2009). Based on these data, we stably overexpressed an NF-κB protein, p65, in Tak1- or/and Tab2-deficient fibroblasts (Fig. 6 D). p65 overexpression increased c-FLIP in Tak1-deficient fibroblasts (Fig. S5). As anticipated, p65 overexpression blocked TNF-induced cell death in Tak1-deficient fibroblasts (Fig. 6 E). Thus, enforced activation of NF-κB can block TNF-induced apoptosis in Tak1-deficient fibroblasts. Importantly, we found that p65 overexpression did not at all rescue TNF-induced cell death in Tab2-deficient fibroblasts (Fig. 6 E). Thus, enforced activation of NF-κB blocks only Tak1 deficiency–caused apoptosis but does not affect RIPK3-dependent cell death in Tab2-deficient fibroblasts, and this allows us to investigate the role of Tak1 in RIPK3-dependent cell death by blocking apoptosis caused by Tak1 deficiency. TNF-induced RIPK3-dependent cell death may be through hyperactivation of TAK1 as we hypothesized or through TAK1-independent mechanisms. If hyperactivation of TAK1 is the mechanism, deletion of Tak1 should rescue TNF-induced cell death in p65 overexpressing Tab2-deficient fibroblasts. Conversely, if Tab2-deficient fibroblasts died upon TNF treatment not solely through a TAK1-dependent mechanism, deletion of Tak1 would not effectively rescue TNF-induced cell death in p65 overexpressing Tab2-deficient fibroblasts. We found that Tak1 and Tab2 double-deficient fibroblasts with a stable overexpression of p65 were completely resistant to TNF-induced cell death (Fig. 6 E). This result clearly indicates that TAK1 mediates TNF-induced cell death in Tab2-deficient fibroblasts.
Figure 6.
Tak1 deletion rescues TNF-induced cell death in Tab2-deficient fibroblasts when apoptosis is blocked. (A) To generate Tab2 KO, Tak1 Het, and Tab2 Tak1 DKO fibroblasts, Rosa26-CreERT Tab2flox/flox, Rosa26-CreERT Tak1flox/+Tab2flox/flox, and Rosa26-CreERT Tak1flox/floxTab2flox/flox dermal fibroblasts were treated with 0.1 µM 4-hydroxytamoxifen to induce the deletion of Tak1 and Tab2. Rosa26-CreERT Tab2flox/flox dermal fibroblasts were treated with vehicle (EtOH) and studied in parallel as Tab2 WT controls. Fibroblasts were stimulated with 200 ng/ml TNF for 24 h and cell viability was determined by the crystal violet assay (three independent experiments; mean ± SD; *, P = 0.029; ***, P = 0. 00056). (B) Tab2 KO Tak1 Het and Tab2 Tak1 DKO fibroblasts were pretreated with vehicle (DMSO) or Z-VAD (20 µM) for 1 h and stimulated with 200 ng/ml TNF for 6 h. Caspase-3 was analyzed by immunoblotting. Immunoblots of TAK1, TAB2, and β-actin are shown as controls. The asterisk indicates a nonspecific band. Caspase-8 activity in cellular extracts from samples treated with the same procedure was measured. Data are shown as caspase-8 activity relative to that in unstimulated Tab2 KO Tak1 Het fibroblasts (three independent experiments; mean ± SD; **, P = 0.0019). (C) Tab2 WT and Tab2 KO fibroblasts were pretreated with vehicle (DMSO) or 5Z-7-Oxozeaenol (Zeaenol; 1 µM) for 1 h and stimulated with TNF (200 ng/ml) for 6 h. Immunoblots of TAB2 and β-actin are shown as controls. The asterisk indicates a nonspecific band. Caspase-8 activity in cellular extracts from samples treated with the same procedure was measured. Data are shown as caspase-8 activity relative to that in unstimulated Tab2 WT fibroblasts (three independent experiments; mean ± SD; ***, P < 0.001; P = 0.00038 and P = 0.000014 from the left). (D and E) Tak1 WT, Tak1 KO, Tab2 WT, Tab2 KO, Tak1 Tab2 WT, and Tak1 Tab2 DKO fibroblasts were stably transfected with p65. Immunoblots of p65, TAK1, TAB2, and β-actin are shown (D). Fibroblasts were stimulated with 200 ng/ml TNF for 24 h, and cell viability was determined by the crystal violet assay (E) (three independent experiments; mean ± SD; ***, P < 0.001; N.S., not significant; P = 3E–05, P = 0.58, and P = 0.00015 from the left).
TAK1 is hyperactivated by inhibition of caspases
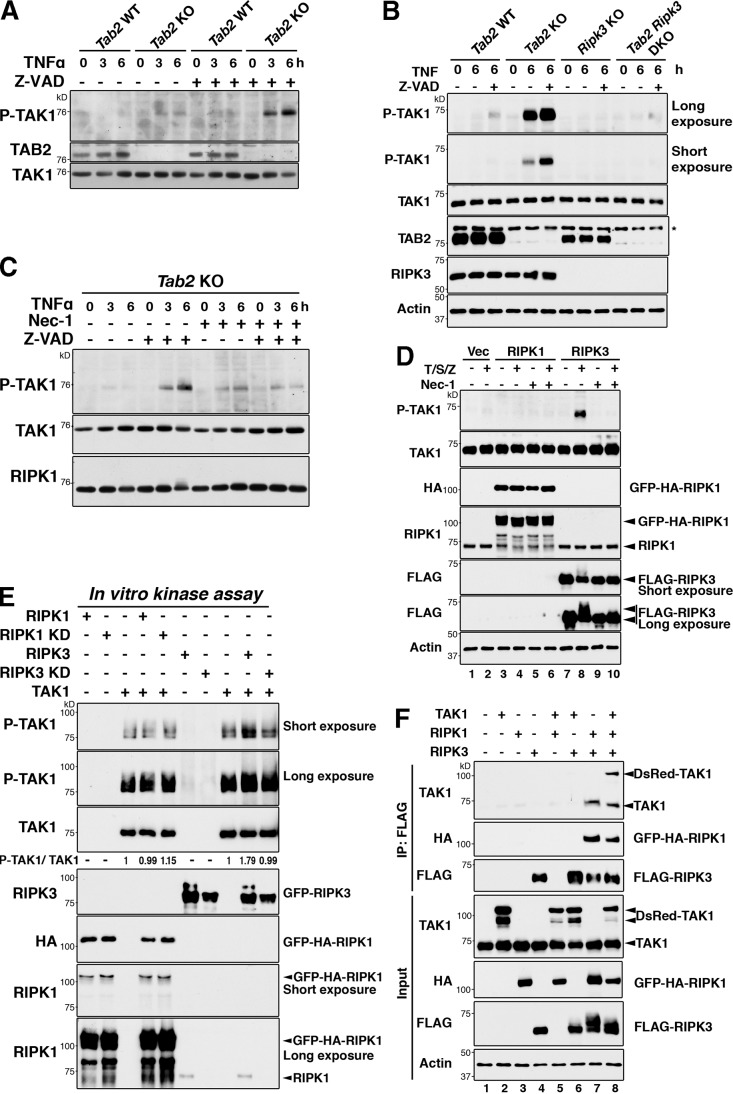
We asked whether TAK1 is hyperactivated under any physiological conditions other than Tab2 deletion. Based on the results showing that TAK1 promotes RIPK3-dependent cell death, we hypothesized that necrosis-inducing conditions might be associated with hyperactivation of TAK1. The most well established condition to promote TNF-induced necrosis is inhibition of caspases, which occurs when viral-derived caspase inhibitors are introduced into cells. We examined whether TAK1 was activated by inhibition of caspases in fibroblasts. Inhibition of caspases (Z-VAD treatment) slightly activated TAK1 at 3–6 h after TNF treatment in wild-type fibroblasts, and inhibition of caspases in Tab2-deficient fibroblasts further up-regulated TAK1 activity (Fig. 7, A and B; 1–6 lanes from left). Thus, hyperactivation of TAK1 is associated with caspase inhibition and necrotic cell death. We next examined the mechanism of TAK1 hyperactivation. Because caspase inhibition upon TNF stimulation activates RIPK1 and RIPK3, we hypothesized that RIPK1 or RIPK3 or both are involved in the hyperactivation of TAK1. We first examined whether RIPK3 is required for TAK1 activation by using Ripk3-deficient dermal fibroblasts. Ripk3 deficiency reduced TNF- and Z-VAD–induced TAK1 activation in Tab2-deficient fibroblasts (Fig. 7 B). This indicates that RIPK3 mediates the TAK1 hyperactivation. To examine the involvement of RIPK1, Nec-1 was used and we found that inhibition of RIPK1 also reduced TNF- and Z-VAD–induced TAK1 activation in Tab2-deficient fibroblasts (Fig. 7 C). These results suggest that both RIPK1 