Abstract
The rugged protein sequence–function landscape complicates efforts, both in nature and in the laboratory, to evolve protein function. Protein library diversification must strike a balance between sufficient variegation to thoroughly sample alternative functionality versus the probability of mutant destabilization below an expressible threshold. In this work, we explore the sequence–function landscape in the context of screening for molecular recognition from an Ig scaffold library. The fibronectin type III domain is used to explore the impact of two sequence diversification strategies: (a) partial wild-type conservation at structurally important positions within the paratope region and (b) tailored amino acid composition mimicking antibody binding-site composition at putative paratope positions. Structurally important positions within the paratope region were identified through stability, structural, and phylogenetic analyses and partially or fully conserved in sequence. To achieve tailored antibody-like diversity, we designed a set of skewed nucleotide mixtures yielding codons approximately matching the distribution observed in antibody complementarity-determining regions without incurring the expense of triphosphoramidite-based construction. These design elements were explored via comparison of three library designs: a random library, a library with wild-type bias in the DE loop only and tyrosine–serine diversity elsewhere, and a library with wild-type bias at 11 positions and the antibody-inspired amino acid distribution. Using pooled libraries for direct competition in a single tube, selection and maturation of binders to seven targets yielded 19 of 21 clones that originated from the structurally biased, tailored-diversity library design. Sequence analysis of the selected clones supports the importance of both tailored compositional diversity and structural bias. In addition, selection of both well and poorly expressed clones from two libraries further elucidated the impact of structural bias.
Keywords: fibronectin type III domain (Fn3), protein engineering, synthetic library, molecular recognition
Introduction
The design and construction of synthetic combinatorial libraries are critical for the development of alternative scaffolds for molecular recognition1 as well as high-throughput approaches to antibody engineering such as those required for proteomic applications.2 The immensity of protein sequence space and the limited capacity of laboratory selection methods necessitates efficient library design in which the diversities at each position combine to yield a population of clones that maintain structural integrity while imparting a wide array of binding specificities. Study of library design and construction enable more efficient selection of high-affinity binders from a variety of scaffolds.
A particularly effective alternative scaffold is the tenth type III domain of human fibronectin (Fn3).3,4 Fn3 is a small (10 kDa), stable β-sandwich devoid of cysteines that can be readily produced in bacteria, thereby providing numerous advantages over antibodies and other scaffolds that lack these attributes. The BC, DE, and FG loops of Fn3, which are structurally analogous to the complementarity-determining regions (CDRs) of antibodies, have proven to be an effective region to diversify for the generation of molecular recognition domains. We sought to develop an improved Fn3 library design through incorporation of two key features: wild-type conservation of residues that are structurally critical and/or are less likely to contribute to the desired binding interaction, and tailored amino acid diversity biased to functional amino acids.
Despite their location in the BC–DE–FG loop region of Fn3, some residues may be critical to the conformational stability of the protein fold. Thus, conservation at structurally critical positions may (a) increase the quantity of potentially functional clones by reducing the frequency of unfolded or highly unstable clones and (b) increase the quality of functional clones by enabling diversity to be focused where it is more likely to contribute to the binding interaction, yielding a more efficient search of sequence space. Moreover, conservation of such critical positions may produce a library population with higher average stability. Stabilization increases the robustness of binders in biotechnology applications such as the stringent washing steps of purification and detection. Stability can impede degradation and aggregation of in vivo diagnostics and therapeutics, thereby maintaining potency and aiding in the prevention of an immune response. Also, stabilization enhances the tolerance to mutation, which increases the capacity for evolution.5 Lastly, enthalpic stabilization may reduce excessive paratope flexibility, which could otherwise diminish the favorable free-energy change upon binding due to a higher entropic cost upon complex formation. Here, we use stability, structural, and sequence analyses to identify conserved sites in Fn3 that benefit library design.
Early library designs commonly used NNB or NNS/NNK randomized codons to approximate an equal distribution of all amino acids.6 Yet not all amino acids are equivalent in their ability to provide physicochemical complementarity for molecular recognition, and so a tailored amino acid composition may be more effective. Sidhu and colleagues have investigated this hypothesis and demonstrated the utility of a tyrosine/serine library as well as the unique efficacy of tyrosine to mediate molecular recognition in antibody fragments.7–9 A tailored antibody library with elevated tyrosine, glycine, and serine and low levels of all other amino acids except cysteine was superior to a tyrosine/serine library in the isolation of binders to human vascular endothelial growth factor.10 A 40% Y, 20% S, 10% G, and 5% each A, D, H, L, N, and R library was used with the Fn3 scaffold to yield a 6 nM binder to maltose-binding protein11 and a novel “affinity clamp” for peptide recognition,12 although the effectiveness of this library was not directly compared to alternate designs. In a comparison of single clones, this maltose-binding protein binder exhibits 5.3±1.3-fold higher affinity than the top tyrosine/serine clone, and structural comparison to a similar tyrosine/serine clone reveals the benefit of conformational flexibility achieved through expanded diversity.11 Direct competition of full diversity and tyrosine/serine diversity libraries in the Fn3 domain was found to be dominated by a full diversity library for selection of high-affinity binders to goat and rabbit immunoglobulin G.13 Thus, although tyrosine/serine may provide ample diversity for binding, an expanded repertoire enables higher complementarity. The expanded repertoire can be effectively utilized with an efficient library design and/or affinity maturation scheme. The aforementioned biased distributions were created by oligonucleotide synthesis with custom trimer phosphoramidite mixtures.14 The current study investigates the ability to create a desired distribution via inexpensive skewed nucleotide mixtures. In particular, the amino acid distribution in human and mouse CDR-H3 loops is effectively mimicked. We demonstrate, using selection to seven targets, that a new library incorporating selective conservation and tailored diversity is superior to both an unbiased library with approximately equal amino acid diversity and a tyrosine/serine binary code library. This library enabled the generation of binders to a multitude of targets with potential applied utility.
Results
Fn3 surface display and stability
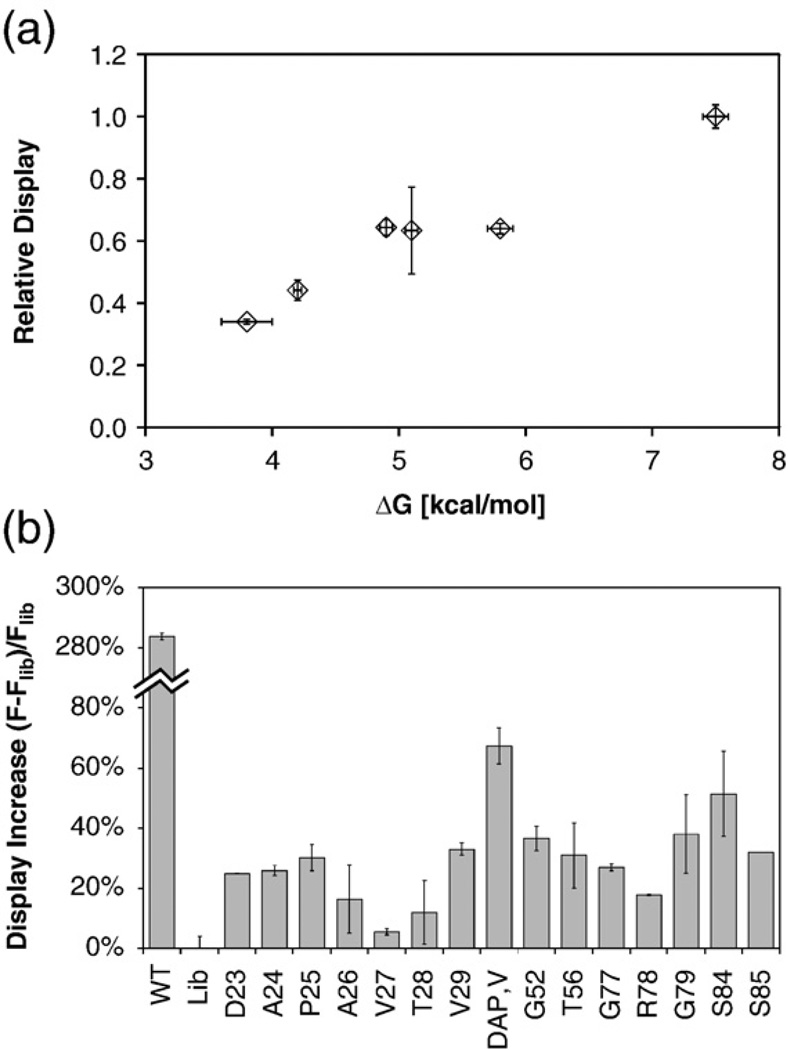
We used yeast surface display for efficient stability analysis of Fn3 clones. Although multiple factors, including stability, solubility, and gene expression, can impact protein expression, it has been demonstrated that the number of displayed single-chain T-cell receptors15 per yeast cell and the yield of yeast-secreted bovine pancreatic trypsin inhibitor16,17 correlate with protein stability. To validate the display–stability correlation for Fn3, we created yeast surface display vectors of binders to vascular endothelial growth factor receptor 2 spanning a range of stabilities: free energies of unfolding from 3.8 to 7.5 kcal/mol and midpoints of thermal denaturation of 42 to 84 °C.18 Clonal cultures of yeast were grown at 30 °C, Fn3 expression was induced at 37 °C, and surface expression of Fn3 was quantified by flow cytometry (Supplementary Fig. 1a). The clones exhibit a positive correlation between display and stability spanning a substantial display range between the least and most stable clones (Fig. 1a), thereby validating this technique for stability comparison.
