Abstract
Although numerous factors have been found to modulate hTERT transcription, the mechanism of its repression in certain leukemias remains unknown. We show here that DEK represses hTERT transcription through its enrichment on the hTERT promoter in cells from chronic and acute myeloid leukemias, chronic lymphocytic leukemia, but not acute lymphocytic leukemias where hTERT is overexpressed. We isolated DEK from the hTERT promoter incubated with nuclear extracts derived from fresh acute myelogenous leukemia (AML) cells and from cells expressing Tax, an hTERT repressor encoded by the human T cell leukemia virus type 1. In addition to the recruitment of DEK, the displacement of two potent known hTERT transactivators from the hTERT promoter characterized both AML cells and Tax-expressing cells. Reporter and chromatin immunoprecipitation assays permitted to map the region that supports the repressive effect of DEK on hTERT transcription, which was proportionate to the level of DEK-promoter association but not with the level of DEK expression. Besides hTERT repression, this context of chromatin redistribution of DEK was found to govern about 40% of overall transcriptional modifications, including those of cancer-prone genes. In conclusion, DEK emerges as an hTERT repressor shared by various leukemia subtypes and seems involved in the deregulation of numerous genes associated with leukemogenesis.
Introduction
The majority of human tumor cells possess shorter telomeres than their normal counterparts, suggesting that abnormal telomere shortening is frequently involved in cancer [1–3]. The cellular reverse transcriptase telomerase counteracts telomere shortening. This enzyme is composed of a catalytic protein subunit, telomerase reverse transcriptase (hTERT), and an RNA template (hTR). Malignant cells regularly bypass replicative senescence and paradoxically combine high telomerase activity with short telomere length [1]. However, the positive correlation between hTERT overexpression, increased telomerase activity, and oncogenesis does not seem to be mandatory in all tumor cases because hTERT underexpression has been shown at some stages of chronic myeloid leukemia (CML) [4,5], adult T cell leukemia/lymphoma [6–8], chronic lymphocytic leukemia (CLL) [9], and acute myelogenous leukemia (AML) [10]. To date, hTERT transcriptional repression has been considered as a tumor-suppressor pathway [11], and in contrast to transcriptional hTERT activation, very little is known about how hTERT is transcriptionally repressed in some hematological malignancies.
The protein DEK was originally identified as a fusion with the CAN/NUP214 nucleoporin in a subset of AML patients who harbored the (6;9)(p23;q64) translocation and was subsequently found overexpressed in most AMLs, as in numerous solid tumors [12]. DEK-CAN induces leukemia in mouse models [13], while its role of DEK in transcription varies on the basis of cell type, gene target, and developmental context. DEK enhances the transcription capacity of AP-2 in human malignant glioblastoma [14] and acts as a co-activator of the nuclear splicing factor U2AF in HeLa cells [15]. However, DEK acts as a co-repressor on p65/nuclear factor κB [16]. More recently, Koleva et al. showed that through their chromatin redistribution, DEK and C/EBPα cooperate together to coordinately activate myeloid gene expression and thereby regulate the differentiation capacity of hematopoietic progenitors [17].
We conducted the present study to assess how hTERT is transcriptionally repressed in certain leukemias. We designed a magnetic promoter precipitation assay coupled with mass spectrometry (MPP-MS) to identify proteins bound to the hTERT promoter in various cell types. We and others previously found that the oncoprotein Tax encoded by the human T cell leukemia virus type 1 (HTLV-1) represses hTERT transcription [6,7,18,19] in proliferating cells, whereas it activates hTERT expression in quiescent cells [7,20]. We therefore hypothesized that leukemic cells with low hTERT expression and Tax-expressing cells might share similar mechanisms of hTERT repression. Using a Tax-based system of hTERT transcriptional repression [6], we first demonstrated that Tax displaces transactivators from the hTERT promoter, where it recruits DEK that we subsequently characterized as an hTERT transcriptional repressor. Furthermore, the data indicate that Tax-expressing cells and fresh AML cells shared numerous common changes of the hTERT promoter proteome including DEK recruitment. Given that DEK is a chromatin protein deregulated in leukemias, we then begun to investigate whether or not DEK was involved in the deregulation of additional Tax-targeted genes as in the repression of hTERT in HTLV-1-unrelated leukemias.
Materials and Methods
The detailed materials and methods are described in the Supplementary Materials and Methods.
Cell Material
After consent was obtained in accordance with the Declaration of Helsinki and institutional guidelines, bone marrow (BM) cells were obtained from 6 donors and 20 patients (Table W1). CD34+ cells were isolated from mononuclear cells using immunomagnetic microbeads and the Dynal CD34 progenitor cell selection system (Dynal Biotechnologies, Oslo, Norway). B-lymphocytes were purified by negative selection using the RosetteSep human B cell enrichment cocktail (STEMCELL Technologies, Grenoble, France). HeLa cells were obtained from the European Collection of Cell Cultures (ECCAC, Salisbury, United Kingdom).
Plasmids, Transient Transfection, Immunoprecipitation, and Western Blot Analysis
HeLa and Jurkat cells were transiently transfected using the calcium phosphate precipitation method (CalPhos Transfection Kit; Clontech, Shiga, Japan) and SuperFect reagent (Qiagen, Courtaboeuf, France), respectively. Lipofectamine RNAi Max (Invitrogen, Carlsbad, CA) was used as the small interfering RNA (siRNA) delivery system. The plasmid vectors pTERTluc800, pCMV-Tax, pCMV-USF2a, and pNGLV3-DEK were previously described [6,21–24]. The corresponding empty vectors were used as controls. DEK siRNA was purchased from Dharmacon (Lafayette, CO). The co-immunoprecipitation experiments were carried out using NP-40 incubation buffer (150 mM NaCl, 50 mM Tris-HCl (pH 7.5), 1% NP-40, 50 mM N-ethylmaleimide, 2 mM EDTA, and protease inhibitors, 1:200, P8340 Sigma, St Louis, MO) and Protein G Sepharose Fast Flow (Sigma). Antibodies are detailed in the Supplementary Materials and Methods.
Quantitative Chromatin Immunoprecipitation and Quantitative Reverse Transcription-Polymerase Chain Reaction
The isolation, sonication, and chromatin immunoprecipitation (ChIP) analysis of nucleoprotein complexes of HeLa cells and leukemic cells are detailed in the Supplementary Materials and Methods. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) assays were performed on a LightCycler 2.0 system (Roche Applied Science, Indianapolis, IN) using the SYBR Green qPCR SuperMix UDG kit as detailed in the Supplementary Materials and Methods. The expression of each gene of interest was normalized against two housekeeping genes, Gus (NM_000181) and HPRT (NM_000194). All controls or samples were analyzed in duplicate. Primer sequences are provided in the Supplementary Materials and Methods.
Array Hybridization and Processing
After quality control, total RNA was amplified and biotin-labeled by a round of in vitro transcription. It was then fragmented and hybridized. Slides were scanned and the image files were analyzed using CodeLink expression software. The microarray analyses consisted of statistical comparison and filtering using GeneSpring software 7.3.1 (Agilent Technologies, Santa Clara, CA). See Supplementary Materials and Methods.
Proteomic Analysis of hTERT Promoter Occupancy In Vivo
Biotinylated hTERT core promoter (hCP) was amplified by PCR from the pTERTLuc800 plasmid [6]. The control template corresponded to a PCR-generated fragment of the HTLV-1 provirus pX region. Streptavidin beads were incubated with biotinylated templates. The beads/DNA complexes were washed, and immobilized templates were freshly prepared before each experiment. Finally, reaction components were incubated for 40 minutes at room temperature with 400 µg of dialyzed nuclear extracts (NEs). After washing, DNA template-specific proteins were eluted and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). See Supplementary Materials and Methods.
Nano-Liquid Chromatography Coupled to Tandem Mass Spectrometry (LC-MS/MS) Analysis
The method consisted of a 60-minute gradient at a flow rate of 200 nl/min, using a gradient of two solvents. MS and tandem mass spectrometry (MS/MS) data were acquired and processed automatically. Consecutive searches against, first, a contaminant database and then against the SwissProt and TREMBL databases were performed for each sample. See Supplementary Materials and Methods.
Statistics
Associations between categorical variables were analyzed by Fisher exact tests. The central tendency differences between groups were compared with the Mann-Whitney or Kruskal-Wallis tests. Non-parametric linear correlations between characteristics were analyzed by the Spearman rank test. All P values were two-sided.
