Rho GTPase signaling establishes a planar polarized actomyosin network within which the actin-binding protein Shroom enhances myosin activity locally to generate robust mechanical forces during axis elongation.
Abstract
Actomyosin contraction generates mechanical forces that influence cell and tissue structure. During convergent extension in Drosophila melanogaster, the spatially regulated activity of the myosin activator Rho-kinase promotes actomyosin contraction at specific planar cell boundaries to produce polarized cell rearrangement. The mechanisms that direct localized Rho-kinase activity are not well understood. We show that Rho GTPase recruits Rho-kinase to adherens junctions and is required for Rho-kinase planar polarity. Shroom, an asymmetrically localized actin- and Rho-kinase–binding protein, amplifies Rho-kinase and myosin II planar polarity and junctional localization downstream of Rho signaling. In Shroom mutants, Rho-kinase and myosin II achieve reduced levels of planar polarity, resulting in decreased junctional tension, a disruption of multicellular rosette formation, and defective convergent extension. These results indicate that Rho GTPase activity is required to establish a planar polarized actomyosin network, and the Shroom actin-binding protein enhances myosin contractility locally to generate robust mechanical forces during axis elongation.
Introduction
Actomyosin contractility generates mechanical forces that determine cell and tissue structure through its role in regulating cell division, cell shape, and cell rearrangement. Spatially and temporally controlled contractile forces during development are achieved in large part by the regulated activity of protein kinases that phosphorylate myosin and promote its activity. Increasing evidence indicates that Rho-kinase, the primary regulator of myosin activity in epithelial tissues, is a critical determinant of where and when myosin is active within the cell (Riento and Ridley, 2003; Quintin et al., 2008; Amano et al., 2010). Rho-kinase is localized to the apical cell cortex in epithelia (Wang and Riechmann, 2007) and recruits myosin apically to induce apical constriction during epithelial bending and cell invagination (Dawes-Hoang et al., 2005; Mason et al., 2013). A distinct pattern of Rho-kinase localization at cell boundaries promotes cell rearrangements during tissue remodeling. Rho-kinase accumulates in a planar polarized fashion at cell contacts that are disassembled during axis elongation in Drosophila melanogaster (Simões et al., 2010). Rho-kinase activity influences the orientation of cell rearrangements during axis elongation by promoting localized actomyosin contractility and inhibiting cell adhesion by excluding the Baz/Par-3 junctional protein (Simões et al., 2010). Rho-kinase planar polarity has been observed in several epithelial tissues, and Rho-kinase activity is required for planar polarized cell rearrangements and cell shape changes during development (Walters et al., 2006; Nishimura et al., 2012; Robertson et al., 2012; Bulgakova et al., 2013). However, although planar polarized Rho-kinase activity plays a critical role in regulating localized actomyosin contractility, the mechanisms that generate Rho-kinase planar polarity are not well understood.
Rho GTPases are conserved upstream regulators of actomyosin organization and contractility (Riento and Ridley, 2003; Jaffe and Hall, 2005). Several lines of evidence suggest that apical myosin activity is regulated by localized Rho GTPase signaling. Rho and its activator RhoGEF2 are required for apical constriction and apical myosin localization in the Drosophila embryo, and RhoGEF2 protein is apically localized (Barrett et al., 1997; Häcker and Perrimon, 1998; Nikolaidou and Barrett, 2004; Dawes-Hoang et al., 2005; Simões et al., 2006; Kölsch et al., 2007). A fluorescent probe that specifically binds to the active, GTP-bound form of Rho is apically localized in epithelial cells, and this distribution requires two apical Rho activators and a basolateral Rho inhibitor (Simões et al., 2006). These results suggest that apically localized Rho GTPase activity promotes myosin contractility at the apical cell cortex during apical constriction. However, it is not known whether localized Rho signaling is involved in planar polarized Rho-kinase localization and myosin activity. The Rho activators RhoGEF2 in Drosophila and its chick homologue, PDZ-RhoGEF, are present in a planar polarized distribution during axis elongation and neural tube closure (Levayer et al., 2011; Nishimura et al., 2012; Warrington et al., 2013). However, the defects in RhoGEF2 mutants are weaker than those caused by loss of Rho-kinase or myosin II (Häcker and Perrimon, 1998; Levayer et al., 2011), suggesting that additional mechanisms regulate actomyosin contractility in the plane of the tissue.
Shroom family proteins are actin-associated proteins that bind to Rho-kinase and target it to the apical cell cortex (Hildebrand and Soriano, 1999; Haigo et al., 2003; Hildebrand, 2005; Nishimura and Takeichi, 2008; Bolinger et al., 2010). Shroom proteins are required for neural tube closure in the mouse, frog, and chick (Hildebrand and Soriano, 1999; Haigo et al., 2003; Nishimura and Takeichi, 2008; Massarwa and Niswander, 2013). These defects are thought to be caused by a role for Shroom in apical constriction, as ectopic Shroom is sufficient to promote apical Rho-kinase and myosin localization and induce apical constriction in epithelial cells (Haigo et al., 2003; Hildebrand, 2005; Nishimura and Takeichi, 2008). Moreover, Shroom is required for many developmental processes that involve apical constriction, such as epithelial bending at neural tube hinge points in Xenopus laevis (Haigo et al., 2003; Lee et al., 2007), invagination of the mammalian lens placode (Plageman et al., 2010), and apical constriction of cells in the zebrafish lateral line (Ernst et al., 2012). However, although the effects of ectopic Shroom have been well characterized in culture, the effects of loss of Shroom on cell behavior and myosin dynamics are less well understood.
In the Drosophila embryo, axis elongation is driven by planar polarized cell rearrangements that require spatially regulated actomyosin contractility. Actin and myosin II are enriched at cell boundaries that are progressively disassembled during elongation (Bertet et al., 2004; Zallen and Wieschaus, 2004; Blankenship et al., 2006), and adherens junction proteins stabilize cell contacts in the complementary cellular domain (Blankenship et al., 2006; Simões et al., 2010; Levayer et al., 2011; Tamada et al., 2012). These spatially regulated processes of contraction and adhesion both require Rho-kinase activity, but the mechanisms that generate planar polarized Rho-kinase localization are not known. Here, we identify distinct roles for Rho GTPase and Shroom in regulating Rho-kinase localization and actomyosin contractility during Drosophila axis elongation. Rho recruits Rho-kinase to adherens junctions and generates a planar polarized distribution of the Shroom actin-binding protein. Shroom in turn is required to amplify and maintain Rho-kinase and myosin planar polarity and junctional localization. These results demonstrate that Rho and Shroom regulate planar polarized Rho-kinase localization and actomyosin contractility to generate sustained mechanical forces during axis elongation.
Results
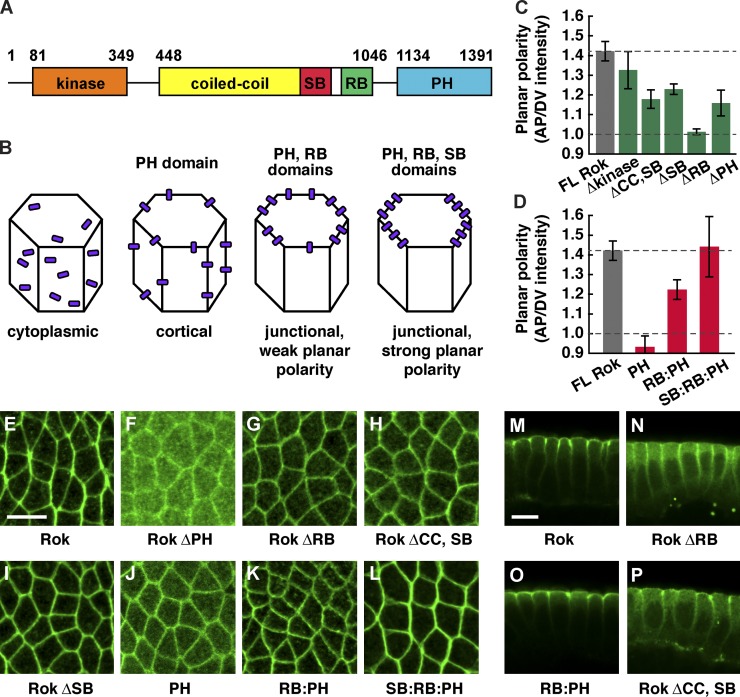
Sequences required for Rho-kinase localization along apical–basal and planar axes
During axis elongation, Rho-kinase is concentrated at adherens junctions between anterior and posterior cells, where it promotes junctional disassembly and oriented cell rearrangement (Simões et al., 2010). To identify the mechanisms that generate localized Rho-kinase activity, we performed structure/function analysis. Rho-kinase has a conserved N-terminal serine/threonine kinase domain, a central coiled-coil (CC) domain that binds to Rho GTPase and the Shroom actin-binding protein, and a C-terminal pleckstrin homology (PH) domain (Fig. 1 A). As overexpression of active Rho-kinase in wild type disrupts embryo morphology, we generated deletions in the context of Venus-tagged, catalytically inactive Rho-kinase that recapitulates the localization of the wild-type protein (Fig. 1 and Fig. S1; Simões et al., 2010).
Figure 1.
The RB, SB, and PH domains regulate distinct aspects of Rho-kinase localization. (A) Drosophila Rho-kinase (Rok) contains an N-terminal kinase domain (aa 81–349), central coiled-coil domain (CC; aa 448–1,046), Shroom-binding domain (SB; aa 834–938), Rho-binding domain (RB; aa 966–1,046), and a C-terminal pleckstrin homology domain (PH; aa 1,134–1,391). (B) Domains required for Rho-kinase cortical, junctional, and planar polarized localization. (C and D) Planar polarized enrichment of Venus:RokK116A proteins at AP cell boundaries (oriented at 75–90° with respect to the AP axis) relative to DV cell boundaries (oriented at 0–15°). Δ denotes deletion of a domain. Dotted line at 1.0 indicates no polarity, and top dotted line indicates wild-type polarity. (E–P) Images of Venus:RokK116A proteins in stage 7 wild-type embryos. Full-length RokK116A (FL Rok; E and M), Rok ΔPH (F), Rok ΔRB (G and N), Rok ΔCC,SB (H and P), Rok ΔSB (I), Rok PH (J), Rok RB:PH (K and O), and Rok SB:RB:PH (L) are shown. (E–L) Anterior is left, and ventral is down. (M–P) Cross sections, with apical up. Rok ΔRB, Rok ΔCC,SB, Rok ΔSB, Rok PH, and RB:PH were less planar polarized than full-length RokK116A (P < 0.003). Rok ΔPH was slightly less planar polarized (P = 0.013). SB:RB:PH planar polarity was similar to full-length RokK116A (P = 0.87). A single value was obtained for each image by averaging 100–200 edges/image; 4–15 images in 4–8 embryos were analyzed/genotype. Means ± SEM between images are shown. Bars, 10 µm.
We identified distinct sequences required for Rho-kinase cortical, junctional, and planar polarized localization (Fig. 1 B). Deletion of the N-terminal and catalytic domains had no effect on Rho-kinase localization (Fig. 1 C). In contrast, Rho-kinase lacking the PH domain (Rok ΔPH) was predominantly cytoplasmic (Fig. 1 F), consistent with previous findings that the PH domain is required for cortical localization (Miyazaki et al., 2006). The PH domain alone was sufficient for cortical localization but displayed no asymmetry along apical–basal or planar cell axes (Fig. 1, D and J), indicating that other domains are required for Rho-kinase junctional localization and planar polarity.