Fig. 1.
Yeast surface display of Fn3 clones and analytical libraries. Yeast clones or libraries were grown to logarithmic growth phase at 30 °C. Expression of Aga2p–Fn3 was induced at 37 °C. Fn3 display level was quantified by flow cytometry using mouse anti-c-myc antibody and anti-mouse antibody–R-phycoerythrin conjugate. (a) The extent of Fn3 display of VEGF-R2 binders was measured, normalized to wild-type Fn3, and compared to the previously determined free energy of unfolding.18 (b) Libraries were created with full diversity in the BC, DE, and FG loops except maintenance of wild type at the position indicated. The fractional improvement in display was calculated as the mean phycoerythrin fluorescence of the singly conserved library minus that of the fully diversified library and normalized to the fully diversified fluorescence.
This validated approach was used to explore domain stabilization via single-site wild-type conservation in the context of a diverse library. To quantify this impact, we constructed a series of libraries: one library with fully diversified BC, DE, and FG loops and multiple libraries of this same design except for wild-type conservation at a single position of interest. The libraries were transformed into a yeast surface display system and the amount of Fn3 displayed upon induction at 37 °C was quantified by flow cytometry. Eleven of 14 positions studied, as well as a multisite library, exhibit improved display with wild-type conservation. Conservation of amino acids A26, V27, and T28 increased display but not to a statistically significant degree (Fig. 1b).
Solvent-accessible surface area
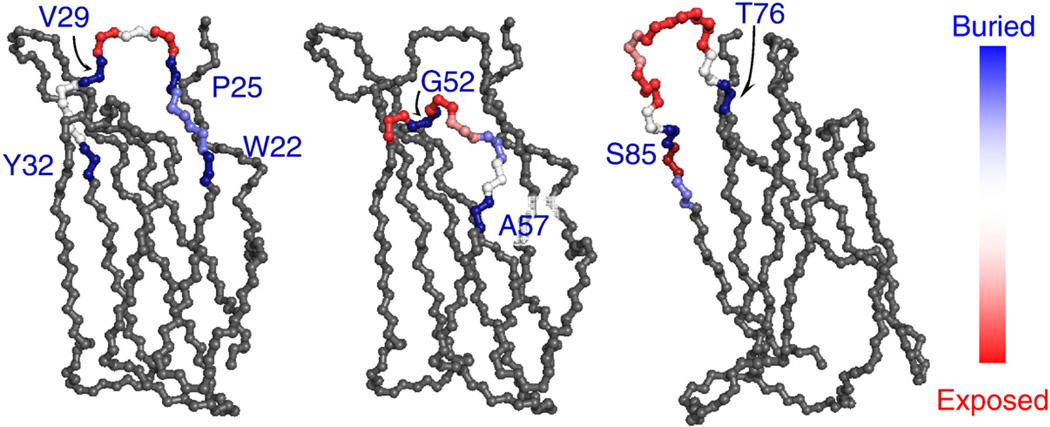
The solvent-accessible surface area (SASA) of each candidate diversified position was calculated with GetArea19 for wild-type Fn3 (solution structure 1TTG20 and crystal structure 1FNA21) and an engineered binder (2OBG22). Despite their presence in previously diversified loop regions, the side chains of D23, A24, P25, V29, G52, and S85 are relatively inaccessible; peripheral residues W22, Y32, A57, T76, and P87 are also buried (Fig. 2). Conversely, the amino acids in the middle of each loop are relatively exposed, supporting the ability of these sites to be diversified while maintaining the correct fold. This information was combined with the phylogenetic analysis described below and surface expression studies described above to identify sites for diversification.
Fig. 2.
Side-chain solvent accessibility. The SASA was calculated for each residue using GetArea19 with a 1.4 Å probe. The SASA of each side chain in the solution (1TTG20) and crystal (1FNA21) structures of wild-type Fn3 and an engineered binder (2OBG22) were normalized by the SASA of the side chain in a random coiled peptide. The fibronectin domain is presented using 1TTG with the residues in the loop regions (W22–Y32, P51–A57, T76–P87) color-coded according to the mean value of the accessibility ratios for the three cases.
Fn3 phylogenetic analysis
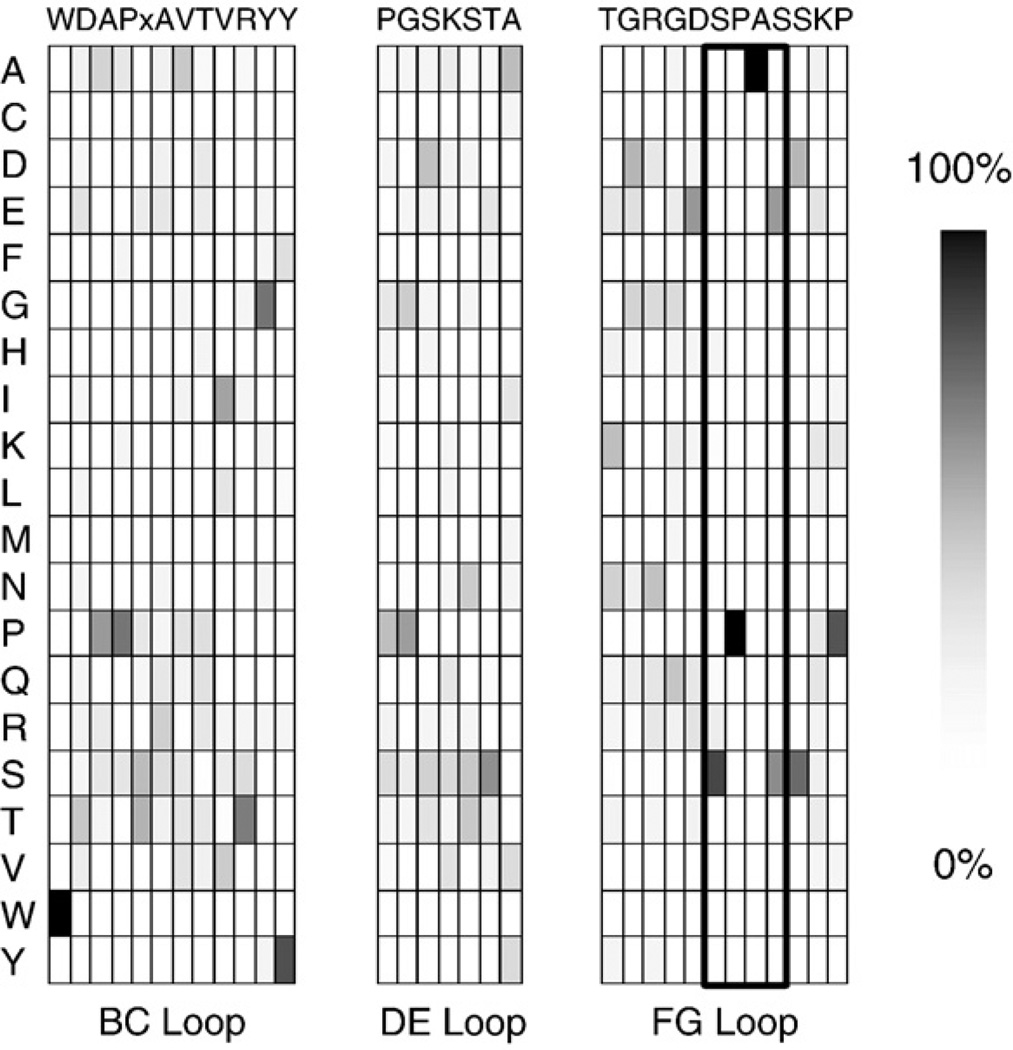
The mutational flexibility of each position was further explored through phylogenetic sequence analysis. The type III domains of fibronectin in chimpanzee, cow, dog, horse, human, mouse, opossum, platypus, rat, and rhesus monkey were aligned, and the relative frequency of each amino acid was determined (Fig. 3). The peripheral residues W22, Y32, P51, A57, and P87 are well conserved; however, T76 is variable. Other sites exhibiting conservation threefold above random are A24 (22%), P25 (62%), V29 (25% as well as 43% isoleucine), G52 (25%), S53 (23%), S55 (27%), G77 (21%), G79 (19%), and S85 (66%); also note that T56 is 12% conserved with 51% of the homolog serine. Thus, the BC loop exhibits conservation of its peripheral hydrophobic residues except Y31. The DE loop, except for the central lysine, is well-conserved. The FG loop has a trend toward glycine from G77 to G79 and two highly conserved sites near the C-terminus.
Fig. 3.
Phylogenetic sequence analysis. The amino acid sequences for the type III domains of fibronectin in chimpanzee, cow, dog, horse, human, mouse, opossum, platypus, rat, and rhesus monkey were aligned. The amino acid frequency at each position is presented in an intensity scale in each column. The wild-type human sequence for the tenth type III domain is presented at the top of each column (W22–Y32, P51–A57, T76–P87). The x in the BC loop corresponds to an amino acid present in other domains that is not present in the human tenth type III domain. The outline around S81–S84 represents rare positions because most type III domains contain shorter FG loops.