and RIPK3 are essential for TAK1 hyperactivation. To further investigate the contribution of RIPK1 and RIPK3 to TAK1 activation, we examined whether RIPK1 and/or RIPK3 can activate TAK1 using the overexpression system in HeLa cells (Fig. 7 D). As mentioned above, HeLa cells expressed endogenous RIPK1 (Fig. 7 D, fourth panel) but not RIPK3 (He et al., 2009). Treatment of TNF, Smac mimetic, and Z-VAD activated overexpressed RIPK3 (Fig. 7 D, sixth panel, lane 8) and endogenous TAK1 (Fig. 7 D, top panel, lane 8). However, RIPK1 overexpression did not activate TAK1 even with treatment with TNF, Smac mimetic, and Z-VAD (Fig. 7 D, top panel, lane 4). This demonstrates that an active RIPK1 alone cannot activate TAK1. Interestingly, TNF-, Smac mimetic–, and Z-VAD–induced activation of TAK1 in RIPK3-expressing cells was completely blocked by Nec-1 (Fig. 7 D, top panel, lane 10). Nec-1 completely blocked activation of RIPK3 (Fig. 7 D, sixth panel, lanes 8 and 10). These data indicate that RIPK3 may be a direct activator and RIPK1 is required for activation of RIPK3. To further evaluate whether RIPK3 is a direct activator of TAK1, we incubated TAK1 immunopurified from TAK1-overexpressing HEK293 cells with RIPK1 or RIPK3, which was separately prepared from RIPK1- or RIPK3-overexpressing HEK293 cells. We found that the in vitro incubation of TAK1 with RIPK3 but not with RIPK1 elevated TAK1 activity (Fig. 7 E). These data confirm that RIPK3 is a direct activator of TAK1. We note that endogenous RIPK1 was co-precipitated with RIPK3 and was present in the in vitro kinase mixture (Fig. 7 E, bottom panel). This raises the possibility that RIPK1 may still participate in RIPK3-induced TAK1 activation through some mechanisms but not through direct activation of TAK1. We examined interaction among TAK1, RIPK1, and RIPK3 using a HEK293 overexpression system (Fig. 7 F). TAK1 and RIPK3 were not detectably associated when both proteins were overexpressed in HEK293 cells (Fig. 7 F, lane 6), but overexpression of RIPK1 induced an interaction between TAK1 and RIPK3 (Fig. 7 F, lane 8). Overexpression of RIPK1 also induced an interaction between RIPK3 and endogenous TAK1 (Fig. 7 F, lane 7). Thus, RIPK1 mediates interaction between TAK1 and RIPK3. These results demonstrate that RIPK1 contributes to activation of TAK1 by facilitating interaction between TAK1 and RIPK3 as well as activating RIPK3 in response to TNF, Smac mimetic, and Z-VAD. We describe that TAK1 activated RIPK3 in Fig. 5, in that endogenous RIPK1 was found to be present in the assay mixture. Thus, RIPK1 may also enhance TAK1-induced activation of RIPK3 by facilitating interaction between TAK1 and RIPK3. Collectively, TAK1 activates RIPK3, RIPK3 activates TAK1, and RIPK1 activates RIPK3 and facilitates interaction between TAK1 and RIPK3. We propose that TAK1, RIPK1, and RIPK3 are activated through a positive feedforward loop mechanism.
Figure 7.
Inhibition of caspases hyperactivates TAK1 through RIPK1 and RIPK3. (A) Tab2 WT and Tab2 KO fibroblasts were pretreated with vehicle (DMSO) or Z-VAD (20 µM) for 1 h and stimulated with TNF (20 ng/ml) for the indicated time periods, and TAK1 activation was determined by immunoblotting with anti–phospho-TAK1. TAB2 and TAK1 are shown as controls. (B) Tab2 WT, Tab2 KO, and Ripk3-deficient (Ripk3 KO) and Tab2 and Ripk3-deficinet (Tab2 Ripk3 DKO) dermal fibroblasts were pretreated with vehicle (DMSO) or Z-VAD (20 µM) for 1 h and stimulated with TNF (20 ng/ml) for 6 h. TAK1 activation was determined by immunoblotting with anti–phospho-TAK1. TAK1, TAB2, RIPK3, and β-actin are shown as controls. The asterisk indicates a nonspecific band. (C) Tab2 KO fibroblasts were pretreated with 20 µM Z-VAD and 30 µM Nec-1 for 1 h as indicated, then stimulated with 20 ng/ml TNF. TAK1 activation was determined by immunoblotting with anti–phospho-TAK1. Immunoblots of total TAK1 and RIPK1 are shown as controls. (D) HeLa cells were transfected with GPF-HA-RIPK1 (RIPK1), FLAG-RIPK3 (RIPK3), or control vectors (Vec) and pretreated with 30 µM Nec-1 for 1 h, then stimulated with 200 ng/ml TNF alone or together with 1 µM Smac mimetic and 20 µM Z-VAD (S/Z). TAK1 activation was determined by immunoblotting with anti–phospho-TAK1. Total amounts of TAK1, overexpressed RIPK1, endogenous RIPK1, and overexpressed RIPK3 and β-actin are shown. (E) The Flag-tagged TAK1 wild-type (TAK1), GFP- and HA-tagged RIPK1 wild-type (RIPK1), or kinase-dead (K45A; RIPK1 KD), GFP-tagged RIPK3 wild-type (RIPK3), or kinase-dead (D161N; RIPK3 KD) were separately expressed in HEK293 cells. TAK1, RIPK1, and RIPK3 were immunoprecipitated with anti-Flag, anti-HA, or anti-RIPK3 antibody, respectively. TAK1 was released from antibody beads and then mixed with RIPK1, RIPK1 KD, RIPK3, or RIPK3 KD immunoprecipitates as indicated. TAK1 activation was determined by immunoblotting with anti–phospho-TAK1. Protein amounts of TAK1, RIPK1, and RIPK3, and endogenous RIPK1 coprecipitated in RIPK3 precipitates were determined by immunoblotting. Density measurement was conducted using Photoshop CS3 and the relative ratio between phospho-TAK1 and total TAK1 to that in TAK1 expression alone (third lane from left) is shown. (F) HEK293 cells were transfected with DsRedMT7-TAK1 (TAK1), GFP-HA-RIPK1 (RIPK1), and Flag-RIPK3 (RIPK3) or control vectors, and total cell lysates were immunoprecipitated with anti-Flag antibody. RIPK3-binding proteins were detected by immunoblotting using anti-HA and anti-TAK1 antibodies. Immunoprecipitated RIPK3 was detected by anti-Flag.
