Abstract
Traditionally large-scale chromatin structure has been studied by microscopic approaches, providing direct spatial information but limited sequence context. In contrast, newer 3C (Chromosome Capture Conformation) methods provide rich sequence context but uncertain spatial context. Recent demonstration of large, topologically linked DNA domains, hundreds to thousands of kb in size, may now link 3C data to actual chromosome physical structures, as visualized directly by microscopic methods. Yet, new data suggesting that 3C may measure cytological rather than molecular proximity prompts a renewed focus on understanding the origin of 3C interactions and dissecting the biological significance of long-range genomic interactions.
Introduction
Traditionally, large-scale chromatin organization has been studied by microscopy, providing spatial information but limited DNA sequence context. In contrast, genomic methods now allow a new, orthogonal approach to investigating large-scale chromatin organization, providing rich sequence context but uncertain spatial context. Both approaches have their own limitations, assumptions, and technical concerns, and both typically are applied by different scientific communities possessing different expertise and posing somewhat different questions. While these new genomic methods have dramatically elevated interest in large-scale chromatin organization, there remains a notable gap that must be bridged to relate genomic results to actual physical models for large-scale chromatin organization.
Here I attempt to provide a conceptual framework for possibly bridging this gap while reviewing, from a microscopist's perspective, key advances from the previous two years. In particular, I focus on the recent demonstration of large, topologically linked DNA domains, hundreds to thousands of kb in size, which may represent constitutive units of chromosome folding.
Folding of chromatin fibers into large-scale chromatin domains
Local chromatin structure, corresponding to the positioning and composition of nucleosomes and the higher-order folding of nucleosome arrays into 30 nm chromatin fibers, has been reviewed recently elsewhere [1-3]. Emerging developments include the concept of a family of "30 nm" structures, replacing a single, canonical 30 nm fiber structure, as well the proposal that the 30 nm chromatin fiber might exist predominantly in regions of low chromatin density, whereas in areas of higher chromatin density intermingling of nucleosomes from adjacent chromatin fibers forms a "polymer melt" [4,5]. More difficult to resolve is to what extent mixtures of 10 and 30 nm chromatin fibers may co-exist within interphase chromosome regions and be distinguished experimentally from regions of polymer melt.
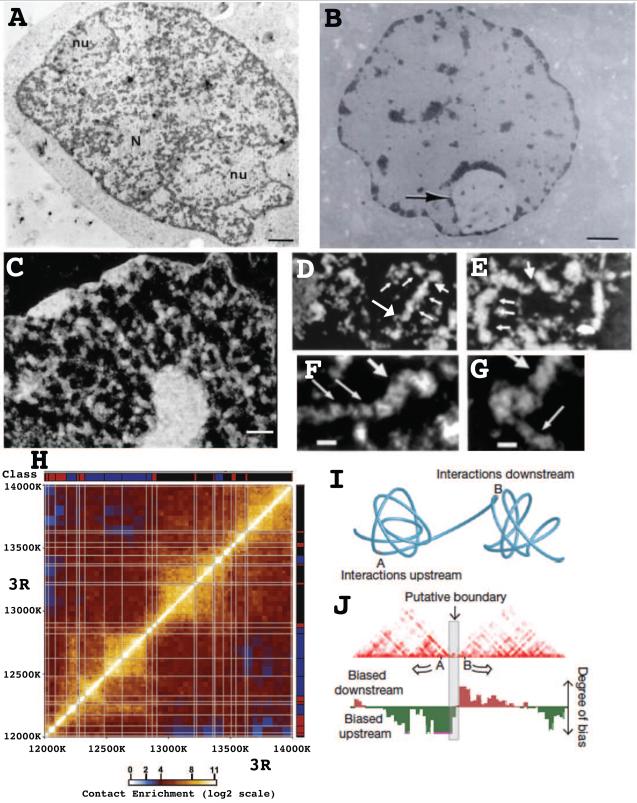
Easier to address are the types of differential chromatin compaction that might exist through the genome. The old "heterochromatin" / "euchromatin" division, derived largely from nonspecific, heavy metal staining of nuclei using transmission electron microscopy (TEM), has been misleading. Multiple DNA specific TEM stains have revealed that the so called "euchromatin" compartment, lightly stained by heavy metals, contains little DNA (Fig. 1A-B); instead most of the genome in the typical mammalian nucleus is packaged into large-scale chromatin structures with higher diameters than 30 nm [6-12] (Fig. 1A-G).
Fig. 1. Large-scale chromatin domains in search of genomic context- genomic interactions in search of structural context.
(A-C) Three staining methods demonstrate large-scale chromatin domains in thin-section TEM of freeze-substitution (A) or conventional (B-C) glutaraldehyde fixed samples: (A) DNA post-embedding immunostaining (dark, positive staining) of TEM thin section reveals most nuclear DNA is present in large-scale, frequently fiber-like structures. Scale bar = 1 μm. Reprinted from Fig. 4a, reference [7], Copyright 1994 J Histochem Cytochem. (B) DNA post-embedding staining by osmium ammine of rat liver nucleus shows no "euchromatin" texture. Large regions of nucleus are DNA free. Staining sensitivity is shown by visualization of decondensed chromatin within nucleolus (arrow). Reprinted from Fig. 1, reference [9], Copyright 1989 J Histochem Cytochem. (C) Uranyl and lead staining of detergent permeabilized Hela nucleus in buffer preserving large-scale chromatin structure. Staining is white in negative image. Extraction of nucleoplasmic protein results in absence of "euchromatin" texture after fixation and reveals large-scale chromatin structures. (D-G) Thin section, serial sections of CHO nuclei reveal (D-E) fiber-like, large-scale structures (small arrows) consisting of ~Mbp size segments punctuated by less condensed chromatin (large arrows). (F-G) Abrupt transitions from thick (large arrows) (~100-130 nm) to thinner (~60-80 nm) diameter size are seen, with local discontinuities by decondensed chromatin (thin arrows). Reprinted from Fig. 6 (D-E) and Fig. 7 (F-G), reference [16] , Copyright JCB 1994. Scale bars = 1 μm (A,B) and 500 nm (C), 120 nm (F-G). (H) Hi-C scatterplot showing long-distance DNA interactions in Drosophila embryonic nuclei over region of chromosome 3R. Off-diagonal, box-like, yellow regions define topological domains within which 3C interactions are favored over neighboring regions. X-Y axes show regions of 3R (bp) with associated chromatin average states. Reprinted from Fig. 3F, reference [37], Copyright 2012, Cell. (I) Cartoon illustrating that topological domains detected by 3C methods correspond to local regions of compact, but irregular chromatin folding separated by extended chromatin. (J) Boundary between topological domains (grey bar) is defined by asymmetry in DNA interactions up versus downstream of boundary region, with DNA within a topological domain interacting preferentially. (I-J) reprinted from Fig. 1b, reference [38], Copyright 2012 Nature.