Published sequences of engineered binders were analyzed similarly, although in this analysis amino acid frequencies must be compared to expected frequencies on the basis of variable library designs (Supplementary Fig. 2). Wild type is present at least twice as often in binders as in the naïve library at three positions: P25 (15% in binders versus 5% in libraries), G52 (26% versus 13%), and G79 (17% versus 5%). In addition, three positions yield substantial enrichment of homologs: alanine at V29 (20% versus 6%), threonine at S55 (25% versus 6%), and serine at T56 (28% versus 11%).
Library design
Stability, accessibility, and sequence analyses (summarized in Table 1) were used to determine the degree of diversification desired at each position. For example, proline at position 25 significantly stabilizes the library, is essentially inaccessible to solvent, and is highly conserved in the type III fibronectin domains of mammals. Thus, the new library was heavily biased toward proline at this position. Conversely, the adjacent alanine at position 26 does not significantly stabilize the library, is highly accessible, and exhibits essentially no conservation. As a result, this position was fully diversified in the new library design.
Table 1.
Fn3 library design summary
| Sequences |
||||||
|---|---|---|---|---|---|---|
| Pos. | WT | Access. | Stability | Native | Binders | Library design |
| BC loop | ||||||
| 22 | W | 1 | — | 100% W | — | Wild type |
| 23 | D | 35 | 25±0% | 5% D+14% E | 1.0× | Ab div. (10% D) |
| 24 | A | 32 | 26±2% | 22% A+12% S | 0.4× | 8% A+Ab div. |
| 25 | P | 10 | 30±4% | 62% P | 2.7× | 42% P+Ab div. |
| 26 | A | 75 | 16±11% | 7% A | 0.7× | Ab div.* |
| 27 | V | 57 | 5±1% | 14% V | 0.4× | Ab div.* |
| 28 | T | 75 | 12±11% | 12% T | 1.6× | Ab div.* |
| 29 | V | 3 | 33±2% | 25% V+56% I+L | 0.6× (3.2×A) | 25% A|L|S|V |
| 30 | R | 49 | — | 6% R | 1.1× | Ab div. |
| 31 | Y | 43 | — | 6% Y, 62% G | — | 50% S, 50% Y |
| 32 | Y | 1 | — | 75% Y | — | Wild type |
| DE loop | ||||||
| 51 | P | 79 | — | 31% P | — | Wild type |
| 52 | G | 12 | 37±4% | 25% G | 2.0× | 49% G |
| 53 | S | 83 | — | 23% S+14% T | 1.0× | 50% S |
| 54 | K | 64 | — | 6% K | 3.6× | NNB div.* |
| 55 | S | 41 | — | 27% S+26% T | 1.6× (4.2×T) | 50% S |
| 56 | T | 48 | 31±11% | 12% T+51% S | 0.8× (2.6×S) | 49% T |
| 57 | A | 8 | — | 32% A | — | Wild type |
| FG loop | ||||||
| 76 | T | 8 | — | 7% T | — | Wild type |
| 77 | G | 48 | 27±1% | 21%G | 0.7× | 12%G+Ab div |
| 78 | R | 81 | 18±0% | 12% R | 1.5× | Ab div. |
| 79 | G | 77 | 38±13% | 19% G | 2.7× | 12% G+Ab div. |
| 80 | D | 74 | — | 7% D+48% E | 1.9× | Ab div.* |
| 81 | S | 69 | — | Rare | 0.7× | Ab div.* |
| 82 | P | 76 | — | Rare | 1.5× | Ab div.* |
| 83 | A | 81 | — | Rare | 0.6× | Ab div.* |
| 84 | S | 54 | 52±14% | Rare | 0.5× | Ab div.* |
| 85 | S | 14 | 32±0% | 66% S | 1.2× | 100% S |
| 86 | K | 88 | — | 12% K | 1.5× | Ab div. |
| 87 | P | 40 | — | 74% P | — | Wild type |
“Pos.” and “WT” are the amino acid position and residue in the human wild-type 10th type III domain, respectively. “Access.” is the ratio of SASA for the residue in the fibronectin domain compared to the residue in a random coiled peptide. “Stability” is the relative increase in yeast surface display level of a library with wild-type conservation at the position of interest. “Native” indicates the frequencies of the indicated amino acids in type III fibronectin domains of 10 species. “Binders” indicates the enrichment of wild-type (or homolog as indicated) in engineered binders relative to the naïve frequency. “Library design” indicates the intended amino acid distribution in the new library. “Ab div.” is the designed amino acid distribution that mimics antibody CDR-H3.
An asterisk indicates the location of loop length variability.
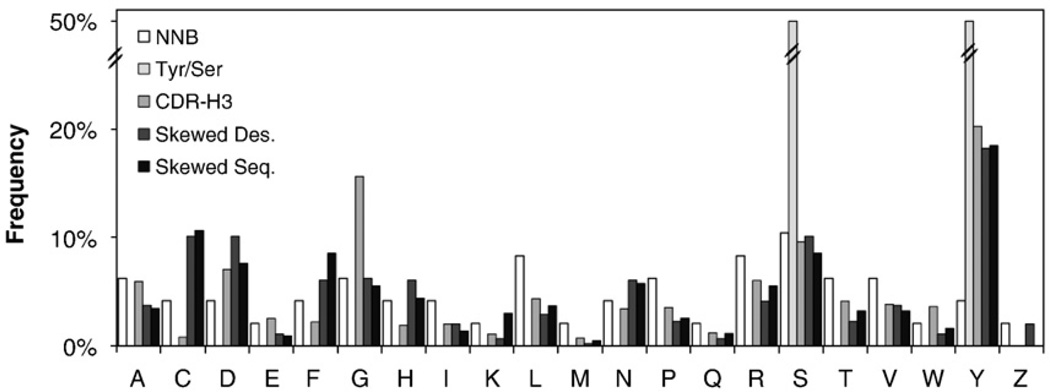
Along with conservation bias to maintain structural integrity and focus diversity on positions better suited for molecular recognition, it was desired to bias the diversity to amino acids of potential functional significance in molecular recognition. Tyrosine has demonstrated unique utility in molecular recognition.7–9 Glycine provides conformational flexibility. Serine and alanine are valuable as small, neutral side chains. Acidic residues, arginine, and lysine provide charge, although recognition utility is unclear.23 Other side chains may provide ideal complementarity in less frequent situations. Thus, it would appear that the ideal diversity contains high levels of tyrosine, glycine, and serine and/or alanine as well as small levels of all other amino acids. For the particular amino acid distribution, we sought guidance from natural molecular recognition. The amino acid distribution in CDR-H3 matches the desired diversity and was used as the library design model (Fig. 4). Each position was designed to incorporate the desired level of wild-type conservation and to match the antibody CDR-H3 repertoire in the nonconserved portion of the distribution. The DE loop is a slight exception because a very similar design was previously validated as effective.13 In this loop, G52, S53, S55, and T56 are highly conserved with wild type at 50% frequency and unbiased distribution of all other amino acids. The lack of antibody-inspired bias in this loop is of limited detriment because of the high degree of conservation of the wild-type amino acids. Multiple loop lengths, selected on the basis of phylogenetic occurrence,25 are included in each loop. The resultant library design is summarized in Table 1.
Fig. 4.
Amino acid distributions. The frequencies of each amino acid in multiple distributions are presented. NNB refers to a degenerate codon with 25% of each nucleotide at the first two positions and 33% of C, T, and G at the third position. Tyr/Ser refers to an even mix of tyrosine and serine. CDR-H3 refers to the expressed human and mouse CDR-H3 sequences.24 Skewed Des. Skewed Seq. refers to the distribution refers to the theoretical distribution attainable using skewed oligonucleotides. attained experimentally using skewed nucleotides.