Discussion
TAK1 kinase switches between apoptosis and necrosis
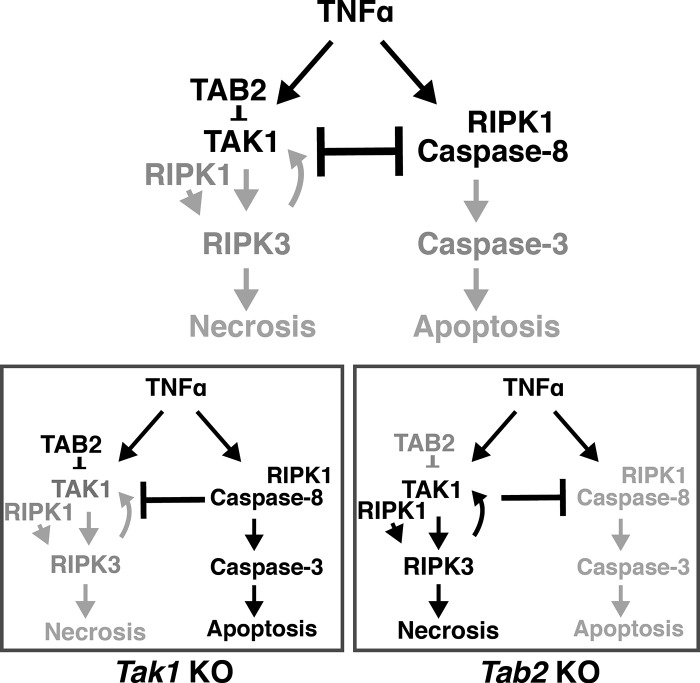
As summarized in Fig. 8, the current study reveals that diminished TAK1 activity activates caspase-8, whereas hyperactivation of TAK1 promotes RIPK3-dependent necrosis. It has been established that a transient activation of TAK1 in response to TNF engages cell survival through induction of NF-κB and antioxidant enzymes (Shim et al., 2005; Omori et al., 2008). Thus, the level and timing of TAK1 activation determine whether cells are alive or undergo apoptosis or necrosis after TNF stimulation.
Figure 8.
Models. In wild-type cells, TAK1 is transiently activated upon TNF stimulation and inhibits caspase-8, which blunts the apoptosis pathway. Tak1 deficiency activates TNF-induced caspase-8 apoptosis pathway, which is inhibitory to the RIPK3-dependent necrosis pathway. Hyperactivation of TAK1 blocks TNF-induced apoptosis and activates the RIPK1–RIPK3 necrotic pathway. TAK1–RIPK1–RIPK3 is activated by a positive feedforward loop mechanism.
Relationship between caspase-8 deficiency and Tak1 deficiency
Inhibition of caspases by gene deletion of either caspase-8 or its adaptor protein FADD causes RIPK1–RIPK3-dependent cell death (Kaiser et al., 2011; Oberst et al., 2011; Zhang et al., 2011). Interestingly, mice having deletion of Tak1, caspase-8, or Fadd genes share similarity. Epidermal-specific deletion of caspase-8 causes severe inflammatory conditions in the skin in neonatal mice (Kovalenko et al., 2009), which resembles the skin in mice having epidermal-specific deletion of Tak1 (Omori et al., 2006). Caspase-8 deficiency is associated with abnormal vascular development during embryogenesis (Kaiser et al., 2011), which is also seen in embryos with endothelial-specific deletion of Tak1 (Morioka et al., 2012). Furthermore, intestinal epithelial-specific deletion of Fadd or caspase-8 causes ileitis (Günther et al., 2011; Welz et al., 2011), which is identical to intestinal epithelial-specific deletion of Tak1 (Kajino-Sakamoto et al., 2008). These might suggest that TAK1 is involved in maintenance of caspase-8 activity, which is pro-cell survival. However, as shown in the current study, caspases are activated after TNF stimulation in Tak1-deficient fibroblasts. Thus, the mechanisms of cell death in Tak1 deficiency and caspase-8 deficiency are different. Indeed, abnormalities in embryogenesis by deletion of caspase-8 or Fadd are rescued by Ripk3 gene deletion (Kaiser et al., 2011; Oberst et al., 2011), but TNF-induced cell death in Tak1-deficient cells is not rescued by Ripk3 deletion in vivo and in vitro as shown in the current study. Vucur et al. (2013) also recently reported that Ripk3 deletion did not rescue liver damage in Tak1-deficient liver. Thus, caspase-8 or Fadd deficiency kills cells through RIPK3-dependent necrosis, whereas TAK1 deficiency causes caspase-induced apoptosis. TNF is expressed at some levels in tissues, and kills cells, if caspase-8, Fadd, or Tak1 gene is deleted, through two different mechanisms leading to similar outcomes.