We next tested the effect of deleting the Rho-binding (RB) domain that interacts with Rho GTPase, an upstream regulator of Rho-kinase activity. Rho-kinase lacking its RB domain (Rok ΔRB) localized to the cortex but was no longer enriched at adherens junctions, resulting in increased levels at the lateral membrane (Fig. 1, M and N). In addition, Rok ΔRB protein at adherens junctions did not display planar asymmetry (Fig. 1, C and G). Conversely, a fusion protein containing the RB and PH domains of Rho-kinase (RB:PH) was sufficient for junctional localization (Fig. 1 O) but displayed only a partial asymmetry in the plane of the tissue (Fig. 1, D and K). These results indicate that binding to Rho GTPase recruits Rho-kinase to adherens junctions but is not sufficient for full Rho-kinase planar polarity.
To identify additional sequences in Rho-kinase that are required for its planar polarized localization, we turned our attention to the CC domain. A protein lacking most of the CC domain (Rok ΔCC,SB) displayed significantly less planar polarity, despite the presence of the PH and RB domains, and had slightly reduced accumulation at adherens junctions (Fig. 1, C, H, and P). The CC domain contains a small region that interacts with the Shroom actin-binding protein (Nishimura and Takeichi, 2008; Bolinger et al., 2010). Removing the Shroom-binding (SB) domain alone (Rok ΔSB) disrupted Rho-kinase planar polarity (Fig. 1, C and I). Similar results were obtained when this domain was deleted in the context of wild-type, active Rho-kinase expressed in a Rho-kinase mutant (Fig. S2, B, C, and I). Moreover, the SB, RB, and PH domains together fully reproduced the planar asymmetry of full-length Rho-kinase (Fig. 1, D and L). Together, these results indicate that Rho-kinase cortical localization is mediated by its PH domain, Rho-kinase enrichment at adherens junctions requires its RB domain, and the RB and SB domains are required for Rho-kinase planar polarity.
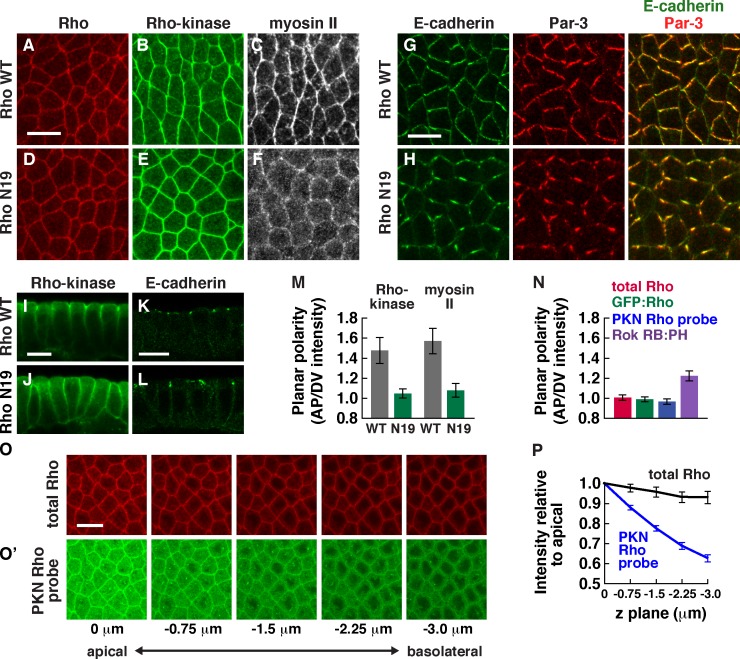
Rho GTPase is required for Rho-kinase junctional localization and planar polarity
Rho GTPase binds directly to Rho-kinase and triggers a conformational change that promotes Rho-kinase activity to phosphorylate its substrates (Riento and Ridley, 2003; Jaffe and Hall, 2005). To ask whether Rho is required for Rho-kinase planar polarity, we inhibited Rho activity during axis elongation. Because Rho is required for early development (Strutt et al., 1997; Magie et al., 1999), we injected embryos in late cellularization with mRNAs encoding wild-type Rho, dominant-negative RhoN19, or constitutively active RhoV14 to express these proteins during axis elongation. Embryos expressing wild-type Rho displayed the proper planar polarized enrichment of Rho-kinase and myosin II at interfaces between anterior and posterior cells (AP interfaces; Fig. 2, A–C). Rho-kinase and myosin II planar polarity were significantly reduced in embryos expressing dominant-negative RhoN19 (Fig. 2, D–F and M). In addition, Rho-kinase failed to concentrate at adherens junctions and was more uniformly distributed along the lateral membrane (Fig. 2, I and J). Adherens junction proteins were correctly localized in embryos expressing dominant-negative RhoN19 (Fig. 2, K and L) and were concentrated at interfaces between dorsal and ventral cells (DV interfaces) as in wild type (Fig. 2, G and H), indicating that the failure of Rho-kinase to localize apically is not caused by a loss of adherens junctions. However, E-cadherin and Baz/Par-3 formed aberrant aggregates (Fig. 2 H), reminiscent of embryos injected with the Rho-kinase inhibitor Y-27632 (Simões et al., 2010), indicating that RhoN19 alters E-cadherin distribution along junctions. Conversely, activated RhoV14 frequently enhanced Rho-kinase and myosin localization and planar polarity ectopically in the basolateral domain (Fig. S3, E and F). Together, these results indicate that regulated Rho GTPase signaling is required for Rho-kinase planar polarity and adherens junction localization.
Figure 2.
Rho GTPase is required for Rho-kinase localization and planar polarity in intercalating cells. (A–F) Localization of HA:Rho, Venus:RokK116A, and Myo:GFP in stage 7 embryos expressing wild-type Rho (Rho WT; A–C) or dominant-negative Rho (RhoN19; D–F). (G and H) Localization of E-cadherin and Baz/Par-3 in stage 7 embryos expressing wild-type Rho (G) or RhoN19 (H). Anterior is left, and ventral is down. (I–L) Localization of Venus:RokK116A (I and J) or E-cadherin (K and L) in stage 7 embryos expressing wild-type Rho (I and K) or RhoN19 (J and L). Cross sections are shown, with apical up. (M) Planar polarized enrichment of Rho-kinase and myosin II at AP cell boundaries (75–90° with respect to the AP axis) relative to DV cell boundaries (0–15°). Rho-kinase and myosin II were significantly less planar polarized in embryos expressing RhoN19 compared with embryos expressing wild-type Rho (P < 0.01). (N) Rok RB:PH was significantly more enriched at AP cell boundaries relative to DV cell boundaries than total Rho protein (P < 0.001). No planar polarized enrichment was detected for total Rho, GFP:Rho, or the PKN Rho probe. (O) Localization of total Rho protein (red) and the PKN Rho probe (green) at different z planes (0 µm is at the level of adherens junctions, and −3 µm is basal to the junctions). Anterior is left, and ventral is down. (P) Total Rho and the PKN Rho probe cortical levels normalized to the apical junctional intensity. PKN Rho probe signal at the basolateral membrane was reduced compared with total Rho (P < 0.002 at −0.75 µm; P < 0.0001 at −1.5, −2.75, and −3.0 µm). A single value was obtained for each image by averaging 100–200 edges/image; 4–16 images in 4–8 embryos were analyzed for each genotype at each z plane. Means ± SEM between images are shown. Bars, 10 µm.
If Rho plays an instructive role in directing Rho-kinase planar polarity, Rho activity is predicted to be spatially localized. However, total Rho protein visualized with an antibody specific to Rho (Magie et al., 2002) was uniformly distributed along apical–basal and planar cell axes (Fig. 2, N–P). To determine whether an active subpopulation of Rho is spatially localized, we expressed a Venus fusion to the RB domains of protein kinase N (PKN). This fusion specifically interacts with active, GTP-bound Rho in vitro and in culture (Simões et al., 2006). Unlike total Rho protein, which is distributed throughout the lateral membrane, the PKN Rho probe was enriched at adherens junctions, in addition to cytoplasmic signal likely caused by unbound protein (Fig. 2, O and P). However, this fusion did not display obvious asymmetry in the plane of the tissue (Fig. 2 N). In an alternative method to detect the pool of active Rho that binds to Rho-kinase, we expressed Venus fused to the RB and PH domains of Rho-kinase (RB:PH). The Rho-kinase RB domain preferentially interacts with GTP-bound Rho in Drosophila and mammals (Fujisawa et al., 1996; Mizuno et al., 1999), and the PH domain is required for strong cortical localization (Fig. 1 F). The Rho-kinase RB:PH fusion displayed a small but reproducible asymmetry in the plane of the tissue (Fig. 1, D and K; and Fig. 2 N). These results indicate that Rho activity is increased at the apical cell cortex and that the active population of Rho that binds to Rho-kinase is at most subtly polarized in the plane of the tissue.
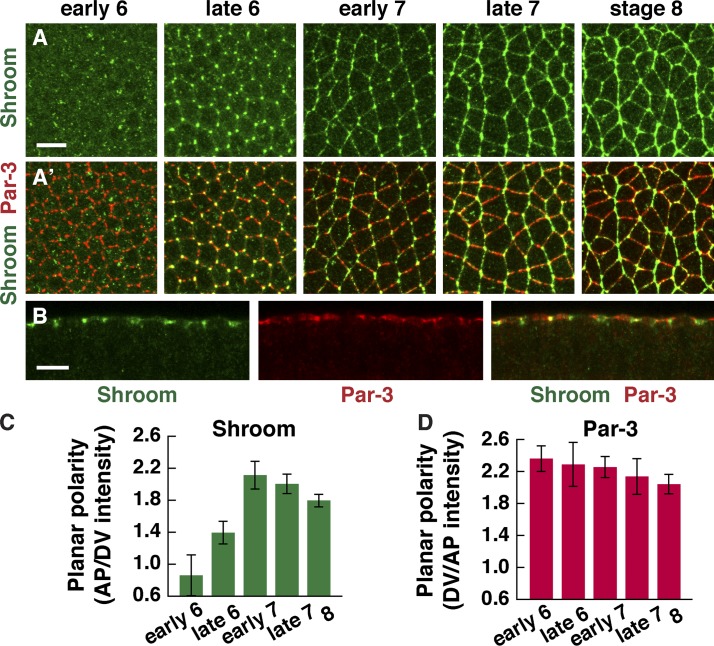
The Shroom actin-binding protein is planar polarized during axis elongation
Because the SB domain is required for Rho-kinase planar polarity, we hypothesized that Shroom enhances Rho-kinase planar polarity downstream of a subtle asymmetry in Rho signaling. To investigate this possibility, we analyzed the localization of ShrmA, the predominant Shroom isoform in the Drosophila embryo. Shroom localizes to adherens junctions at three-cell vertices before axis elongation (Fig. 3 A, stage 6), consistent with a previous study (Bolinger et al., 2010). In addition, we found that Shroom has a striking planar polarized distribution during axis elongation. During stages 7 and 8, when cells undergo active cell rearrangement, Shroom was strongly enriched in adherens junctions at AP interfaces, colocalizing with Rho-kinase and myosin II (Fig. 3, A–C). In contrast to the Baz/Par-3 junctional protein, which is enriched in the complementary domain in stage 6 before elongation (Blankenship et al., 2006), Shroom planar polarity was most evident during cell rearrangement in stage 7 (Fig. 3, C and D). This distribution is consistent with a potential role for Shroom in regulating planar polarity during axis elongation.
Figure 3.
The Shroom actin-binding protein is planar polarized in intercalating cells. (A and B) Localization of Shroom (A–B) and Baz/Par-3 (A′ and B) in wild-type embryos at the indicated stages. (A) Anterior is left, and ventral is down. (B) Cross section of a stage 8 embryo, with apical up. (C) Planar polarized enrichment of Shroom at AP cell boundaries (75–90° with respect to the AP axis) relative to DV cell boundaries (0–15°). (D) Planar polarized enrichment of Par-3 at DV cell boundaries relative to AP cell boundaries at the indicated stages. Note that Shroom planar polarity appears later than Par-3. A single value was obtained for each image by averaging 100–200 edges/image; 5–13 images in 2–7 embryos were analyzed/stage, except early stage 6 (three images in one embryo). Means ± SEM between images are shown. Bars, 10 µm.