Library construction
Although trimer phosphoramidite library construction enables precise creation of unique amino acid distributions, this approach is expensive with the inclusion of multiple specialty codon mixtures. As an inexpensive alternative, we employed standard oligonucleotide synthesis using custom mixtures of skewed nucleotides at each position. The optimal set of three nucleotide mixtures was determined for each codon as follows. All possible sets of nucleotide mixtures with each component at 5% increments were filtered to select only those that closely match the desired levels of wild type and tyrosine and reasonably match glycine, serine, aspartic acid, alanine, and arginine; these amino acids are the most frequent in antibody CDR-H324 and are functionally diverse. Sample protein libraries were then produced in silico from the amino acid probability distributions resulting from the sets of nucleotide mixtures. The library calculated to be most likely to be produced from the intended distribution (i.e., the antibody repertoire with the appropriate wild-type bias) was selected as optimal. This process was repeated for each position in the library. In general, these skewed nucleotide mixtures provide good matches to the desired amino acid distribution (Fig. 4). The two exceptions are decreased levels of glycine and elevated cysteine. Since the latter two positions in a cysteine codon (TGT or TGC) are shared by glycine (GGN), it is not possible to create high levels of glycine without also yielding high levels of cysteine unless TNN codons are depleted, which depletes tyrosine. Thus, a compromise is reached with 6% glycine and 10% cysteine. Although this incorporates a relatively high level of cysteine, library design still yields many cysteine-free clones; moreover, interloop disulfide bonds are a potentially advantageous element.26
Fn3 genes were constructed by overlap extension PCR of partially degenerate oligonucleotides. Transformation into yeast by electroporation with homologous recombination yielded 2.5×108 transformants. Sequencing and flow cytometry analysis indicate 60% of clones encode for full-length Fn3, resulting in 1.5×108 Fn3 clones. Sequence analysis reveals that the skewed nucleotides accurately match their intended distribution (Fig. 4). The library is termed G4, as it is the fourth generation Fn3 library created in our laboratory after the two-loop, single-length BF14 library,26 the three-loop, length-diversified NNB library,25 and the three-loop, DE-conserved tyrosine/serine library YS.13
Library comparison
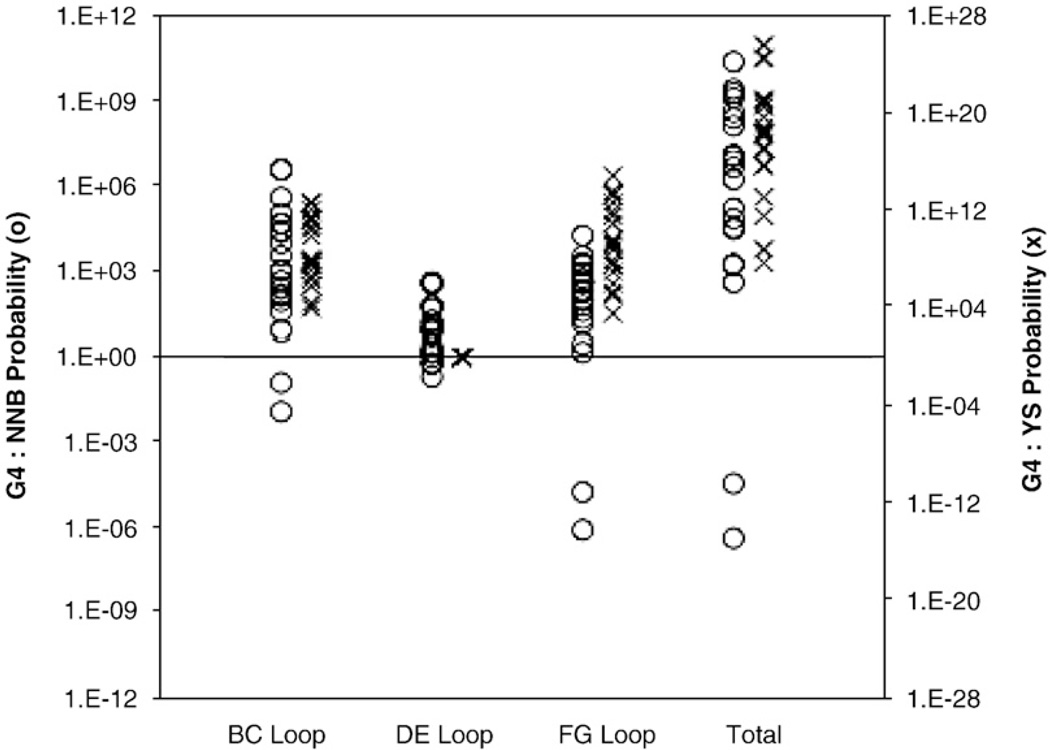
The new G4 library design was compared to a nonconserved, full-diversity library (NNB)25 and a library with wild-type conservation in the DE loop only and tyrosine/serine diversity (YS)13 (Table 2). The libraries were pooled for comparison and tested for their ability to generate binders to seven targets: human A33, mouse A33, epidermal growth factor receptor (EGFR), Fcγ receptors IIA and IIIA (FcγRIIA and FcγRIIIA), mouse immunoglobulin G (mIgG), and human serum albumin (HSA). The naïve library was sorted by magnetic bead selections,27 and lead clones were diversified by error-prone PCR on the full Fn3 gene and shuffling of mutagenized Fn3 loops.25 Multiple rounds of diversification and selection, by magnetic beads and ultimately flow cytometry (Supplementary Fig. 1b), were performed to yield binders to each target, as described previously.25 Sequence analysis of each binding population revealed that 19 of 21 binders originated from the G4 library, while two clones were likely of NNB origin and no YS clones were identified (Table 3 and Fig. 5). Given the comparable number of clones in the naïve libraries, this result indicates that G4 is a library design superior to both NNB and YS for the selection of protein binders. In other words, the abundance of selectable sequences with the desired functionality is significantly higher in the G4 population, by direct experimental comparison to the NNB and YS populations.
Table 2.
Library design
| Library | Loop diversity | Biased positions | Full-length Fn3s |
|---|---|---|---|
| NNB | Full diversity (NNB codons) | None | 0.7 ×108 |
| YS | 50% Y, 50% S | 52, 53, 55, 56 | 1.5 ×108 |
| G4 | Antibody based (18% Y, 10% S, …) | 23, 24, 25, 29, 31, 52, 53, 55, 56, 77, 79, 85 | 1.5 ×108 |
“Loop diversity” indicates the library of codons included at positions without wild-type bias. “Biased positions” indicates positions within the diversified loops (23–31, 52–56, 77–86) that are biased toward wild-type. “Full-length Fn3s” indicates the library size, that is, the number of yeast transformants that encode for full-length Fn3 domains.
Table 3.
Engineered binder sequences
| Name | Target | 23 | 24 | 25 | 26 | 27 | 28 | — | 29 | 30 | 31 | 52 | 53 | 54 | 55 | 56 | 77 | 78 | 79 | 80 | 81 | 82 | 83 | 84 | 85 | 86 | Framework | Kd (nM) | Tm (°C) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | — | D | A | P | A | V | T | — | V | R | Y | G | S | K | S | T | G | R | G | D | S | P | A | S | S | K | — | — | 85.7 |
| E4.2.1 | EGFR | Y | G | F | S | L | — | — | A | S | S | R | S | P | W | F | S | N | D | F | S | N | R | Y | S | G | — | 30±3 | 56.2 |
| E6.2.6 | EGFR | F | D | Y | A | — | — | — | V | T | Y | G | W | I | S | T | D | N | S | H | W | P | F | R | S | T | I90T | 0.26±0.13 | 65.7 |
| E6.2.10 | EGFR | Y | L | R | D | P | R | Y | V | D | Y | W | Y | L | P | E | Y | D | G | Y | R | E | S | T | P | L | — | 0.96±0.11 | — |
| EI1.4.1 | EGFR | Y | G | P | F | Y | Y | V | A | H | S | R | S | P | W | F | S | K | C | Y | D | G | — | — | S | V | — | 0.85±0.50 | 52.8 |
| EI2.4.6 | EGFR | Y | H | P | F | Y | Y | V | A | H | S | R | S | P | W | F | D | S | N | G | — | — | — | — | S | H | — | 2.9±0.3 | 63.9 |
| EI3.4.2 | EGFR | Y | G | S | S | Y | — | — | A | S | Y | R | S | P | W | F | P | S | G | I | — | — | — | — | S | A | T58I | 9.5±3.5 | — |
| EI3.4.3 | EGFR | L | H | H | R | S | D | — | V | R | S | G | S | R | S | L | W | G | S | Y | C | C | S | N | E47K | 0.25±0.05 | 72.8 | ||
| EI4.4.2 | EGFR | Y | F | R | D | P | R | Y | V | D | Y | W | Y | L | P | E | G | D | D | Q | N | A | — | — | G | L | V45A | 1.4±0.2 | 70.2 |
| IIa8.2.6 | Fcγlla | C | T | H | L | H | — | — | W | D | Y | A | L | C | P | G | V | G | G | D | — | — | — | — | D | W | R6G, T35F, V72A, I88S, K98E | 850 | — |
| IIIa6.2.6 | FcγlIIa | D | M | P | F | — | — | — | S | D | S | G | T | D | S | L | S | S | G | S | N | — | — | — | S | Y | A12V, S21N, T35A | 530 | — |
| hA2.2.1 | hA33 | Y | C | P | D | G | C | H | S | Y | Y | R | S | I | S | S | F | R | W | P | — | — | — | — | S | F | — | — | — |
| hA2.2.2 | hA33 | N | T | Y | F | S | F | — | L | Y | Y | S | S | L | H | T | G | T | W | P | — | — | — | — | S | Y | — | — | — |
| hA3.2.1 | hA33 | S | Y | S | S | Y | N | S | W | D | S | N | S | D | C | I | R | D | C | D | F | Y | — | — | S | Y | Y32F | — | — |
| hA3.2.2 | hA33 | Y | Y | H | L | R | G | — | L | D | S | R | S | Y | S | T | V | N | D | Y | I | — | — | — | S | Y | S21G Q46K T49A | — | — |
| mA3.2.1 | mA33 | S | S | S | L | Y | N | — | S | A | Y | V | W | D | C | T | P | N | Y | S | F | — | — | — | S | L | Y32F | — | — |
| mA3.2.2 | mA33 | C | C | L | F | F | — | — | S | G | Y | G | L | V | Y | W | D | N | V | G | — | — | — | — | S | N | I90V | — | — |
| mA3.2.3 | mA33 | S | F | P | C | V | — | — | S | S | S | G | D | T | T | S | S | T | C | Y | P | — | — | — | S | Y | — | — | — |
| mA3.2.4 | mA33 | S | C | P | I | C | P | R | A | T | S | A | T | — | S | S | D | Q | G | Y | D | D | — | — | S | A | I34V | — | — |
| mA3.2.5 | mA33 | Q | C | H | Y | Y | Y | — | A | Q | S | S | S | K | S | T | Y | N | W | F | L | D | S | V | S | I | A12V | — | — |
| Alb3.2.1 | hAlb | G | A | P | A | C | — | — | A | A | Y | G | S | G | T | S | S | R | Y | Y | Y | C | — | — | S | E | — | — | — |
| mI2.2.1 | mlgG | C | C | S | D | N | C | — | S | N | S | R | S | C | F | M | D | S | N | G | — | — | — | — | P | H | V72A | 4.1±0.7 | — |
“Name” is the name of each clone. “Target” is the cognate protein bound by the Fn3 clone. 23–31,52–56, and 77–86 refer to the amino acid positions diversified in the naive library. “Framework” refers to amino acid mutations outside of the diversified loops. A dash indicates no amino acid. Underline indicates wild-type amino acid.