In this context, old autoradiography studies showing concentrated sites of transcription at "heterochromatin /euchromatin boundaries" can now be reinterpreted as suggesting transcription at the edges of large-scale chromatin domains [13]. This interpretation is supported by demonstration of transcription on a condensed template in BAC transgene arrays [14] and by recent super-resolution light microscopy showing Br-UTP incorporation, RNA pol II immunostaining, and enrichment of some active chromatin marks at the periphery of large-scale chromatin domains [15].
Thus the old dichotomy of "heterochromatin" versus "euchromatin" now needs replacement with differentiation between multiple types of chromatin. From a structural perspective, TEM shows a range or continuum of large-scale chromatin compaction. In less compact chromosomal regions, large-scale chromatin domains form discontinuous fibers which can be traced in late G1 and S phase nuclei for up to several microns in selected regions; these fibers frequently fold back and supercoil on themselves to form plectonemic chromosome loops [16]. A range of large-scale chromatin domain diameters is seen within the same nucleus and these diameters can vary through the cell cycle [11,16,17]. Pericentric heterochromatin versus the facultative heterochromatin of the inactive X chromosome are easily distinguished, with the condensed Barr body of the inactive X showing large-scale chromatin domains comparable in diameter to more condensed regions elsewhere in the nucleus [18].
Sharp transitions between different compaction states can be visualized at the TEM level (Fig. 1D-G). A simple interpretation is that discontinuous fiber segments Mbp in size are connected by sharp transitions to less folded states; these fiber segments may correspond to structurally stable chromatin domains of similar, ~Mbp size consisting of clusters of DNA replication origins which initiate DNA replication synchronously [19-21]. While it has been assumed that these apparent fibers form from hierarchical folding of chromatin, topological looping of DNA may co-exist within these fiber-like structures [22].
The major problem with these structural results has been the difficulty testing whether these distinct structural states are reproducible for specific DNA sequences and mapping structure to sequence. Visualization of engineered chromosome regions by light and electron microscopy has correlated large-scale chromatin compaction with transcriptional activity, but this has relied on transgene arrays that may not completely recapitulate normal chromosome structure [14,22-24]. Fluorescent in situ hybridization (FISH) has demonstrated variable large-scale compaction correlated with gene transcriptional activity over specific gene loci [25-27] but this approach is low resolution, given both limitations of light microscopy and possible structural perturbations induced by the hybridization procedure [28-30].
The Good
Enter 3C (chromosome conformation capture) methods, possibly to the rescue. As reviewed recently [31-36], multiple variations on 3C use a proximity- ligation assay, typically together with PCR, to probe long-distance interactions between loci on the same or different chromosomes: the original 3C examines pairwise interactions, 4C examines interactions of one particular "bait" region with all other genomic loci, 5C examines pairwise interactions over a defined genomic interval, and Hi-C examines all pairwise interactions over the entire genome but at lower resolution than 5C. Reassuringly, these 3C methods have revealed general features of large-scale chromatin organization, discovered through decades of microscopy on specific genomic regions by FISH, but now demonstrated in higher throughput and genomic context. This includes folding of DNA into spatially segregated chromosome territories, proximity of arms from the same chromosome, clustering between centromeres and between telomeres, and clustering between transcriptionally active genome regions.
Several independent groups now have demonstrated distinct topological domains in the hundreds of kb to Mbp size range using 5C and Hi-C methods in Drosophila or mammalian systems [37-41]. 3C interactions occur at significantly higher frequency with other sequences lying in these same "topological associated domains" (TADs) [39] relative to sequences lying outside these domains (Fig. 1).
To date the highest resolution Hi-C studies have been in Drosophila, given its ~20 fold smaller genome size. Two studies, one using mixed embryonic nuclei from different developmental stages and lineages [32] and another using Kc tissue culture cells [40], each describe the delineation of roughly 1000 domains, with median and mean values of 60 or 62 kb and 107 or 100 kb, respectively. Half the domain boundaries are the same in both studies with the overall domain pattern even more similar. Thus a large fraction of these domains are common to a range of cell types. This similarity in domain structure is striking, given the differences in starting material and the different computational methods used to extract domain boundaries from still noisy data sets.
These topological domains largely demarcate regions enriched for either repressive or active epigenetic chromatin classifications, as defined by cluster analysis of protein binding and histone modification genomic patterns [42,43]. Larger domains are biased heavily towards repressive epigenetic classes and smaller domains heavily towards transcriptionally active epigenetic classes. Domain borders are significantly enriched in marks of transcriptionally active chromatin, including H3K4me3, DNase I hypersensitivity sites, transcriptional start sites (TSSs) and overall gene density, rather than numbers of active genes. One study concluded that domain borders were determined by multiple insulator sites, particularly those combinations bound by multiple insulator proteins including CP190, that may or may not be associated with transcriptionally active sites [37]. The other study concluded that domain boundaries are particularly enriched in insulators containing sites for three or four of the insulator protein types (+/− Su(Hw)) combined with bound RNA pol II and TSSs [40].
