Abstract
The ability of electrospray to propel large viruses into a mass spectrometer is established and is rationalised by analogy to the atmospheric transmission of the common cold. Much less clear is the fate of membrane embedded molecular machines in the gas phase. Here we show that rotary ATPases/synthases from Thermus thermophilus and Enterococcus hirae can be maintained intact with membrane and soluble subunit interactions preserved in vacuum. Mass spectra reveal subunit stoichiometries and the identity of tightly bound lipids within the membrane rotors. Moreover subcomplexes formed in solution and gas phases reveal the regulatory effects of nucleotide binding on both ATP hydrolysis and proton translocation. Consequently we can link specific lipid and nucleotide binding with distinct regulatory roles.
Rotary ATPases/synthases are membrane associated molecular machines that perform biological energy conversion. Both V-type and F-type complexes consist of two reversible motors: the ion pump/turbine in VO/FO and the chemical motor/generator in V1/F1. The mode of operation is influenced by the ratio of the two fuels (protons:ATP) that drive the two motors. (1-3). The membrane-embedded VO/FO domain mediates the movement of Na+ or protons across the membrane while V1/F1 domains interact with nucleotides and inorganic phosphate either to produce or consume ATP in the case of the eukaryotic F and V-type families respectively. Eubacteria and archaea typically have only one type of rotary ATPases/synthases for both functions. Most bacteria have complexes of the F-type, but some bacteria and all known archaea have complexes closely related to eukaryotic V-type ATPases (also known as A-type ATPases/synthases (4)). Whether of F or V-type, the physiological function of most prokaryotic complexes is ATP synthesis, however many have evolved regulatory functions that allow reversal into ATP-driven proton pumps if required.
F1-FO, and V1-VO, are mechanically coupled by a central rotating shaft and held together by peripheral stalks (Fig. 1). Structural details derive from isolated subcomplexes of F1 and V1 (5-7), and from membrane embedded proteolipid rings of various species (8-11). Despite this wealth of structural information no high resolution structures of any intact rotary ATPases/synthases have been reported. Thus, regulatory allosteric changes that involve both the soluble head and the membrane sector are lost. In addition heterogeneous interactions with lipids and nucleotides are difficult to observe using existing structural biology approaches.
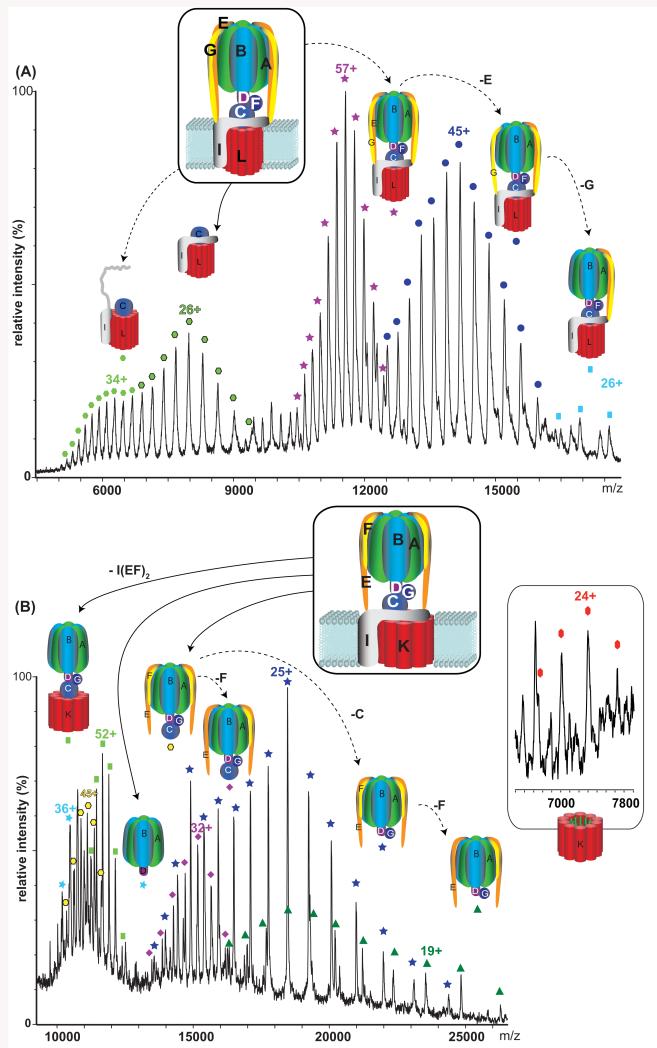
Figure 1. Mass spectra of the intact rotary ATPases from Thermus thermophilus and Enterococuccs hirae.