Results
Tax-Expressing Cells and Fresh Myeloblasts Share Common hTERT Promoter Proteome Modifications
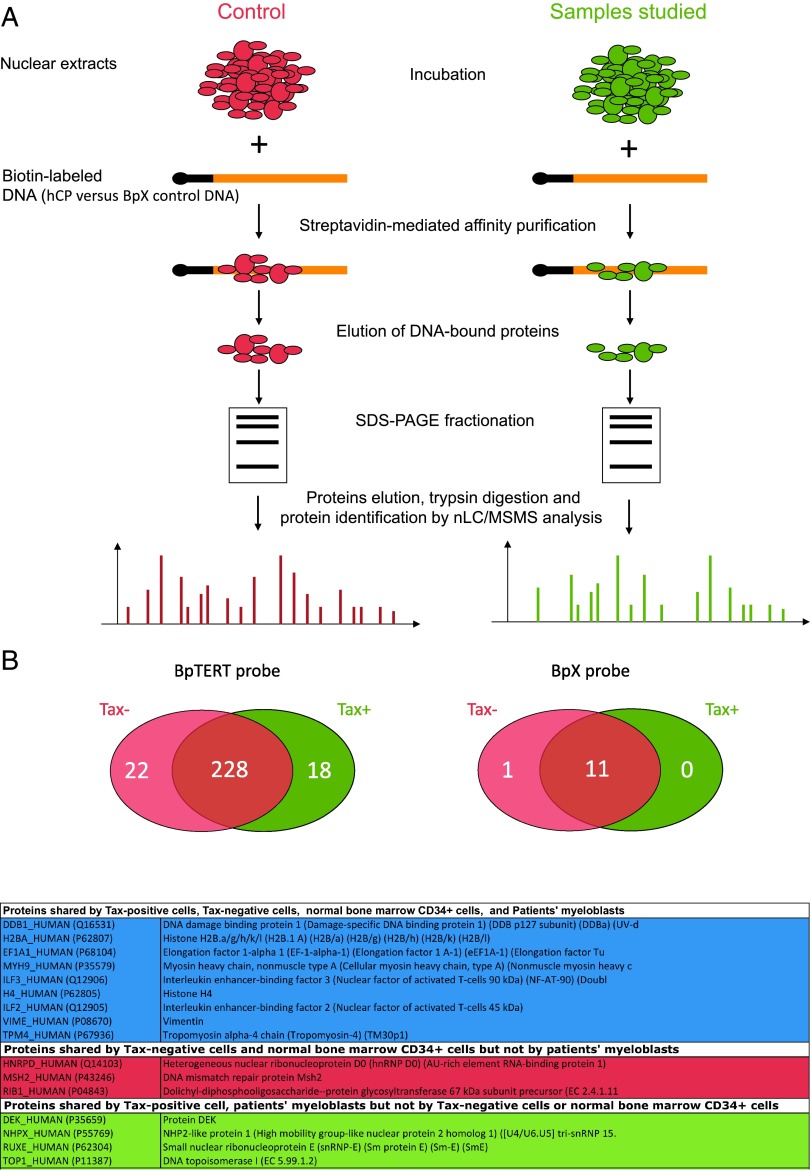
After validation of MPP assays as a reliable tool for isolation of specific hTERT promoter partners (Supplementary Results and Figure W1), our MPP-MS assays (Figure 1A) permitted to isolate 268 hCP-bound proteins from NEs deriving from Tax- or Tax+ cells (Figure 1B and Table W2). Of these, 22 and 18 were identified in NEs derived from Tax- and Tax+ cells, respectively. As shown in Figure 1B (bottom) and Table W2, myeloblast- and normal CD34+-derived NEs shared common hTERT promoter partners with Tax+- and/or Tax--derived NEs. Remarkably DEK, TOP1, NHPX, and RUXE were eluted from hCP incubated with Tax+- and myeloblast- but not from Tax- and normal CD34+-derived NEs (Table W2). In contrast, MSH2, hnRNP D0, and RIB1 were eluted from hCP incubated with Tax-- and CD34+-but not from Tax+- and myeloblast-derived NEs. These similarities suggested that, although occurring in different phenotypic contexts, both leukemic processes, i.e., AML and adult T cell leukemia, might share common mechanisms of hTERT repression.
Figure 1.
Proteomic analysis of hTERT promoter occupancy. (A) Nuclear protein extracts were prepared either from HeLa cells transfected with a Tax-expressing plasmid (right) or from fresh BM AML tumor cells. HeLa cells transfected with the control empty plasmid and normal BM CD34+ cells served as controls (left). Proteins were incubated with biotin-labeled DNA probes that corresponded to the hCP or to a control DNA stretch (BpX), as detailed in the Materials and Methods section. After SDS-PAGE fractionation, eluted products were digested with trypsin and then analyzed by MS. (B) The Venn diagram (top) represents the distribution of proteins detected in Tax+ versus Tax- NEs analyzed as shown in A. Data correspond to proteins recurrently detected in three independent experiments. Isolated proteins are described in Table W2. (Bottom) Distribution of hTERT promoter partners shared between transfected HeLa cells and BM cells derived from donors or from patients with AML.
hTERT Promoter Repression Coincides with the Displacement of hTERT Transactivators and the Recruitment of DEK
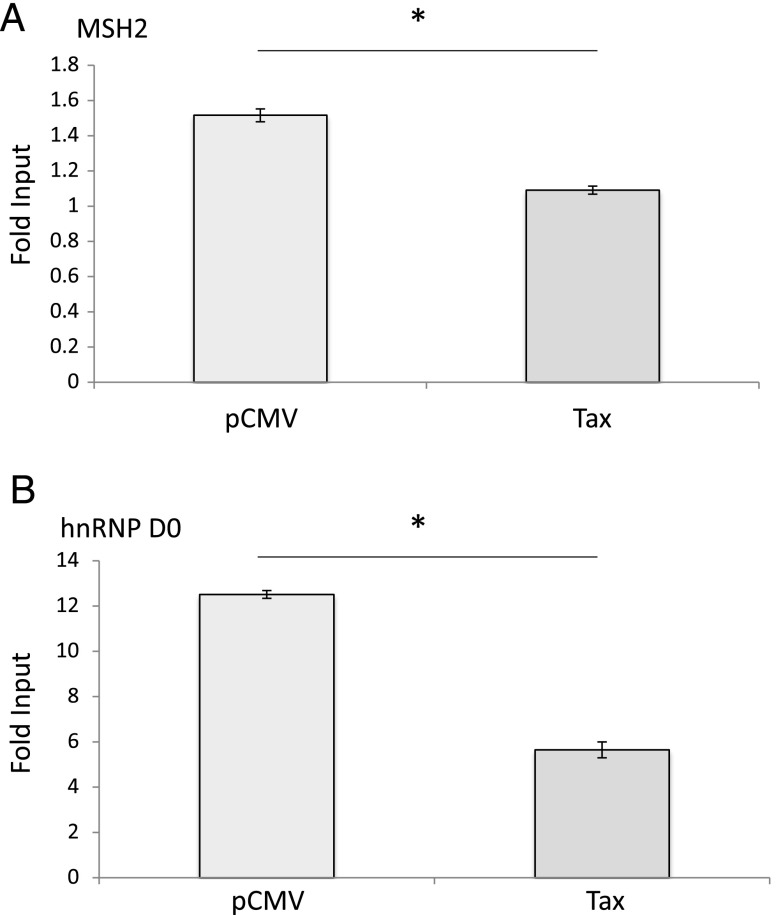
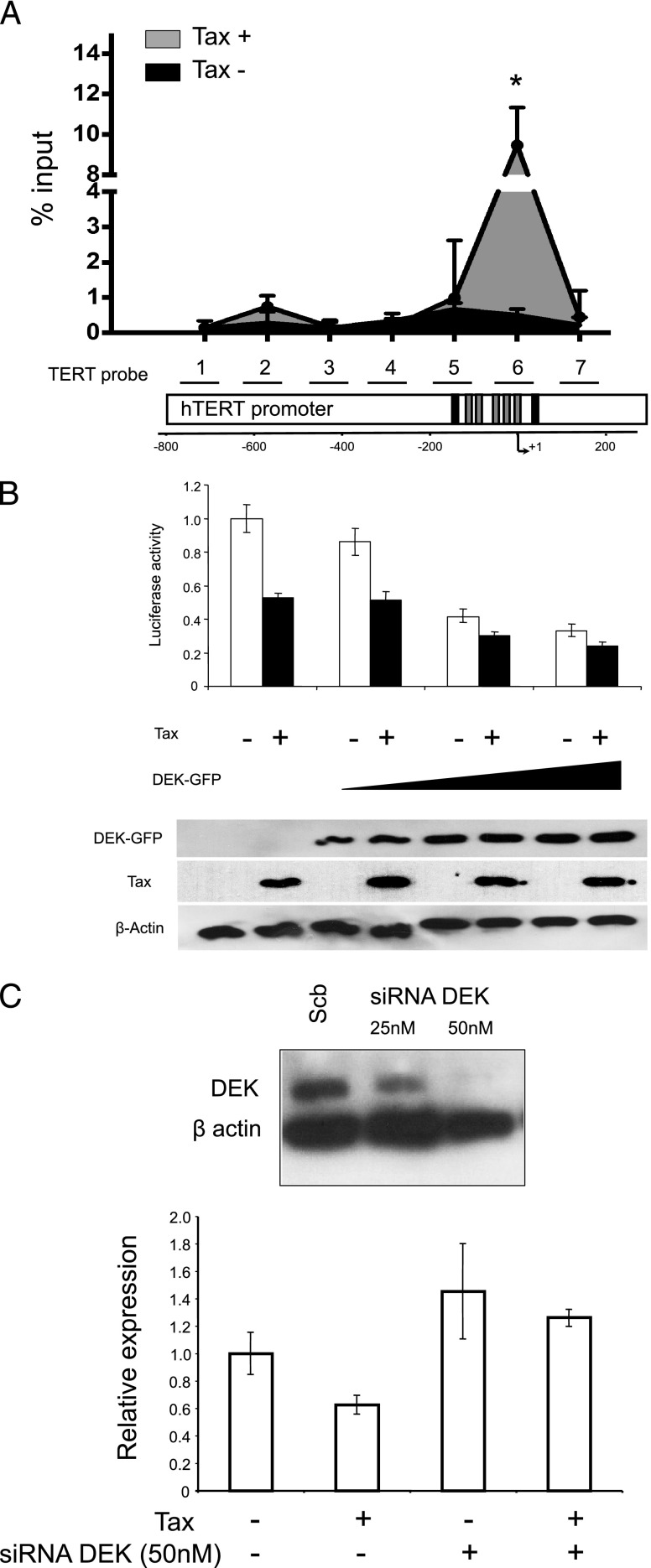
To validate MS data, we assessed the hTERT promoter occupancy in vivo for some MPP-eluted factors. Among those specifically isolated from Tax- and normal CD34+ cells, but not from Tax+ and AML NEs, we selected the proteins MSH2 and hnRNP D0 because they have been previously reported to activate hTERT transcription through binding two distinct regions on the hTERT promoter in oral squamous cell carcinoma cells [25]. Quantitative ChIP (qChIP) assays showed that the relative amounts of immunoprecipitated hTERT promoter fragments were lower in Tax+ than in Tax- cells (2.2- and 1.4-fold for hnRNPD0 and MSH2, respectively; Figure 2). Among the hCP-bound proteins specifically shared by Tax+- and AML-hCP proteomes (Table W2), we paid particular attention to the proto-oncogene DEK due to its important role in modifying the chromatin topology and gene expression during cell differentiation and transformation [26]. DEK occupancy of hTERT promoter was estimated by qChIP with anti-DEK antibody and seven primer sets spanning the hCP (Figure 3A). Figure 3A shows that DEK-DNA association was higher in Tax-expressing cells. In addition, the amount of amplified DNA varied along the immunoprecipitated hTERT promoter from Tax+ cells where the intensity peaked at 9.46% input with the TERT6 primer set that overlaps the transcription start site (Figure 3A). The effect of DEK on hTERT transcription was then assessed through reporter assays. Figure 3B shows that pNGLV3-DEK inhibited TERTLuc800 expression in a concentration-dependent manner and that Tax expression showed an additive effect on this repression. Conversely, siRNA-mediated knockdown of DEK expression increased hTERT transcription and abolished the negative effect of Tax on endogenous hTERT expression in HeLa cells (Figure 3C). Change in DEK expression level was also found to modulate endogenous hTERT expression in Jurkat cell lines (not shown).