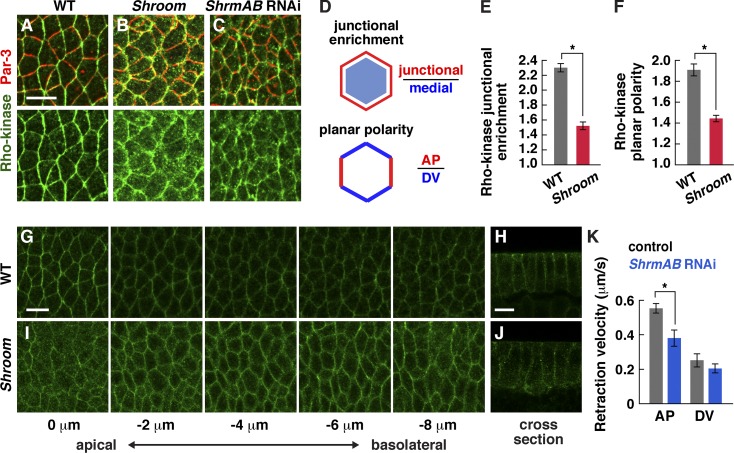
Shroom increases Rho-kinase and myosin II planar polarity and junctional localization
To determine whether Shroom regulates Rho-kinase localization and actomyosin contractility, we used two approaches to disrupt Shroom function. First, we injected embryos with two independent double-stranded RNAs (dsRNAs) that target both embryonic ShrmA and ShrmB isoforms (ShrmAB RNAi) and strongly reduce Shroom protein levels (Fig. S4 A) or a third dsRNA that specifically targets the ShrmA isoform (ShrmA RNAi). In addition, we generated two Shroom mutations, ShrmΔ11 and ShrmΔ13.6, through P element transposase-mediated recombination (Materials and methods). These mutations remove part of the Shroom open reading frame, including the Rho-kinase–binding domain, and show an absence of Shroom protein by immunostaining (Fig. S4, B–D). We characterized embryos lacking maternal and zygotic Shroom (referred to as Shroom mutants), which were the progeny of ShrmΔ11 or ShrmΔ13.6 females transheterozygous for the Df(2R)Exel7131 deficiency that removes the Shroom locus crossed to Shrm/Df or Shrm/+ males. Shrm/Df females were viable, but their progeny had increased embryonic lethality (Materials and methods).
In wild-type embryos, Rho-kinase is enriched at adherens junctions and is present at lower levels on the lateral membrane (Fig. 4, A and G). In contrast, in Shroom mutants, Rho-kinase accumulated less strongly at adherens junctions and was present in ectopic aggregates in the apical–medial cortex and cytoplasm (Fig. 4, B, E, and I). In addition, Rho-kinase had significantly reduced planar polarity in Shroom mutants compared with wild type (Fig. 4 F). Consistent with the decreased Rho-kinase localization at adherens junctions, the apical enrichment of Rho-kinase was slightly reduced in Shroom mutants (Fig. 4, G–J). Similar defects were observed in ShrmAB RNAi embryos (Fig. 4 C). These results demonstrate that Shroom is required to promote Rho-kinase junctional localization and planar polarity during axis elongation.
Figure 4.
Shroom is required for Rho-kinase localization and force generation during convergent extension. (A–C) Localization of Rho-kinaseK116A and Baz/Par-3 in wild-type (WT; A), ShrmΔ13.6 mutant (B), and ShrmAB (C) RNAi embryos at stage 8. Anterior is left, and ventral is down. (D) Junctional enrichment is the ratio of the mean pixel intensity at adherens junctions divided by the mean pixel intensity in the medial–apical cortex and cytoplasm (pixels ≥1 µm from the cell boundary). Planar polarity is the mean intensity of AP edges (75–90° relative to the AP axis) divided by the mean intensity of DV edges (0–15°). (E) Rho-kinase was less junctionally enriched in Shroom mutants in stage 8 (P < 0.0001). (F) Rho-kinase was less planar polarized in Shroom mutants in stage 8 (P = 0.05). Wild-type Rho-kinase planar polarity was slightly higher than in Fig. 1 C because different Rho-kinaseK116A transgenes and fixation methods were used (Materials and methods). (G and H) Z stack (G) and cross section (H) of Rho-kinaseK116A in a stage 8 wild-type embryo (0 µm is at the level of adherens junctions, and −2 to 8–µm images are at the indicated distance basal to the junctions). (I and J) Z stack (I) and cross section (J) of Rho-kinaseK116A in a stage 8 ShrmΔ11 mutant. (K) Peak retraction velocities after laser ablation of control (flp dsRNA injected) and ShrmAB RNAi embryos in early stage 8. ShrmAB RNAi embryos had slower retraction velocities at AP edges (P = 0.003) and no change at DV edges (P = 0.36) compared with controls (19 AP and 8 DV ablations in flp RNAi; 16 AP and 6 DV ablations in ShrmAB RNAi). ShrmΔ11 and ShrmΔ13.6 were combined for the analysis in E and F. A single value was obtained for each image by analyzing 100–200 edges/image, and 8–11 images in 5–6 embryos were analyzed/genotype. *, P ≤ 0.05. Means ± SEM between images are shown. Bars, 10 µm.
To investigate the effect of Rho-kinase mislocalization on myosin activity, we analyzed relative contractile forces using laser ablation to sever single cell–cell interfaces. Laser ablation leads to a local cellular retraction that is proportional to the tension acting on the interface, assuming uniform viscoelastic properties (Hutson et al., 2003). This response is normally strongest at AP cell boundaries (Fernandez-Gonzalez et al., 2009). The response to laser ablation was significantly attenuated in ShrmAB RNAi embryos, and AP edges had lower retraction velocities compared with water-injected controls, with no difference in retraction velocities at DV edges (Fig. 4 K). Together, these results demonstrate that Shroom is required to recruit or stabilize Rho-kinase at adherens junctions and to enhance planar polarized force generation during axis elongation.
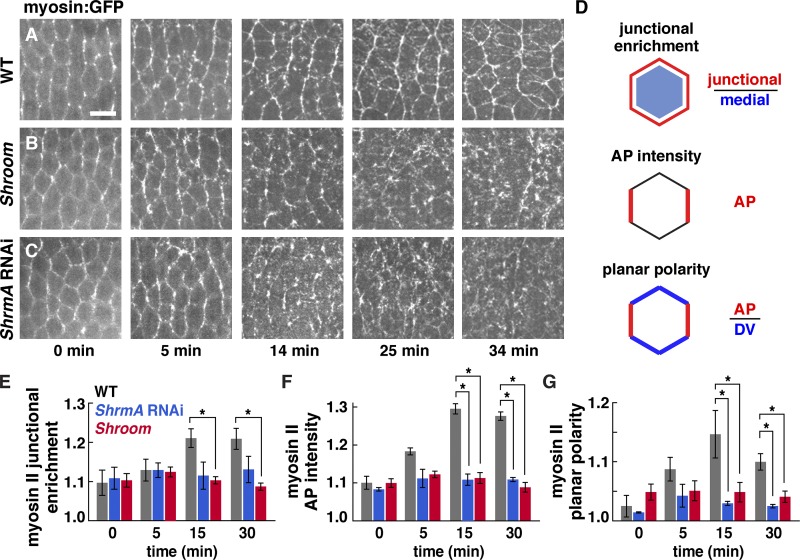
To analyze the effect of Shroom on myosin II localization and dynamics, we performed time-lapse imaging of embryos expressing GFP fused to the myosin regulatory light chain (Myo:GFP; Videos 1–6). Myosin is present in two populations in intercalating cells: a junctional population at cell–cell contacts and a medial population that spans the apical cell cortex (Rauzi et al., 2010; Fernandez-Gonzalez and Zallen, 2011; Sawyer et al., 2011). During axis elongation, myosin becomes increasingly enriched at adherens junctions relative to the medial cortex in wild type (Fig. 5, A and E, 14–34 min). In contrast, myosin junctional enrichment did not increase during elongation in Shroom mutants (Fig. 5, B and E). In addition, the planar polarized enrichment of myosin at AP cell boundaries, which normally increases substantially throughout elongation, plateaued early in elongation and failed to reach wild-type levels in Shroom mutants and ShrmA RNAi embryos (Fig. 5, B–D, F, and G). Par-3 planar polarity occurred normally in Shroom mutants and ShrmA and ShrmAB RNAi embryos (unpublished data). These results demonstrate that Shroom is required to amplify myosin II planar polarity and junctional localization during axis elongation.
Figure 5.
Shroom increases myosin II cortical localization and planar polarity during convergent extension. (A–C) Myo:GFP in time-lapse videos of wild-type (WT; A), ShrmΔ11 mutant (B), and ShrmA RNAi (C) embryos (t = 0 is the onset of elongation; stage 7 [0–10 min]; stage 8 [10–35 min]). Anterior is left, and ventral is down. Bar, 10 µm. (D) Junctional enrichment is the ratio of the mean pixel intensity at adherens junctions divided by the mean pixel intensity in the medial–apical cortex and cytoplasm (pixels ≥1 µm from the cell boundary). AP intensity is the absolute AP edge intensity normalized to the mean pixel intensity for the image. Planar polarity is the mean intensity of AP edges (75–90° relative to the AP axis) divided by the mean intensity of DV edges (0–15°). Myo:GFP intensity was quantified directly in living embryos. (E–G) Myo:GFP junctional enrichment (E), AP intensity (F), and planar polarity (G) in time-lapse videos of wild-type (gray), ShrmA RNAi (blue), and ShrmΔ11 mutant (red) embryos. (E) Myo:GFP junctional enrichment was significantly reduced in Shroom mutants (P < 0.001 at 15 and 30 min) but did not reach statistical significance for ShrmA RNAi. (F) Myo:GFP AP intensity was significantly reduced in Shroom mutant and ShrmA RNAi embryos (P ≤ 0.003 at 15 and 30 min). (G) Myo:GFP planar polarity was significantly reduced in Shroom mutant and ShrmA RNAi embryos (P ≤ 0.02 at 15 and 30 min). A single value was obtained for each image by analyzing 100–200 edges/time point, and four time points/video in three wild-type, seven ShrmΔ11 mutant, and four ShrmA RNAi videos were analyzed. *, P ≤ 0.02. Means ± SEM between images are shown.
Shroom is required for multicellular rosette formation and convergent extension
The failure to maintain Rho-kinase and myosin II localization at specific planar junctions is predicted to impair the generation of sustained actomyosin contractility during cell rearrangement. To test whether Shroom is required for cell behavior, we performed time-lapse imaging of wild-type and Shroom mutant embryos expressing Spider:GFP (Videos 7 and 8). During axis elongation, the Drosophila germband elongates along the AP axis and simultaneously narrows along the DV axis, producing a 4.00 ± 0.32–fold increase in the tissue aspect ratio (length to width ratio), a process known as convergent extension. Shroom mutants had reduced convergent extension, resulting in a smaller increase in the tissue length to width ratio compared with wild type (3.16 ± 0.15–fold, P = 0.02; Fig. 6 C). In contrast, tissue length along the AP axis was only slightly reduced in Shroom mutants (1.99 ± 0.04–fold) compared with wild type (2.13 ± 0.05–fold, P = 0.074). These results show that Shroom is required for convergent extension but does not strongly affect the final length of the tissue.