(A) Peaks are assigned to the intact TtATPase complex (stars), loss of the membrane subcomplex (ICL12) in solution and gas phases (dark green and green hexagons, respectively) and dissociation of subunits E and G from the peripheral stalk (blue circles and squares respectively). (B) For EhATPase the membrane subcomplex is observed in contact with the soluble head (green squares). Inset: mass spectra of the K ring in aqueous solution.
We show using electrospray mass spectrometry (MS) that rotary ATPases/synthases from Thermus thermophilus (TtATPase) and Enterococcus hirae (EhATPase) can remain intact in the gas phase. Previously composite models were assembled of the intact TtATPase using low resolution electron microscopy (EM) data in combination with high resolution X-ray structures of subunits (12) while the first cryo-EM data revealed views of the entire membrane embedded region (13). We compared the MS of the two complexes, the TtATPase with that of the less well-characterised EhATPase. Current models suggest that the EhATPase has only one peripheral stalk (14) and the stoichiometry of the K subunits in the membrane ring was determined as seven from EM (15) and 10 from X-ray analysis (8).
TtATPase was purified as described (16) using DDM for solubilisation since under these conditions the complex is most stable and does not form aggregates. The complex was introduced into a mass spectrometer modified for high mass complexes (17). Well-resolved charge states were assigned to the intact particle consisting of 26 subunits and nine different proteins (Fig. 1A). An experimentally determined mass of 659 202 (+/−131) Da corresponds to that calculated for the intact complex, based on subunit masses determined by MS (Table S1), plus additional mass due to incomplete desolvation, lipid and nucleotide binding (Fig. S1). Gas phase activation is necessary to release the complex from its detergent micelle (18, 19) giving rise to peaks at higher m/z than the intact complex, formed by unfolding and dissociation of highly charged subunits, predominantly subunits E, G and I (20) (Fig S2). At lower m/z charge states are assigned to the membranous subcomplex VO (ICL12), their bimodal distribution implying that they are formed both in solution and gas phases. The corresponding soluble V1 complex is also observed confirming that under these solution conditions a proportion of the complex dissociates spontaneously (21) (Fig. S3).
Using similar experimental parameters we recorded a spectrum for the EhATPase isolated as in DDM where its functional activity has been established previously (22, 23). The membrane embedded rotor for this complex is larger since each K subunit contains four trans-membrane helices as opposed to two for the corresponding L subunits from TtATPase. Under conditions where the complex emerges from the micelle surviving intact, the spectrum is not well-resolved (Fig. S4). Increases in activation energy lead to better desolvation and also appearance of subcomplexes in which either the membrane ring remains but the peripheral stalks and subunit I have dissociated, or the stalk subunits are attached but the membrane region is disrupted (Fig. 1B and S5). The number of peripheral stalks was determined as two and the stoichiometry in the K ring as 10 (175 kDa)(Fig. S6).
The lipid components in the K10 ring were identified as a series of negatively charged cardiolipins (Figs. 2, S7 and Table S2). The stoichiometry of lipid binding was determined as ten from the mass of the membrane ring. We identified six cardiolipin isomers and from quantitative analysis and measurement of the protein concentration we deduce specific binding of one lipid per subunit (Eh K subunit 1: 1.2 ±0.1 cardiolipins (Fig. S8)). Previously, the lipids were located using the atomic structure of the isolated K10 ring, in which peaks of positive density were attributed to 20 bifurcated phosphatidylglycerol lipids (8). We docked ten negatively charged symmetric cardiolipins inside the K10 ring, proximal to the conserved lys-32. The four hydrophobic chains are positioned with two chains emanating from both sides of the polar head, thus providing a hydrophobic lining to the inside of the ring (Fig. 2A).
Figure 2. Lipid binding and its effect on the membrane rings.