Figure 2.
hnRNP D0 and MSH2 are displaced from the hTERT promoter upon Tax expression. Analysis of MSH2 (A) and hnRNP D0 (B) associations with the hTERT promoter was carried out by qChIP with HeLa cells expressing or not Tax, using antibodies against hnRNP D0 and MSH2 and qPCR with PCR primers complementary to the hTERT promoter, as described in the Materials and Methods section. qChIP assays were performed in triplicates with P < .05 (*) tested with the one-tailed Mann-Whitney test.
Figure 3.
DEK binds to and suppresses the hTERT promoter on Tax expression. (A) DEK and hTERT promoter association in Tax-expressing cells. The seven primer pairs used for ChIP and qChIP spanned a 964-bp region of the hTERT promoter. This region encompasses several known critical sites involved in the regulation of hTERT transcription, including the transcription initiation site, the start codon, as well as GC and E boxes. The black boxes represent the E boxes, while the gray boxes represent the five Sp1 binding sites. + 1 is the transcription initiation site. qChIP analysis of DEK association with the hTERT promoter was carried out as described in the Materials and Methods section. Results (means ± SDs) are representative of triplicate experiments. *P < .05, Mann-Whitney test. (B) HeLa cells were co-transfected with WT hTERT promoter-luciferase reporter plasmid TERTLuc800, in combination with the pCMV-Tax plasmid and/or a control vector (pCMV) in the absence or presence of increasing amounts of pNGLV3-DEK. Forty-eight hours after transfection, HeLa cells were collected and transcriptional activity was assayed by luciferase activity (see Materials and Methods section). (Bottom) DEK and Tax expression in transfected HeLa cells were assayed by Western blot analysis. (C) DEK knockdown increased endogenous hTERT expression and prevented its repression by Tax. hTERT expression was quantified through quantitative real-time PCR in HeLa cells transfected with the pCMV-Tax plasmid and/or control vector and/or the DEK siRNA (50 nM) and/or scrambled RNA. siRNAmediated knockdown of DEK expression was checked by Western blot analysis (top). Data shown in B and C are the means (±SDs) of one representative experiment performed in triplicate.
In Fresh Leukemic Cells, DEK Recruitment Parallels hTERT Transcriptional Repression
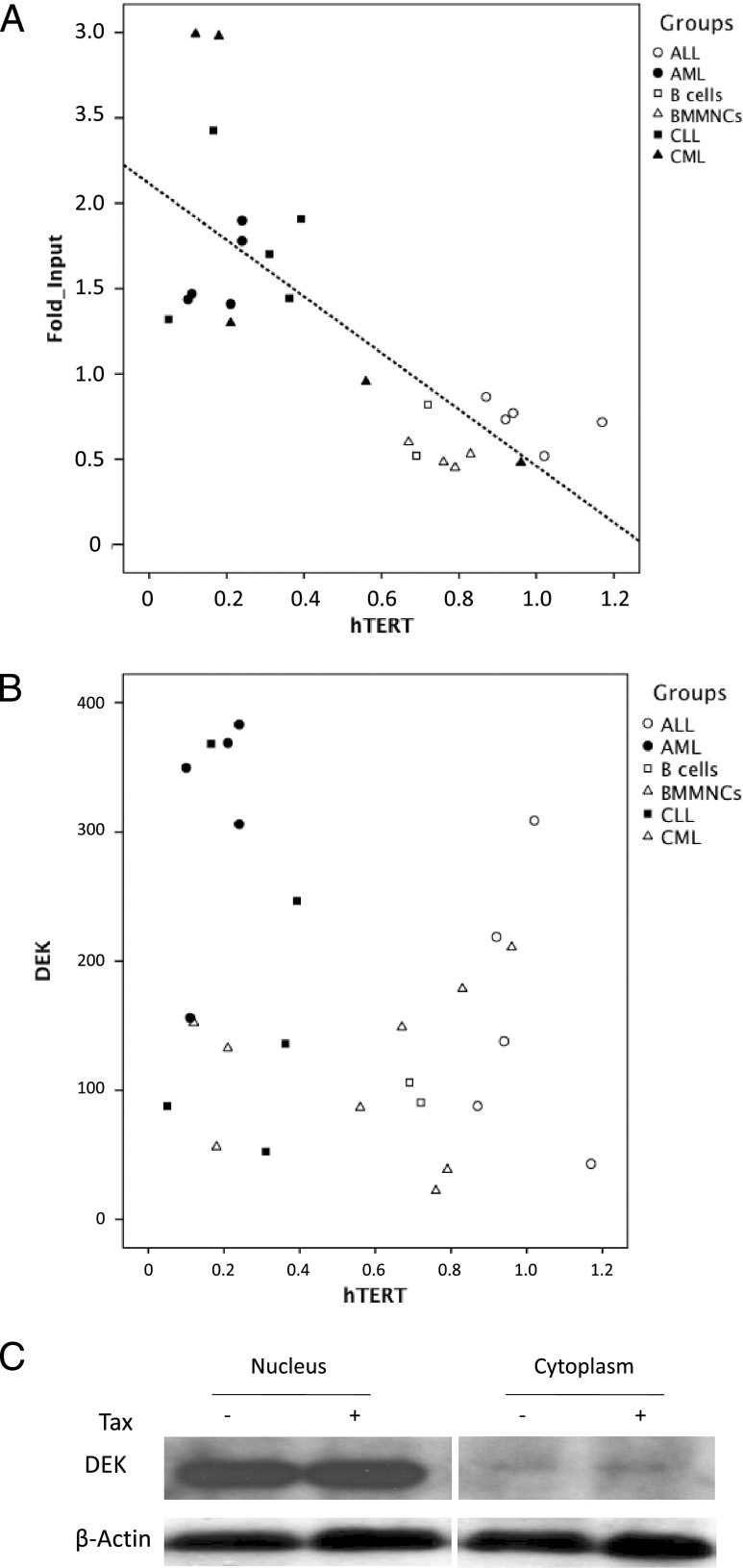
The above results prompted us to investigate whether the DEK-mediated transcriptional repression of hTERT pertains to other leukemic contexts. To this end, hTERT transcripts were quantified by qPCR in cells derived from CML, acute lymphoblastic leukemia (ALL), and CLL patients (Table W1) and in normal BM mononuclear cells (BMMNCs) and purified B cells used as controls. In parallel, the fold enrichment of DEK at the hTERT promoter DNA sequences was assessed by qChIP assays in the same samples (Figure 4). The mean amounts of hTERT transcripts for AML, CML, ALL, and CLL values were 0.28, 0.41, 0.98, and 0.26 arbitrary units, respectively, while those of normal BMMNCs, purified BM CD34+ cells, and purified peripheral B cells were 0.76, 0.73, and 0.72 arbitrary units, respectively. This confirmed previous results showing that hTERT expression is increased in ALL and decreased in CML, CLL, and AML [4–7,9,10,19]. The mean fold enrichment of DEK at the hTERT promoters in normal BMMNCs, normal BM purified CD34+ cells, AML, CML, ALL, normal purified circulating B cells, and CLL samples were 0.44, 0.56, 1.41, 1.74, 0.72, 0.68, and 1.76, respectively (P = .014, Kruskal-Wallis test). Samples with the highest DEK-hTERT association displayed the lowest amounts of hTERT transcripts, and by linear regression analysis, a significant negative correlation linked these two values (Figure 4A; P = .00002, R ∼ -0.75, Spearman rank correlation). In contrast, Figure 4B shows that in vivo, DEK mRNA levels were widely dispersed over cell samples without any statistical correlation between DEK and hTERT mRNA levels (P = .26, R ∼ -0.21, Spearman rank correlation). Similarly, DEK expression remained unchanged upon Tax expression (Figure 4C).
Figure 4.
In vivo and ex vivo, DEK recruitment on the endogenous hTERT promoter correlates with hTERT transcriptional repression but not with DEK expression. (A) Analysis of DEK association with the hTERT promoter was carried out by qChIP using an antibody against DEK and qPCR with the TERT6 primer pair as described in the Materials and Methods section. For each sample, hTERT expression was measured by qRT-PCR. AML, CML, and ALL cells and BMMNCs were derived from the BM, while purified B cells and malignant CLL cells were derived from the blood. All patient samples were collected at the time of diagnosis, after written informed consent. Signals were normalized to input, and background levels in immunoprecipitation (IP) with control IgGs were assigned. (B) DEK expression did not correlate with hTERT transcriptional repression in hematological samples. For each sample, DEK and hTERT mRNA were quantified by qRT-PCR. (C) Tax expression did not modify DEK expression. NEs and cytoplasmic extracts from Tax- versus control empty vector-transfected HeLa cells were analyzed by Western blot analysis with an anti-DEK antibody.