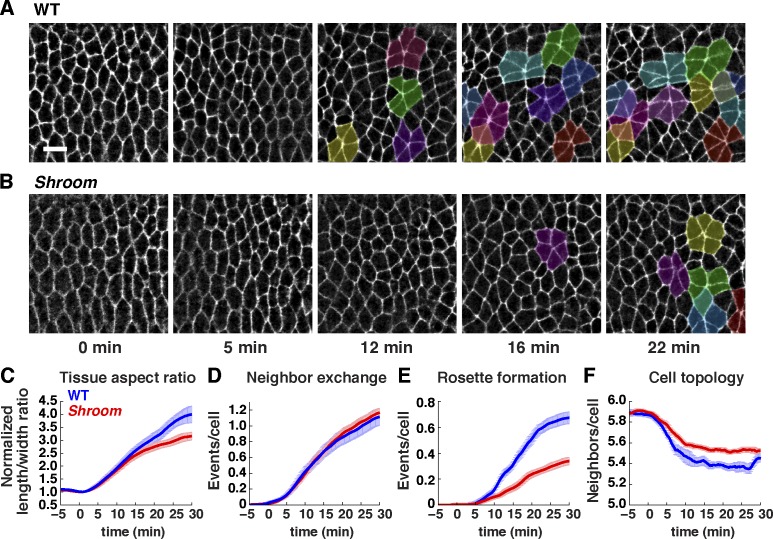
Figure 6.
Shroom is required for convergent extension and multicellular rosette formation. (A and B) Stills of time-lapse videos of wild-type (WT; A) and ShrmΔ11 mutant (B) embryos expressing Spider:GFP (t = 0 is the onset of elongation in early stage 7). Stage 7 (0–10 min); stage 8 (10–30 min). Anterior is left, and ventral is down. Rosettes (groups of five or more cells that meet at a vertex) are highlighted. Bar, 10 µm. (C–F) Cell behavior in wild-type (blue) and ShrmΔ11 mutant (red) embryos. (C) Tissue aspect ratio (tissue length along the AP axis relative to its width along the DV axis) normalized to the value at t = 0. Shroom mutants have a reduced tissue aspect ratio in stage 8 (P = 0.02 at 30 min). (D and E) Neighbors lost per cell through neighbor exchange, also known as a T1 process (resulting from single edge contraction events; D) and rosette formation (resulting from the contraction of multiple, consecutive edges; E). Rosette formation was significantly reduced in Shroom mutants (P < 0.001 at 30 min). Local neighbor exchange was not significantly affected (P = 0.63 at 30 min). (F) Mean number of neighbors per cell. Wild-type cells have progressively fewer neighbors midway through elongation as a result of cell rearrangement. Cells in Shroom mutants have more neighbors on average midway through elongation (P < 0.001 at 20 min), consistent with reduced cell rearrangement. Videos of four wild-type Spider:GFP and eight Shroom mutants were analyzed at 15-s intervals (185–276 cells tracked/embryo). Means ± SEM between embryos are shown.
To investigate the cellular basis of the convergent extension defects in Shroom mutants, we analyzed cell rearrangement, the major mechanism driving convergent extension in Drosophila (Irvine and Wieschaus, 1994). Cell rearrangements occur through local neighbor exchange (resulting from single edge contraction events; Bertet et al., 2004) and multicellular rosette formation (resulting from the contraction of multiple consecutive edges; Blankenship et al., 2006). Myosin II mislocalization in Shroom mutants occurred partway through elongation in stage 8 (t = 15–30 min; Fig. 5, E–G), when multicellular rosette formation is the predominant form of cell rearrangement in the tissue (Fernandez-Gonzalez et al., 2009). Using computational cell-tracking methods, we found that Shroom mutants had a strong reduction in rosette behaviors (Fig. 6, A, B, and E), with little change in local neighbor exchange (Fig. 6 D). Similar defects were observed in ShrmAB RNAi embryos (Fig. S5, A–D). These results indicate that cell rearrangement is significantly decreased in Shroom mutants and can account for the reduction in convergent extension in the absence of Shroom activity.
The loss of Shroom is associated with increased apical cell area in the frog, chick, and mouse (Lee et al., 2007; Nishimura and Takeichi, 2008; Plageman et al., 2010). Consistent with these studies, we found that intercalating cells in Shroom mutants had slightly larger apical cell areas during elongation (6%, P = 0.03 at 20 min), with no difference between wild-type and mutant cells before elongation (P = 0.66 at 0 min). This is suggestive of reduced apical myosin contractility in Shroom mutants. In contrast, mesoderm invagination, which requires apical constriction, occurs with the proper developmental timing in Shroom mutants (Fig. S5 E), although we cannot rule out subtle defects in this tissue. The combined effect of decreased rosette formation and increased apical cell area in Shroom mutants is a significantly reduced tissue aspect ratio but nearly normal tissue length along the AP axis. Together, these results demonstrate that Shroom is required for multicellular rosette formation and convergent extension midway through elongation, coincident with the defects in myosin localization.
Shroom planar polarity requires Rho activity and the actin cytoskeleton
In one model, Shroom could act downstream of a subtle asymmetry in Rho GTPase activity to enhance Rho-kinase and myosin II planar polarity. Alternatively, Rho and Shroom could act independently to regulate planar polarity in Drosophila. Different relationships between Rho and Shroom have been reported in vertebrates, in which Rho and Shroom appear to act independently to regulate myosin localization and apical constriction in Xenopus (Haigo et al., 2003; Hildebrand, 2005; Nishimura and Takeichi, 2008; Nishimura et al., 2012), but Rho is required for Shroom3-mediated apical constriction in the lens placode of the chick and mouse (Plageman et al., 2011). To test whether Shroom acts downstream or independently of Rho signaling in Drosophila, we analyzed Shroom localization in embryos expressing dominant-negative RhoN19. Shroom planar polarity was abolished in RhoN19-expressing embryos (Fig. 7, A, B, and G), whereas depleting both isoforms of Shroom in ShrmAB RNAi embryos had no effect on Rho localization or activity (Fig. S5, F–I). Thus, Shroom localization requires Rho GTPase activity but not vice versa, consistent with a model in which Shroom acts downstream of Rho.
Figure 7.
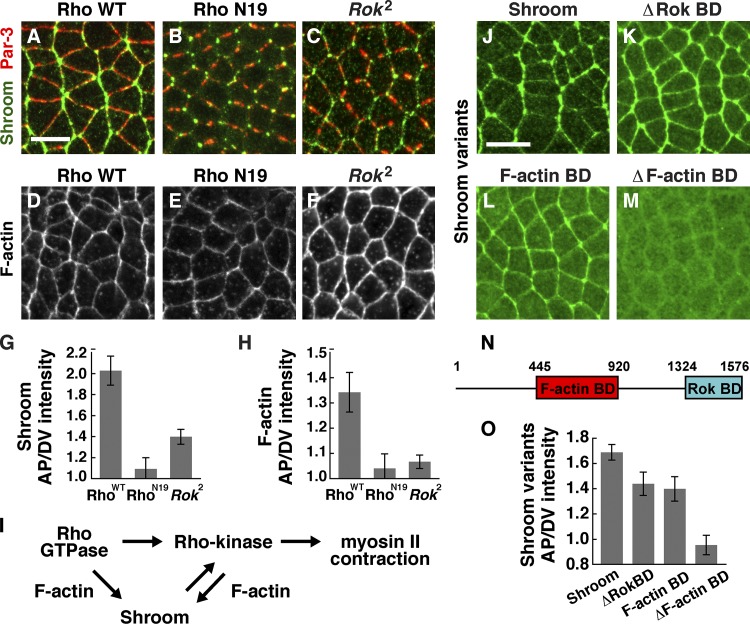
Rho activity, Rho-kinase, and F-actin binding are required for Shroom planar polarity. (A–F) Shroom, Baz/Par-3, and F-actin (phalloidin) in embryos expressing wild-type Rho (Rho WT; A and D), embryos expressing dominant-negative RhoN19 (B and E), and Rok2 maternal (C and F) mutants. Anterior is left, and ventral is down. (G and H) Planar polarized enrichment of Shroom (G) and F-actin (H) at AP cell boundaries (75–90° with respect to the AP axis) relative to DV cell boundaries (0–15°). (G) Shroom planar polarity (visualized with the anti-Shroom antibody) was reduced in embryos expressing RhoN19 compared with embryos expressing wild-type Rho (P < 0.0001) and in Rok2 mutants compared with wild type (P < 0.0001). (H) F-actin planar polarity was strongly reduced in embryos expressing RhoN19 compared with embryos expressing wild-type Rho (P < 0.005) and was slightly reduced (but did not reach statistical significance) in Rok2 mutants compared with wild type (P = 0.08). (I) Model. (J–M) Venus:Shroom proteins in stage 7 wild-type embryos visualized with anti-GFP. Wild-type Shroom (J), Shroom ΔRok–binding domain (BD; K), F-actin–binding domain alone (L), and Shroom ΔF-actin–binding domain (M) are shown. Anterior is left, and ventral is down. (N) Shroom protein (the ShrmA isoform aa 1–1,576) contains an F-actin–binding domain (aa 445–920) and a Rho-kinase–binding domain (Rok BD, aa 1,324–1,576). (O) Planar polarized enrichment of Venus:Shroom proteins at AP cell boundaries relative to DV cell boundaries visualized with anti-GFP antibody. Shroom ΔF-actin–binding domain planar polarity was strongly reduced (P < 0.0001) and Shroom F-actin–binding domain and ΔRok–binding domain planar polarity were moderately reduced (P < 0.05) compared with full-length Shroom. Shroom planar polarity was slightly lower than in Fig. 3 C and Fig. 7 G because different antibodies were used to detect endogenous and ectopic Shroom protein (Materials and methods). A single value was obtained for each image by averaging 100–200 edges/image; 12–26 images in 6–18 embryos were analyzed/genotype. Means ± SEM between images are shown. Bars, 10 µm.
To investigate how Rho influences Shroom localization, we identified the sequences in the Shroom protein that are required for Shroom planar polarity. Shroom does not contain an RB domain, suggesting that the regulation of Shroom localization by Rho signaling occurs indirectly. The ShrmA isoform contains a central domain that associates with F-actin and a C-terminal domain that binds to Rho-kinase (Fig. 7 N; Bolinger et al., 2010). Ectopically expressed Venus-tagged ShrmA recapitulates the localization of the endogenous protein (Fig. 7 J). Shroom lacking the Rho-kinase–binding domain displayed substantial planar polarity, although slightly less than the full-length protein (Fig. 7, K and O). A similar distribution was observed for the F-actin–binding domain alone (Fig. 7, L and O). In contrast, deletion of the F-actin–binding domain completely eliminated Shroom planar polarity and strongly reduced its cortical localization (Fig. 7, M and O). These results suggest that Shroom localization requires a direct interaction with the actin cytoskeleton.
The finding that the Shroom actin-binding domain is required to generate Shroom planar polarity suggests that Rho could influence Shroom through its effects on actin. In wild-type embryos, F-actin is enriched at AP interfaces in a planar polarized fashion (Fig. 7, D and H; Blankenship et al., 2006). F-actin planar polarity was abolished in embryos expressing dominant-negative RhoN19 (Fig. 7, E and H), consistent with a model in which Rho generates Shroom planar polarity by modifying the actin cytoskeleton. Although the Rho-kinase–binding domain is largely dispensable for Shroom localization, Shroom planar polarity was reduced in Rho-kinase mutants (Fig. 7, C and G), suggesting that Shroom and Rho-kinase mutually reinforce each other’s localization. Rho-kinase regulates actin organization (Amano et al., 2001; Verdier et al., 2006), and consistent with this role, we found that Rho-kinase mutants display reduced F-actin planar polarity (Fig. 7, F and H), and Rho-kinase activity is necessary for its own planar polarized localization (Fig. S2, D and J). These results are consistent with a model in which Rho GTPase modulates the actin cytoskeleton to generate a planar polarized localization of Shroom, and Shroom in turn stabilizes Rho-kinase at planar polarized adherens junctions to enhance planar polarized actomyosin contractility during convergent extension (Fig. 7 I).