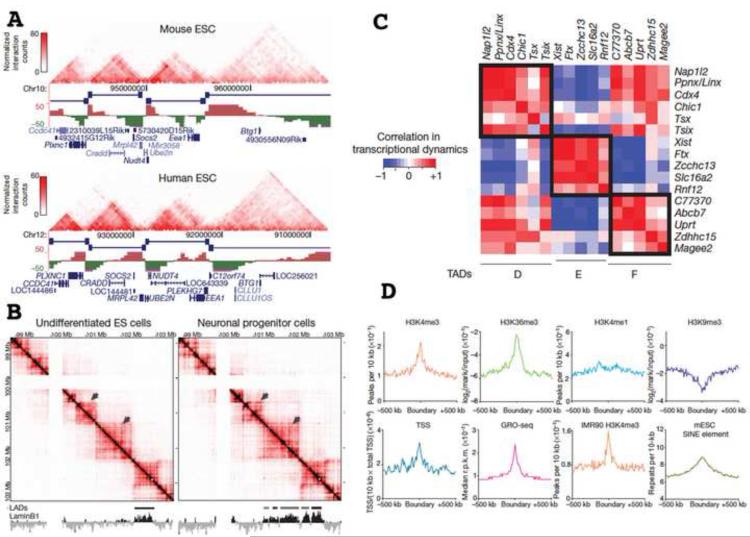
Several Hi-C and 5C studies in mammalian cells strikingly have described quite similar topological domain structure [38,39,41] with median size of ~900 kb. A large fraction of these are conserved between mouse and human and invariant during development (Fig. 2A-B). As in Drosophila, domain boundaries are enriched in insulator (CTCF) binding sites, active promoters, gene bodies, transcription start sites, engaged RNA pol II, housekeeping genes, tRNA genes, and active histone marks (Fig. 2D). Again, domain boundaries appear to demarcate chromatin domains varying in replication timing, lamin B binding, or distinctive histone modifications [38,39]. However, these chromatin domain types can change during development and can be perturbed by mutation or knockdown without changing the TAD domain organization [39].
Fig. 2. An incomplete list of important biological features of 3C defined topological domains.
(A) Conservation of topological domains- comparison of mouse versus human domain organization over syntenic region. Reprinted from Fig. 3g-h, reference [38], Copyright 2012 Nature. (B) Preservation of topological domain organization during differentiation- comparison of domain organization in undifferentiated ES cells versus neuronal progenitor cells. Topological domains are fixed, while lamin associated domains (LADs) change organization. However, topological boundaries appear to mark boundaries of LADs and other epigenetic domains. (C) In topological domains identified in one 5C study, genes within topological domains show higher correlation of gene expression than genes lying in different domains. Reprinted from Fig. 3 (B) and Fig. 4 (C) from reference [39], Copyright 2012 Nature. (D) Topological boundaries are enriched in marks for active chromatin and transcription. Reprinted from Fig. 4a, reference [38], Copyright 2012 Nature.
Therefore many TAD boundaries may act as constitutive boundaries to spreading of chromatin states. Across several neighboring TADs identified by 5C, expression of genes with promoters within a single TAD correlated better than expression of genes in different TADs [39] (Fig. 2C). Focusing on putative enhancer / promoter interactions within single TADs identified a lncRNA likely involved in X chromosome inactivation regulation [39]. More generally, mapping TAD structure may facilitate identification of functional long-range interactions important in gene regulation. Dissecting determinants of TAD organization and long-range interactions within TADs is beginning, leading to suggestions that constitutive, Mbp size TADs are demarcated by CTCF/cohesin while Mediator and cohesin bridge enhancer/promoter interactions over shorter distances [41].
The Surprising
These new Hi-C and 5C results clearly direct a renewed focus on large-scale chromatin architecture. Yet, conceptually there remains a large contrast between the richness of 3C interaction data versus our understanding of how to relate these interaction frequencies to actual large-scale chromatin structures and dynamics. Normalization schemes now correct for technical biases- for instance, varying ligation rates as a function of restriction fragment length and nucleotide composition [44]. A more general correction for "visibility" to Hi-C interactions across the genome corrects for unknown biases that vary over the 200kb-1Mbp bin sizes used now for Hi-C [45].
Less tractable are questions related to variations in fixation efficiency across the genome and, even more basic, whether detected 3C interactions reflect molecular (~1-10 nm) versus cytological (100-1000 nm) proximity. For instance, DNase I HS sites show elevated 3C interaction frequencies [44,46]. But they also show lower cross-linked protein after formaldehyde fixation. Indeed, the FAIRE genomic method for identifying HS sites is based on HS sites purifying as "naked DNA" in a standard phenol / chloroform DNA extraction [47,48]. Likewise, genomic interactions detected by 3C methods are nearly always conceptualized as the result of ligation of two DNA fragments self-associating in vivo through a direct molecular interaction. Yet formaldehyde concentrations much lower than used in 3C protocols result in extensive nucleosome cross-linking within and between chromatin fibers [49], raising the possibility of large networks of cross-linked chromatin fragments.