Predominant lipid molecules, one of the six cardioplipins (left) phosphatidylethanolamine (PE, right) found in intact EhATPase and TtATPase respectively. Main panel: Tandem MS of a subcomplex from TtATPase (ICL12E2G2F (282 740 Da)) leads to disruption of the L12 ring, releasing proteolipids L +/− PE (red/green circle, 8539 Da, red circles 7849 Da) and a stripped-complex ICE2G2F (blue squares, 184 242 Da). Atomic structure of the K10 ring (8) of EhATPase with docking of 10 cardioplipins to show reduction in the inner diameter (A) and after docking subunit C (B). Models for six-fold symmetry of the L12 ring with 6 PE molecules (green) (C) and with subunit C (42) (blue) docked into the ring (D). Lower panel: schematics of the rotor ring with 12 L subunits each having two TMH (red cylinders) and one conserved glu-63 (yellow) as seen in EM of 2D crystals (27) (i). Transformation into a six-fold symmetric ring ((ii) and (iii)). (iv) Comparison of the 6-fold symmetrical model with EM data reported previously for the TtATPase (iv) (16).
For the TtATPase L12 ring we identified the bound lipid as phosphatidylethanolamine (PE) (Fig. 2, S9). Unexpectedly, dissociated L subunits were observed either with bound PE lipid (holo) or without lipid (apo) after tandem MS of a subcomplex containing the membrane subunits and peripheral stalks (Fig. 2). Prior to disruption the mass of the subcomplex is consistent with binding of six lipids. This means that apo and holo subunits coexist within the same complex. We confirmed this observation by quantifying the protein:lipid ratio as one L subunit: 0.55+/−0.1 PE (ie one PE lipid per L subunit dimer) (Fig. S10). Six equivalent binding sites for lipids in protein dimers in a 12-membered ring imply a six-fold symmetrical state. Previously EM studies of the same preparation of the TtATPase revealed a 6-fold symmetric membrane embedded ring (Fig. 2, S11) (16). Both sets of data are consistent with close packing of two neighboring subunits, forming six dimers each with four transmembrane helices (TMH), thus emulating the arrangement of c subunits in eukaryotic V-ATPases in which gene duplication has led to four TMH per subunit (24, 25). From modelling we find that a rotation of the dimers by about 60° relative to their original orientation in the ring effectively locks six glu-63 residues in an occluded position, preserving lipid binding at the dimer interface. Proton transfer can occur at the remaining six active glutamates (Fig. 2 i-iii); consequently the proton:ATP ratio for the rotary enzyme is effectively halved.
Interestingly, in contrast to E. hirae which has both F and V-type ATPases, T. thermophilus has only one type of rotary ATPase that has been shown to operate in both proton pumping and ATP synthesis modes in vitro (26). Given that each of the 12 L subunits in the TtATPase membrane ring contains only two TMH, alternating between synthesis and pumping would involve switching the 12-membered ring from high proton:ATP ratio, primed for synthesis, into a lower one biased for pumping (27). This latter scenario thus mimics the eukaryotic V-type enzyme where gene duplication has provided four TMH each with one active glutamate. Consequently this mechanism for switching between ATP synthesis and ion pumping modes is likely assisted by specific lipid binding for this dual-function rotary ATPase.
These different lipid binding patterns in the two rotary ATPases studied here were found to be invariant from repeat preparations and from different detergent concentrations. Together they provide an explanation as to how the disparate membrane rings L12 and K10 (93.6 kDa and 160 kDa) can interact with their respective C subunits (35.8 and 38.2 kDa) which are likely conserved. Lining with cardiolipin, bound specifically to the inside of the K10 ring, has the effect of reducing the orifice from 54 Å to 38 Å (Fig. 2A, Fig. S12). Similarly converting a 12 to a six-membered ring for TtATPase reduces the orifice from 47 Å to 39 Å, creating two very similar inner diameters (38 Å and 39 Å) (Fig 2B).
To assess the role of nucleotides in changing subunit interactions we compared spectra for the TtATPase, with and without addition of 50 μM ATP. In the presence of ATP the intact complex and membrane embedded subcomplex ICL12 are the predominant species formed in solution (Fig. 3a and S13). When ATP is depleted loss of subunit B leads to extensive dissociation of the soluble head. In addition tandem MS of V1 in the presence of ATP leads predominantly to loss of subunit F, with subsequent loss of subunit D (Fig. 3b). When ATP is depleted, however, loss of subunit D prior to F becomes possible, leaving a subcomplex with F interacting directly with the A3B3 hexamer. Extension of F towards the soluble head has been proposed previously as a ‘braking’ mechanism to prevent unregulated consumption of ATP following in vivo dissociation of the head from the base in yeast V-ATPase (28). In line with this proposal an X-ray structure and cryoEM reconstruction of the isolated A3B3DF complex from TtATPase and yeast V-ATPase respectively, also reveal interactions of subunit F with the A3B3 hexamer (7, 29). An X-ray structure of the auto inhibited F1 head of the E. coli F-ATPase showed interactions between the subunits analogous to F1 ε and β in the soluble head (30). We subjected the E. coli F1 complex to the same tandem MS procedure outlined above. Results showed that subunit ε makes direct interactions with the soluble head (Fig. S14). The similar MS dissociation patterns observed in E. coli F-ATPase, in which the braking mechanism is well-established, and in TtATPase suggest an analogous mechanism to prevent ATP hydrolysis in the uncoupled V1 complex of TtATPase.