Discussion
Rho-kinase is an essential regulator of actomyosin contractility, but the mechanisms that generate Rho-kinase asymmetry to produce spatially regulated forces during development are not well understood. Here, we show that Rho GTPase signaling is required for the planar polarized localization of Rho-kinase and myosin II during Drosophila axis elongation. Direct interaction between Rho and Rho-kinase recruits Rho-kinase to adherens junctions but is not sufficient for full Rho-kinase planar polarity, suggesting that other mechanisms amplify the effects of Rho signaling. We provide evidence that the actin-binding protein Shroom regulates Rho-kinase localization and planar polarized actomyosin contractility to promote sustained cell rearrangements during axis elongation. Shroom is present in a planar polarized distribution at adherens junctions in intercalating cells, consistent with a direct and localized function. Shroom planar polarity requires Rho activity, indicating that Shroom is an effector of Rho signaling. In Shroom mutants, Rho-kinase and myosin II junctional localization and planar polarity initiate normally but fail to be amplified and maintained during axis elongation. Consequently, planar polarized contractile forces and multicellular rosette rearrangements are reduced in Shroom mutants, resulting in decreased convergent extension. These results support a role for Shroom in regulating planar polarized actomyosin contractility and junctional remodeling during convergent extension, expanding the morphogenetic functions of this highly conserved protein beyond its known role in apical constriction.
Our data support a model in which Rho GTPase and Shroom have distinct functions in regulating Rho-kinase localization and planar polarized myosin contractility during convergent extension. Rho GTPase recruits Rho-kinase to adherens junctions and initiates planar polarity, and Shroom plays a modulatory role in enhancing and maintaining planar polarized myosin contractility downstream of Rho signaling. Rho GTPase binds to Rho-kinase and could regulate its localization directly (Riento and Ridley, 2003; Jaffe and Hall, 2005). Rho does not bind to Shroom but may regulate Shroom planar polarity indirectly through its effect on the actin cytoskeleton. We show that Rho-kinase, usually viewed as a downstream effector of Shroom, feeds back to maintain Shroom planar polarity and its own planar polarized localization. Rho-kinase could directly phosphorylate Shroom to reinforce planar cell polarity. Alternatively, Rho-kinase could promote Shroom localization through remodeling of the actin cytoskeleton, as the Shroom actin-binding domain is necessary and sufficient for targeting to planar junctions, and Rho-kinase can phosphorylate known regulators of actin (Amano et al., 2001; Verdier et al., 2006).
These findings may be relevant to neural tube development in vertebrates, which involves a combination of apical constriction, polarized junctional remodeling, and cell shape changes (Nishimura and Takeichi, 2008; Nishimura et al., 2012). Shroom3 is required for neural tube closure in the mouse, frog, and chick (Hildebrand and Soriano, 1999; Haigo et al., 2003; Nishimura and Takeichi 2008), and disrupting the interaction between Shroom and Rho-kinase reduces the number of rosettes in the chick neural plate (Nishimura and Takeichi, 2008). Unlike mutants that have disrupted rosette-based movements caused by defects in cell adhesion (Tamada et al., 2012), the defects in Shroom mutants are likely a result of reduced myosin II activity. Rosette behaviors in Drosophila predominate midway through elongation at stage 8 (Fernandez-Gonzalez et al., 2009), coinciding with the stage when myosin becomes mislocalized in Shroom mutants. A failure to reinforce actomyosin contractility during elongation in Shroom mutants could selectively disrupt later-onset, higher-order cell rearrangements, with no effect on local neighbor exchange events that are more frequent at earlier stages. Alternatively, rosette formation may require more force, as rosettes form through the contraction of multicellular actomyosin cables that are under a higher level of tension and accumulate more myosin (Fernandez-Gonzalez et al., 2009). In Shroom mutants, defects in myosin junctional localization may prevent contractile forces from reaching the levels necessary to produce rosette-based convergent extension movements. It will be interesting to explore whether planar polarized Shroom activity plays a general role in promoting junctional remodeling and enhancing mechanical force generation in processes that require strong actomyosin contractility during development.
Rho GTPase signaling is an excellent candidate to break planar symmetry, as a small fraction of active Rho protein can trigger rapid and dramatic changes in the actin cytoskeleton (Jaffe and Hall, 2005). In one model, a subtle increase in Rho activity at AP cell boundaries could provide an instructive cue, guiding planar cell polarity by recruiting Rho-kinase, modifying the actin cytoskeleton, and facilitating the cortical association of the Rho-kinase regulator Shroom. Alternatively, Rho could regulate Rho-kinase planar polarity indirectly through its role in promoting Rho-kinase apical localization. Although it is challenging to visualize a small and highly dynamic population of active Rho protein in vivo, several findings support the idea that localized Rho activity could play an instructive role in planar polarity. First, myosin planar polarity and directional cell rearrangements occur normally at early stages in Shroom mutants, suggesting that other signals are able to generate localized myosin activity. The partial planar asymmetry of a fragment containing the RB domain of Rho-kinase, which is predicted to interact with the active pool of Rho GTPase, suggests that Rho could contribute to this asymmetry. Second, Rho is required for the planar polarized localization of Shroom, raising the possibility that Rho signaling could provide an essential source of Shroom asymmetry. Third, the upstream Rho activator RhoGEF2 in Drosophila and PDZ-RhoGEF in the chick display a subtle planar asymmetry during epithelial bending and elongation (Levayer et al., 2011; Nishimura et al., 2012; Warrington et al., 2013). Multiple activators and inhibitors of Rho could act together to generate a spatially localized pattern of Rho activity, as is the case for apical constriction (Simões et al., 2006). Notably, although Rho GTPase activity is necessary to establish Rho-kinase and myosin planar polarity, it is not sufficient to maintain their activity at high enough levels to allow sustained force generation and rosette rearrangements in Shroom mutants. We propose that Rho promotes the recruitment of Shroom as part of a positive feed-forward mechanism that reinforces planar polarized actomyosin contractility during convergent extension.
Planar polarized cell rearrangements require the active maintenance of cell polarity in large populations of dynamically moving cells. We show that Shroom and Rho GTPase signaling play distinct roles in the establishment and maintenance of polarized actomyosin contractility during convergent extension. The upstream spatial cues that localize actomyosin contractility to specific planar cellular domains are not known. An asymmetry in the organization of the actin cytoskeleton is the earliest evidence of planar polarity in the Drosophila embryo (Blankenship et al., 2006). Distinct actin-binding domains in different Shroom isoforms have been proposed to target Shroom protein and its effectors to different regions of the cell (Hildebrand and Soriano, 1999; Dietz et al., 2006). Moreover, the actin-binding domain is critical for Shroom planar polarity. These findings support the idea that an asymmetry in the actin cytoskeleton is an essential spatial input that regulates the localization of Shroom, the contractile machinery, and ultimately the forces that control cell rearrangement and tissue structure. The upstream spatial cues that generate these asymmetries could involve an asymmetry in Rho signaling, perhaps through the local activation of upstream signaling proteins that regulate Rho GTPase activity. Alternatively, the critical event in the establishment of planar cell polarity could be a Rho-independent reorganization of the actin cytoskeleton that biases the activity of Shroom, Rho-kinase, and myosin, which in turn modify the cytoskeleton to allow robust and sustained cell polarization. Elucidation of the upstream spatial cues that regulate actomyosin localization and dynamics will provide insight into the mechanisms that direct polarized cell behavior.
Materials and methods
Fly stocks and genetics
Embryos were generated at 25°C. Wild type was yw unless otherwise indicated. Alleles were Rok1, Rok2 (Winter et al., 2001), ShrmΔ11, and ShrmΔ13.6 (this work), and Df(2R)Exel7131 (Bloomington Drosophila Stock Center). The expression of Rho-kinase, Shroom, Rho1, PKN, and Dicer-2 transgenes was driven by the upstream activating sequence (UAS) enhancer under the control of Gal4. Embryos expressing UASp–Rho-kinase transgenes, UASp-Shroom transgenes, UAST-GFP:Rho1, UASp-PKNG58A:Venus, or UAST–Dicer-2 were the F2 progeny of UAS males × matαtub67;15 Gal4 females (gift of D. St Johnston, University of Cambridge, Cambridge, England, UK), except UASp-Venus:ShrmA and UASp-Venus:RBD:PH, which were the F2 progeny of UAS males × matαtub15 Gal4 females.
ShrmΔ11 and ShrmΔ13.6 were generated by P element transposase-mediated male recombination (Chen et al., 1998). The P elements pSuport-pKG04646 (50F1) and p{EPgyp2}CG8613[EY06332] (50F6) were recombined with cn1 and sp1, respectively. P element–induced recombination occurred in males of the following genotype: w+/y; cn1 pSuport-pKG04646/p{EPgyp2}CG8613[EY06332] sp1; Dr Δ2-3 99/+ (source of transposase, Bloomington Drosophila Stock Center). These males were crossed to w+/w+; cn1 sp1/SM6a, cn1 sp1 females (Bloomington Drosophila Stock Center), and the progeny were screened for deletions in Shroom by the presence of cn1 and sp1 and the loss of white+, as both P elements were excised in the deletions. Deletion breakpoints were molecularly mapped by PCR (Fig. S4 B).
Shroom mutants were the progeny of ShrmΔ11/Df(2R)Exel7131 or ShrmΔ13.6/Df(2R)Exel7131 females and males (Fig. 4, B, E, and F; Fig. 5, B and E–G; Fig. 6, B–F; and Fig. S5 E). Similar results were observed in the progeny of ShrmΔ11/Df or ShrmΔ13.6/Df females crossed to ShrmΔ11/+ or ShrmΔ13.6/+ males (Fig. 4, I and J): 6/8 ShrmΔ11 and 8/11 ShrmΔ13.6 maternal/zygotic mutants displayed aberrant Rho-kinaseK116A localization compared with 0/9 wild-type embryos, and 3/3 videos of ShrmΔ11 maternal/zygotic mutants displayed aberrant Myo:GFP localization compared with 0/3 videos of wild-type embryos. In crosses to Shrm/+ males, progeny that lack maternal and zygotic Shroom were identified by the absence of balancer markers. In crosses to Shrm/Df males, all progeny were maternally and zygotically mutant for Shroom, and 25% are predicted to be homozygous for the small Df(2R)Exel7131 deficiency. The defects in the progeny of Shrm/Df females and males were fully penetrant, indicating that these defects are not a result of the zygotic loss of other genes uncovered by the deficiency. Moreover, crosses to Shrm/+ males and injection of three independent dsRNAs to Shroom gave similar results (Fig. 4 C and Fig. S5, A–D), suggesting that these defects are caused by the loss of Shroom.
Embryonic hatch rates were 10% of embryos from ShrmΔ11/Df; GFP:RokK116A females and 23% of embryos from ShrmΔ13.6/Df; GFP:RokK116A females crossed to Shrm/+ males compared with 80% of embryos laid by control GFP:RokK116A females (176–200 embryos/genotype). Weaker but detectable lethality was also observed in Shroom mutants that do not express a GFP marker, and these could not be maintained as homozygous stocks. Rok1 and Rok2 germline clones were generated using the FLP recombinase–dominant female sterile system (Chou and Perrimon, 1992) and ovoD2 FRT19A (gift of N. Tolwinski, Yale-NUS College, Singapore) by heat shocking larvae of the following genotypes and crossing them to wild-type males: Rok1 FRT19A/ovoD2 FRT19A; matαtub67/hs-FLP38 UASp-Venus:Rok transgene and Rok2 FRT19A/ovoD2 FRT19A; hs-FLP38 UASp-Venus:Rok transgene/+; matαtub15/+.