The effect of caspase inhibition in Tak1-deficient cells
To block TNF-induced apoptosis in Tak1-deficient cells, we initially used a pan-caspase inhibitor, Z-VAD. However, Z-VAD failed to rescue cell death in Tak1-deficient cells (Fig. S1 D). We assume that caspase inhibition by Z-VAD activates necrosis in response to TNF in Tak1-deficient cells, consistent with reports showing that Z-VAD does not block but rather enhances TNF-induced necrotic cell death (Hirsch et al., 1997; Degterev et al., 2005; Han et al., 2011). NF-κB is known to up-regulate inhibitory molecules for caspase-8 such as c-FLIP and cIAPs (Micheau and Tschopp, 2003). In contrast to Z-VAD treatment, we show that overexpression of NF-κB could suppress apoptosis without activating necrosis (Fig. 6). What causes these different outcomes of Z-VAD treatment and NF-κB overexpression? TNF stimulation normally induces the formation of a complex containing RIPK1, RIPK3, FADD, c-FLIP, and caspase-8 (Green et al., 2011). Dillon et al. (2012) described that c-FLIP makes a heterodimer with caspase-8 and prevents an active homodimer formation of caspase-8. The c-FLIP–caspase-8 heterodimer inhibits RIPK3, resulting in no activation of either caspase-8 or RIPK3. We found that the basal expression level of c-FLIP was reduced in Tak1-deficient cells, which was restored by NF-κB overexpression (Fig. S5). These data suggest that reduced c-FLIP expression and subsequent caspase-8 homodimer formation may be involved in TNF-induced caspase-8 activation in Tak1-deficient cells. In this context, restoration of c-FLIP by NF-κB overexpression inhibits the homodimer formation of caspase-8 and normalizes the complex composition so that RIPK3 is not activated. On the other hand, caspase-8 inhibition without restoration of c-FLIP in Tak1-deficient cells would activate RIPK3-dependent necrosis.
RIPK1-dependent caspase-8 activation in Tak1-deficient fibroblasts
Tak1-deficient fibroblasts undergo apoptosis upon TNF stimulation. Our results clearly demonstrate that RIPK1 activity is required for this apoptosis. However, RIPK1 is not typically involved in death ligand–induced apoptosis (Ting et al., 1996). Only when polyubiquitination of RIPK1 is blocked by inhibition of ubiquitination enzymes cIAPs or by mutation at the polyubiquitination site of RIPK1, RIPK1 becomes a pro-apoptotic and activates caspase-8 (O’Donnell et al., 2007; Wang et al., 2008). Treatment of a cIAP inhibitor, Smac mimetic, together with TNF is well known to induce this type of apoptosis. Interestingly, we have previously reported that cIAPs are down-regulated by Tak1 deletion in keratinocytes (Morioka et al., 2009). Thus, it is likely that TNF-induced apoptosis in Tak1-deficient fibroblasts is through a mechanism similar to that of Smac mimetic treatment.
Relationship between TAK1 and RIPK1–RIPK3
It has been shown that phosphorylation of RIPK1 and RIPK3 is important for stable RIPK1–RIPK3 complex formation and subsequent execution of necrotic cell death (Cho et al., 2009; He et al., 2009). RIPK1 kinase activity is required for RIPK3 phosphorylation, and reciprocally RIPK3 phosphorylates RIPK1 (Cho et al., 2009). However, the mechanism by which these phosphorylation events are regulated was still elusive. We reveal that hyperactivation of TAK1 induces phosphorylation of RIPK3 even without TNF stimulation. This raises the possibility that TAK1 acts upstream of RIPK3. We also found that RIPK3 activation leads to hyperactivation of TAK1, suggesting that TAK1 is not only upstream of RIPK3 but also a target of RIPK3. Furthermore, we show that RIPK1 facilitates RIPK3-induced TAK1 activation, suggesting that TAK1–RIPK1–RIPK3 is activated in a positive feedforward mechanism. Our results demonstrate that TAK1 promotes RIPK1–RIPK3–necrosis; however, it is still not clear whether TAK1 is entirely required for this type of death. Inhibition of caspase in Tak1-deficient fibroblasts initially blocked but still induced cell death upon TNF stimulation in later time points (Fig. S1, B and D). Because treatment of Z-VAD and TNF normally activates RIPK3, this cell death is presumably RIPK3-dependent necrosis. Thus, TAK1 may not be required for RIPK3 activation under some circumstances. Alternatively, Tak1 deficiency may cause RIPK3-independent cell death when caspases are inhibited. Further studies in cell death in Tak1-deficient cells will clarify contribution of TAK1 to RIPK3-dependent and potentially other types of cell death. In addition, further studies are required to define precise physical interactions and potential phosphorylation sites of TAK1, RIPK1, and RIPK3 using purified proteins. Our results identify TAK1–RIPK1–RIPK3 as a necrosis promoting protein kinase cascade.
Materials and methods
Mice and cell culture
Tab2-floxed (Tab2flox/flox) mice (Sanjo et al., 2003) were backcrossed for a minimum of five generations to C57BL/6 mice. Fibroblasts were isolated from the dermis of the neonatal mice and spontaneously immortalized in culture. Tab2-deficient fibroblasts were generated by infection of a retroviral vector for the expression of Cre recombinase. We isolated more than 10 independent clones of control and Tab2-deficient fibroblasts (Broglie et al., 2010), and we confirmed that all Tab2-deficient fibroblast clones were sensitive to TNF-induced cell death. Several double-deficient fibroblasts were generated by using mice having combinations of Tab2flox/flox, Tak1flox/flox (Sato et al., 2005), and Ripk3−/− mice (a gift from V.M. Dixit, Genentech, Inc., South San Francisco, CA; Newton et al., 2004) and a deleter transgene, Rosa26-CreERT (The Jackson Laboratory; Badea et al., 2003). Gene deletion was induced in vitro by incubation with 0.1 µM 4-hydroxytamoxifen for 3 d. Fibroblasts, HeLa, and HEK293 cells were cultured in DMEM with 10% bovine growth serum (Hyclone) and penicillin/streptomycin at 37°C in 5% CO2. Alb.Cre and Tnfr1−/− C57BL/6 mice were obtained from The Jackson Laboratory (Pfeffer et al., 1993; Postic et al., 1999). Hepatocyte-specific Tak1-deficient Alb.Cre Tak1flox/flox (Tak1LPC-KO) mice, in a background of either Tnfr1−/− or Ripk3−/−, were generated and compared with Alb.Cre Tak1flox/flox in a wild-type background. To induce gene deletion in mice having Rosa26-CreERT transgene, mice were intraperitoneally injected with 50 mg/kg tamoxifen for three consecutive days. Mice were subjected to experiments at least two weeks after tamoxifen treatment to prevent a potential effect from acute Cre toxicity as described previously (Takaesu et al., 2012). All animal experiments were conducted with the approval of the North Carolina State University Institutional Animal Care and Use Committee.