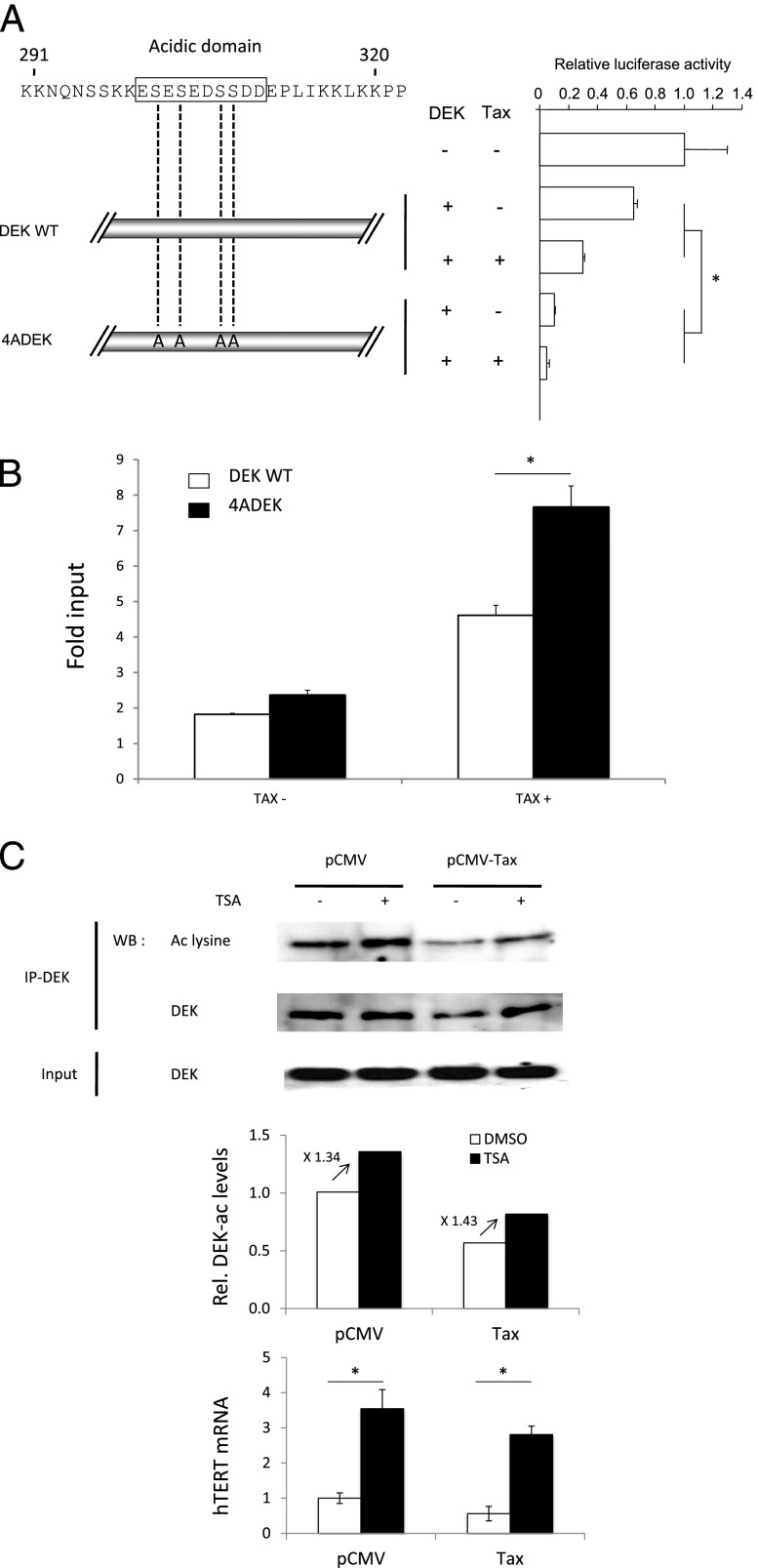
hTERT Repression Is Influenced by DEK Posttranslational Modification
Both phosphorylation and acetylation decrease DEK's DNA affinity [16] and thereby modulate its transcriptional effects [27]. To address the function of DEK phosphorylation on hTERT expression, we generated a DEK phosphorylation mutant, 4ADEK, by introducing alanine substitutions at serines 301, 303, 306, and 307. These residues were chosen because they are involved in DEK-DNA binding [28,29]. After transfection, the overall DEK amounts were unchanged between assays using either 4ADEK- or the wild-type (WT)-DEK-expressing vectors (not shown). In contrast, the suppressive effect of the 4ADEK mutant on the hTERT transcription was three times higher than that of WT-DEK (Figure 5A). In parallel, qChIP revealed that the 4ADEK-hTERT DNA association was higher than that obtained with the WT-DEK plasmid (Figure 5B). These results strongly supported that DEK-DNA association governed DEK-dependent hTERT transcriptional repression and suggested that DEK phosphorylation was critical for these processes. To investigate the relationship between DEK acetylation and hTERT expression, we assessed the amount of acetylated DEK in Tax- and Tax+ NEs. Figure 5C shows that Tax decreased DEK acetylation without modifying its expression. In contrast, the exposure of HeLa cells to the deacetylase inhibitor trichostatin A (TSA) led to a decrease in DEK occupancy at the hTERT promoter (Figure W2), while it increased the amounts of both acetylated DEK and hTERT mRNA in HeLa cells expressing or not Tax (Figure 5C). This suggests that DEK acetylation could counteract the transcriptional repression of hTERT. Typically, modifications in DEK acetylation could result from either Tax-induced deacetylase activity or Tax-induced inhibition of histone acetyltransferase activity. However, Figure 5C shows that TSA exposure did not alleviate the negative effect of Tax on DEK acetylation (Figure 5C), thereby ruling out the Tax-dependent deacetylation possibility. Therefore, it was possible to propose that the decreased amount of acetylated DEK mainly relies on a Tax-triggered histone acetyltransferase inhibition rather than on a Tax-triggered deacetylase effect. Taken together, these results demonstrated that DEK-dependent hTERT repression does not rely on increased DEK expression but rather on a greater DEK-DNA association, the level of which seems to be modulated at the post-translational level through DEK phosphorylation and acetylation.
Figure 5.
Transcriptional repression of hTERT depend on posttranslational modifications of DEK. (A) Mutations of the DEK phosphorylation site increase the repressive effect of DEK and Tax on hTERT promoter activity. HeLa cells were co-transfected with theWT hTERT promoter-luciferase reporter plasmid TERTLuc800, in combination with either the pCMV-Tax plasmid, the pNGLV3-DEK WT, or the pNGLV3-4ADEK mutant. Transcriptional activity was assayed by luciferase activity (see Materials and Methods section). (B) Mutations of the DEK phosphorylation site increased the recruitment of DEK on the hTERT promoter. qChIP was carried out with an anti-DEK antibody and the TERT6 primer pair, and nuclear proteins were derived from HeLa cells co-transfected with either the pCMV-Tax plasmid, the pCMV control vector, the pNGLV3-DEK WT, or the pNGLV3-4ADEK mutant. (C) Tax decreased DEK acetylation in a TSA-independent manner. HeLa cells were transfected with Tax or the control empty pCMV plasmid in the presence of TSA or DMSO and subjected to immunoprecipitation and Western blot analysis for DEK. The Western blot analysis membrane was stripped and reprobed with an anti-acetylated lysine antibody. The endogenous expression of hTERT was measured by qRT-PCR. Error bars represent the S.D. in triplicate experiments. Data shown in B and C are the means (±SDs) of one representative experiment performed in triplicate. *P < .05, Mann-Whitney test.
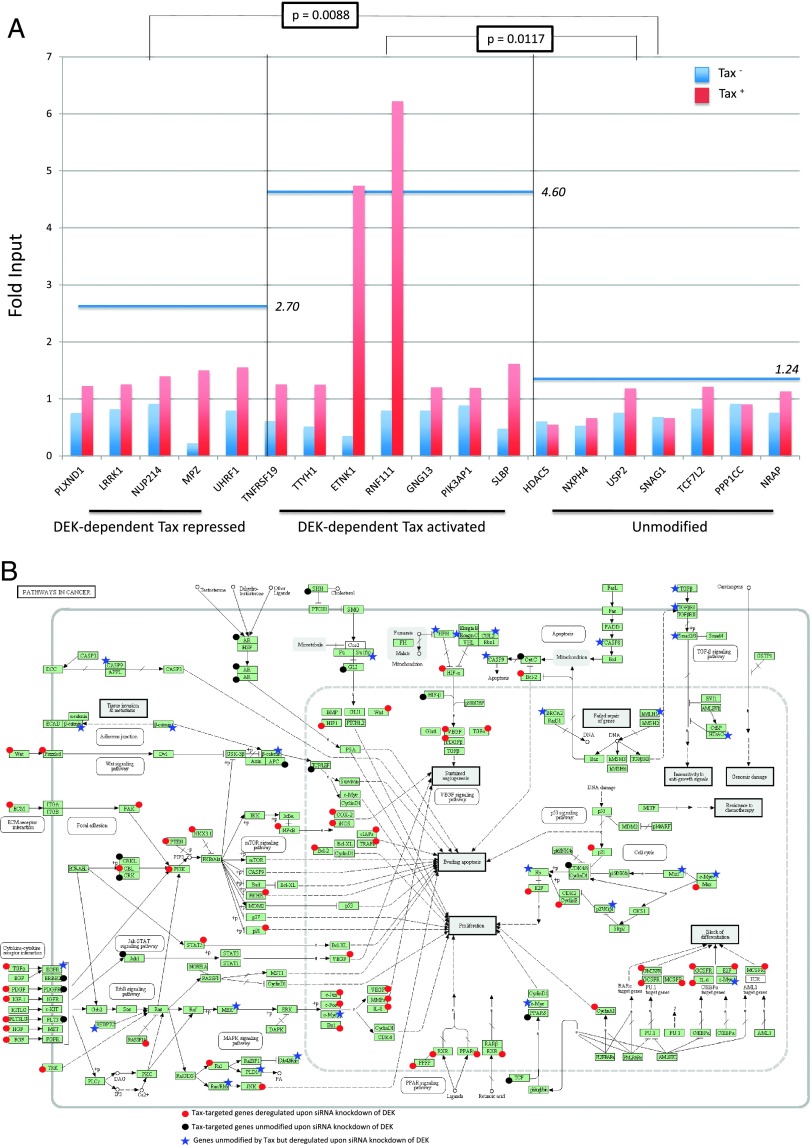
Tax-Associated Cellular Gene Transcriptional Deregulation Frequently Depends on DEK
Given that DEK is an abundant chromatin protein in human cells, we investigated whether Tax-mediated chromatin redistribution of DEK might influence the transcription of other genes than hTERT. As detailed in the Supplementary Results and shown in Tables 1, W3, and W4, DEK was found involved in the transcriptional deregulation of 42% of Tax-targeted genes including 44% of Tax-repressed genes and 40% of Tax-activated genes. qChIP assays showed that Tax expression strengthened DEK recruitment on the promoter of 12/12 genes found to be transcriptionally modified by Tax in a DEK-dependent manner, including 5/5 Tax-repressed and 7/7 Tax-activated genes (Figure 6A). Furthermore, qChIP revealed that the average level of DEK-hTERT promoter association was higher for Tax-activated (P = .0117) and Tax-repressed (P = .0088) genes than for unmodified genes. By using the DAVID bioinformatics resources (http://david.abcc.ncifcrf.gov/), ontological analysis of the Tax-targeted genes showed that the two top-ranked term categories “Pathways in cancer” and “Cytokine-cytokine receptor interaction” were DEK-dependent (Supplementary Results, Tables 1, 2, and W4). In contrast, the terms “Neuroactive ligand-receptor interaction” and “MAPK signaling pathway” appeared independent of DEK. Figure 6B represents the “Pathways in cancer” and shows that the vast majority of Tax-targeted genes in these pathways are DEK-dependent genes.
Table 1.