Transgenic lines
Transgenes were UASp-Venus:RokK116A, UASp-Venus:RokWT (Simões et al., 2010), sqh-GFP:RokK116A, UASp-Venus:RokΔCAT (Rok Δaa 1–412), UASp-Venus:RokK116AΔPH (Rok Δaa 1,134–1,391), UASp-Venus:RokK116AΔRB (Rok K948M L955A Δaa 963–1,046), UASp-Venus:RokK116AΔCC,SB, UASp-Venus:RokWTΔCC,SB (Δaa 547–923), UASp-Venus:RokK116AΔSB, UASp-Venus:RokWTΔSB (Δaa 835–937), UASp-Venus:RB:PH (Rok aa 900–1,391), UASp-Venus:SB:RB:PH (Rok aa 700–1,391), UASp-Venus:ShrmA (full-length ShrmA isoform, aa 1–1,576), UASp-Venus:ShrmAΔRokBD (Δaa 1,324–1,576), UASp-Venus:ShrmAΔF-actinBD (Δaa 445–920), and UASp-Venus:ShrmA F-actinBD (aa 445–920).
UASp-Venus:Rok and UASp-Venus:Shroom constructs were made by recombining pENTR clones into the pUASp-W-attB destination vector (gift of M. Buszczak, University of Texas Southwestern Medical Center, Dallas, TX) using the Gateway system (Invitrogen) and were inserted in attp40 (chromosome II). UASp-PKNG58A:Venus was modified from Simões et al. (2006) using the UASp promoter, and sqh-GFP:RokK116A (Fig. 4, B and E–J) was modified from Simões et al. (2010) using pENTR-RokK116A recombined into the pSqh-GFP-W-attB destination vector (gift of F. Wirtz-Peitz, Harvard Medical School, Boston, MA); both were inserted in attp2 (chromosome III). Other transgenic fly stocks were UAST-Rho1, UAST-GFP:Rho1 (Bloomington Drosophila Stock Center), and UAS–Dicer-2 (X) (gift of P. Smibert, Mount Sinai Hospital, New York, NY).
Immunohistochemistry
Tagged Rho-kinase and Shroom proteins were visualized with anti-GFP antibody in embryos fixed 5 min in 1:1 37% FA in PBS/heptane (Fig. 1 and Fig. 7) or 1 h in 1:1 3.7% FA in PBS/heptane (Fig. 2 and Fig. 4) and manually devitellinized. Myo:GFP (sqh:GFP; Royou et al., 2004), F-actin, Rho, PKNG48A:Venus, HA:RhoWT, HA:RhoN19, and HA:Rok:PH were visualized in embryos fixed 1 h in 1:1 3.7% FA in PBS/heptane and manually devitellinized. Endogenous Shroom protein (the ShrmA isoform) was detected in embryos boiled 10 s in 0.03% Triton X-100/0.4% NaCl, cooled on ice, and devitellinized in heptane/methanol. Antibodies were rabbit GFP (1:100; Torrey Pines Biolabs, Inc.), rat HA (1:500; Roche), guinea pig Baz/Par-3 (1:500; Blankenship et al., 2006), mouse Rho1 (1:50; Developmental Studies Hybridoma Bank), rat ShrmA R20 (1:100; Bolinger et al., 2010), and mouse Arm/β-catenin (1:50; Developmental Studies Hybridoma Bank). Rhodamine-conjugated phalloidin and secondary antibodies conjugated to Alexa Fluor 488, Alexa Fluor 568, or Alexa Fluor 647 (Molecular Probes) were used at 1:500. PKNG48A:Venus was visualized after fixation without antibody. Embryos were mounted in ProLong gold (Molecular Probes) and imaged on a confocal microscope (LSM 700; Carl Zeiss) with a Plan Neofluor 40×, 1.3 NA objective; 1.0-µm z slices were acquired at 0.5-µm steps. Maximum intensity projections of 2–3 µm in the apical junctional domain were analyzed.
In vitro transcription and mRNA injection
To synthesize mRNA encoding HA:RhoWT, HA:RhoN19, and HA:RhoV14, coding sequences were PCR amplified from pGEX-RhoWT (gift of J. Settleman, Genentech, San Francisco, CA), pUAST-RhoN19, and pUAST-RhoV14 (gifts of D. Strutt, University of Sheffield, Sheffield, England, UK). To synthesize mRNA encoding HA:Rok PH, a PCR fragment corresponding to Rok aa 1,084–1,391 was amplified from wild-type genomic DNA. Products were cloned into pCS2+ using BamH1 and XhoI restriction sites. pCS2+HA:RhoWT, pCS2+HA:RhoN19, pCS2+HA:RhoV14, and pCS2+HA:Rok PH were digested with NotI and used as templates for in vitro transcription to generate mRNAs encoding HA:RhoWT, HA:RhoN19, HA:RhoV14, and HA:Rok PH, respectively, with the mMESSAGE mMACHINE SP6 kit (Ambion). Embryos were collected for 20 min, aged 2 h and 20 min at 25°C, injected ventrally during mid-to-late cellularization with 0.1 nl of 2-mg/ml mRNA, incubated 45 min at 25°C in a humidified chamber, and processed for immunostaining.
dsRNA injection
Templates to produce dsRNA against ShrmA or both ShrmA and ShrmB isoforms were generated by PCR from the LP13775 EST. PCR products were used as templates for the T7 transcription reactions with the 5× MEGAscript T7 kit (Ambion). dsRNA was injected ventrally (for immunostaining) or dorsally (for live imaging) with 0.1 nl of 2-mg/ml dsRNA into 0–1-h embryos that were the F2 progeny of UAS–Dicer-2 (X); UAS-Venus:RokK116A × matαtub67;15 (Fig. 4, A and C), UAS–Dicer-2; sqh-sqh:GFP × matαtub67;15 (Fig. 4 K and Fig. 5 C), UAS–Dicer-2; Spider:GFP × matαtub67;15 (Fig. S5 E), and UAS–Dicer-2; UAS-RhoWT UAS-PKNG58A:Venus × matαtub67;15 (Fig. 2, O and P; and Fig. S5, F–I). All other injections were without Dicer-2. With Dicer-2, similar effects were observed at a greater distance from the site of injection. Embryos were incubated 2 h and 30 min at 25°C in a humidified chamber and processed for immunostaining or live imaging.
Planar polarity analysis
Using the SIESTA (Scientific Image Segmentation and Analysis) algorithm (Fernandez-Gonzalez and Zallen, 2011), user-drawn, 3-pixel-wide lines for all edges in a 50 × 50–µm region were analyzed to obtain the mean pixel intensity and orientation for each edge. Intensities were averaged for all edges in each angular range, and the mean cytoplasmic intensity of all pixels >1 µm from user-drawn lines was subtracted for background correction. The mean, background-subtracted edge intensity for all edges in the 75–90° angular range was normalized to the mean, background-subtracted intensity in the 0–15° bin for Rho-kinase, myosin, Shroom, and F-actin. The inverse ratio was calculated for Par-3. A single value was obtained for each embryo, and error bars indicate the SEMs between embryos. P-values were calculated using the F test followed by the appropriate t test.
Time-lapse imaging
Time-lapse imaging was performed with Spider:GFP (gift of A. Debec, Insitut Jacques Monod, Paris, France) and Myo:GFP (sqh:GFP; Royou et al., 2004). Embryos were dechorionated 2 min in 50% bleach, washed in water, mounted on an oxygen-permeable membrane (YSI Incorporated) with halocarbon oil 27 (Sigma-Aldrich), and imaged on a spinning-disk confocal microscope (UltraView RS5; PerkinElmer) with Plan Neofluor 40×, 1.3 NA or 63×, 1.4 NA objectives (Carl Zeiss). Z stacks were acquired at 1-µm steps (40× objective) or 0.5-µm steps (63× objective) at 30-, 15-, or 5-s intervals as indicated. Maximum intensity projections of 2–3 µm in the apical junctional domain were analyzed. Videos 7 and 8 were background noise corrected by multiplying pixel intensities in three consecutive z planes and normalizing to occupy the full intensity range. Cell behavior was analyzed as previously described (Simões et al., 2010; Tamada et al., 2012). Tissue aspect ratio was the ratio of the long axis to the short axis of an ellipse fit to a group of cells that were tracked ≥30 min after t = 0. Cumulative cell rearrangement events for cells tracked ≥12.5 min after t = 0 were used for analysis of neighbor exchange and rosette formation. Cell area and topology (number of neighbors) were calculated for all cells at each time point. A single value was obtained for each embryo at each time point, and error bars indicate the SEMs between embryos. The p-values were calculated using the F test followed by the appropriate t test using the t = 30 min value as the test statistic.
Laser ablation
Embryos injected with dsRNAs targeting ShrmA, both ShrmA and ShrmB isoforms, or flp were mounted in halocarbon oil 27 and imaged on a spinning-disk confocal (UltraView RS5) with a Plan Neofluor 63×, 1.4 NA oil immersion lens that was also used to focus the MicroPoint laser. An N2 laser (MicroPoint; Photonics Instruments) tuned to 365 nm was used to ablate cell boundaries labeled with Myo:GFP. Z stacks of three optical slices at 0.5-µm steps were acquired at 2-s intervals. The two vertices attached to the cut edge were manually identified and analyzed in SIESTA (Fernandez-Gonzalez and Zallen, 2011).