Fig. 5.
Library source probability. For each binding clone sequence, the probability of origination from each library was calculated on the basis of library design. The relative preferences for G4 versus NNB (○) or G4 versus YS (×) are presented for each loop as well as the total domain. Each symbol indicates a sequenced clone.
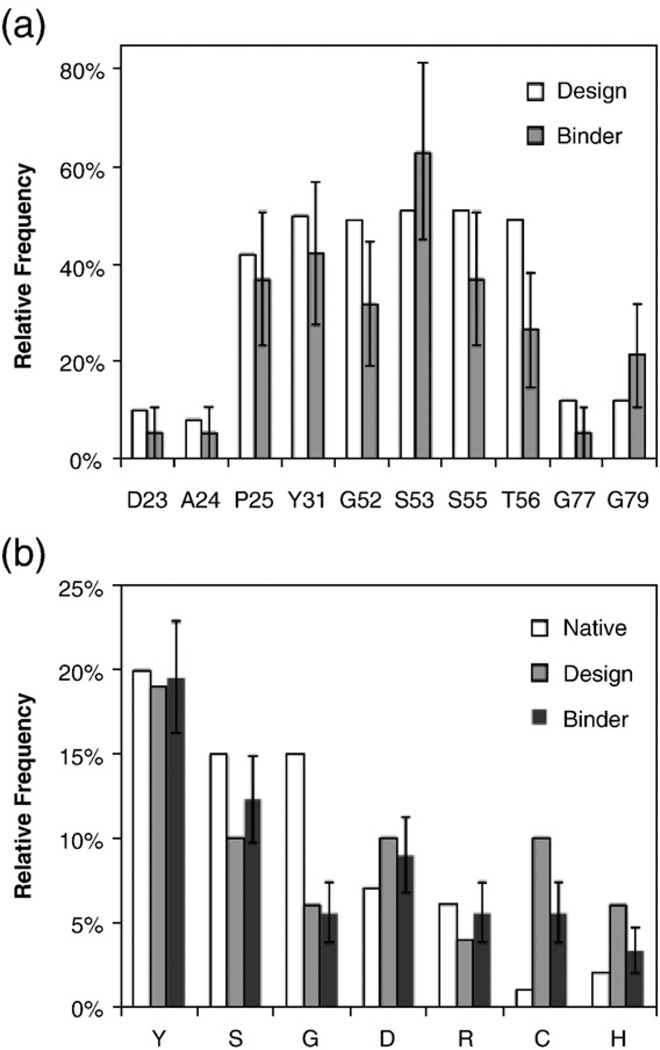
Sequence analysis reveals that wild-type bias is approximately maintained or perhaps slightly reduced in the BC and FG loops of binders, while the strong bias at G52, S55, and T56 is slightly reduced but still highly frequent (Fig. 6a). It is noteworthy that in addition to 21% occurrence at G79, glycine is present at 16% at position 80. At position 29, equal amounts of alanine, leucine, serine, and wild-type valine were included in the naïve library; in binders, the smallest available side-chain, alanine, is present at 37%, while the largest side-chain, leucine, occurs with only 11% frequency. Cumulative analysis of amino acid frequency at positions without wild-type bias indicates maintenance of the preferentially high levels of tyrosine, serine, glycine, aspartic acid, and arginine (Fig. 6b). Conversely, cysteine and histidine, which were included at higher frequency than intended because of their codon similarity to tyrosine, are present at reduced levels in binders. Eight of 19 (42%) G4-based binders are cysteine-free as compared to 19% in the naïve library. Interestingly, only three clones (15%) have a single cysteine as compared to a naïve 33%, whereas seven clones (35%) contain two cysteines (26% in the naïve library). A single clone has four cysteines. Thus, a strong selective pressure exists against unpaired cysteines; this occurs despite the potential for lone cysteine side chains to covalently bind the target protein. Of particular interest, six of the seven two-cysteine clones contain cysteine residues in identical or adjacent loops at proximal positions, suggesting feasible disulfide bonding, which can stabilize the domain.26 Thus, both wild-type bias and tailored diversity were effective in producing an effective library. Additional engineering campaigns and sequence analysis will improve the statistical significance of these trends and guide further library improvement.
Fig. 6.
Analysis of binder sequences for presence of biased amino acids. The 19 binders from the G4 library were aligned and analyzed. (a) The wild-type frequency at each position with wild-type bias is indicated. (b) The amino acid frequency at positions without wild-type bias is indicated. Native indicates phylogenetic frequency in CDR-H3. Design indicates the frequency in the G4 library design. Binder indicates the frequency in sequenced binders. The error bars represent a single standard deviation calculated as the square root of the counted amino acid occurrences divided by the total number of sequences.
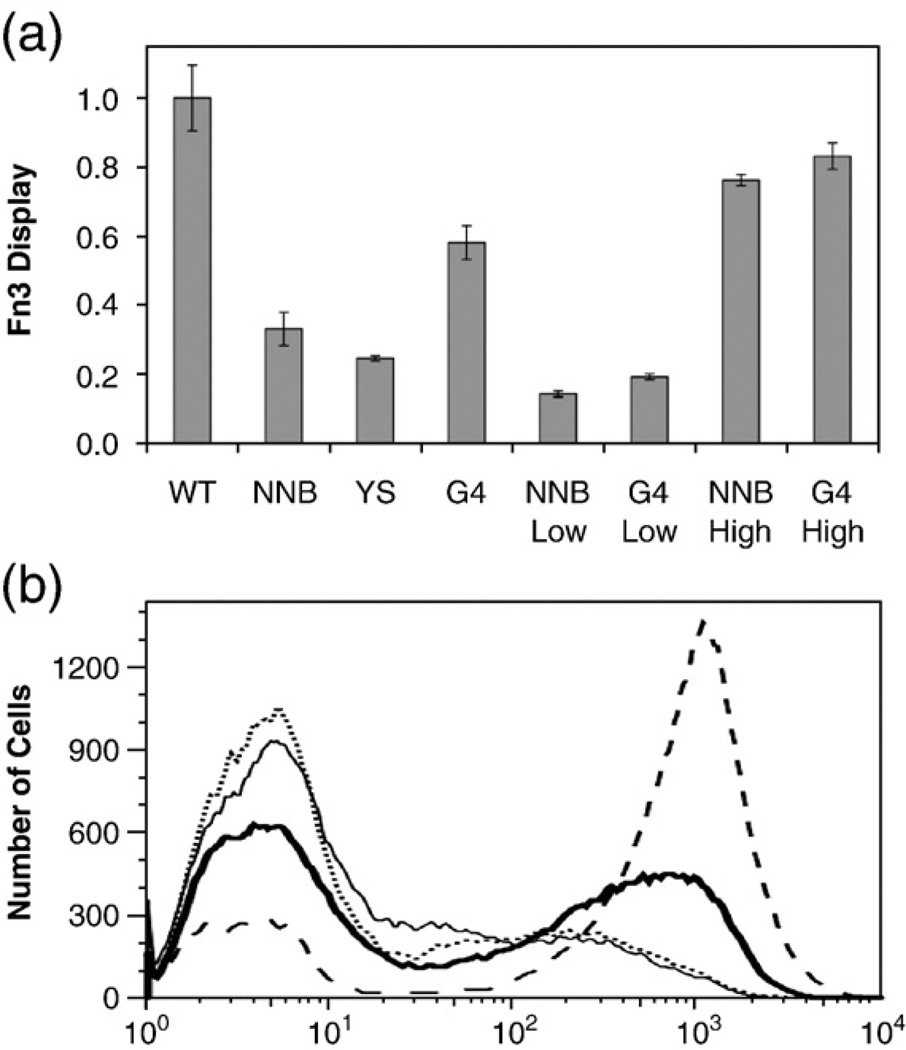
The effect of wild-type bias and tailored diversity on domain stability was analyzed. The NNB, YS, and G4 libraries were independently induced for yeast surface display at elevated temperature (37 °C). The G4 library exhibits 76±40% higher average display than the NNB library (Fig. 7), indicating higher average stability (p<0.001). Conversely, the YS library exhibits 26±13% lower display than the NNB library (p<0.005). The NNB and G4 libraries were then sorted by flow cytometry to identify clones of low and high stability. About 50 clones were sequenced from each resultant population and the amino acid frequencies in low- and high-stability clones were compared (Table 4). The biased positions in the BC loop were not enriched by stability sorting in this analysis except position 29. As observed in binder sequence analysis, the small side-chain alanine is preferred, whereas the larger side-chain leucine is destabilizing. Wild-type amino acids at the four biased positions in the DE loop are stabilizing, especially S53 and S55. While G77 is perhaps mildly stabilizing, G79 is present at substantially higher frequency in stable clones. The complete conservation of S85 in the G4 library is justified by the preferential occurrence of S85 in stable clones from the NNB library. At positions without wild-type bias, none of the preferred amino acids are substantially destabilizing, thereby validating their inclusion at elevated levels.