Antibodies, reagents, and plasmids
Anti–phospho-TAK1 (T187) rabbit polyclonal antibody (Cell Signaling Technology) was used to detect the phosphorylated form of TAK1. Anti-TAK1, -TAB1, and -TAB2 rabbit polyclonal antibodies were described previously (Ninomiya-Tsuji et al., 1999; Kishimoto et al., 2000; Takaesu et al., 2000). Anti-RIPK3 (R4277, rabbit polyclonal; Sigma-Aldrich), anti-RIPK1 (mouse monoclonal, BD), anti–caspase-3 (8G10, rabbit polyclonal; Cell Signaling Technology), anti-p65 (C20, rabbit polyclonal; Santa Cruz Biotechnology, Inc.), anti–β-actin (C5, mouse monoclonal; Sigma-Aldrich), anti-HA (HA.11, mouse monoclonal; Covance), and anti-FLAG (M2, mouse monoclonal; Sigma-Aldrich) were used. The reagents used were TNF (PeproTech), Z-VAD-fmk (Z-VAD; Enzo Life Sciences), and Necrostatin-1 (Nec-1; Enzo Life Sciences). Smac mimetic (C2-symmetric compound; Li et al., 2004) was a gift from X. Wang (University of Texas Southwestern Medical Center, Dallas, TX). Full-length TAK1 cDNA was subcloned into pCMV-DsRedMonomer-T7 vector to generate pCMV-DsRedMT7-TAK1. pCMV-TAB1 and pCMV-TAK1 (Ninomiya-Tsuji et al., 1999; Inagaki et al., 2008) and HA-tagged RIPK3 expression plasmid (pCI-neo-RIPK3-HA, a gift from X. Wang) were used. GFP-tagged wild-type and kinase-dead (D161N) RIPK3 expression plasmid (pEGFP-N1-GFP-RIP3 and pEGFP-N1-GFP-RIP3 (D161N)) and GFP- and HA-tagged wild-type and kinase-dead (K45A) RIPK1 (pEGFP-N1-GFP-RIP1 and pEGFP-N1-GFP-RIP1 (K45A)) were a gift from F. Chan (University of Massachusetts Medical School, Worcester, MA; Addgene plasmids).
Crystal violet assay
Cells were plated on 24-well dishes overnight at 37°C in 5% CO2 at a concentration of 104 cells per well. Cells were either pretreated with Z-VAD or Nec-1 for 1 h and exposed to TNF for 24 h. Cells were fixed in 10% formalin and stained with 0.1% crystal violet. The dye was eluted and analyzed at 595 nm.
Caspase assay
Cell extracts were mixed with the substrate mixture (Caspase-Glo; Promega) for caspase-8 or caspase-3, and the cleaved substrates were determined according to the manufacturer’s instructions.
Immunoblotting
Whole-cell extracts were prepared using an extraction buffer containing 20 mM Hepes, pH 7.4, 150 mM NaCl, 12.5 mM β-glycerophosphate, 1.5 mM MgCl2, 2 mM EGTA, 10 mM NaF, 2 mM DTT, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 20 µM aprotinin, and 0.5% Triton X-100. Cell extracts were resolved on SDS-PAGE and transferred to Hypond-P membranes (GE Healthcare). The membranes were immunoblotted with various antibodies, and the bound antibodies were visualized with horseradish peroxidase–conjugated antibodies against rabbit or mouse IgG using the ECL Western blotting system (GE Healthcare).
Transmission electron microscopy
Fibroblasts were suspended in McDowell and Trump’s 4F:1G fixative. The samples were then suspended in 1% osmium tetroxide in 0.1 M phosphate buffer (pH 7.4) for 1 h, dehydrated, and infiltrated with Spurr resin. 90-nm ultra-thin sections were stained with uranyl acetate and analyzed through an electron microscope (FEI/Philips EM 208S/Morgagni) at 80 kV.
Ripk3 knockdown
Two different Ripk3 and nontargeting control siRNAs were generated (Ripk3 siRNA-1 [#1], 5′-GAGCTGTTATTTGATGTCAACCTGA-3′; Ripk3 siRNA-2 [#2], 5′-TGGCACTCCTCAGATTCCACATACT-3′; and non-targeting siRNA, 5′-UUCUCCGAACGUGUCACGU-3′). Fibroblasts were transfected with the siRNAs using Lipofectamine 2000 (Invitrogen).
Dephosphorylation of RIPK3
Cell lysates from HeLa cells were incubated with lambda protein phosphatase (New England Biolabs, Inc.) for 4 h at 30°C in a buffer containing 1 mM MnCl2 according to the manufacturer’s instructions, and analyzed by immunoblotting.
Liver damage analysis
Spontaneous liver injury was examined at 1–2 months of age. For acute liver injury model, 4–7-mo-old mice were intraperitoneally injected with LPS at 10 mg/kg. Liver specimens were fixed in 4% paraformaldehyde, and paraffin sections were subjected to TUNEL (Promega) staining according to the manufacturer’s protocol. Serum ALT levels were measured using an ALT assay kit (BioVision).
Intradermal TNF injection
Mice were anesthetized by intraperitoneal injection of 100 mg/kg ketamine and 10 mg/kg xylazine. After shaving hair, 1 µg of TNF was intradermally injected to the skin. After 6 h, skin samples were collected.
Wound surgery
Mice were anesthetized and a skin wound was generated using a disposable biopsy punch (6.0 mm; Sklar Instruments). The wound area was monitored and measured every day, and wound closure was expressed as a percentage of initial wound size.
Microscopy and image analysis
TUNEL staining was observed with 20× (UPlanFI; Olympus) and 40× (UPlanFLN; Olympus) objectives, and images were acquired using a fluorescence microscope (model BX41; Olympus) and camera (model XM10; Olympus) at room temperature with a DAPI or FITC filter. 3–10 immunofluorescent images per mouse were randomly photographed, and at least 1,000 DAPI-stained cells per mouse were counted. Quantitative results were generated from the counted numbers in 3–6 mice from independent experiments. Bright-field images were acquired with a 20× (LCAchN; Olympus) objective using an inverted microscope (model CKS41; Olympus) at room temperature. Image analysis was performed using cellSens Standard (Olympus) image analysis software.
Flow cytometric analysis
HeLa cells and dermal fibroblasts were transfected with 1 µg of plasmids using Lipofectamine 2000. At 48 h after transfection, cells were stimulated with TNF, Smac mimetic, and Z-VAD for 6 h. Cell death was analyzed by fixable viability dye eFluor 780 (eBioscience) at 1 µl/ml and Pacific Blue–annexin V (Invitrogen). Stained cells were analyzed by a flow cytometer (LSR II; BD). Data were analyzed using FlowJo software (Tree Star).
Immunoprecipitation and in vitro kinase assay
HEK293 cells were transfected with plasmids using the standard calcium phosphate method. Cell lysates were prepared in the extraction buffer described above. Flag-tagged TAK1, GFP-HA-tagged RIPK1, and GFP-tagged RIPK3 were immunoprecipitated with specific antibodies for 3 h at 4°C (kinase assay) or overnight at 4°C. The resulting immune complexes were washed with IP washing buffer (20 mM Hepes, pH 7.4, 10 mM MgCl2, and 500 mM NaCl) three times followed by a wash with kinase buffer (10 mM Hepes, pH 7.4, 1 mM DTT, and 5 mM MgCl2). TAK1 complex was released from antibody beads by incubation with 100 µM Flag peptide. For the kinase assay using RIPK3 as a substrate, TAK1 was pre-activated by incubation in kinase buffer with 1 mM ATP for 30 min at 30°C as described previously (Kishimoto et al., 2000) and washed once with kinase butter. The kinase and substrate were mixed and incubated in kinase buffer supplemented with 200 µM ATP and 5 µCi γ-[32P]ATP for 30 min at 30°C. Samples were resolved on SDS-PAGE and exposed to autoradiographic films or subjected to immunoblotting.