Distribution of Gene Expression according to Tax Expression and DEK Knockdown.
| Effect of DEK Knockdown | Tax-Activated (%) | Tax-Repressed (%) | Tax-Unmodified (%) |
| Activation | 539 (13) | 2029 (44) | 1991 (9) |
| Repression | 1613 (40) | 644 (14) | 3235 (14) |
| None | 1898 (47) | 1895 (42) | 18101 (77) |
Figure 6.
qChIP analysis of DEK-promoter association upon Tax expression for genes other than hTERT. (A) HeLa cells were transfected with either pCMV-Tax or control pCMV empty vector, and qChIP was carried out with a DEK antibody and PCR primers complementary to a promoter region encompassing the transcription start site of 19 genes. For each gene, the signal was normalized to input for Tax+ (blue) and Tax- HeLa cells (red). As indicated at the bottom, these genes included five DEK-dependent/Tax-repressed (left), seven DEK-dependent/Tax-activated (center), and seven DEK-independent/Tax-unmodified genes (right). For each gene category, a horizontal blue line represents the mean DEK-promoter association ratio of Tax+ to Tax- signals. (B) Distribution of “cancer” pathways according to the DEK-dependent (red circles) versus DEK-independent (black circles) nature of Tax-targeted genes. A blue asterisk identifies Tax-independent genes that were deregulated on siRNA knockdown of DEK.
Table 2.
Ontological Analysis of DEK-Dependent and DEK-Independent Tax-Targeted Genes.
| Terms | Gene Count | P |
| Enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of genes dysregulated by Tax expression in a DEK-dependent or DEK-independent manner | ||
| hsa04080: neuroactive ligand-receptor interaction | 63 | 9.193E-06 |
| hsa04010: MAPK signaling pathway | 59 | 4.504E-04 |
| hsa05200: pathways in cancer | 59 | 4.257E-02 |
| hsa04060: cytokine-cytokine receptor interaction | 52 | 1.052E-02 |
| hsa04020: calcium signaling pathway | 48 | 7.685E-06 |
| hsa04514: cell adhesion molecules (CAMs) | 26 | 7.960E-02 |
| hsa04270: vascular smooth muscle contraction | 25 | 2.403E-02 |
| hsa05416: viral myocarditis | 19 | 9.047E-03 |
| hsa05414: dilated cardiomyopathy | 19 | 9.702E-02 |
| hsa05412: arrhythmogenic right ventricular cardiomyopathy (ARVC) | 18 | 3.614E-02 |
| hsa05410: hypertrophic cardiomyopathy (HCM) | 18 | 8.922E-02 |
| hsa04730: long-term depression | 17 | 3.043E-02 |
| hsa05218: melanoma | 17 | 3.881E-02 |
| hsa04260: cardiac muscle contraction | 17 | 8.103E-02 |
| hsa02010: ABC transporters | 11 | 8.656E-02 |
| Enriched KEGG pathways of genes dysregulated by Tax expression in a DEK-independent manner | ||
| hsa04080: neuroactive ligand-receptor interaction | 34 | 1.833E-04 |
| hsa04010: MAPK signaling pathway | 26 | 5.523E-02 |
| hsa04020: calcium signaling pathway | 23 | 3.228E-03 |
| hsa04514: cell adhesion molecules (CAMs) | 15 | 5.862E-02 |
| hsa04340: hedgehog signaling pathway | 9 | 3.217E-02 |
Discussion
The study identified and characterized hTERT promoter partners involved in hTERT transcriptional repression in leukemias. Some of these partners such as MSH2 and hnRNP D0 were found displaced from the hTERT promoter in both AML cells and Tax-expressing cells, while other factors including DEK were found recruited to this promoter in both cell subtypes. Given its known implication in leukemias, we further detailed the role of DEK in hTERT transcription. DEK was found recruited on hTERT promoter not only in AML and Tax-expressing cells but also in CML and CLL cells. We found that DEK repressed hTERT transcription and that this repression relied on the recruitment of DEK on the hTERT promoter but not on the amounts of DEK protein in tumoral cells. Posttranslational DEK modifications were found to influence its interplays with hTERT. In addition to hTERT, DEK/promoter enrichment was also found to regulate more than 40% of Tax-controlled genes, including those having critical roles in cancer.
DEK has emerged as a novel class of DNA topology modulators that can be both targets and effectors of tumor initiation and addiction [12,16]. The proto-oncogene DEK is involved in chromatin remodeling, transcriptional repression or activation, and mRNA maturation [12,16]. DEK is capable to associate with numerous promoters for modulating their occupancy in transcriptional factors and chromatin modifiers such as hDaxx [30], P/CAF, p300, and p65/nuclear factor κB [26,31]. DEK prefers structured DNA, such as supercoiled and four-way junctions, to specific nucleic acid sequence [28]. Such structured DNA forms are enriched in the vicinity of promoter regions [32]. DEK and other cruciform-binding proteins such as p53, BRCA1, MSH2, PARP1, 14-3-3, and topoisomerase 1 are frequently involved in transcriptional regulation as well as in DNA repair and replication [33]. Interestingly, our results show that in addition to enrichment in the cruciform-binding protein DEK at the vicinity of the hTERT transcription start site, hTERT repression also included changes in occupancy of additional cruciform-binding proteins such as MSH2, hnRNPs, and topoisomerase 1 (Figure 1).
AML, CML, and CLL cells display significantly lower amounts of hTERT transcripts than their normal counterparts [4,5,9,10]. In contrast, ALL cells overexpress hTERT when compared to normal BMMNCs or to normal B or T lymphocytes [10,34]. These results were confirmed in the present series of 20 additional patients (Figure 5), where there was no significant difference in the level of DEK expression between AML, ALL, CML, CLL, and normal BM samples or normal B cells. In contrast, qChIP revealed a significantly higher DEK-hTERT promoter association in AML, CML, and CLL than in control cells (Figure 4). In ALL cells, which express elevated amounts of hTERT mRNA, the hTERT promoter was found depleted of DEK. A strong negative correlation linked DEK-hTERT promoter association and hTERT expression supporting an identical interplay between DEK-hTERT promoter association and hTERT expression in fresh leukemic cells and Tax-expressing cells. Knowing now that Tax represses hTERT in a concentration-dependent manner [6] through the parallel recruitment of DEK on the hTERT promoter (present results), it could be speculated that, in AML, CML, and CLL cells, a Tax surrogate that reproduces the same telomere effects as those observed in Tax-expressing cells exists. Accordingly, deciphering the DEK interactome in leukemic cells will help identify the important factors involved in telomere-dependent chromosomal instability. Given that DEK is overexpressed in numerous tumor types, the effects of DEK on gene expression, cell differentiation, and transformation have hitherto mainly been studied through DEK overexpression and/or genetic depletion. Here, DEK was found to repress hTERT ex vivo in a concentration-dependent manner. However, in Tax+ cells as in AML, CML, and CLL cells, our results did not support any correlation between the cellular amounts of DEK and the expression level of its target genes. Rather, our study pinpoints DEK redistribution along the chromatin as the main process governing its target gene transcription. Consistent with the known relationships between DEK acetylation or phosphorylation and its DNA affinity [16], the present results strongly suggest that through influencing DEK-DNA affinity and thereby target gene expression, qualitative and quantitative posttranslational changes of the DEK protein play an important role in tumorigenesis.
In conclusion, DEK was found here to repress hTERT through its redistribution on the hTERT promoter, thus permitting to explain the recently shown suboptimal hTERT expression in CLL [9], CML [4,5], and AML [10,34] as in certain Tax-expressing cells, such as activated HTLV-1-positive but untransformed CD4+ T cells [8]. Preventing telomere elongation in proliferating premalignant cells is assumed to promote genetic instability leading to tumor initiation. In overt transformed cells, such telomere defects are assumed to sustain genetic plasticity, permitting immune escape, resistance to treatment, and relapse. DEK redistribution appears to be involved in more than 40% of Tax-targeted genes and helps explain the transcriptional pleiotropic effect of Tax, notably in the field of oncogenesis (Table 2 and Figure 6B). Whether DEK redistribution leads to such pleiotropic transcriptional effects in other HTLV-1-unrelated leukemias remains to be investigated.
Supplementary Materials and Methods
Samples Studied
After consent was obtained in accordance with the Declaration of Helsinki and institutional guidelines, BM cells were obtained from 6 donors and 20 patients (at the Edouard Herriot University Hospital, Lyon, France) with acute leukemias (ALs) and having more than 85% BM blasts. BM samples were obtained from patients before treatment. The diagnosis of leukemia was based on routine morphologic evaluation, immunophenotyping, cytochemical smears, and karyotyping. Mononuclear cell fractions from donor BM were separated using a Ficoll-Hypaque gradient. CD34+ cells were then isolated with immunomagnetic microbeads and the Dynal CD34 progenitor cell selection system (Dynal Biotechnologies), according to the manufacturer's instructions. The kits contain Dynabeads and DETACHaBEAD for isolation and release of pure CD34+ cells with an intact antigen profile. Purity was controlled by FACS with CD34+-fluorescein isothiocyanate antibody (Dako, Trappes, France).
Isolation of Normal Peripheral B Cells
B lymphocytes were purified by negative selection using the RosetteSep human B cell enrichment cocktail (STEMCELL Technologies). The percentage of CD19+ cells was determined by cytometric assay, using an α-CD19-phycoerythrin (PE) antibody (Amersham-Biosciences, Uppsala, Sweden).
Plasmids and siRNA
The hTERT promoter-Luc reporter plasmid, pTERTluc800, contains the proximal 800 bp of the hTERT promoter placed upstream of the firefly luciferase coding region [1,2]. The Renilla luciferase reporter construct, pHRG-TK, used as an internal control in the luciferase-based reporter gene assays, was purchased from Promega (Madison, WI). Expression vectors pCMV-Tax, pCMV-USF2a, and pNGLV3-DEK plasmids were previously described [3–5]. The corresponding empty vectors were used as controls. DEK siRNA oligonucleotides (M-003881-01) and their respective nontargeting siRNA (D-001210-04) were purchased from Dharmacon.