Fig. 7.
Yeast surface display of Fn3 libraries. Yeast containing the indicated Fn3 populations were grown to logarithmic growth phase at 30 °C. Expression of Aga2p–Fn3 was induced at 37 °C. The mean Fn3 display level for each population was quantified by flow cytometry using mouse anti-c-myc antibody and anti-mouse antibody– Alexa Fluor 488 conjugate. (a) Quantified Fn3 display. WT is wild-type Fn3. NNB, YS, and G4 are the naïve libraries. Low and High indicate the populations sorted for low and high display, respectively. Display levels are normalized to the wild-type value. (b) Raw flow cytometry data. WT (dashed line), NNB (dotted line), YS (continuous thin line), G4 (continuous thick line).
Table 4.
Stability analysis
| High–low (stability) |
|||
|---|---|---|---|
| AA | G4 design | NNB | G4 |
| Positions with wild-type bias | |||
| D23 | 10% D | 0% | +3% |
| A24 | 8% A | −4% | +1% |
| P25 | 42% P | 0% | +4% |
| V29 | A|L|S|V | +10|−19|+18|+5 | + 29|−11|−27|+12 |
| Y31 | S|Y | — | 0% |
| G52 | 49% G | +7% | +10% |
| S53 | 51% S | +5% | +20% |
| S55 | 51% S | +18% | +44% |
| T56 | 49% T | +10% | +8% |
| G77 | 12% G | +1% | +5% |
| G79 | 12% G | +17% | +17% |
| S85 | 100% S | +18% | — |
| Positions without wild-type bias | |||
| Y | 19% | −1% | −4% |
| S | 10% | +5% | +2% |
| G | 6% | 0% | +1% |
| D | 10% | +2% | +5% |
| R | 4% | −1% | +2% |
| C | 10% | 1% | −2% |
| H | 6% | 0% | 0% |
The NNB and G4 libraries were independently sorted for clones of low stability and high stability. Sequences of about 50 clones from each sorted population were analyzed. “AA” indicates the wild-type amino acid at positions with wild-type bias or amino acids of elevated frequency at positions without wild-type bias. “G4” design indicates the designed frequency of the indicated amino acid. “NNB” and “G4” indicate the difference in amino acid frequency between the high- and low-stability populations from the indicated library, respectively.
Stability analysis
The engineered binders are active in soluble form, as the EGFR binders, produced and purified from Escherichia coli, effectively bind EGFR ectodomain (B.J.H., unpublished data). These clones were also induced in the yeast surface display system at elevated temperature (37 °C) to investigate stability. The engineered binders exhibit moderate to high display levels indicative of stable clones (Supplementary Fig. 3a). Moreover, to further corroborate the display–stability correlation from Fig. 1, the clones were analyzed by circular dichroism thermal denaturation. The midpoints of thermal denaturation range from 53 °C to 73 °C (Table 3), indicative of stable clones, and agree with the display–stability correlation (Supplementary Figs. 3b and 4). The display–stability correlation for the 12 clones analyzed has a Pearson correlation coefficient of 0.86 and a Spearman rank-order correlation coefficient of 0.78 (p<0.005) demonstrative of a robust correlation. In addition, wavelength scans exhibit spectra indicative of beta sheet structure (Supplementary Fig. 4).
Discussion
Two elements of binding repertoire diversity are examined in this work in the context of the Fn3 framework: amino acid composition bias, and sitewise conservation of structural elements in binding loops. Considerable insight has recently been gained into the most functional amino acid compositions for antibody repertoires,7–9 and we also find that mimicking the natural antibody CDR composition bias produces greater functionality in Fn3 scaffold-based repertoires. Construction of scaffold libraries often treats loop sequences as completely unconstrained by structural requirements. However, incorporation of destabilizing substitutions would significantly decrease the screenable functional diversity of a repertoire. Applying phylogenetic analyses and direct measurements, conserved sites in the Fn3 scaffold were identified and diversification at these locations was reduced in constructed libraries, leading to a more highly functional repertoire for selection of binders.
The current study demonstrates that tailored CDR-like diversity is superior for library construction to nearly fully random (e.g., NNB) or overly constrained (e.g., YS) diversity. This is evidenced by the dominant selection of clones from the G4 library as well as the maintenance of the favored amino acids in binder sequences (Fig. 6b). Tailored diversity improves the search of sequence space by increasing the frequency of functional binders. This results both through improving the likelihood of beneficial contacts, largely by elevation of tyrosine, and reducing detrimental constraints. The latter element is achieved through reduction of hydrophobic isoleucine, leucine, methionine, proline, threonine, and valine as well as the large, positively charged arginine and lysine, in deference to small, neutral serine. Yet, a binary code of tyrosine and serine constrains sequence space such that it often lacks high-affinity binders. Thus, through modest incorporation of other amino acids in the library and a broad, yet efficient mutagenesis approach, tailored diversity yields a vastly improved hybrid of the two extremes of NNB and YS.
The inclusion of wild-type bias is also an important element of the G4 library design. This bias increases the frequency of functional clones both by enabling diversity to be used at positions with more impact on binding and by reducing the number of misfolded clones that result from detrimental mutation of a structurally critical residue. Moreover, the improved stability of G4 clones (Fig. 7) improves evolvability,5 allowing otherwise unstable sequence motifs to be explored. This improved stability is also beneficial in a variety of applications as outlined in the Introduction.
The methodology and techniques in the current study are directly applicable to other protein engineering efforts. While the designed skewed nucleotide mixtures for particular sites are unique to Fn3, the antibody mimic mixture should be generally applicable to nonstabilizing, solvent-exposed sites in molecular recognition scaffolds. Moreover, the mixture design algorithm may be reapplied to any design distribution. The identification of positions most likely to benefit from wild-type bias can be readily applied to other scaffolds through high throughput stability analysis in the context of protein libraries, demonstrated here using yeast surface display. When available, sequence and structural data provide additional avenues of analysis. The relative efficacy of each of these approaches will be elucidated as continued analyses expand the sequence data set and evolved library designs are tested.
Although the thrust of this work entails study of sequence–structure–function relationships and library design, the panel of binders generated provides useful reagents for a variety of applications from tumor targeting (EGFR, human A33, and mouse A33) to biotechnology (HSA and mouse IgG) to immunology (FcγRIIa and FcγRIIIa).
Materials and Methods
Stability–display relationship
Yeast surface display plasmids were created for six Fn3 domains of previously published stabilities18: wild type, 159, 159(wt DE), 159(Q8L), 159(A56E), and 159(Q8L, A56E). Genes were constructed by overlap extension PCR of eight oligonucleotides (IDT, Coralville, IA) and transformed into EBY100 yeast as described.25 Gene construction was verified by DNA sequencing. Clonal populations were grown at 30 °C in SD-CAA medium [0.07 M sodium citrate (pH 5.3), yeast nitrogen base (6.7 g/L), casamino acids (5 g/L), and glucose (20 g/L)] and induced at 37°C in SG-CAA [0.1 M sodium phosphate (pH 6.0), yeast nitrogen base (6.7 g/L), casamino acids (5 g/L), galactose (19 g/L), and glucose (1 g/L)]. Yeast were labeled with mouse anti-c-myc antibody (clone 9E10, Covance, Denver, PA) followed by phycoerythrin-conjugated goat anti-mouse antibody (Invitrogen, Carlsbad, CA). Yeast were washed and phycoerythrin fluorescence was analyzed with an Epics XL flow cytometer (Beckman Coulter, Fullerton, CA).
Library stability comparison
A library was constructed in which positions 23–30 (DAPAVTVR), 52–55 (GSKST), and 77–86 (GRGDSPASSK) were diversified with NNB codons. The library was constructed by overlap extension PCR of eight oligonucleotides and transformed into EBY100 yeast. Fourteen similar libraries were constructed with identical design except a single codon of interest was maintained as wild type within the otherwise diversified regions. Separate libraries were constructed for D23, A24, P25, A26, V27, T28, V29, G52, T56, G77, R78, G79, S84, and S85; in addition, a library was constructed that maintained D23, A24, P25, and V29. These libraries, as well as wild-type Fn3, were grown at 30 °C and induced at 37 °C; Fn3 expression was analyzed by flow cytometry as indicated above. The fractional improvement in display was calculated as the mean phycoerythrin fluorescence of the singly conserved library minus that of the fully diversified library and normalized to the fully diversified fluorescence.