Retroviral infection
Retroviral vectors for p65 (pMXpuro-p65) were generated by inserting p65 cDNA into the retroviral vector pMX-puro. PLAT-E cells (Morita et al., 2000) were transiently transfected with pMX-puro-p65. After 48 h culture, the growth medium containing the retrovirus was collected and filtered with a 0.45-µm cellulose acetate membrane to remove packaging cells. Fibroblasts were incubated with the collected virus-containing medium with 10 µg/ml polybrene for 24 h. Uninfected cells were removed by puromycin selection.
Statistical analyses
Statistical analyses were performed using two-tailed Student’s t test or one-way ANOVA for comparing the means of two or multiple groups, respectively. *, P < 0.05; **, P < 0.01; ***, P < 0.001; N.S., not significant when P > 0.05.
Online supplemental material
Fig. S1 A shows TNF-induced hyperactivation of TAK1 in Tab2-deficient fibroblasts. Fig. S1 B shows bright-field images of Tak1- and Tab2-deficient fibroblasts after TNF stimulation. Fig. S1 C shows TEM images of Tak1- and Tab2-deficient fibroblasts after TNF stimulation. Fig. S1 D shows that Z-VAD does not inhibit TNF-induced cell death in Tak1-deficient fibroblasts. Fig. S2 A shows that Nec-1 blocks TNF-induced cell death in Tak1-deficient fibroblasts. Fig. S2 B shows that Tab2 and Ripk3 double-deficient fibroblasts are completely resistant to TNF-induced cell death. Fig. S3 shows that Ripk3 deletion does not rescue chronic and acute liver injury in liver-specific Tak1-deficient mice. Fig. S4 shows that overexpression of an active TAK1 induces RIPK3-dependent cell death in fibroblasts. Fig. S5 shows that cFLIP is reduced in Tak1-deficient fibroblasts and is restored by p65 overexpression. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201305070/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Dixit for providing Ripk3−/− mice; Dr. Wang for providing Smac mimetic and RIPK3 expression plasmids; Dr. Chan for providing GFP-tagged RIPK3 plasmids; Dr. Kitamura for PLAT-E cells; Dr. Dykstra, A. Wood, and J. Shipley-Phillips at the North Carolina State University Laboratory for Advanced Electron and Light Optical Methods core facility for TEM support; and the North Carolina State University Histology Laboratory for preparation of paraffin sections.
This work was supported by National Institutes of Health grant GM068812 (to J. Ninomiya-Tsuji).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- ALT
- alanine aminotransferase
- c-FLIP
- cellular FLICE-like inhibitory protein
- cIAP
- cellular inhibitor of apoptosis protein
- DISC
- death-inducing signaling complex
- FADD
- Fas-associated death domain
- IKK
- IκB kinase
- NF-κB
- nuclear factor-κB
- PP6
- protein phosphatase 6
- RIPK
- receptor-interacting protein kinase
- TAB
- TAK1-binding protein
- TAK
- TGF-β–activated kinase
- TNF-α
- tumor necrosis factor-α
- TNFR
- TNF receptor
- TRAF
- TNF receptor-associated factor
- Z-VAD
- Z-VAD(OMe)-FMK
References
- Arslan S.C., Scheidereit C. 2011. The prevalence of TNFα-induced necrosis over apoptosis is determined by TAK1-RIP1 interplay. PLoS ONE. 6:e26069 10.1371/journal.pone.0026069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea T.C., Wang Y., Nathans J. 2003. A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J. Neurosci. 23:2314–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettermann K., Vucur M., Haybaeck J., Koppe C., Janssen J., Heymann F., Weber A., Weiskirchen R., Liedtke C., Gassler N., et al. 2010. TAK1 suppresses a NEMO-dependent but NF-kappaB-independent pathway to liver cancer. Cancer Cell. 17:481–496 10.1016/j.ccr.2010.03.021 [DOI] [PubMed] [Google Scholar]
- Broglie P., Matsumoto K., Akira S., Brautigan D.L., Ninomiya-Tsuji J. 2010. Transforming growth factor β-activated kinase 1 (TAK1) kinase adaptor, TAK1-binding protein 2, plays dual roles in TAK1 signaling by recruiting both an activator and an inhibitor of TAK1 kinase in tumor necrosis factor signaling pathway. J. Biol. Chem. 285:2333–2339 10.1074/jbc.M109.090522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y.S., Challa S., Moquin D., Genga R., Ray T.D., Guildford M., Chan F.K. 2009. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 137:1112–1123 10.1016/j.cell.2009.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degterev A., Huang Z., Boyce M., Li Y., Jagtap P., Mizushima N., Cuny G.D., Mitchison T.J., Moskowitz M.A., Yuan J. 2005. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat. Chem. Biol. 1:112–119 10.1038/nchembio711 [DOI] [PubMed] [Google Scholar]
- Degterev A., Hitomi J., Germscheid M., Ch’en I.L., Korkina O., Teng X., Abbott D., Cuny G.D., Yuan C., Wagner G., et al. 2008. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat. Chem. Biol. 4:313–321 10.1038/nchembio.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon C.P., Oberst A., Weinlich R., Janke L.J., Kang T.B., Ben-Moshe T., Mak T.W., Wallach D., Green D.R. 2012. Survival function of the FADD-CASPASE-8-cFLIP(L) complex. Cell Rep. 1:401–407 10.1016/j.celrep.2012.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondelinger Y., Aguileta M.A., Goossens V., Dubuisson C., Grootjans S., Dejardin E., Vandenabeele P., Bertrand M.J. 2013. RIPK3 contributes to TNFR1-mediated RIPK1 kinase-dependent apoptosis in conditions of cIAP1/2 depletion or TAK1 kinase inhibition. Cell Death Differ. 20:1381–1392 10.1038/cdd.2013.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feoktistova M., Geserick P., Kellert B., Dimitrova D.P., Langlais C., Hupe M., Cain K., MacFarlane M., Häcker G., Leverkus M. 2011. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol. Cell. 43:449–463 10.1016/j.molcel.2011.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green D.R., Oberst A., Dillon C.P., Weinlich R., Salvesen G.S. 2011. RIPK-dependent necrosis and its regulation by caspases: a mystery in five acts. Mol. Cell. 44:9–16 10.1016/j.molcel.2011.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther C., Martini E., Wittkopf N., Amann K., Weigmann B., Neumann H., Waldner M.J., Hedrick S.M., Tenzer S., Neurath M.F., Becker C. 2011. Caspase-8 regulates TNF-α-induced epithelial necroptosis and terminal ileitis. Nature. 477:335–339 10.1038/nature10400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Zhong C.Q., Zhang D.W. 2011. Programmed necrosis: backup to and competitor with apoptosis in the immune system. Nat. Immunol. 12:1143–1149 10.1038/ni.2159 [DOI] [PubMed] [Google Scholar]