Antibodies
Antibodies to DEK were purchased either from Santa Cruz Biotechnology (Santa Cruz, CA; H-300) or BD Biosciences (San Jose, CA; 610948). Antibodies to MSH2 (clone 3A2) and hnRNP D0 (#07-260) were purchased from Cell Signaling Technology (Danvers, MA) and Upstate Biotechnology (Charlottesville, VA), respectively. The anti-Tax monoclonal antibody was HY474 [1]. The mouse monoclonal anti-acetylated lysine antibody was AKL5C1 from Santa Cruz Biotechnology, while the monoclonal anti-actin (AC-40 or AC-15) antibody was obtained from Sigma. Rabbit polyclonal antibodies recognizing USFs, USF1 and USF2a, were previously described [5]. Antirabbit and anti-mouse IgG-peroxidase antibodies were used for Western blot analysis. The antibodies used in immunofluorescence staining experiments were goat anti-rabbit IgG2a conjugated to fluorescein isothiocyanate and goat anti-mouse IgG conjugated to Alexa Fluor 546.
Cells, Transfections, and Reporter Assays
HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (Invitrogen), penicillin (100 units/ml), and streptomycin (100 µg/ml) at 37°C in a 5% CO2 incubator. Jurkat cells were grown in RPMI 1640. The calcium phosphate precipitation method (CalPhos Transfection Kit; Clontech) was used for transfecting HeLa cells with expression plasmids, while Lipofectamine RNAi Max (Invitrogen) was used as the siRNA delivery system. For luciferase assays, cells were seeded in 24-well plates at 75% confluence and co-transfected with 500 ng of pTERTLuc800, 20 ng of the internal control Renilla luciferase reporter construct pHRG-TK, and the indicated recombinant plasmids. Cells were washed three times with ice-cold phosphate-buffered saline (PBS) and harvested to analyze luciferase activity 48 hours after transfection using a luminometer and adding a luciferase assay reagent (Promega). Jurkat cells were transfected using SuperFect (Qiagen), according to the manufacturer's protocol. Data are presented as means ± SD. Data shown in Figures 3, A–C, and 5, B, D, E, and G, are the means (±SDs) of one representative experiment performed in triplicate.
Co-Immunoprecipitation Experiments
Whole-cell extracts were prepared from about 5 x 105 HeLa cells transfected with either pCMV-Tax or the corresponding empty vector. Twenty-four hours after transfection, cells were washed once with PBS, lysed in 300 µl of NP-40 incubation buffer [150 mM NaCl, 50 mM Tris-HCl (pH 7.5), 1% NP-40, 50 mM N-ethylmaleimide, 2 mM EDTA, and protease inhibitors, 1:200, P8340; Sigma], and clarified by centrifugation at 15,000g for 10 minutes. Proteins from cell lysates were incubated with DEK antibody and 50 µl of Protein G Sepharose Fast Flow (Sigma) overnight at 4°C in incubation buffer. Beads were collected by centrifugation, washed once in NP-40 incubation buffer and twice in PBS, and resuspended in Laemmli sample buffer for Western blot analysis.
Western Blot
Cells were lysed and protein concentration was assayed using the Bio-Rad DC Protein Assay Kit (Bio-Rad, Hercules, CA). Equal amounts of proteins were subjected to 10% SDS-PAGE. Fractionated proteins were transferred to polyvinylidene fluoride (PVDF) membranes (Immobilon-P Transfer Membranes; Millipore, Billerica, MA). Membranes were blocked in PBS containing 5% nonfat milk and 0.2% Tween 20, then probed with the appropriate antibody, followed by secondary IgG HRP-linked antibody (Cell Signaling Technology). Blots were then developed using an enhanced chemiluminescence detection system (Lumi-Lightplus Western Blotting Substrate; Roche).
Quantitative Reverse Transcription-Polymerase Chain Reaction
RNA was isolated with TRIzol reagent (Invitrogen). Before RT, RNA was treated with DNase (Invitrogen) to prevent DNA contamination. First-strand cDNA was synthesized from 0.5 µg of RNA using random primers (Promega) and Superscript II reverse transcriptase (Invitrogen). RNA concentration and purity were determined by UV spectrophotometry (Nanodrop, Montchanin, DE). Sequences of primers are available on request. Each primer set used to quantify gene expression was first tested by PCR using a control cDNA to ensure specific amplification, as shown by the presence of a unique specific signal after agarose gel electrophoresis (not shown). PCR assays were performed on a LightCycler 2.0 system (Roche Applied Science). All reactions were performed using a Platinum SYBR Green qPCR SuperMix UDG kit. The reaction mixture contained 5 µl of water, 10 µl of SYBR Green MasterMix, 1 µl of BSA (20x), 2 µl of primers at 10 µM, and 2 µl of a 1/10 dilution of cDNA. The final reaction volume was 20 µl. Water was used as a negative control. All samples were kept at 4°C during preparation. Thermocycling conditions were as previously described [6]. All controls or samples were analyzed in duplicate. All fluorescence data were analyzed by LightCycler 4.0 software (Roche) and the Ct results were exported to Excel sheets. Amplified DNA with primer sets used to quantify gene expression was diluted and aliquoted. The 10-7 dilution served as the calibrator for all the qPCR runs. For relative quantification and normalization, the comparative Ct (or E-ΔΔC, where E was the primer-dependent efficiency of the PCR) method was used [7]. The expression of each gene of interest was normalized against two housekeeping genes, Gus (NM_000181) and HPRT (NM_000194), which had been validated with BestKeeper software tool to adjust for variations in RNA levels and cDNA synthesis.
Quantitative Chromatin Immunoprecipitation
HeLa cells were fixed in 1% formaldehyde for 10 minutes at room temperature and then for 40 minutes at +4°C. Cross-linking was stopped by adding 125 mM glycine for 5 minutes. Cells were washed in hypotonic buffer and resuspended in 1 to 2 ml of SDS lysis buffer (1% SDS, 10 mM EDTA, and 50 mM Tris-HCl, pH 8). Nucleoprotein complexes derived from HeLa cells expressing Tax or not were sonicated to reduce the length of DNA fragments to 200 to 300 bp. For nucleoprotein complexes derived from patients and donors, DNA fragments ranging from 300 to 1000 bp were obtained after sonication. Insoluble material was removed and the supernatant was collected. Thirty microliters of this fraction were preserved as an input control, and the rest was diluted 1:10 in ChIP dilution buffer (ChIP Assay Kit; Upstate Biotechnology). The chromatin solution was precleared for 1 hour by incubation with 80 µl of salmon sperm DNA-protein A-agarose beads (Upstate Biotechnology). The soluble fraction was collected and 15 µl of anti-DEK and anti-mouse IgG (Dakocytomation, Trappes, France) antibodies were added and incubated overnight. Then, for ChIP monoclonal antibodies, 1 µg of rabbit polyclonal anti-mouse antibody (Dakocytomation) was added and incubated for 1 hour. After immunoprecipitation, immune complexes were collected by adding 60 µl of salmon sperm DNA-protein A-agarose beads for 1 hour. The supernatant corresponding to the unbound fraction was collected. After washing (according to the manufacturer's instructions), complexes were eluted from the beads in 1% SDS and 0.1 M NaHCO3. This fraction corresponded to the bound (anti-DEK) or the non-DEK-specific antibody (anti-mouse IgG) fractions. Cross-links were reversed by heating samples at 65°C in 200 mM NaCl. DNA was recovered by proteinase K digestion, phenol extraction, and ethanol precipitation. Finally, DNA samples from the input, unbound, non-DEK-specific antibody, and bound fractions were quantified by real-time PCR with specific promoter primers.
Array Hybridization and Processing
HeLa cells were transfected with either pCMV-Tax or an empty vector. At 24-hour post-transfection, Tax-positive cells received either scrambled siRNA or DEK siRNA, while Tax-negative HeLa cells were transfected with a scrambled siRNA as a control. Cells were harvested 24 hours later, and Tax expression and DEK knockdown were confirmed by Western blot analysis of whole-cell extracts. Total RNA was extracted using an RNeasy Mini Kit (Qiagen), including DNAse treatment, and according to the manufacturer's instructions. Total RNA yield was measured by OD260, with an A260/A280 ratio of 1.9 to 2.1 demonstrating purity. Quality was evaluated on nanochips with the Agilent 2100 Bioanalyzer (Agilent Technologies), according to the manufacturer's protocol. Total RNA (1 µg) was amplified and biotin-labeled by a round of in vitro transcription with the MessageAmp II a RNA Amplification Kit (Ambion, Huntingdon, United Kingdom), following the manufacturer's protocol. Before amplification, spikes of synthetic mRNA at different concentrations were added to all samples; these positive controls were used to ascertain the quality of the process. The aRNA yield was measured with a Nanodrop and quality checked on nanochips with the Agilent 2100 Bioanalyzer (Agilent Technologies). Biotin-labeled aRNA was fragmented and hybridized following the manufacturer's protocol (Applied Microarrays, Branchburg, NJ). Briefly, 10 µg of aRNA was fragmented using fragmentation buffer, then mixed with hybridization solution (Applied Microarrays), and injected onto CodeLink Human Whole Genome bioarrays. After overnight hybridization at 37°C, arrays were washed and stained with a streptavidin-cy5 solution (GE Healthcare, Freiburg, Germany). Slides were scanned using a Genepix 4000B scanner (Axon, Union City, CA) and Genepix software, with the laser set at 635 mm, the laser power at 100%, and the photomultiplier tube voltage at 60%. The scanned image files were analyzed using CodeLink expression software, version 5.0 (GE Healthcare), which produces both raw and normalized hybridization signals for each spot on the array. The microarray analyses consisted of statistical comparison and filtering, using GeneSpring software 7.3.1 (Agilent Technologies). Pairwise comparisons were performed between each cell category (Tax-, Tax+, and Tax+DEK-). Only genes showing a fold variation of ≥1.3 were retained.