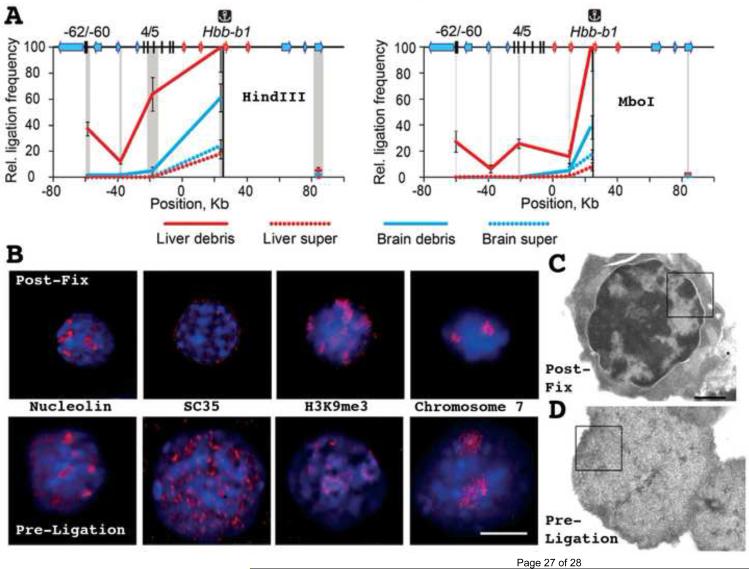
Indeed, Razin and colleagues now have shown that 3C signals are predominately, if not exclusively, generated from the insoluble fraction of the "lysed nuclei" fraction (Fig. 3A) [50]. This insoluble fraction is comprised of moderately swollen nuclei with basic architectural features- nucleoli, SC35 speckles, heterochromatic domains, and chromosome territories- largely preserved at the light microscope level (Fig. 3C) but severely perturbed as seen by TEM (Fig. 3D). Using 3C interactions within the mouse beta-globin locus as their assay, nearly all long-range 3C interactions were demonstrated as arising from the insoluble fraction, with disruption of nuclear structure by sonication decreasing these interactions proportionally. These results raise the intriguing possibility that 3C interactions might measure cytological rather than molecular proximity (Fig. 4).
Fig. 3. 3C Interactions predominately derive from insoluble fraction corresponding to swollen nuclei.
(A) Comparison of 3C interactions between beta-globin promoter region and flanking regulatory sequences in beta-globin locus from soluble (dotted lines) versus insoluble (solid lines) fractions. Data is normalized for DNA amount in each fraction. Red lines correspond to fetal liver where gene is active, while blue lines correspond to brain where gene is inactive. 3C peaks from known long-distance interactions in beta-globin locus derive from insoluble fraction containing swollen nuclei. (B) Nuclear structure immediately after fixation (top) versus just prior to 3C ligation step (previously assumed to act on soluble, small, DNA-protein molecular complexes). Nuclei enlarge with SDS and other treatments prior to ligation step, chromosome territories swell, but nuclear bodies such as nucleoli, SC35 domains, and patches of epigenetically marked chromatin domains are still recognizable. DNA (DAPI) staining blue, immunofluorescence or FISH signals red. (C,D) TEM images of nuclei after fixation (C) or after fixation but just prior to ligation step of 3C procedure (D)- 3C procedures result in obvious disruption of condensed chromatin masses in fetal liver nuclei. Scale bars = 5000 nm (B), 1000 nm in (C-D). Reprinted from Fig. 2C (A), Fig. 3 (B), and Fig. 4 (C-D) from reference [50], Copyright 2013 Nucleic Acids Research.
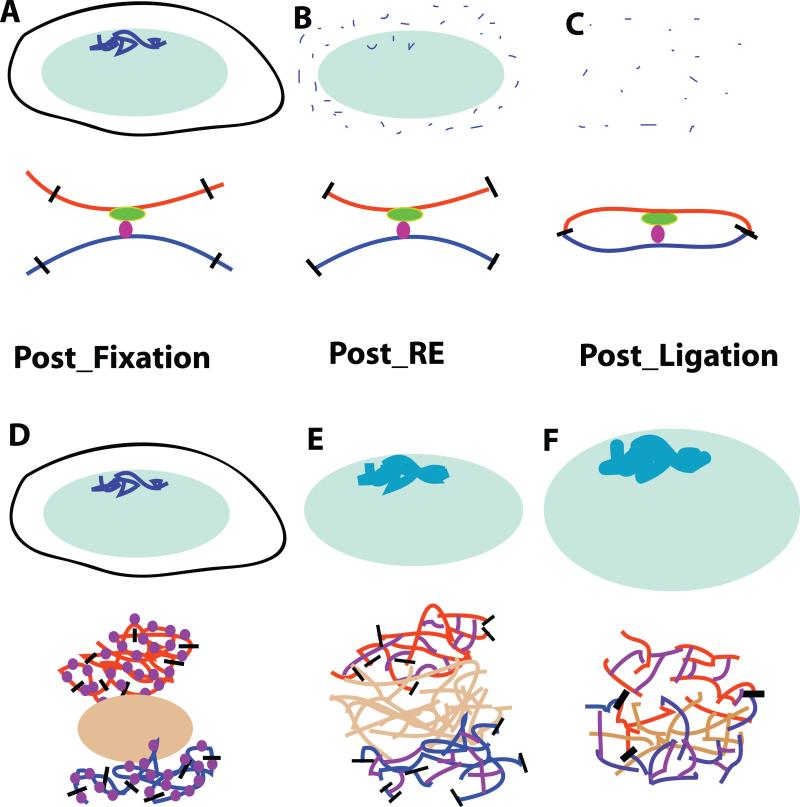
Fig. 4. Do long-range cis and trans 3C interactions imply molecular versus cytological proximity?