- Hayden M.S., Ghosh S. 2008. Shared principles in NF-kappaB signaling. Cell. 132:344–362 10.1016/j.cell.2008.01.020 [DOI] [PubMed] [Google Scholar]
- He S., Wang L., Miao L., Wang T., Du F., Zhao L., Wang X. 2009. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-α. Cell. 137:1100–1111 10.1016/j.cell.2009.05.021 [DOI] [PubMed] [Google Scholar]
- Hirsch T., Marchetti P., Susin S.A., Dallaporta B., Zamzami N., Marzo I., Geuskens M., Kroemer G. 1997. The apoptosis-necrosis paradox. Apoptogenic proteases activated after mitochondrial permeability transition determine the mode of cell death. Oncogene. 15:1573–1581 10.1038/sj.onc.1201324 [DOI] [PubMed] [Google Scholar]
- Hitomi J., Christofferson D.E., Ng A., Yao J., Degterev A., Xavier R.J., Yuan J. 2008. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 135:1311–1323 10.1016/j.cell.2008.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki M., Omori E., Kim J.Y., Komatsu Y., Scott G., Ray M.K., Yamada G., Matsumoto K., Mishina Y., Ninomiya-Tsuji J. 2008. TAK1-binding protein 1, TAB1, mediates osmotic stress-induced TAK1 activation but is dispensable for TAK1-mediated cytokine signaling. J. Biol. Chem. 283:33080–33086 10.1074/jbc.M807574200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inokuchi S., Aoyama T., Miura K., Osterreicher C.H., Kodama Y., Miyai K., Akira S., Brenner D.A., Seki E. 2010. Disruption of TAK1 in hepatocytes causes hepatic injury, inflammation, fibrosis, and carcinogenesis. Proc. Natl. Acad. Sci. USA. 107:844–849 10.1073/pnas.0909781107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser W.J., Upton J.W., Long A.B., Livingston-Rosanoff D., Daley-Bauer L.P., Hakem R., Caspary T., Mocarski E.S. 2011. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature. 471:368–372 10.1038/nature09857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajino T., Ren H., Iemura S., Natsume T., Stefansson B., Brautigan D.L., Matsumoto K., Ninomiya-Tsuji J. 2006. Protein phosphatase 6 down-regulates TAK1 kinase activation in the IL-1 signaling pathway. J. Biol. Chem. 281:39891–39896 10.1074/jbc.M608155200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajino-Sakamoto R., Inagaki M., Lippert E., Akira S., Robine S., Matsumoto K., Jobin C., Ninomiya-Tsuji J. 2008. Enterocyte-derived TAK1 signaling prevents epithelium apoptosis and the development of ileitis and colitis. J. Immunol. 181:1143–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.I., Kwak J.H., Wang L., Choi M.E. 2008. Protein phosphatase 2A is a negative regulator of transforming growth factor-β1-induced TAK1 activation in mesangial cells. J. Biol. Chem. 283:10753–10763 10.1074/jbc.M801263200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto K., Matsumoto K., Ninomiya-Tsuji J. 2000. TAK1 mitogen-activated protein kinase kinase kinase is activated by autophosphorylation within its activation loop. J. Biol. Chem. 275:7359–7364 10.1074/jbc.275.10.7359 [DOI] [PubMed] [Google Scholar]
- Kovalenko A., Kim J.C., Kang T.B., Rajput A., Bogdanov K., Dittrich-Breiholz O., Kracht M., Brenner O., Wallach D. 2009. Caspase-8 deficiency in epidermal keratinocytes triggers an inflammatory skin disease. J. Exp. Med. 206:2161–2177 10.1084/jem.20090616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamothe B., Lai Y., Xie M., Schneider M.D., Darnay B.G. 2013. TAK1 is essential for osteoclast differentiation and is an important modulator of cell death by apoptosis and necroptosis. Mol. Cell. Biol. 33:582–595 10.1128/MCB.01225-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist M., Gantner F., Bohlinger I., Tiegs G., Germann P.G., Wendel A. 1995. Tumor necrosis factor-induced hepatocyte apoptosis precedes liver failure in experimental murine shock models. Am. J. Pathol. 146:1220–1234 [PMC free article] [PubMed] [Google Scholar]
- Li L., Thomas R.M., Suzuki H., De Brabander J.K., Wang X., Harran P.G. 2004. A small molecule Smac mimic potentiates TRAIL- and TNFalpha-mediated cell death. Science. 305:1471–1474 10.1126/science.1098231 [DOI] [PubMed] [Google Scholar]
- Micheau O., Tschopp J. 2003. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 114:181–190 10.1016/S0092-8674(03)00521-X [DOI] [PubMed] [Google Scholar]
- Morioka S., Omori E., Kajino T., Kajino-Sakamoto R., Matsumoto K., Ninomiya-Tsuji J. 2009. TAK1 kinase determines TRAIL sensitivity by modulating reactive oxygen species and cIAP. Oncogene. 28:2257–2265 10.1038/onc.2009.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morioka S., Inagaki M., Komatsu Y., Mishina Y., Matsumoto K., Ninomiya-Tsuji J. 2012. TAK1 kinase signaling regulates embryonic angiogenesis by modulating endothelial cell survival and migration. Blood. 120:3846–3857 10.1182/blood-2012-03-416198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S., Kojima T., Kitamura T. 2000. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7:1063–1066 10.1038/sj.gt.3301206 [DOI] [PubMed] [Google Scholar]
- Newton K., Sun X., Dixit V.M. 2004. Kinase RIP3 is dispensable for normal NF-κ Bs, signaling by the B-cell and T-cell receptors, tumor necrosis factor receptor 1, and Toll-like receptors 2 and 4. Mol. Cell. Biol. 24:1464–1469 10.1128/MCB.24.4.1464-1469.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya-Tsuji J., Kishimoto K., Hiyama A., Inoue J., Cao Z., Matsumoto K. 1999. The kinase TAK1 can activate the NIK-I kappaB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature. 398:252–256 10.1038/18465 [DOI] [PubMed] [Google Scholar]