Proteomic Analysis of hTERT Promoter Occupancy In Vivo
hTERT promoter occupancy was investigated in HeLa cells expressing or not Tax (triplicate assays) and in BM samples derived from three healthy donors or from patients with AML. NEs were prepared using the Nuclear Extract Kit (Active Motif, Carlsbad, CA) according to the manufacturer's instructions and dialyzed overnight at 4°C against dialysis buffer [20 mM Hepes, 10% glycerol, 50 mM KCl, 1 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT (pH 7.5), and protease inhibitor] using Slide-A-Lyser MINI Dialysis Unit Plus Float 3500 MWCO (Pierce, Rockford, IL). Protein concentrations were measured by colorimetric assay (Bio-Rad Assay DC Protein; Bio-Rad). Biotinylated templates consisting of the hCP and an irrelevant 720-bp DNA fragment (Bpx) which one derived from the pX region of the HTLV-1 provirus and served as an internal control for unspecific DNA binding. Biotinylated hCP was amplified by PCR from the pTERTLuc800 plasmid 1 using biotinylated primers BpTERT-F (5′-Biot-AAAAATTTAAATGGATCCAAGCTCAGATCC) and BpTERT-F (5′-Biot-AAAATTTAAATACAGTACCGGAATGCCAAG). The control template corresponded to a PCR-generated fragment of the HTLV-1 provirus pX region with BpX-F (Biot-AAAAATTTAAATCACCTGTCCAGAGCATCAGA) and BpX-R (Biot-AAAAATTTAAATGTGGTAGGCCTTGGTTTGAA). PCR conditions were 95°C for 10 minutes, followed by 30 cycles of 95°C for 1 minute and 58°C for 1 minute performed with 50 pmol of each primer, 250 µM dNTP, 4% DMSO, and 2.5 units of Herculase Hotstart DNA Polymerase (Stratagene, La Jolla, CA). Amplified products were extracted with phenol/chloroform (1:1) and ethanol precipitated before DNA affinity precipitation assay. Streptavidin beads (Dynabeads M-280 Streptavidin; Dynal Biotechnologies) were washed twice in Buffer T [10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 1 M NaCl] and concentrated at 10 mg/ml in Buffer T with 0.003% NP-40, using the magnetic particle concentrator (Dynal Biotechnologies). Streptavidin beads were incubated with biotinylated template (20 fmol/µg bead) in Buffer T for 30 minutes at room temperature with constant agitation and washed three times with Buffer T. Immobilized templates were blocked 15 minutes at room temperature in Buffer T to which 60 µg/ml casein (Sigma; C-5890), 5 µg/ml polyvinylpyrrolidone, and 2.5 mM DTT were added. The beads/DNA complexes were washed four times in transcription buffer and concentrated to 10 µg/ml in Buffer T. Immobilized templates were freshly prepared before each experiment. Finally, reaction components were incubated for 40 minutes at room temperature with 400 µg of dialyzed NEs in Buffer T to which 2.5 mM DTT, 0.1% NP-40, and protease inhibitors were added. After washing with Buffer T containing 2.5 mM DTT and 0.05% NP-40, DNA template-specific proteins were eluted with Laemmli buffer and resolved by 12% SDS-PAGE.
Protein Digestion
Protein bands were manually excised from the gels and transferred to 96-well microtitration plates. Sample preparation was carried out automatically (EVO150; Tecan, Crailsheim, Germany). Excised gel samples were washed several times by incubating in 25 mM NH4HCO3 for 15 minutes and then in 50% (vol/vol) acetonitrile containing 25 mM NH4HCO3 for 15 minutes. Gel pieces were then dehydrated with 100% acetonitrile and incubated with 7% H2O2 for 15 minutes before being washed again with the destaining solutions described above. Then, 0.15 µg of modified trypsin (Promega; sequencing grade) in 25 mM NH4HCO3 was added to the dehydrated gel spots, depending on protein amount. After a 30-minute incubation at room temperature, 20 µl of 25 mM NH4HCO3 was added to gel pieces before incubating overnight at 37°C. Peptides were then extracted from gel pieces in three 15-minute sequential extraction steps in 30 µl of 50% acetonitrile, 30 µl of 5% formic acid, and finally 30 µl of 100% acetonitrile. The pooled supernatants were then transferred to microcentrifuge tubes and dried under vacuum.
Nano-LC-MS/MS Analysis
For nano-LC-MS/MS analysis, the dried extracted peptides were resuspended in water containing 2.5% acetonitrile and 2.5% trifluoroacetic acid, before being transferred to vials compatible with nano-LC-MS/MS analysis (CapLC, Waters and ESI-Q-TOF Ultima; Micromass UK, Manchester, United Kingdom). The method consisted of a 60-minute gradient at a flow rate of 200 nl/min, using a gradient of two solvents: A (5% acetonitrile and 0.1% formic acid in water) and B (80% acetonitrile and 0.08%formic acid in water). The system included a 300 µm x 5 mm PepMap C18 pre-column to pre-concentrate peptides and a 75 µm x 150 mm C18 column (Gemini C18 phase for in-house built columns) used for peptide elution. Spectra were calibrated by fragmentation of glufibrino-peptide in MS/MS mode. MS and MS/MS data were acquired and processed automatically using MassLynx 4.0 software (Waters, Milford, MA). Consecutive searches against, first, a contaminant database and then against the SwissProt and TREMBL databases were performed for each sample using Mascot 2.0. Peptide modifications allowed during the search were N-acetyl (protein), dioxidation (M), oxidation (M), cysteic acid (C), and sulfone (M). The other parameters were peptide tolerance = 0.4 Da, MS/MS tolerance = 0.4 Da, and one missed cleavage site by trypsin allowed. Proteins showing two peptides with a score above 40 were automatically validated using homemade software (IRMa, CEA/DSV/iRTSV/LEDyP). Each protein identified by only one peptide was checked manually using standard fragmentation rules (five consecutive y ions, proline rule, b ions complementary to y ions, major peaks assigned). IRMa was then able to transform a manually validated Mascot data file into a result file in Excel format.
Statistics
Associations between categorical variables were analyzed by Fisher exact tests. The central tendency differences between groups were compared with the Mann-Whitney or Kruskal-Wallis tests. Non-parametric linear correlations between characteristics were analyzed by the Spearman rank test. All P values were two-sided.
Acknowledgments
We thank W.C. Greene for the pCMV-Tax plasmid, B. Viollet for the pCR3-USF2a plasmid and anti-USF antibodies, and D.M. Markovitz for the pNGLV3-DEK plasmid. The authors thank the genomic platform facility ProfileXpert (Université Lyon I, Bron, France) and the PLATIM (Plateau Technique Imagerie/Microscopie, Université Lyon I, Lyon France) for microarray and microscopy analyses, respectively.
Footnotes
This work was supported by the Ligue Nationale Contre le Cancer (Comités de l'Ain et du Rhône), Fondation de France, Association Laurette Fugain, Association pour la Recherche sur le Cancer (ARC), Association Guillaume Espoir, Agence Nationale pour la Recherche, AMGEN, Celgene, Novartis, Centre Léon Bérard, Hospices Civils de Lyon, University Lyon I, Centre National pour la Recherche Scientifique, and Institut National de la Santé et de la Recherche Médicale (INSERM). M.K. was supported by bursaries from the Société Française d'Hématologie, ARC, and the Guillaume Espoir Association. F.M. is supported by INSERM and by the Hospices Civils de Lyon (AVIESAN CHRT program 2010). E.W. is supported by Hospices Civils de Lyon and Lyon I University. The authors declare no conflict of interest.
This article refers to supplementary materials, which are designated by Tables W1 to W4 and Figures W1 and W2 and are available online at www.neoplasia.com.
References
- 1.Deng Y, Chan SS, Chang S. Telomere dysfunction and tumour suppression: the senescence connection. Nat Rev Cancer. 2008;8:450–458. doi: 10.1038/nrc2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christodoulidou A, Raftopoulou C, Chiourea M, Papaioannou GK, Hoshiyama H, Wright WE, Shay JW, Gagos S. The roles of telomerase in the generation of polyploidy during neoplastic cell growth. Neoplasia. 2013;15:156–168. doi: 10.1593/neo.121398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuzyk A, Mai S. Selected telomere length changes and aberrant three-dimensional nuclear telomere organization during fast-onset mouse plasmacytomas. Neoplasia. 2012;14:344–351. doi: 10.1593/neo.12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell LJ, Fidler C, Eagleton H, Peniket A, Kusec R, Gal S, Littlewood TJ, Wainscoat JS, Boultwood J. hTERT, the catalytic component of telomerase, is downregulated in the haematopoietic stem cells of patients with chronic myeloid leukaemia. Leukemia. 2006;20:671–679. doi: 10.1038/sj.leu.2404141. [DOI] [PubMed] [Google Scholar]
- 5.Drummond MW, Hoare SF, Monaghan A, Graham SM, Alcorn MJ, Keith WN, Holyoake TL. Dysregulated expression of the major telomerase components in leukaemic stem cells. Leukemia. 2005;19:381–389. doi: 10.1038/sj.leu.2403616. [DOI] [PubMed] [Google Scholar]
- 6.Gabet AS, Mortreux F, Charneau P, Riou P, Duc-Dodon M, Wu Y, Jeang KT, Wattel E. Inactivation of hTERT transcription by Tax. Oncogene. 2003;22:3734–3741. doi: 10.1038/sj.onc.1206468. [DOI] [PubMed] [Google Scholar]
- 7.Hara T, Matsumura-Arioka Y, Ohtani K, Nakamura M. Role of human T-cell leukemia virus type I Tax in expression of the human telomerase reverse transcriptase (hTERT) gene in human T-cells. Cancer Sci. 2008;99:1155–1163. doi: 10.1111/j.1349-7006.2008.00798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zane L, Sibon D, Capraro V, Galia P, Karam M, Delfau-Larue MH, Gilson E, Gessain A, Gout O, Hermine O, et al. HTLV-1 positive and negative T cells cloned from infected individuals display telomerase and telomere genes deregulation that predominate in activated but untransformed CD4+ T cells. Int J Cancer. 2011;131:821–833. doi: 10.1002/ijc.26270. [DOI] [PubMed] [Google Scholar]