Figure 3. Nucleotide binding and its effects on intact TtATPase and the membrane embedded and soluble head complexes.
(a) Depletion of ATP leads to dissociation of the B subunit from the head and subunit I from ICL12. (b) Tandem MS of the soluble head (A3B3DF) reveals sequential loss of subunits F and D in the presence of 50 μM ATP. In TtATPase solutions containing 50 μM ADP two conformations of subunit F are evident from the bimodal distribution of charge states formed for the V1 stripped of F (Δ6+ and Δ12+) together with a direct loss of subunit D. The Δ6+ series is similar to that formed from the ATP-bound complex. The Δ12+ series is consistent with an extended conformation of subunit F. Insert schematic representation of effects of ATP/ADP on the membrane ICL12 complex and the movement of an extended subunit F in the A3B3DF complex.
We observed further sensitivity of TtATPase to low ATP levels, notably loss of subunit I from ICL12 to form CL12 (Fig. 3a). Expansion of the peaks assigned to ICL12 is consistent with binding of up to six lipids and up to two nucleotides (ATP or ADP) (Fig. S15). This is in line with proposals that a eukaryotic functional equivalent of subunit I senses cellular nucleotide levels by binding selectively to ADP and undergoing conformational change (31). To investigate this conformational change we applied ion mobility MS (IM-MS) (32) to the intact TtATPase, ICL12 and CL12 subcomplexes. Since ions with multiple conformations result in broad arrival time distributions (ATDs) we conclude that the relatively compact ATDs for both the intact ATPase and CL12 are consistent with one predominant conformation (S16 and tables S3, S4). By contrast the ATDs for the ICL12 complex are much broader than those of the intact TtATPase and CL12, consistent with multiple conformations of subunit I in the isolated VO complex (Fig. 4, table S4). The lack of conformational heterogeneity in the intact complex is rationalised by the ‘tethering’ of subunit I by forces exerted by the peripheral stalks (subunits E and G) as suggested recently (13). Once released from the intact complex, subunit I in VO is not constrained and flexibility of the hinge domain, located between the soluble and transmembrane domains, likely leads to its conformational heterogeneity.
Figure 4. Conformational heterogeneity and dissociation of subunit I from ICL12 implies a mechanism for closing the H+ channel.
(a) IM-MS of the transmembrane ICL12 and CL12 complexes formed in solution from intact TtATPase. Charge states used for IM measurement are labelled grey and red (b,c). Broader arrival time distributions for ICL12 than for CL12 are consistent with conformational heterogenity in subunit I. (d) Possible mechanism to ‘close’ the H+ channel following lateral movement of subunit I within the membrane. Surrounding lipids effectively block the channel. The direction of proton pumping (green) and rotation of the ring (yellow) for ATP hydrolysis, as seen from the top (site of interaction with V1). (e) Coarse grain and atomic model of the ICL12 complex generated according to IM restraints and homology modelling (see supplementary methods). The proposed nucleotide binding area in the hinge region of I (orange) and Glu63:Arg563 from subunits L and I are shown yellow and green respectively.
The soluble domain of I in EM density maps of the intact TtATPase is at 90° to the proton channel (12, 13). Our IM data for the VO complex, based on modeling of the ICL12 and CL12 complexes, (Fig. S17) are consistent with a range of conformational states with angles from 90° to 135° (Fig. S18). We propose therefore that the conformational dynamics evidenced here, together with preferential binding of ADP to the proposed site near the hinge region (31), destabilize interactions between I and CL12 as evidenced by the facile loss of subunit I under low ATP conditions.