- O’Donnell M.A., Legarda-Addison D., Skountzos P., Yeh W.C., Ting A.T. 2007. Ubiquitination of RIP1 regulates an NF-kappaB-independent cell-death switch in TNF signaling. Curr. Biol. 17:418–424 10.1016/j.cub.2007.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell M.A., Perez-Jimenez E., Oberst A., Ng A., Massoumi R., Xavier R., Green D.R., Ting A.T. 2011. Caspase 8 inhibits programmed necrosis by processing CYLD. Nat. Cell Biol. 13:1437–1442 10.1038/ncb2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberst A., Green D.R. 2011. It cuts both ways: reconciling the dual roles of caspase 8 in cell death and survival. Nat. Rev. Mol. Cell Biol. 12:757–763 10.1038/nrm3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberst A., Dillon C.P., Weinlich R., McCormick L.L., Fitzgerald P., Pop C., Hakem R., Salvesen G.S., Green D.R. 2011. Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature. 471:363–367 10.1038/nature09852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori E., Matsumoto K., Sanjo H., Sato S., Akira S., Smart R.C., Ninomiya-Tsuji J. 2006. TAK1 is a master regulator of epidermal homeostasis involving skin inflammation and apoptosis. J. Biol. Chem. 281:19610–19617 10.1074/jbc.M603384200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori E., Morioka S., Matsumoto K., Ninomiya-Tsuji J. 2008. TAK1 regulates reactive oxygen species and cell death in keratinocytes, which is essential for skin integrity. J. Biol. Chem. 283:26161–26168 10.1074/jbc.M804513200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasparakis M. 2009. Regulation of tissue homeostasis by NF-kappaB signalling: implications for inflammatory diseases. Nat. Rev. Immunol. 9:778–788 10.1038/nri2655 [DOI] [PubMed] [Google Scholar]
- Pfeffer K., Matsuyama T., Kündig T.M., Wakeham A., Kishihara K., Shahinian A., Wiegmann K., Ohashi P.S., Krönke M., Mak T.W. 1993. Mice deficient for the 55 kd tumor necrosis factor receptor are resistant to endotoxic shock, yet succumb to L. monocytogenes infection. Cell. 73:457–467 10.1016/0092-8674(93)90134-C [DOI] [PubMed] [Google Scholar]
- Postic C., Shiota M., Niswender K.D., Jetton T.L., Chen Y., Moates J.M., Shelton K.D., Lindner J., Cherrington A.D., Magnuson M.A. 1999. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic β cell-specific gene knock-outs using Cre recombinase. J. Biol. Chem. 274:305–315 10.1074/jbc.274.1.305 [DOI] [PubMed] [Google Scholar]
- Sanjo H., Takeda K., Tsujimura T., Ninomiya-Tsuji J., Matsumoto K., Akira S. 2003. TAB2 is essential for prevention of apoptosis in fetal liver but not for interleukin-1 signaling. Mol. Cell. Biol. 23:1231–1238 10.1128/MCB.23.4.1231-1238.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Sanjo H., Takeda K., Ninomiya-Tsuji J., Yamamoto M., Kawai T., Matsumoto K., Takeuchi O., Akira S. 2005. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat. Immunol. 6:1087–1095 10.1038/ni1255 [DOI] [PubMed] [Google Scholar]
- Shim J.H., Xiao C., Paschal A.E., Bailey S.T., Rao P., Hayden M.S., Lee K.Y., Bussey C., Steckel M., Tanaka N., et al. 2005. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev. 19:2668–2681 10.1101/gad.1360605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaesu G., Kishida S., Hiyama A., Yamaguchi K., Shibuya H., Irie K., Ninomiya-Tsuji J., Matsumoto K. 2000. TAB2, a novel adaptor protein, mediates activation of TAK1 MAPKKK by linking TAK1 to TRAF6 in the IL-1 signal transduction pathway. Mol. Cell. 5:649–658 10.1016/S1097-2765(00)80244-0 [DOI] [PubMed] [Google Scholar]
- Takaesu G., Inagaki M., Takubo K., Mishina Y., Hess P.R., Dean G.A., Yoshimura A., Matsumoto K., Suda T., Ninomiya-Tsuji J. 2012. TAK1 (MAP3K7) signaling regulates hematopoietic stem cells through TNF-dependent and -independent mechanisms. PLoS ONE. 7:e51073 10.1371/journal.pone.0051073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenev T., Bianchi K., Darding M., Broemer M., Langlais C., Wallberg F., Zachariou A., Lopez J., MacFarlane M., Cain K., Meier P. 2011. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol. Cell. 43:432–448 10.1016/j.molcel.2011.06.006 [DOI] [PubMed] [Google Scholar]
- Ting A.T., Pimentel-Muiños F.X., Seed B. 1996. RIP mediates tumor necrosis factor receptor 1 activation of NF-kappaB but not Fas/APO-1-initiated apoptosis. EMBO J. 15:6189–6196 [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele P., Melino G. 2012. The flick of a switch: which death program to choose? Cell Death Differ. 19:1093–1095 10.1038/cdd.2012.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele P., Galluzzi L., Vanden Berghe T., Kroemer G. 2010. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat. Rev. Mol. Cell Biol. 11:700–714 10.1038/nrm2970 [DOI] [PubMed] [Google Scholar]
- Vanlangenakker N., Vanden Berghe T., Bogaert P., Laukens B., Zobel K., Deshayes K., Vucic D., Fulda S., Vandenabeele P., Bertrand M.J. 2011. cIAP1 and TAK1 protect cells from TNF-induced necrosis by preventing RIP1/RIP3-dependent reactive oxygen species production. Cell Death Differ. 18:656–665 10.1038/cdd.2010.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlangenakker N., Vanden Berghe T., Vandenabeele P. 2012. Many stimuli pull the necrotic trigger, an overview. Cell Death Differ. 19:75–86 10.1038/cdd.2011.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucur M., Reisinger F., Gautheron J., Janssen J., Roderburg C., Cardenas D.V., Kreggenwinkel K., Koppe C., Hammerich L., Hakem R., et al. 2013. RIP3 inhibits inflammatory hepatocarcinogenesis but promotes cholestasis by controlling caspase-8- and JNK-dependent compensatory cell proliferation. Cell Rep. 4:776–790 10.1016/j.celrep.2013.07.035 [DOI] [PubMed] [Google Scholar]
- Wang L., Du F., Wang X. 2008. TNF-α induces two distinct caspase-8 activation pathways. Cell. 133:693–703 10.1016/j.cell.2008.03.036 [DOI] [PubMed] [Google Scholar]
- Welz P.S., Wullaert A., Vlantis K., Kondylis V., Fernández-Majada V., Ermolaeva M., Kirsch P., Sterner-Kock A., van Loo G., Pasparakis M. 2011. FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature. 477:330–334 10.1038/nature10273 [DOI] [PubMed] [Google Scholar]
- Werner S., Grose R. 2003. Regulation of wound healing by growth factors and cytokines. Physiol. Rev. 83:835–870 [DOI] [PubMed] [Google Scholar]
- Xiao Y., Li H., Zhang J., Volk A., Zhang S., Wei W., Zhang S., Breslin P., Zhang J. 2011. TNF-α/Fas-RIP-1-induced cell death signaling separates murine hematopoietic stem cells/progenitors into 2 distinct populations. Blood. 118:6057–6067 10.1182/blood-2011-06-359448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Kroemer G. 2010. Alternative cell death mechanisms in development and beyond. Genes Dev. 24:2592–2602 10.1101/gad.1984410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhou X., McQuade T., Li J., Chan F.K., Zhang J. 2011. Functional complementation between FADD and RIP1 in embryos and lymphocytes. Nature. 471:373–376 10.1038/nature09878 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.