- 9.Poncet D, Belleville A, t'kint de Roodenbeke C, Roborel de Climens A, Ben Simon E, Merle-Beral H, Callet-Bauchu E, Salles G, Sabatier L, Delic J, et al. Changes in the expression of telomere maintenance genes suggest global telomere dysfunction in B-chronic lymphocytic leukemia. Blood. 2008;111:2388–2391. doi: 10.1182/blood-2007-09-111245. [DOI] [PubMed] [Google Scholar]
- 10.Capraro V, Zane L, Poncet D, Perol D, Galia P, Preudhomme C, Bonnefoy-Berard N, Gilson E, Thomas X, El-Hamri M, et al. Telomere deregulations possess cytogenetic, phenotype, and prognostic specificities in acute leukemias. Exp Hematol. 2011;39:195–202. e192. doi: 10.1016/j.exphem.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 11.Lin SY, Elledge SJ. Multiple tumor suppressor pathways negatively regulate telomerase. Cell. 2003;113:881–889. doi: 10.1016/s0092-8674(03)00430-6. [DOI] [PubMed] [Google Scholar]
- 12.Waldmann T, Scholten I, Kappes F, Hu HG, Knippers R. The DEK protein—an abundant and ubiquitous constituent of mammalian chromatin. Gene. 2004;343:1–9. doi: 10.1016/j.gene.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 13.Oancea C, Ruster B, Henschler R, Puccetti E, Ruthardt M. The t(6;9) associated DEK/CAN fusion protein targets a population of long-term repopulating hematopoietic stem cells for leukemogenic transformation. Leukemia. 2010;24:1910–1919. doi: 10.1038/leu.2010.180. [DOI] [PubMed] [Google Scholar]
- 14.Campillos M, García MA, Valdivieso F, Vázquez J. Transcriptional activation by AP-2α is modulated by the oncogene DEK. Nucleic Acids Res. 2003;31:1571–1575. doi: 10.1093/nar/gkg247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soares LM, Zanier K, Mackereth C, Sattler M, Valcárcel J. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science. 2006;312:1961–1965. doi: 10.1126/science.1128659. [DOI] [PubMed] [Google Scholar]
- 16.Riveiro-Falkenbach E, Soengas MS. Control of tumorigenesis and chemoresistance by the DEK oncogene. Clin Cancer Res. 2010;16:2932–2938. doi: 10.1158/1078-0432.CCR-09-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koleva RI, Ficarro SB, Radomska HS, Carrasco-Alfonso MJ, Alberta JA, Webber JT, Luckey CJ, Marcucci G, Tenen DG, Marto JA. C/EBPα and DEK coordinately regulate myeloid differentiation. Blood. 2012;119:4878–4888. doi: 10.1182/blood-2011-10-383083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhlmann AS, Villaudy J, Gazzolo L, Castellazzi M, Mesnard JM, Duc Dodon M. HTLV-1 HBZ cooperates with JunD to enhance transcription of the human telomerase reverse transcriptase gene (hTERT) Retrovirology. 2007;4:92. doi: 10.1186/1742-4690-4-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terme JM, Mocquet V, Kuhlmann AS, Zane L, Mortreux F, Wattel E, Duc Dodon M, Jalinot P. Inhibition of the hTERT promoter by the proto-oncogenic protein TAL1. Leukemia. 2009;23:2081–2089. doi: 10.1038/leu.2009.131. [DOI] [PubMed] [Google Scholar]
- 20.Sinha-Datta U, Horikawa I, Michishita E, Datta A, Sigler-Nicot JC, Brown M, Kazanji M, Barrett JC, Nicot C. Transcriptional activation of hTERT through the NF-κB pathway in HTLV-I-transformed cells. Blood. 2004;104:2523–2531. doi: 10.1182/blood-2003-12-4251. [DOI] [PubMed] [Google Scholar]
- 21.Hausmann S, Biddison WE, Smith KJ, Ding YH, Garboczi DN, Utz U, Wiley DC, Wucherpfennig KW. Peptide recognition by two HLA-A2/Tax11–19-specific T cell clones in relationship to their MHC/peptide/TCR crystal structures. J Immunol. 1999;162:5389–5397. [PubMed] [Google Scholar]
- 22.Cleary J, Sitwala KV, Khodadoust MS, Kwok RP, Mor-Vaknin N, Cebrat M, Cole PA, Markovitz DM. p300/CBP-associated factor drives DEK into interchromatin granule clusters. J Biol Chem. 2005;280:31760–31767. doi: 10.1074/jbc.M500884200. [DOI] [PubMed] [Google Scholar]
- 23.Smith MJ, Charron-Prochownik DC, Prochownik EV. The leucine zipper of c-Myc is required for full inhibition of erythroleukemia differentiation. Mol Cell Biol. 1990;10:5333–5339. doi: 10.1128/mcb.10.10.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Viollet B, Lefrancois-Martinez AM, Henrion A, Kahn A, Raymondjean M, Martinez A. Immunochemical characterization and transacting properties of upstream stimulatory factor isoforms. J Biol Chem. 1996;271:1405–1415. doi: 10.1074/jbc.271.3.1405. [DOI] [PubMed] [Google Scholar]
- 25.Kang X, Chen W, Kim RH, Kang MK, Park NH. Regulation of the hTERT promoter activity by MSH2, the hnRNPs K and D, and GRHL2 in human oral squamous cell carcinoma cells. Oncogene. 2009;28:565–574. doi: 10.1038/onc.2008.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sammons M, Wan SS, Vogel NL, Mientjes EJ, Grosveld G, Ashburner BP. Negative regulation of the RelA/p65 transactivation function by the product of the DEK proto-oncogene. J Biol Chem. 2006;281:26802–26812. doi: 10.1074/jbc.M600915200. [DOI] [PubMed] [Google Scholar]
- 27.Kappes F, Damoc C, Knippers R, Przybylski M, Pinna LA, Gruss C. Phosphorylation by protein kinase CK2 changes the DNA binding properties of the human chromatin protein DEK. Mol Cell Biol. 2004;24:6011–6020. doi: 10.1128/MCB.24.13.6011-6020.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waldmann T, Baack M, Richter N, Gruss C. Structure-specific binding of the proto-oncogene protein DEK to DNA. Nucleic Acids Res. 2003;31:7003–7010. doi: 10.1093/nar/gkg864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waldmann T, Eckerich C, Baack M, Gruss C. The ubiquitous chromatin protein DEK alters the structure of DNA by introducing positive supercoils. J Biol Chem. 2002;277:24988–24994. doi: 10.1074/jbc.M204045200. [DOI] [PubMed] [Google Scholar]
- 30.Hollenbach AD, McPherson CJ, Mientjes EJ, Iyengar R, Grosveld G. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J Cell Sci. 2002;115:3319–3330. doi: 10.1242/jcs.115.16.3319. [DOI] [PubMed] [Google Scholar]
- 31.Ko SI, Lee IS, Kim JY, Kim SM, Kim DW, Lee KS, Woo KM, Baek JH, Choo JK, Seo SB. Regulation of histone acetyltransferase activity of p300 and PCAF by proto-oncogene protein DEK. FEBS Lett. 2006;580:3217–3222. doi: 10.1016/j.febslet.2006.04.081. [DOI] [PubMed] [Google Scholar]
- 32.van Holde K, Zlatanova J. Unusual DNA structures, chromatin and transcription. Bioessays. 1994;16:59–68. doi: 10.1002/bies.950160110. [DOI] [PubMed] [Google Scholar]
- 33.Brazda V, Laister RC, Jagelská EB, Arrowsmith C. Cruciform structures are a common DNA feature important for regulating biological processes. BMC Mol Biol. 2011;12:33. doi: 10.1186/1471-2199-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cogulu O, Kosova B, Gunduz C, Karaca E, Aksoylar S, Erbay A, Karapinar D, Vergin C, Vural F, Tombuloglu M, et al. The evaluation of hTERT mRNA expression in acute leukemia children and 2 years follow-up of 40 cases. Int J Hematol. 2008;87:276–283. doi: 10.1007/s12185-008-0054-y. [DOI] [PubMed] [Google Scholar]
Supplementary References
- 1.Gabet AS, Mortreux F, Charneau P, Riou P, Duc-Dodon M, Wu Y, Jeang KT, Wattel E. Inactivation of hTERT transcription by Tax. Oncogene. 2003;22(24):3734–3741. doi: 10.1038/sj.onc.1206468. Prepublished on 2003/06/13 as doi:10.1038/sj.onc.1206468. [DOI] [PubMed] [Google Scholar]
- 2.Hausmann S, Biddison WE, Smith KJ, Ding YH, Garboczi DN, Utz U, Wiley DC, Wucherpfennig KW. Peptide recognition by two HLA-A2/Tax11-19-specific T cell clones in relationship to theirMHC/peptide/TCR crystal structures. J Immunol. 1999;162(9):5389–5397. [PubMed] [Google Scholar]
- 3.Cleary J, Sitwala KV, Khodadoust MS, Kwok RP, Mor-Vaknin N, Cebrat M, Cole PA, Markovitz DM. p300/CBP-associated factor drives DEK into interchromatin granule clusters. J Biol Chem. 2005;280(36):31760–31767. doi: 10.1074/jbc.M500884200. Prepublished on 2005/07/01 as doi:10.1074/jbc.M500884200. [DOI] [PubMed] [Google Scholar]
- 4.Smith MR, Greene WC. Identification of HTLV-I tax trans-activator mutants exhibiting novel transcriptional phenotypes. Genes Dev. 1990;4(11):1875–1885. doi: 10.1101/gad.4.11.1875. Prepublished on 1990/11/01 as doi:10.1101/gad.4.11.1875. [DOI] [PubMed] [Google Scholar]
- 5.Viollet B, Lefrancois-Martinez AM, Henrion A, Kahn A, Raymondjean M, Martinez A. Immunochemical characterization and transacting properties of upstream stimulatory factor isoforms. J Biol Chem. 1996;271(3):1405–1415. doi: 10.1074/jbc.271.3.1405. Prepublished on 1996/01/19 as doi:10.1074/jbc.271.3.1405. [DOI] [PubMed] [Google Scholar]
- 6.Zane L, Sibon D, Capraro V, Galia P, Karam M, Delfau-Larue MH, Gilson E, Gessain A, Gout O, Hermine O, et al. HTLV-1 positive and negative T cells cloned from infected individuals display telomerase and telomere genes deregulation that predominate in activated but untransformed CD4+ T cells. Int J Cancer. 2011 doi: 10.1002/ijc.26270. Prepublished on 2011/07/01 as doi:10.1002/ijc.26270. [DOI] [PubMed] [Google Scholar]
- 7.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26(6):509–515. doi: 10.1023/b:bile.0000019559.84305.47. Prepublished on 2004/05/07 as doi:10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 8.Chang JT, Yang HT, Wang TC, Cheng AJ. Upstream stimulatory factor (USF) as a transcriptional suppressor of human telomerase reverse transcriptase (hTERT) in oral cancer cells. Mol Carcinog. 2005;44(3):183–192. doi: 10.1002/mc.20129. Prepublished on 2005/07/13 as doi:10.1002/mc.20129. [DOI] [PubMed] [Google Scholar]
- 9.Jiang S, Galindo MR, Jarrett HW. Purification and identification of a transcription factor, USF-2, binding to E-box element in the promoter of human telomerase reverse transcriptase (hTERT) Proteomics. 2010;10(2):203–211. doi: 10.1002/pmic.200800693. Prepublished on 2009/11/10 as doi:10.1002/pmic.200800693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.