Given that cellular nucleotide levels likely affect proton translocation in the isolated VO complex it might also be anticipated that changes in the proton gradient would induce similar regulatory effects. To test this hypothesis we increased the pH of the ATPase-containing solution to mimic reduction of the proton concentration. Mass spectra of the TtATPase incubated at pH 9.0 lead to subcomplexes formed by loss of IGE (Fig. S19). This observation, together with the dissociation of subunit I from VO, implicates subunit I in a regulatory role, sensing both proton and ATP levels. Interestingly previous proposals invoked a locking together of the membrane portion of subunit I with the CL12 membrane ring preventing relative movement and hence explaining the absence of passive H+ translocation in isolated VO (33, 34). Since we observe facile loss of subunit I under both low [H+] and low [ATP] conditions, and given the lack of extensive interactions between I and the membrane ring observed in cryo-EM data (13), our results point to a mechanism in which subunit I moves away from the ring, and the resulting gap could then be sealed with membrane lipids, (Fig. 4d).
Our results show that lipids with two and four hydrophobic chains associate with subunits with two and four TMH, subunits L and K respectively. Interestingly in both ATPases the lipids identified in situ are not the most prevalent in the cell, (35, 36) implying that lipids are selected from the available pool for specific structural roles and metabolic regulation. This lends further support to the proposal that membrane proteins possess specific lipid binding sites (37) and demonstrates the ability of lipids to fine tune subunit interactions by defining the conformations and inner dimensions of the membrane rings. Our nucleotide binding experiments show that a drop in cellular ATP levels is sensed by V1, not only with the movement of subunit F but also with changes in interactions at the A:B interface. Unexpectedly VO is also sensitive to low [ATP] and [H+], both of which promote displacement of subunit I. We suggest that membrane lipids subsequently seal the proton conducting channel. Consequently both ATP and proton/ion gradients are conserved when reversible dissociation takes place in vivo.
The existence of intact rotary ATPases/synthases in vacuum has been demonstrated previously using LILBID MS (38, 39). At its current resolution however it is not possible to identify bound lipids/nucleotides or to probe their effects on subunit interactions. Moreover previous ES-MS experiments gave well-resolved mass spectra for the V1 domain (40, 41) but lack of an intact VO domain, or any interactions between V1 and VO, is attributed to dissociation of the complex in the absence of the protective micelle (18, 19). By contrast the ES approach used here enables interrogation of subunit interactions within intact rotary ATPase/synthases and allows us to probe the synergistic effects of lipid and nucleotide binding.
Supplementary Material
Acknowledgements
Funding from the Wellcome Trust and by the PROSPECTS (HEALTHF4-2008-201648) grant within the Research Framework of the European Union together with funding from the Royal Society (CVR), the Australian NH&MRC grant 573712 (DS) and the FONDECYT 1100515 (NB) is acknowledged.
References
- 1.Jefferies KC, Cipriano DJ, Forgac M. Arch Biochem Biophys. 2008;476:33. doi: 10.1016/j.abb.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.von Ballmoos C, Wiedenmann A, Dimroth P. Annu Rev Biochem. 2009;78:649. doi: 10.1146/annurev.biochem.78.081307.104803. [DOI] [PubMed] [Google Scholar]
- 3.Muench SP, Trinick J, Harrison MA. Q Rev Biophys. 2011:1. [Google Scholar]
- 4.Muller V, Gruber G. Cell Mol Life Sci. 2003;60:474. doi: 10.1007/s000180300040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abrahams JP, Leslie AG, Lutter R, Walker JE. Nature. 1994;370:621. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 6.Okazaki K, Takada S. Structure. 2011;19:588. doi: 10.1016/j.str.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 7.Numoto N, Hasegawa Y, Takeda K, Miki K. EMBO Rep. 2009;25:25. doi: 10.1038/embor.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murata T, Yamato I, Kakinuma Y, Leslie AG, Walker JE. Science. 2005;308:654. doi: 10.1126/science.1110064. [DOI] [PubMed] [Google Scholar]