(A-C) Conventional models interpret 3C interactions as demonstrating molecular proximity. Top: After fixation (A), chromosomes (dark blue) within nuclei (light green) are digested and solubilized by restriction enzyme digestion (B), followed by dilution of soluble fraction prior to ligation to favor intramolecular ligation events (C). Bottom: DNA strands from distant chromosome sites (blue or red) are brought into proximity through specific binding of trans factors (green and purple) binding to different cis elements; this proximity is fixed by formaldehyde cross-linking of trans and cis factors (A). Restriction digest cuts restriction sites (black lines) (B) which then ligate DNA fragments together from different genomic regions (C). (D-F) Alternative model suggests 3C interactions correspond instead to proximity over cytological rather than distances. Top: Interphase chromosome structures (blue) after fixation (D) are disrupted progressively during 3C procedure. Restriction digest (E) results in fragments that remain cross-linked to insoluble, cross-linked nuclear structures and ligation (F) occurs within these partially disrupted chromosome residual structures. Bottom: Chromosome regions (red and blue) adjacent to nuclear bodies (brown) are densely cross-linked by formaldehyde fixation (D). SDS treatment and heat prior to restriction digest denatures histone and nonhistone proteins (purple) that act as linkers creating a protein-DNA network anchoring restriction fragments after restriction digest (E). Ligation events (black lines) connect fragments over large distances within this disordered network (F). In this model, long-range 3C interactions might derive for instance from co-association of two gene loci to the same nuclear body, even if they are separated in the live cell by cytological (100-1000 nm) distances.
The Still Perplexing
Many examples now exist in which 3C interactions bridging LCR, enhancer, and/or promoter sequences depend on the recruitment of specific transcription factors or co-factors to these cis-regulatory sequences. Enhancer-promoter looping is the dominant model for enhancer function. The demonstration of transcription factor dependent 3C interactions connecting enhancer and promoter elements therefore appears to confirm this looping model while demonstrating the molecular origin of 3C interactions.
An excellent example is the 3C interaction between DNase I HS site 2 of the beta-globin LCR and the beta-globin promoter [51]. This interaction is dependent on the Lbd1 protein and the hematopoietic-restricted factors KLF1, GATA1, and the GATA1 cofactor FOG1. Ldb1 is present at both the LCR and promoter and GATA1 is required for Lbd1 recruitment at the promoter. In one model, Ldb1 homo-dimerization or oligomer formation anchors a loop between the LCR and the promoter, generating a 3C detectable interaction [52]. In an elegant experiment, the GATA1 dependence of this 3C interaction was bypassed by targeted tethering of Lbd1 directly to the promoter via a zinc finger-Lbd1 fusion protein or the same zinc finger protein fused to the self-interacting domain of Lbd1 [52]. Transcriptional activation via these Lbd1 fusion proteins was within 20% of that seen after GATA1 activation.
And yet there also exist numerous published examples where validation of 3C interactions of very long-distance cis and trans interactions by FISH showed adjacent (~200-1000 nm) rather than overlapping signals. These adjacent but non-overlapping signals suggest cytological versus molecular interactions, connected by extensive cross-linking consistent with the origin of 3C interactions from the insoluble rather than soluble template. Such cytological interactions would also explain how genes on different chromosomes that co-localize at nuclear bodies, such as transcription factories, are identified by 3C interactions [53].
Thus there is strong evidence that 3C interactions connecting regulatory elements in trans or over very long-distance in cis arise from cytological rather than molecular interactions. Higher frequency 3C interactions over ~100 kb distances are most simply explained by molecular looping interactions, but this has not yet been clearly established by the high-resolution imaging approaches needed to distinguish molecular interactions from longer-range interactions generated by extensive cross-linking. Interestingly, in the beta-globin example, Lbd1 is also required for the migration of the beta-globin locus from the nuclear periphery to the interior [54]. One can imagine therefore an alternative model in which Lbd1 at both the LCR and promoter recruit the beta-globin locus to a transcription factory or nuclear speckle.
Regardless of the origin of shorter-range, enhancer-promoter interactions, the demonstration that many Mbp-scale 3C interactions arise from cytological rather than molecular interactions, and from the insoluble rather than soluble fraction, supports the idea that TADs may correspond to distinct large-scale chromatin folding motifs. Structural perturbations induced by the 3C protocol might even increase 3C interactions across the TAD, facilitating their detection.
Certainly the rough similarity between the ~1000 number of Drosophila TADs and the 3286 Drosophila polytene band/interband units estimated by electron microscopy is tantalizing [55]. Like TADs, polytene banding patterns are also highly conserved between different tissues [56]. The ~1000 TAD number is almost certainly an underestimate given the still limited Hi-C resolution. Indeed, internal structure and sub-TAD domains exist within larger TAD domains, as revealed more clearly by higher resolution 5C analysis [38,39,41].
Yet this analogy of TADs with polytene banding patterns raises several provocative questions. FISH of polytene chromosomes have generally always shown distinct stripes of target sequences within individual bands and interbands with no evidence for long-range looping interactions; also at best very few highly reproducible, long-range interactions between different chromosome regions were seen in polytene nuclei [57]. What then is the biological meaning of the very long-range (inter-TAD) and trans interactions detected by 3C, whose ligation frequencies are orders of magnitude lower than shorter-range 3C interactions? Most Drosophila larval tissues are polytene and carry out the vast majority of biological functions required of diploid nuclei. Might this argue for limited functional significance in diploid cells for these long-range interactions for most genes?
For example, while 3C long-range interactions connect the alpha-globin locus with distant chromosome sites, comparison of 22 species revealed an ~120kb kb syntenic region sufficient to drive normal developmental expression patterns [58]. Perhaps very long-range interactions rather have evolved to play highly specialized roles for a small number of more complex examples of gene regulation. An extreme example of the latter might be the mono-allelic expression selected from the thousands of olfactory receptor genes in mammals in which clustering of inactive alleles into heterochromatic foci in the nuclear interior may be key to their silencing [59]. A more typical developmental example might be the transient Mbp enhancer-promoter looping at the Shh locus seen in tissues where Shh expression is lower and more transient than in tissues regulated by short-range enhancers [60].
Alternatively, might these inter-TAD and trans interactions in diploid nuclei fine-tune gene regulation for a larger number of genes, either through small changes in gene expression or more robust regulation of expression levels? Here an interesting example might be the fine-tuning of expression of the haploinsufficient genes PTHLH (in cis) and Sox9 (in trans) by a cis-regulatory region 24 Mbp downstream of the PTHLH gene coding for a lncRNA, with loss of this fine-tuning leading to brachydactyly [61].