Molecular biology
Full-length pENTR-Venus:Rok and pENTR-Venus:RokK116A plasmids were generated as previously described (Simões et al., 2010). For pENTR-Venus:RokΔkinase (Δaa 1–412), a PCR fragment was amplified with the primers Rok413f, 5′-CACCATGGACTATCAGCTGCTGTCTAGCGAC-3′, and Rok1391r, 5′-TCATTTCAGCGATGAATTGGCTGG-3′, and cloned into pENTR/D-topo (Invitrogen). N-terminal Venus was inserted into the NotI site (PCR amplified with the primers NotIVenusf, 5′-ATGCGGCCGCCACCATGGTGAGCAAGGGCGAGGAGCTG-3′, and NotIVenusr, 5′-ATGCGGCCGCCGTGGACCGGTGCTTGTACAGCTC-3′. For pENTR-Venus:RokK116AΔPH (Δaa 1,134–1,391), a stop codon was introduced in pENTR-Venus:RokK116A at position S1134 by site-directed mutagenesis (QuikChange; Agilent Technologies) with the primers Rok S1134stopf, 5′-CCTCACACAGGACTAGGTCTTCGAGGGCTG-3′, and Rok S1134stopr, 5′-CAGCCCTCGAAGACCTAGTCCTGTGTGAGG-3′. For pENTR-Venus:RokK116AΔRB (K948M L955A Δaa 967–1,046), an internal deletion was created by amplifying a PCR product from pENTR-Venus:RokK116A with the 5′ phosphorylated primers: Rok966r, 5′-CAGCTTCTTGTGCAGTTCGTTTTCGGCCTC-3′, and Rok1047f, 5′-GAGAAGGAGATGCGTCGGCTGCAG-3′. The product was self-ligated, and two point mutations (K948M and L955A) were added sequentially by site-directed mutagenesis using the primers RokK948Mf, 5′-CGAGATCAACGCCATGGAGGCGGCCCTAGC-3′, RokK948Mr, 5′-GCTAGGGCCGCCTCCATGGCGTTGATCTCG-3′, RokL955Af, 5′-GGCGGCCCTAGCCACGGCCAAGGAGGCCG-3′, and RokL955Ar, 5′-CGGCCTCCTTGGCCGTGGCTAGGGCCGCC-3′. K948 and L955 are conserved sites that directly interact with Rho in mammals (Fujisawa et al., 1996). For pENTR-Venus:RokΔCC and pENTR-Venus:RokK116AΔCC (Δaa 547–923), an internal deletion was created by amplifying a PCR product from pENTR-Venus:Rok or pENTR-Venus:RokK116A with the 5′ phosphorylated primers Rok546r, 5′-CTTTTGCTCGTTTTCCAGATTG-3′, and Rok924f, 5′-GAAAAGGAGAAGACTATCAAGGAGC-3′. The product was self-ligated. For pENTR-Venus:RokΔSBD and pENTR-Venus:RokK116AΔSBD (Δaa 835–937), an internal deletion was created by amplifying a PCR product from pENTR-Venus:Rok or pEntr-Venus:RokK116A with the 5′ phosphorylated primers Rok938f, 5′-ATGAAGCATCGCAACGAGATCAACG-3′, and Rok834r, 5′-CTCCTCGAAGCGTCGCTTCGCCTC-3′. The product was self-ligated. For pENTR-Venus:RBD:PH (aa 900–1,391), a PCR product was amplified with the primers Rok900f, 5′-CACCATGCAGGTGGCCGTTGCCCGTGCAGAC-3′, and Rok1391r, 5′-TCATTTCAGCGATGAATTGGCTGG-3′, and cloned into pENTR. N-terminal Venus was inserted in the NotI site. For pENTR-Venus:SBD:RBD:PH (aa 700–1,391), a PCR product was amplified with the 5′ phosphorylated primers Rok700f, 5′-CAGGAGGTCAAGGCACACCAAGAG-3′, and Venusr, 5′-CGCCGTGGACCGGTGCTTGTACAG-3′. The product was self-ligated. For pENTR-Venus:ShrmA (full length, aa 1–1,576), a PCR product was amplified from the LP13775 EST with the primers ShrmAf1, 5′-CACCATGAAAATGCGCAATCACAAGGAGAACGG-3′, and ShrmAr1, 5′-CTAACAATCGCTTTGGACTAGCGC-3′, and cloned into pENTR. N-terminal Venus was inserted in the NotI site. For pENTR-Venus:ShrmA ΔRokBD (Δaa 1,324–1,576), a stop codon was introduced at position 1,324 in pENTR-Venus:ShrmA by site-directed mutagenesis with the primers ShrmA 1324stopf, 5′-GGAGCTGGTTAAGCTCCTATGACCTGGTGACAAGATCGCCG-3′, and ShrmA 1324stopr, 5′-CGGCGATCTTGTCACCAGGTCATAGGAGCTTAACCAGCTCC-3′. For pENTR-Venus:ShrmA F-actin BD (aa 445–920), a PCR product was amplified from the LP13775 EST with the primers ShrmA445f, 5′-CACCATGGCACCACAACCGCCAGCTGGTAAG-3′, and ShrmA920r, 5′-CTAATTCGATCGTTTCTGCTGGTTGC-3′, and cloned into pENTR. For pENTR-Venus:ShrmA ΔF-actin BD (Δaa 445–920), a PCR product was amplified from pENTR-Venus:ShrmA with the 5′ phosphorylated primers ShrmAaa920f, 5′-AATTCCAAGGCCTCATACTTGCCG-3′, and ShrmAaa445r, 5′-TGCAATGGTGACCACGGCCTCGCTGC-3′. The product was self-ligated. For pUASp-PKNG58A:Venus, a PCR product was amplified from pUAST-PKNG58A-EGFP (Simões et al., 2006) with the primers PKNf, 5′-CACCATGTCGGATTCGTATTATCAGGG-3′, and PKNr, 5′-TAGCTGGACGATCTGCAGCTCCG-3′, and cloned into pENTR. C-terminal Venus was inserted in the AscI site (PCR amplified with the primers AscIVenusf, 5′-TGGGCGCGCCGGAGGTGGAGGAGGTATGGTGAGCAAGGGCGAGGAGCTG-3′, and AscIVenusr, 5′-TGGGCGCGCCTCACGTGGACCGGTGCTTGTACAG-3′).
Shroom mutant breakpoint analysis
ShrmΔ11 and ShrmΔ13.6 both delete exons within the Shroom open reading frame that encode the Rho-kinase domain, assessed by the failure to amplify PCR products with the following primer pairs: (a) Shroomf, 5′-GATGTCGGCGCACCTGTGTGATGC-3′, and Shroomr, 5′-GCTCCTCGCTCGGCTCTGCCAGCTG-3′, and (b) seq12f, 5′-TGAAGTCCATCACGTCGTCTGCC-3′, and seq12r, 5′-CTTTGGACTAGCGCATCGCTGAGG-3′. In addition, ShrmΔ13.6 (but not ShrmΔ11) failed to amplify the following band in the middle of the Shroom coding region: (c) seq14f, 5′-TTGGCCCACCAGCAGCACCACCCGC-3′, and seq14r, 5′-GATTGCTGGCAATCACATTGTCATGG-3′. Both ShrmΔ11 or ShrmΔ13.6 mutants retained sequences in the 5′ coding region and just outside of the P elements on either side, assessed by the ability to amplify PCR products with the following primer pairs: (d) CG8613f, 5′-GAGAAGATGCTCTATGATCAGGG-3′, and CG8613r, 5′-CACACCATAGCACGAACGGATGAG-3′; (e) tejf, 5′-TCGTCAGAAGACGCGTAGTGTAAGC-3′, and tejr, 5′-CCGTCCATACGCCTCATATTGACC-3′; (f) seq13f, 5′-CAGGAGAGCGATCTGCTGTCGCG-3′, and seq13r, 5′-CTCGTTGTTGTGCAGATACAGCACG-3′; and (g) seq15f, 5′-CTCATAAACCTTGTCTGTCTGACCC-3′, and seq15r, 5′-CGACCGATGCGGAACTGAAGCTGGC-3′.
Primers for dsRNA injection, in vitro transcription, and mRNA injection
Templates to produce dsRNA against ShrmA or both ShrmA and ShrmB isoforms were generated by PCR from the LP13775 EST using the following primer pairs that contain a 5′ T7 promoter sequence (5′-TAATACGACTCACTATAGGGAGACCAC-3′): ShrmA T7f, 5′-CACAGTGCAGCCCACCTGGCACTCG-3′, and ShrmA T7r, 5′-CTTCGTAGGTGGTGCGACGTGTGG-3′; ShrmAB1 T7f, 5′-GCCTCATACTTGCCGCGTCAGAG-3′, and ShrmAB1 T7r, 5′-CTCTGGCTGCTTGTCGTGCACATC-3′; and ShrmAB2 T7f, 5′-GAGGAACTGCAGCTGATGCAGCGC-3′, and ShrmAB2 T7r, 5′-CGCATCGCTGAGGGAGCTAAGCTG-3′. The template to produce control dsRNA against flp was generated by PCR from hs-FLP38–bearing flies, using the following primers that contain a 5′ T7 promoter sequence: Flp T7f, 5′-CAGCAATCAAGAGAGCCACATTC-3′, and Flp T7r, 5′-TCCCACAACATTAGTCAACTCCG-3′.
To synthesize mRNA encoding HA:RhoWT and HA:RhoN19, coding sequences were PCR amplified from pGEX-RhoWT (gift of J. Settleman) and pUAST-RhoN19 (gift of D. Strutt) with the following primers: BamHIHARho1f, 5′-ATGGATCCCACCATGTACCCATATGATGTTCCAGATTACGCTGGAATGACGACGATTCGCAAGAAATTGG-3′, and XhoIRho1r, 5′-ATCTCGAGTTAGAGCAAAAGGCATCTGGTCTTC-3′. To synthesize mRNA encoding HA:Rok PH, a PCR fragment corresponding to Rok aa 1,084–1,391 was amplified using the following primers: BamHIHARok1084f, 5′-ATGGATCCCACCATGTACCCATATGATGTTCCAGATTACGCTGGACTCAAGCAGAAGATGGTCATGGAG-3′, and XhoIRok1391r, 5′-ATCTCGAGTCATTTCAGCGATGAATTGGCTGGC-3′.
Online supplemental material
Fig. S1 shows a description of Rho-kinase transgenes. Fig. S2 provides an analysis of Rho-kinase localization in Rho-kinase mutants. Fig. S3 shows an analysis of activated RhoV14 expression. Fig. S4 provides validation of Shroom mutants and RNAi treatment. Fig. S5 shows additional characterization of Shroom mutant and ShrmAB RNAi embryos. Videos 1 and 2 show Myosin II junctional and medial populations in a wild-type embryo (Video 1) and Shroom mutant embryo (Video 2). Videos 3–6 show Myosin II dynamics in a wild-type embryo (Video 3), Shroom mutant embryo (Video 4), a control injected embryo (Video 5), and ShrmA RNAi embryo (Video 6). Videos 7 and 8 show cell behavior in wild-type (Video 7) and Shroom mutant (Video 8) embryos expressing Spider:GFP. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201307070/DC1.
Supplementary Material
Acknowledgments
We are grateful to Chris Fincher for excellent technical assistance, Karen Kasza for help with laser ablation experiments, Dene Farrell for help with computational analysis, and Chris Fincher, Karen Kasza, Emily Marcinkevicius, German Sabio, Masako Tamada, Athea Vichas, and Richard Zallen for insightful discussions and comments on the manuscript. We thank Rodrigo Fernandez-Gonzalez for the sqh-GFP:RokK116A flies, Frederik Wirtz-Peitz for the pSqh-GFP-W-attB plasmid, Mike Buszczak for the pUASp-W-attB plasmid, Jeff Hildebrand for the ShrmA antibody, and Nick Tolwinski for the ovoD2 FRT19A flies.