- 9.Meier T, Polzer P, Diederichs K, Welte W, Dimroth P. Science. 2005;308:659. doi: 10.1126/science.1111199. [DOI] [PubMed] [Google Scholar]
- 10.Dautant A, Velours J, Giraud MF. J Biol Chem. 2010;285:29502. doi: 10.1074/jbc.M110.124529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watt IN, Montgomery MG, Runswick MJ, Leslie AG, Walker JE. Proc Natl Acad Sci U S A. 2010;107:16823. doi: 10.1073/pnas.1011099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee LK, Stewart AG, Donohoe M, Bernal RA, Stock D. Nat Struct Mol Biol. 2010;17:373. doi: 10.1038/nsmb.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lau WC, Rubinstein JL. Proc Natl Acad Sci U S A. 2010;107:1367. doi: 10.1073/pnas.0911085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto M, et al. J Biol Chem. 2008;283:19422. doi: 10.1074/jbc.M801772200. [DOI] [PubMed] [Google Scholar]
- 15.Murata T, et al. J Biol Chem. 2003;278:21162. doi: 10.1074/jbc.M301620200. [DOI] [PubMed] [Google Scholar]
- 16.Bernal RA, Stock D. Structure. 2004;12:1789. doi: 10.1016/j.str.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez H, Robinson CV. Nat Protoc. 2007;2:715. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 18.Barrera NP, Di Bartolo N, Booth PJ, Robinson CV. Science. 2008;321:243. doi: 10.1126/science.1159292. [DOI] [PubMed] [Google Scholar]
- 19.Barrera NP, et al. Nat Methods. 2009;6:585. doi: 10.1038/nmeth.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benesch JL, Robinson CV. Curr Opin Struct Biol. 2006;16:245. doi: 10.1016/j.sbi.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 21.Yokoyama K, et al. J Biol Chem. 2003;278:42686. doi: 10.1074/jbc.M305853200. [DOI] [PubMed] [Google Scholar]
- 22.Takase K, et al. J Biol Chem. 1994;269:11037. [PubMed] [Google Scholar]
- 23.Murata T, Takase K, Yamato I, Igarashi K, Kakinuma Y. J Biochem. 1999;125:414. doi: 10.1093/oxfordjournals.jbchem.a022302. [DOI] [PubMed] [Google Scholar]
- 24.Muller EG, Snydsman BE, Novik I, Hailey DW, Gestaut DR, Niemann CA, O’Toole ET, Giddings TH, Jr., Sundin BA, Davis TN. Mol Biol Cell. 2005;16:3341. doi: 10.1091/mbc.E05-03-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clare DK, et al. Structure. 2006;14:1149. doi: 10.1016/j.str.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 26.Yokoyama K, et al. J Biol Chem. 2000;275:13955. doi: 10.1074/jbc.275.18.13955. [DOI] [PubMed] [Google Scholar]
- 27.Toei M, et al. Proc Natl Acad Sci U S A. 2007;104:20256. doi: 10.1073/pnas.0706914105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kane PM, Smardon AM. J Bioenerg Biomembr. 2003;35:313. doi: 10.1023/a:1025724814656. [DOI] [PubMed] [Google Scholar]
- 29.Hildenbrand ZL, Molugu SK, Stock D, Bernal RA. PLOS one. 2010;5:e12588. doi: 10.1371/journal.pone.0012588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cingolani G, Duncan TM. Nat Struct Mol Biol. 2011;18:701. doi: 10.1038/nsmb.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Armbruster A, et al. FEBS Lett. 2005;579:1961. doi: 10.1016/j.febslet.2005.02.042. [DOI] [PubMed] [Google Scholar]
- 32.Uetrecht C, Rose RJ, van Duijn E, Lorenzen K, Heck AJ. Chem Soc Rev. 2010;39:1633. doi: 10.1039/b914002f. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, Myers M, Forgac M. J Biol Chem. 1992;267:9773. [PubMed] [Google Scholar]
- 34.Beltran C, Nelson N. Acta Physiol Scand Suppl. 1992;607:41. [PubMed] [Google Scholar]
- 35.Carson D, Pieringer RA, Daneo-Moore L. Biochim Biophys Acta. 1979;575:225. doi: 10.1016/0005-2760(79)90024-9. [DOI] [PubMed] [Google Scholar]
- 36.Ray PH, White DC, Brock TD. J Bacteriol. 1971;108:227. doi: 10.1128/jb.108.1.227-235.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hunte C, Richers S. Curr Opin Struct Biol. 2008;18:406. doi: 10.1016/j.sbi.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 38.Meier T, et al. Mol Microbiol. 2007;65:1181. doi: 10.1111/j.1365-2958.2007.05857.x. [DOI] [PubMed] [Google Scholar]
- 39.Hoffmann J, et al. Phys Chem Chem Phys. 2010;12:13375. doi: 10.1039/c0cp00733a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esteban O, et al. J Biol Chem. 2008;283:2595. doi: 10.1074/jbc.M704941200. [DOI] [PubMed] [Google Scholar]
- 41.Kitagawa N, Mazon H, Heck AJ, Wilkens S. J Biol Chem. 2008;283:3329. doi: 10.1074/jbc.M707924200. [DOI] [PubMed] [Google Scholar]
- 42.Iwata M, et al. Proc Natl Acad Sci U S A. 2004;101:59. doi: 10.1073/pnas.0305165101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.