Most BAC transgenes express within several fold of endogenous genes in mammalian systems [62]. Those that don't would identify gene loci in which long-range interactions are essential. But exploring the basis for the several fold reduction in expression relative to endogenous genes would be one means to explore the fine-tuning role played by very long-range interactions.
Moving forward
Through their identification of topological domains, Hi-C, 5C, and other 3C approaches open new avenues, where none existed previously, for dissecting the molecular basis for long-range genome organization and gene regulation. A major goal for the future will be understanding how this topological organization revealed by 3C methods relates to actual large-scale chromatin ultrastructure directly visualized by high-resolution microscopy. Meanwhile a combination of sequence and gene knockdown manipulations should lead to rapid progress in unraveling the determinants of this level of genome organization and their functional impact on gene regulation.
Acknowledgements
This work was supported by National Institutes of Health grants R01 GM58460 and R01 GM98319 to A.S.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bian Q, Belmont AS. Revisiting higher-order and large-scale chromatin organization. Current opinion in cell biology. 2012;24:359–366. doi: 10.1016/j.ceb.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghirlando R, Felsenfeld G. Chromatin structure outside and inside the nucleus. Biopolymers. 2013;99:225–232. doi: 10.1002/bip.22157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grigoryev SA, Woodcock CL. Chromatin organization - the 30 nm fiber. Experimental cell research. 2012;318:1448–1455. doi: 10.1016/j.yexcr.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 4.Maeshima K, Hihara S, Eltsov M. Chromatin structure: does the 30-nm fibre exist in vivo? Current opinion in cell biology. 2010;22:291–297. doi: 10.1016/j.ceb.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Nishino Y, Eltsov M, Joti Y, Ito K, Takata H, Takahashi Y, Hihara S, Frangakis AS, Imamoto N, Ishikawa T, et al. Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm chromatin structure. The EMBO journal. 2012;31:1644–1653. doi: 10.1038/emboj.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belmont AS, Braunfeld MB, Sedat JW, Agard DA. Large-scale chromatin structural domains within mitotic and interphase chromosomes in vivo and in vitro. Chromosoma. 1989;98:129–143. doi: 10.1007/BF00291049. [DOI] [PubMed] [Google Scholar]
- 7.Bohrmann B, Kellenberger E. Immunostaining of DNA in electron microscopy: an amplification and staining procedure for thin sections as alternative to gold labeling. J. Histochem. Cytochem. 1994;42:635–643. doi: 10.1177/42.5.7512586. [DOI] [PubMed] [Google Scholar]
- 8.Testillano PS, Sanchez-Pina MA, Olmedilla A, Ollacarizqueta MA, Tandler CJ, Risueno MC. A specific ultrastructural method to reveal DNA: the NAMA-Ur. J Histochem Cytochem. 1991;39:1427–1438. doi: 10.1177/39.10.1719069. [DOI] [PubMed] [Google Scholar]
- 9.Olins AL, Moyer BA, Kim SH, Allison DP. Synthesis of a more stable osmium ammine electron-dense DNA stain. J. Histochem. Cytochem. 1989;37:395–398. doi: 10.1177/37.3.2465337. [DOI] [PubMed] [Google Scholar]
- 10.Bazett-Jones DP, Hendzel MJ. Electron spectroscopic imaging of chromatin. Methods. 1999;17:188–200. doi: 10.1006/meth.1998.0729. [DOI] [PubMed] [Google Scholar]
- 11.Kireev I, Lakonishok M, Liu W, Joshi VN, Powell R, Belmont AS. In vivo immunogold labeling confirms large-scale chromatin folding motifs. Nat Methods. 2008;5:311–313. doi: 10.1038/nmeth.1196. [DOI] [PubMed] [Google Scholar]
- 12.Skaer RJ, Whytock S. The fixation of nuclei and chromosomes. J. Cell Sci. 1976;20:221–231. doi: 10.1242/jcs.20.1.221. [DOI] [PubMed] [Google Scholar]
- 13.Fakan S, Bernhard W. Localization of rapidly and slowly labeled nuclear RNA as visualized by high resolution autoradiography. Exp. Cell Res. 1971;67:129–141. doi: 10.1016/0014-4827(71)90628-8. [DOI] [PubMed] [Google Scholar]
- 14.Hu Y, Kireev I, Plutz MJ, Ashourian N, Belmont AS. Large-scale chromatin structure of inducible genes- transcription on a linear template J. Cell Biol. 2009;185:87–100. doi: 10.1083/jcb.200809196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Markaki Y, Gunkel M, Schermelleh L, Beichmanis S, Neumann J, Heidemann M, Leonhardt H, Eick D, Cremer C, Cremer T. Functional nuclear organization of transcription and DNA replication: a topographical marriage between chromatin domains and the interchromatin compartment. Cold Spring Harbor symposia on quantitative biology. 2010;75:475–492. doi: 10.1101/sqb.2010.75.042. [DOI] [PubMed] [Google Scholar]
- 16.Belmont AS, Bruce K. Visualization of G1 chromosomes: a folded, twisted, supercoiled chromonema model of interphase chromatid structure. J Cell Biol. 1994;127:287–302. doi: 10.1083/jcb.127.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kireeva N, Lakonishok M, Kireev I, Hirano T, Belmont AS. Visualization of early chromosome condensation: a hierarchical folding, axial glue model of chromosome structure. J Cell Biol. 2004;166:775–785. doi: 10.1083/jcb.200406049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rego A, Sinclair PB, Tao W, Kireev I, Belmont AS. The facultative heterochromatin of the inactive X chromosome has a distinctive condensed ultrastructure. J. Cell Sci. 2008;121:1119–1127. doi: 10.1242/jcs.026104. [DOI] [PubMed] [Google Scholar]
- 19.Sparvoli E, Levi M, Rossi E. Replicon clusters may form structurally stable complexes of chromatin and chromosomes. Journal of cell science. 1994;107(Pt 11):3097–3103. doi: 10.1242/jcs.107.11.3097. [DOI] [PubMed] [Google Scholar]