This work was funded by National Institutes of Health/National Institute of General Medical Sciences R01 grants GM079340 and GM102803 to J.A. Zallen. J.A. Zallen is an Early Career Scientist of the Howard Hughes Medical Institute.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CC
- coiled coil
- dsRNA
- double-stranded RNA
- PH
- pleckstrin homology
- PKN
- protein kinase N
- RB
- Rho binding
- SB
- Shroom binding
- UAS
- upstream activating sequence
References
- Amano M., Nakayama M., Kaibuchi K. 2010. Rho-kinase/ROCK: A key regulator of the cytoskeleton and cell polarity. Cytoskeleton (Hoboken). 67:545–554 10.1002/cm.20472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano T., Tanabe K., Eto T., Narumiya S., Mizuno K. 2001. LIM-kinase 2 induces formation of stress fibres, focal adhesions and membrane blebs, dependent on its activation by Rho-associated kinase-catalysed phosphorylation at threonine-505. Biochem. J. 354:149–159 10.1042/0264-6021:3540149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett K., Leptin M., Settleman J. 1997. The Rho GTPase and a putative RhoGEF mediate a signaling pathway for the cell shape changes in Drosophila gastrulation. Cell. 91:905–915 10.1016/S0092-8674(00)80482-1 [DOI] [PubMed] [Google Scholar]
- Bertet C., Sulak L., Lecuit T. 2004. Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature. 429:667–671 10.1038/nature02590 [DOI] [PubMed] [Google Scholar]
- Blankenship J.T., Backovic S.T., Sanny J.S., Weitz O., Zallen J.A. 2006. Multicellular rosette formation links planar cell polarity to tissue morphogenesis. Dev. Cell. 11:459–470 10.1016/j.devcel.2006.09.007 [DOI] [PubMed] [Google Scholar]
- Bolinger C., Zasadil L., Rizaldy R., Hildebrand J.D. 2010. Specific isoforms of Drosophila shroom define spatial requirements for the induction of apical constriction. Dev. Dyn. 239:2078–2093 10.1002/dvdy.22326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgakova N.A., Grigoriev I., Yap A.S., Akhmanova A., Brown N.H. 2013. Dynamic microtubules produce an asymmetric E-cadherin–Bazooka complex to maintain segment boundaries. J. Cell Biol. 201:887–901 10.1083/jcb.201211159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T.-B., Perrimon N. 1992. Use of a yeast site-specific recombinase to produce female germline chimeras in Drosophila. Genetics. 131:643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Chu T., Harms E., Gergen J.P., Strickland S. 1998. Mapping of Drosophila mutations using site-specific male recombination. Genetics. 149:157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes-Hoang R.E., Parmar K.M., Christiansen A.E., Phelps C.B., Brand A.H., Wieschaus E.F. 2005. folded gastrulation, cell shape change and the control of myosin localization. Development. 132:4165–4178 10.1242/dev.01938 [DOI] [PubMed] [Google Scholar]
- Dietz M.L., Bernaciak T.M., Vendetti F., Kielec J.M., Hildebrand J.D. 2006. Differential actin-dependent localization modulates the evolutionarily conserved activity of Shroom family proteins. J. Biol. Chem. 281:20542–20554 10.1074/jbc.M512463200 [DOI] [PubMed] [Google Scholar]
- Ernst S., Liu K., Agarwala S., Moratscheck N., Avci M.E., Dalle Nogare D., Chitnis A.B., Ronneberger O., Lecaudey V. 2012. Shroom3 is required downstream of FGF signalling to mediate proneuromast assembly in zebrafish. Development. 139:4571–4581 10.1242/dev.083253 [DOI] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R., Zallen J.A. 2011. Oscillatory behaviors and hierarchical assembly of contractile structures in intercalating cells. Phys. Biol. 8:045005 10.1088/1478-3975/8/4/045005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R., Simoes S.M., Röper J.C., Eaton S., Zallen J.A. 2009. Myosin II dynamics are regulated by tension in intercalating cells. Dev. Cell. 17:736–743 10.1016/j.devcel.2009.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa K., Fujita A., Ishizaki T., Saito Y., Narumiya S. 1996. Identification of the Rho-binding domain of p160ROCK, a Rho-associated coiled-coil containing protein kinase. J. Biol. Chem. 271:23022–23028 10.1074/jbc.271.38.23022 [DOI] [PubMed] [Google Scholar]
- Häcker U., Perrimon N. 1998. DRhoGEF2 encodes a member of the Dbl family of oncogenes and controls cell shape changes during gastrulation in Drosophila. Genes Dev. 12:274–284 10.1101/gad.12.2.274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigo S.L., Hildebrand J.D., Harland R.M., Wallingford J.B. 2003. Shroom induces apical constriction and is required for hingepoint formation during neural tube closure. Curr. Biol. 13:2125–2137 10.1016/j.cub.2003.11.054 [DOI] [PubMed] [Google Scholar]
- Hildebrand J.D. 2005. Shroom regulates epithelial cell shape via the apical positioning of an actomyosin network. J. Cell Sci. 118:5191–5203 10.1242/jcs.02626 [DOI] [PubMed] [Google Scholar]
- Hildebrand J.D., Soriano P. 1999. Shroom, a PDZ domain-containing actin-binding protein, is required for neural tube morphogenesis in mice. Cell. 99:485–497 10.1016/S0092-8674(00)81537-8 [DOI] [PubMed] [Google Scholar]
- Hutson M.S., Tokutake Y., Chang M.S., Bloor J.W., Venakides S., Kiehart D.P., Edwards G.S. 2003. Forces for morphogenesis investigated with laser microsurgery and quantitative modeling. Science. 300:145–149 10.1126/science.1079552 [DOI] [PubMed] [Google Scholar]
- Irvine K.D., Wieschaus E. 1994. Cell intercalation during Drosophila germband extension and its regulation by pair-rule segmentation genes. Development. 120:827–841 [DOI] [PubMed] [Google Scholar]
- Jaffe A.B., Hall A. 2005. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21:247–269 10.1146/annurev.cellbio.21.020604.150721 [DOI] [PubMed] [Google Scholar]
- Kölsch V., Seher T., Fernandez-Ballester G.J., Serrano L., Leptin M. 2007. Control of Drosophila gastrulation by apical localization of adherens junctions and RhoGEF2. Science. 315:384–386 10.1126/science.1134833 [DOI] [PubMed] [Google Scholar]
- Lee C., Scherr H.M., Wallingford J.B. 2007. Shroom family proteins regulate gamma-tubulin distribution and microtubule architecture during epithelial cell shape change. Development. 134:1431–1441 10.1242/dev.02828 [DOI] [PubMed] [Google Scholar]
- Levayer R., Pelissier-Monier A., Lecuit T. 2011. Spatial regulation of Dia and Myosin-II by RhoGEF2 controls initiation of E-cadherin endocytosis during epithelial morphogenesis. Nat. Cell Biol. 13:529–540 10.1038/ncb2224 [DOI] [PubMed] [Google Scholar]
- Magie C.R., Meyer M.R., Gorsuch M.S., Parkhurst S.M. 1999. Mutations in the Rho1 small GTPase disrupt morphogenesis and segmentation during early Drosophila development. Development. 126:5353–5364 [DOI] [PubMed] [Google Scholar]
- Magie C.R., Pinto-Santini D., Parkhurst S.M. 2002. Rho1 interacts with p120ctn and α-catenin, and regulates cadherin-based adherens junction components in Drosophila. Development. 129:3771–3782 [DOI] [PubMed] [Google Scholar]
- Mason F.M., Tworoger M., Martin A.C. 2013. Apical domain polarization localizes actin-myosin activity to drive ratchet-like apical constriction. Nat. Cell Biol. 15:926–936 10.1038/ncb2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massarwa R., Niswander L. 2013. In toto live imaging of mouse morphogenesis and new insights into neural tube closure. Development. 140:226–236 10.1242/dev.085001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki K., Komatsu S., Ikebe M. 2006. Dynamics of RhoA and ROKα translocation in single living cells. Cell Biochem. Biophys. 45:243–254 10.1385/CBB:45:3:243 [DOI] [PubMed] [Google Scholar]
- Mizuno T., Amano M., Kaibuchi K., Nishida Y. 1999. Identification and characterization of Drosophila homolog of Rho-kinase. Gene. 238:437–444 10.1016/S0378-1119(99)00351-0 [DOI] [PubMed] [Google Scholar]
- Nikolaidou K.K., Barrett K. 2004. A Rho GTPase signaling pathway is used reiteratively in epithelial folding and potentially selects the outcome of Rho activation. Curr. Biol. 14:1822–1826 10.1016/j.cub.2004.09.080 [DOI] [PubMed] [Google Scholar]
- Nishimura T., Takeichi M. 2008. Shroom3-mediated recruitment of Rho kinases to the apical cell junctions regulates epithelial and neuroepithelial planar remodeling. Development. 135:1493–1502 10.1242/dev.019646 [DOI] [PubMed] [Google Scholar]
- Nishimura T., Honda H., Takeichi M. 2012. Planar cell polarity links axes of spatial dynamics in neural-tube closure. Cell. 149:1084–1097 10.1016/j.cell.2012.04.021 [DOI] [PubMed] [Google Scholar]
- Plageman T.F., Jr, Chung M.I., Lou M., Smith A.N., Hildebrand J.D., Wallingford J.B., Lang R.A. 2010. Pax6-dependent Shroom3 expression regulates apical constriction during lens placode invagination. Development. 137:405–415 10.1242/dev.045369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plageman T.F., Jr, Chauhan B.K., Yang C., Jaudon F., Shang X., Zheng Y., Lou M., Debant A., Hildebrand J.D., Lang R.A. 2011. A Trio-RhoA-Shroom3 pathway is required for apical constriction and epithelial invagination. Development. 138:5177–5188 10.1242/dev.067868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintin S., Gally C., Labouesse M. 2008. Epithelial morphogenesis in embryos: asymmetries, motors and brakes. Trends Genet. 24:221–230 10.1016/j.tig.2008.02.005 [DOI] [PubMed] [Google Scholar]
- Rauzi M., Lenne P.F., Lecuit T. 2010. Planar polarized actomyosin contractile flows control epithelial junction remodelling. Nature. 468:1110–1114 10.1038/nature09566 [DOI] [PubMed] [Google Scholar]
- Riento K., Ridley A.J. 2003. Rocks: multifunctional kinases in cell behaviour. Nat. Rev. Mol. Cell Biol. 4:446–456 10.1038/nrm1128 [DOI] [PubMed] [Google Scholar]
- Robertson F., Pinal N., Fichelson P., Pichaud F. 2012. Atonal and EGFR signalling orchestrate rok- and Drak-dependent adherens junction remodelling during ommatidia morphogenesis. Development. 139:3432–3441 10.1242/dev.080762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royou A., Field C., Sisson J.C., Sullivan W., Karess R. 2004. Reassessing the role and dynamics of nonmuscle myosin II during furrow formation in early Drosophila embryos. Mol. Biol. Cell. 15:838–850 10.1091/mbc.E03-06-0440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer J.K., Choi W., Jung K.C., He L., Harris N.J., Peifer M. 2011. A contractile actomyosin network linked to adherens junctions by Canoe/afadin helps drive convergent extension. Mol. Biol. Cell. 22:2491–2508 10.1091/mbc.E11-05-0411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões S., Denholm B., Azevedo D., Sotillos S., Martin P., Skaer H., Hombría J.C., Jacinto A. 2006. Compartmentalisation of Rho regulators directs cell invagination during tissue morphogenesis. Development. 133:4257–4267 10.1242/dev.02588 [DOI] [PubMed] [Google Scholar]
- Simões S.M., Blankenship J.T., Weitz O., Farrell D.L., Tamada M., Fernandez-Gonzalez R., Zallen J.A. 2010. Rho-kinase directs Bazooka/Par-3 planar polarity during Drosophila axis elongation. Dev. Cell. 19:377–388 10.1016/j.devcel.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt D.I., Weber U., Mlodzik M. 1997. The role of RhoA in tissue polarity and Frizzled signalling. Nature. 387:292–295 10.1038/387292a0 [DOI] [PubMed] [Google Scholar]
- Tamada M., Farrell D.L., Zallen J.A. 2012. Abl regulates planar polarized junctional dynamics through β-catenin tyrosine phosphorylation. Dev. Cell. 22:309–319 10.1016/j.devcel.2011.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier V., Guang-Chao-Chen, andSettleman J. 2006. Rho-kinase regulates tissue morphogenesis via non-muscle myosin and LIM-kinase during Drosophila development. BMC Dev. Biol. 6:38 10.1186/1471-213X-6-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters J.W., Dilks S.A., DiNardo S. 2006. Planar polarization of the denticle field in the Drosophila embryo: roles for Myosin II (zipper) and fringe. Dev. Biol. 297:323–339 10.1016/j.ydbio.2006.04.454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Riechmann V. 2007. The role of the actomyosin cytoskeleton in coordination of tissue growth during Drosophila oogenesis. Curr. Biol. 17:1349–1355 10.1016/j.cub.2007.06.067 [DOI] [PubMed] [Google Scholar]
- Warrington S.J., Strutt H., Strutt D. 2013. The Frizzled-dependent planar polarity pathway locally promotes E-cadherin turnover via recruitment of RhoGEF2. Development. 140:1045–1054 10.1242/dev.088724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter C.G., Wang B., Ballew A., Royou A., Karess R., Axelrod J.D., Luo L. 2001. Drosophila Rho-associated kinase (Drok) links Frizzled-mediated planar cell polarity signaling to the actin cytoskeleton. Cell. 105:81–91 10.1016/S0092-8674(01)00298-7 [DOI] [PubMed] [Google Scholar]
- Zallen J.A., Wieschaus E. 2004. Patterned gene expression directs bipolar planar polarity in Drosophila. Dev. Cell. 6:343–355 10.1016/S1534-5807(04)00060-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.