Solvent-accessible surface area
The relative SASA of positions 22–32, 51–57, and 76–87 were calculated for wild-type Fn3 (solution structure 1TTG20 and crystal structures 1FNA21) and an engineered binder (2OBG22). The area accessible to a 1.4 Å sphere was determined for each side chain in each structure and compared to the accessible area in a G-X-G random coiled peptide with GetArea.19
Phylogenetic sequence alignment
The following fibronectin sequences were used: chimpanzee (XP_516072), cow (P07589), dog (XP_536059), horse (XP_001489154), human (NP_997647), mouse (NP_034363), opossum (XP_001368449), platypus (XP_001509150), rat (NP_062016), and rhesus monkey (XP_001083548). The sequences were aligned with ClustalW.28 The relative frequency of each amino acid was calculated at each position.
A similar analysis was conducted with engineered binder sequences. Engineered Fn3 domain sequences3,11–13,18,22,25,26,29–32 were aligned; identical loop sequences in related clones were only counted once to avoid bias. The amino acid frequency at each position was calculated and compared to the expected amino acid frequency as determined from a weighted average of theoretical library designs (e.g., NNS, NNB, serine/tyrosine, etc.).
Library construction
Degenerate oligonucleotides were designed to provide the desired amino acid distribution at each position. All three-site combinations of skewed nucleotide mixtures within 5% increments were considered (e.g., 20% A, 5% C, 35% G, 40% T at the first position, 15% A, 45% C, 10% G, 30% T at the second position, and 35% A, 25% C, 30% G, 10% T at the third position). The sets were filtered to identify those with good tyrosine matching and reasonable matching of alanine, aspartic acid, glycine, argininine, and serine. Specifically, tyrosine was required to occur at 0.5–2 times the intended frequency; alanine, aspartic acid, glycine, arginine, and serine were required to occur at 0.33–3 times the intended frequency. The sets that fulfilled these criteria were then used to produce numerous in silico protein libraries on the basis of their amino acid probability distribution. For each clone, the probability of occurrence from a library that precisely matched the desired distribution was calculated. The sum of probabilities for each sample library was used as a metric of library fitness. The skewed nucleotide designs were selected on the basis of fitness and the ability to use identical mixtures at multiple sites (e.g., 45% C, 10% G, 45% T at the wobble position of multiple codons). Nucleotide designs are included in Supplementary Table 1.
Degenerate oligonucleotides were synthesized with skewed nucleotides at diversified positions and nucleotides encoding wild-type Fn3 at fully conserved positions. The library design, summarized in Table 1, includes four, three, and four loop lengths in the BC, DE, and FG loops, respectively.25 Separate oligonucleotides were synthesized to yield each length. Overlap extension PCR of eight oligonucleotides was performed to construct complete Fn3 genes. Separate reactions were conducted for each loop length to avoid bias toward shorter loops. The gene libraries were transformed into yeast by homologous recombination with linearized yeast surface display vector, which includes the Aga2p protein fusion, N-terminal hemagglutinin (HA) epitope, and C-terminal c-myc epitope. The fraction of clones that produce full-length Fn3 was determined by flow cytometry as the fraction displaying the N-terminal HA tag that also contained the C-terminal c-myc epitope; these results were corroborated by sequence analysis.
Binder selections
Human and mouse A33 extracellular domains were both produced with His6 epitope tags in human embryonic kidney cells and purified by metal-affinity chromatography. Protein was biotinylated either on free amines with the sulfo-NHS biotinylation kit (Pierce, Rockford, IL) or by site-specific sortase-based conjugation of GGGGG-biotin to an LPETG C-terminal epitope.33 EGFR mutant 404SG34 was produced in Saccharomyces cerevisiae yeast, purified by metal-affinity chromatography and anti-EGFR antibody affinity chromatography, and biotinylated on free amines with the sulfo-NHS biotinylation kit. Biotinylated FcγRIIA and FcγRIIIA were a kind gift from Jeffrey Ravetch (Rockefeller University). Biotinylated mIgG was purchased from Rockland Immunochemicals (Gilbertsville, MD). Human serum albumin (Sigma, St. Louis, MO) was biotinylated with the sulfo-NHS biotinylation kit. The NNB, YS, and G4 libraries were pooled for direct competition. The libraries were sorted for binding to the seven protein targets and affinity-matured as described.13 Yeast were grown and induced to display Fn3. Binders to streptavidin-coated magnetic Dynabeads (Invitrogen) were removed.35 Biotinylated protein was loaded on streptavidin-coated magnetic Dynabeads and incubated with the remaining yeast. The beads were washed with phosphate-buffered saline with bovine serum albumin (PBSA), and the beads with attached cells were grown for further selection. After two magnetic bead sorts, full-length Fn3 clones were selected by fluorescence-activated cell sorting with the C-terminal c-myc epitope for identification of full-length clones. Plasmid DNA was zymoprepped from the cells and mutagenized by error-prone PCR of the entire Fn3 gene or the BC, DE, and FG loops. Mutants were transformed into yeast by electroporation with homologous recombination and requisite shuffling of the loop mutants. The lead clones and their mutants were pooled for further cycles of selection and mutagenesis. Once significant binder enrichment was observed during magnetic bead sorts, fluorescence-activated cell sorting was used. Yeast displaying Fn3 were incubated with biotinylated target protein and anti-c-myc antibody (clone 9E10 or chicken anti-c-myc, Invitrogen). Cells were washed and incubated with Alexa Fluor 488-, phycoerythrin-, or Alexa Fluor 647-conjugated streptavidin (Invitrogen) and fluorophore-conjugated anti-mouse or anti-chicken antibody (Invitrogen). Cells were washed and cells with the highest target to c-myc labeling ratio were selected on a FACS Aria (Becton Dickinson, Franklin Lakes, NJ) or MoFlo (Dako Cytomation, Carpinteria, CA) flow cytometer. Plasmids from binding populations were zymoprepped and transformed into E. coli; transformants were grown, miniprepped, and sequenced.
Library source determination
For each clone, the probabilities that it originated from the NNB, YS, or G4 library were calculated with the designed nucleotide distributions at each position as well as the probability of mutation by error-prone PCR.
Fn3 production
The Fn3 gene was digested with NheI and BamHI and transformed to a pET vector containing a HHHHHHK-GSGK-encoding C-terminus. The six histidines enable metal-affinity purification, and the pentapeptide provides two additional amines for chemical conjugation. The plasmid was transformed into Rosetta (DE3) E. coli, which was grown in LB medium with kanamycin (100 mg/L) and chloramphenicol (34 mg/L) at 37 °C. Two hundred microliters of overnight culture was added to 100 mL of LB medium, grown to an optical density of 0.2–1.5 units, and induced with 0.5 mM IPTG for 3–24 h. Cells were pelleted, resuspended in lysis buffer [50 mM sodium phosphate (pH 8.0), 0.5 M NaCl, 5% glycerol, 5 mM CHAPS, 25 mM imidazole, and 1× complete EDTA (ethylenediaminetetraacetic acid)-free protease inhibitor cocktail], and exposed to four freeze-thaw cycles. The soluble fraction was clarified by centrifugation at 15,000g for 10 min, and Fn3 was purified by metal-affinity chromatography on TALON resin.
Affinity titration
The equilibrium dissociation constants of select clones were determined by titration of soluble antigen for binding to yeast surface displayed Fn3 as described.36 Affinities for FcγRIIA and FcγRIIIA were determined by surface plasmon resonance. Affinities for EGFR were also measured with soluble Fn3 and EGFR-expressing A431 cells. Purified Fn3 was buffer-exchanged into PBS and biotinylated with NHS-LC-biotin according to the manufacturer’s instructions. A431 cells were washed in PBSA and incubated with various concentrations of biotinylated Fn3 on ice. The number of cells and sample volumes were selected to ensure excess Fn3 relative to EGFR. Cells were incubated on ice for sufficient time to ensure that the approach to equilibrium was at least 98% complete. Cells were then pelleted, washed with 1 mL PBSA, and incubated in PBSA with streptavidin-R-phycoerythrin (10 mg/L) for 10–30 min. Cells were washed and resuspended with PBSA and analyzed by flow cytometry. The minimum and maximum fluorescence and the Kd value were determined by minimizing the sum of squared errors assuming a 1:1 binding interaction.
Circular dichroism
EGFR binders were produced as described and purified by metal-affinity chromatography and high-performance liquid chromatography on a C18 column. Protein was lyophilized and resuspended in PBS. Ellipticity was measured from 260 to 205 nm on a Jasco 815 spectrometer by means of a quartz cuvette with a 1 -mm path length. Thermal denaturation was conducted by measuring ellipticity at 216 nm from 20 to 98 °C and calculating Tm from a standard two-state unfolding curve.
Library stability selection and analysis
The NNB and G4 libraries were independently grown at 30 °C and induced at 37 °C. Yeast were labeled with mouse anti-HA antibody (clone 16B12, Covance) and chicken anti-c-myc antibody to label the N- and C-terminal epitopes. Cells were washed, incubated with phycoerythrin-conjugated goat anti-mouse antibody and Alexa Fluor 488-conjugated goat anti-chicken antibody, and sorted by flow cytometry. Only cells with comparable signals for each epitope were considered to avoid selecting epitope mutants. Approximately 1% of the cells with the lowest or highest display fluorescence were collected and grown for an additional induction and selection. Plasmids were isolated and transformed into E. coli. About 50 clones from each resultant population (both low and high stability for both NNB and G4) were miniprepped and sequenced. Sequences were aligned and the amino acid frequencies at each position were determined.