- 20.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma H, Samarabandu J, Devdhar RS, Acharya R, Cheng P-c, Meng C, Berezney R. Spatial and Temporal Dynamics of DNA Replication Sites in Mammalian Cells. J. Cell Biol. 1998;143:1415–1425. doi: 10.1083/jcb.143.6.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belmont AS, Hu Y, Sinclair PB, Wu W, Bian Q, Kireev I. Insights into interphase large-scale chromatin structure from analysis of engineered chromosome regions. Cold Spring Harbor symposia on quantitative biology. 2010;75:453–460. doi: 10.1101/sqb.2010.75.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller WG, Walker D, Hager GL, McNally JG. Large-scale chromatin decondensation and recondensation regulated by transcription from a natural promoter. J Cell Biol. 2001;154:33–48. doi: 10.1083/jcb.200011069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tumbar T, Sudlow G, Belmont AS. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J. Cell Bio. 1999;145:1341–1354. doi: 10.1083/jcb.145.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gierman HJ, Indemans MH, Koster J, Goetze S, Seppen J, Geerts D, van Driel R, Versteeg R. Domain-wide regulation of gene expression in the human genome. Genome Res. 2007;17:1286–1295. doi: 10.1101/gr.6276007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christova R, Jones T, Wu PJ, Bolzer A, Costa-Pereira AP, Watling D, Kerr IM, Sheer D. P-STAT1 mediates higher-order chromatin remodelling of the human MHC in response to IFNgamma. J Cell Sci. 2007;120:3262–3270. doi: 10.1242/jcs.012328. [DOI] [PubMed] [Google Scholar]
- 27.Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markaki Y, Smeets D, Fiedler S, Schmid VJ, Schermelleh L, Cremer T, Cremer M. The potential of 3D-FISH and super-resolution structured illumination microscopy for studies of 3D nuclear architecture: 3D structured illumination microscopy of defined chromosomal structures visualized by 3D (immuno)-FISH opens new perspectives for studies of nuclear architecture. BioEssays : news and reviews in molecular, cellular and developmental biology. 2012;34:412–426. doi: 10.1002/bies.201100176. [DOI] [PubMed] [Google Scholar]
- 29.Solovei I, Cavallo A, Schermelleh L, Jaunin F, Scasselati C, Cmarko D, Cremer C, Fakan S, Cremer T. Spatial preservation of nuclear chromatin architecture during three-dimensional fluorescence in situ hybridization (3D-FISH) Exp Cell Res. 2002;276:10–23. doi: 10.1006/excr.2002.5513. [DOI] [PubMed] [Google Scholar]
- 30.Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont AS. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dostie J, Bickmore WA. Chromosome organization in the nucleus - charting new territory across the Hi-Cs. Current opinion in genetics & development. 2012;22:125–131. doi: 10.1016/j.gde.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 32.Tanay A, Cavalli G. Chromosomal domains: epigenetic contexts and functional implications of genomic compartmentalization. Current opinion in genetics & development. 2013;23:197–203. doi: 10.1016/j.gde.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 33.Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nature reviews. Genetics. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes & development. 2012;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanizawa H, Noma K. Unravelling global genome organization by 3C-seq. Seminars in cell & developmental biology. 2012;23:213–221. doi: 10.1016/j.semcdb.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell. 2013;152:1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 37.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- **.3C analysis of Drosophila embryonic nuclei using a modified Hi-C protocol divides genome into ~1000 topological domains. Domain boundaries correlate with boundaries between distinct epigenetic domains and are enriched in binding of multiple insulator proteins and active chromatin marks. Active domains interact more frequently with other active domains and show increased interchromosomal contacts relative to inactive domains.
- 38.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **.Hi-C analysis of genome interactions of mammalian cells in different differentiated states reveals Mbp size topological domains in which DNA sequences show increased interactions with other intra-domain sequences. These domains are conserved between mouse and human and largely invariant during differentiation. Domain boundaries are enriched in CTCF, active chromatin marks, and housekeeping genes, and appear to demarcate domains with distinct epigenetic marks.
- 39.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **.5C analysis of 4.5 Mbp region encompassing the mouse X-inactivation center reveals 9 topologically associating domains (TADs) 200 kb to 1 Mbp in size which remain constant through cell differentiation. TADs correspond to blocks of chromatin with distinct histone marks, but TAD organization does not require histone modifications. Expression of genes within individual TADs show increased correlation during cell differentiation.
- 40.Hou C, Li L, Qin ZS, Corces VG. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Molecular cell. 2012;48:471–484. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **.Hi-C analysis defines ~1000 topological domains in Drosophila Kc tissue culture cells similar to those identified in in previous study of Drosophila embryonic nuclei. Domain boundaries are enriched in active chromatin and gene dense regions as well as binding of multiple insulator proteins. Domain boundaries are hotspots for transposon insertion, suggesting DNA is more accessible, and reporter genes inserted near these boundaries are less likely to show variegated expression.
- 41.Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **.HIgh resolution 5C maps across seven genomic loci define additional organization, or subTADS, within TADs previously defined by lower resolution Hi-C methods. CTCF and cohesin were observed to anchor constituitive long-range interactions while Mediator and cohesin appeared to bridge shorter-range enhancer-promoter interactions.
- 42.Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yaffe E, Tanay A. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nature genetics. 2011;43:1059–1065. doi: 10.1038/ng.947. [DOI] [PubMed] [Google Scholar]
- *.Protocol for renormalizing Hi-C data is described to correct for biases created by factors including restriction fragment length, GC content, and sequence uniqueness. Application of this correction to Hi-C data sets reveals clustering of genomic regions bearing active chromatin marks and DNase I HS sites.
- 45.Imakaev M, Fudenberg G, McCord RP, Naumova N, Goloborodko A, Lajoie BR, Dekker J, Mirny LA. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nature methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.An iterative protocol is described for nomalizing Hi-C data to correct for biases including "visibility" of genomic regions for ligation events and mapping and sequencing. Algorithm assumes all genomic regions should have same visibility and corrects for known and unknown biases using this assumption. The method, however, can not correct for biases within a given bin size used for analysis of the Hi-C data.
- 46.Hakim O, Sung MH, Voss TC, Splinter E, John S, Sabo PJ, Thurman RE, Stamatoyannopoulos JA, de Laat W, Hager GL. Diverse gene reprogramming events occur in the same spatial clusters of distal regulatory elements. Genome research. 2011;21:697–706. doi: 10.1101/gr.111153.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.4C analysis reveals that genomic regions contacting glucocorticoid receptor (GR)-responsive gene loci are not enriched for GR responsive genes or any other particular functional group of genes but are highly enriched for regions containing DNase I hypersensitivity sites.
- 47.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome research. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simon JM, Giresi PG, Davis IJ, Lieb JD. Ausubel Frederick M., et al., editors. A detailed protocol for formaldehyde-assisted isolation of regulatory elements (FAIRE) Current protocols in molecular biology. 2013 doi: 10.1002/0471142727.mb2126s102. Chapter 21:Unit21 26. [DOI] [PubMed] [Google Scholar]
- 49.Grigoryev SA, Arya G, Correll S, Woodcock CL, Schlick T. Evidence for heteromorphic chromatin fibers from analysis of nucleosome interactions. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13317–13322. doi: 10.1073/pnas.0903280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gavrilov AA, Gushchanskaya ES, Strelkova O, Zhironkina O, Kireev II, Iarovaia OV, Razin SV. Disclosure of a structural milieu for the proximity ligation reveals the elusive nature of an active chromatin hub. Nucleic acids research. 2013;41:3563–3575. doi: 10.1093/nar/gkt067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **.This study demonstrates that the predominant source of 3C interactions comes from the insoluble sample fraction and shows that this insoluble fraction corresponds to swollen nuclei that retain chromatin territory organization and nuclear body architecture. These results challenge the standard explanation of 3C interactions arising from molecular interactions directly bridging two different DNA regions and instead suggest the possibility that 3C interactions can result from ligation of two DNA fragments separated by an extensive network of cross-linked DNA and protein.
- 51.Krivega I, Dean A. Enhancer and promoter interactions-long distance calls. Current opinion in genetics & development. 2012;22:79–85. doi: 10.1016/j.gde.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149:1233–1244. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **.An elegant approach uses a zinc finger - Ldb1 fusion protein directed to the beta-globin promoter region to restore an apparent looping interaction between the LCR and the promoter which is normally dependent on the GATA1 transcription factor. The authors propose that dimerization or multimerization of Ldb1 bridges the LCR and promoter region resulting in significant transcriptional activation even in the absence of GATA1.
- 53.Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nature genetics. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song SH, Kim A, Ragoczy T, Bender MA, Groudine M, Dean A. Multiple functions of Ldb1 required for beta-globin activation during erythroid differentiation. Blood. 2010;116:2356–2364. doi: 10.1182/blood-2010-03-272252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sorsa V. Chromosome Maps of Drosophila. CRC Press; 1988. [Google Scholar]
- 56.Hochstrasser M. Chromosome structure in four wild-type polytene tissues of Drosophila melanogaster. The 87A and 87C heat shock loci are induced unequally in the midgut in a manner dependent on growth temperature. Chromosoma. 1987;95:197–208. doi: 10.1007/BF00330351. [DOI] [PubMed] [Google Scholar]
- 57.Hochstrasser M, Sedat JW. Three-dimensional organization of Drosophila melanogaster interphase nuclei. II. Chromosome spatial organization and gene regulation. The Journal of cell biology. 1987;104:1471–1483. doi: 10.1083/jcb.104.6.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wallace HA, Marques-Kranc F, Richardson M, Luna-Crespo F, Sharpe JA, Hughes J, Wood WG, Higgs DR, Smith AJ. Manipulating the mouse genome to engineer precise functional syntenic replacements with human sequence. Cell. 2007;128:197–209. doi: 10.1016/j.cell.2006.11.044. [DOI] [PubMed] [Google Scholar]
- 59.Clowney EJ, LeGros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou EC, Myllys M, Barnea G, Larabell CA, Lomvardas S. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151:724–737. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **.Monoallelic expression of a single olfactory receptor (OR) gene is linked to clustering of the thousands of inactive OR genes in a small number of heterochromatin foci. This clustering is facilitated by downregulation of lamin B receptor expression and upregulation of lamin B receptor expression is sufficient to block OR gene clustering and disrupt monoallelic expression.
- 60.Amano T, Sagai T, Tanabe H, Mizushina Y, Nakazawa H, Shiroishi T. Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev Cell. 2009;16:47–57. doi: 10.1016/j.devcel.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 61.Maass PG, Rump A, Schulz H, Stricker S, Schulze L, Platzer K, Aydin A, Tinschert S, Goldring MB, Luft FC, et al. A misplaced lncRNA causes brachydactyly in humans. The Journal of clinical investigation. 2012;122:3990–4002. doi: 10.1172/JCI65508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Chromosome translocation causing brachydactyly leads to discovery by 3C of a long-range regulatory element interacting in cis across 24 Mbp with the PTHLH gene and in trans with the SOX9 gene. This regulatory element contains a lncRNA which was upregulated by the chromosome translocation and at the same time showed reduced binding to the PTHLH gene.
- 62.Heintz N. Analysis of mammalian central nervous system gene expression and function using bacterial artificial chromosome-mediated transgenesis. Human molecular genetics. 2000;9:937–943. doi: 10.1093/hmg/9.6.937. [DOI] [PubMed] [Google Scholar]