Supplementary Material
Acknowledgements
Steve Sazinsky (MIT) provided EGFR mutant 404SG. Jeffrey Ravetch (Rockefeller University) provided biotinylated FcγRIIA and FcγRIIIA. The work was supported by the MIT-Portugal program and NIH grants CA101830 and CA96504.
Abbreviations used
- CDR
complementarity-determining region
- EGFR
epidermal growth factor receptor
- FcγR
Fcγ receptor
- Fn3
tenth type III domain of human fibronectin
- HA
hemagglutinin
- HSA
human serum albumin
- mIgG
mouse immunoglobulin G
- PBSA
phosphate-buffered saline with bovine serum albumin
- SASA
solvent-accessible surface area
Footnotes
Supplementary Data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2010.06.004
References
- 1.Binz H, Amstutz P, Plückthun A. Engineering novel binding proteins from nonimmunoglobulin domains. Nat. Biotechnol. 2005;23:1257–1268. doi: 10.1038/nbt1127. [DOI] [PubMed] [Google Scholar]
- 2.Sidhu S, Fellouse F. Synthetic therapeutic antibodies. Nat. Chem. Biol. 2006;2:682–688. doi: 10.1038/nchembio843. [DOI] [PubMed] [Google Scholar]
- 3.Koide A, Bailey CW, Huang X, Koide S. The fibronectin type III domain as a scaffold for novel binding proteins. J. Mol. Biol. 1998;284:1141–1151. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- 4.Koide A, Koide S. Monobodies: antibody mimics based on the scaffold of the fibronectin type III domain. Methods Mol. Biol. 2007;352:95–109. doi: 10.1385/1-59745-187-8:95. [DOI] [PubMed] [Google Scholar]
- 5.Bloom JD, Labthavikul ST, Otey CR, Arnold FH. Protein stability promotes evolvability. Proc. Natl Acad. Sci. USA. 2006;103:5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbas CF, Bain JD, Hoekstra DM, Lerner RA. Semisynthetic combinatorial antibody libraries: a chemical solution to the diversity problem. Proc. Natl Acad. Sci. USA. 1992;89:4457–4461. doi: 10.1073/pnas.89.10.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fellouse F, Wiesmann C, Sidhu S. Synthetic antibodies from a four-amino-acid code: a dominant role for tyrosine in antigen recognition. Proc. Natl Acad. Sci. USA. 2004;101:12467–12472. doi: 10.1073/pnas.0401786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fellouse F, Li B, Compaan DM, Peden AA, Hymowitz SG, Sidhu S. Molecular recognition by a binary code. J. Mol. Biol. 2005;348:1153–1162. doi: 10.1016/j.jmb.2005.03.041. [DOI] [PubMed] [Google Scholar]
- 9.Fellouse F, Barthelemy PA, Kelley RF, Sidhu S. Tyrosine plays a dominant functional role in the paratope of a synthetic antibody derived from a four amino acid code. J. Mol. Biol. 2006;357:100–114. doi: 10.1016/j.jmb.2005.11.092. [DOI] [PubMed] [Google Scholar]
- 10.Fellouse F, Esaki K, Birtalan S, Raptis D, Cancasci VJ, Koide A, et al. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J. Mol. Biol. 2007;373:924–940. doi: 10.1016/j.jmb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Gilbreth RN, Esaki K, Koide A, Sidhu S, Koide S. A dominant conformational role for amino acid diversity in minimalist protein–protein interfaces. J. Mol. Biol. 2008;381:407–418. doi: 10.1016/j.jmb.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang J, Koide A, Makabe K, Koide S. Design of protein function leaps by directed domain interface evolution. Proc. Natl Acad. Sci. USA. 2008;105:6578–6583. doi: 10.1073/pnas.0801097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hackel BJ, Wittrup KD. The full amino acid repertoire is superior to serine/tyrosine for selection of high affinity immunoglobulin G binders from the fibronectin scaffold. Protein Eng. Des. Select. 2010;23:211–219. doi: 10.1093/protein/gzp083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Virnekäs B, Ge L, Plückthun A, Schneider KC, Wellnhofer G, Moroney SE. Trinucleotide phosphoramidites: ideal reagents for the synthesis of mixed oligonucleotides for random mutagenesis. Nucleic Acids Res. 1994;22:5600–5607. doi: 10.1093/nar/22.25.5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shusta EV, Kieke MC, Parke E, Kranz DM, Wittrup KD. Yeast polypeptide fusion surface display levels predict thermal stability and soluble secretion efficiency. J. Mol. Biol. 1999;292:949–956. doi: 10.1006/jmbi.1999.3130. [DOI] [PubMed] [Google Scholar]
- 16.Kowalski JM, Parekh RN, Mao J, Wittrup KD. Protein folding stability can determine the efficiency of escape from endoplasmic reticulum quality control. J. Biol. Chem. 1998;273:19453–19458. doi: 10.1074/jbc.273.31.19453. [DOI] [PubMed] [Google Scholar]
- 17.Kowalski JM, Parekh RN, Wittrup KD. Secretion efficiency in Saccharomyces cerevisiae of bovine pancreatic trypsin inhibitor mutants lacking disulfide bonds is correlated with thermodynamic stability. Biochemistry. 1998;37:1264–1273. doi: 10.1021/bi9722397. [DOI] [PubMed] [Google Scholar]
- 18.Parker M, Chen Y, Danehy F, Dufu K, Ekstrom J, Getmanova E, et al. Antibody mimics based on human fibronectin type three domain engineered for thermostability and high-affinity binding to vascular endothelial growth factor receptor two. Protein. Eng. Des. Select. 2005;18:435–444. doi: 10.1093/protein/gzi050. [DOI] [PubMed] [Google Scholar]
- 19.Fraczkiewicz R, Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for …. J. Comput. Chem. 1998;19:319–333. [Google Scholar]
- 20.Main AL, Harvey TS, Baron M, Boyd J, Campbell ID. The three-dimensional structure of the tenth type III module of fibronectin: an insight into RGD-mediated interactions. Cell. 1992;71:671–678. doi: 10.1016/0092-8674(92)90600-h. [DOI] [PubMed] [Google Scholar]
- 21.Dickinson CD, Veerapandian B, Dai XP, Hamlin RC, Xuong NH, Ruoslahti E, Ely KR. Crystal structure of the tenth type III cell adhesion module of human fibronectin. J. Mol. Biol. 1994;236:1079–1092. doi: 10.1016/0022-2836(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 22.Koide A, Gilbreth RN, Esaki K, Tereshko V, Koide S. High-affinity single-domain binding proteins with a binary-code interface. Proc. Natl Acad. Sci. USA. 2007;104:6632–6637. doi: 10.1073/pnas.0700149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Birtalan S, Zhang Y, Fellouse F, Shao L, Schaefer G, Sidhu S. The intrinsic contributions of tyrosine, serine, glycine and arginine to the affinity and specificity of antibodies. J. Mol. Biol. 2008;377:1518–1528. doi: 10.1016/j.jmb.2008.01.093. [DOI] [PubMed] [Google Scholar]
- 24.Zemlin M, Klinger M, Link J, Zemlin C, Bauer K, Engler JA, et al. Expressed murine and human CDR-H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range of structures. J. Mol. Biol. 2003;334:733–749. doi: 10.1016/j.jmb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 25.Hackel B, Kapila A, Wittrup K. Picomolar affinity fibronectin domains engineered utilizing loop length diversity, recursive mutagenesis, and loop shuffling. J. Mol. Biol. 2008;381:1238–1252. doi: 10.1016/j.jmb.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lipovsek D, Lippow S, Hackel B, Gregson MW, Cheng P, Kapila A, Wittrup K. Evolution of an interloop disulfide bond in high-affinity antibody mimics based on fibronectin type III domain and selected by yeast surface display: molecular convergence with single-domain camelid and shark antibodies. J. Mol. Biol. 2007;368:1024–1041. doi: 10.1016/j.jmb.2007.02.029. [DOI] [PubMed] [Google Scholar]
- 27.Ackerman M, Levary D, Tobon G, Hackel B, Orcutt KD, Wittrup KD. Highly avid magnetic bead capture: an efficient selection method for de novo protein engineering utilizing yeast surface display. Biotechnol. Prog. 2009;25:774–783. doi: 10.1002/btpr.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 29.Koide A, Abbatiello S, Rothgery L, Koide S. Probing protein conformational changes in living cells by using designer binding proteins: application to the estrogen receptor. Proc. Natl Acad. Sci. USA. 2002;99:1253–1258. doi: 10.1073/pnas.032665299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu L, Aha P, Gu K, Kuimelis RG, Kurz M, Lam T, et al. Directed evolution of high-affinity antibody mimics using mRNA display. Chem. Biol. 2002;9:933–942. doi: 10.1016/s1074-5521(02)00187-4. [DOI] [PubMed] [Google Scholar]
- 31.Karatan E, Merguerian M, Han Z, Scholle MD, Koide S, Kay BK. Molecular recognition properties of FN3 monobodies that bind the Src SH3 domain. Chem. Biol. 2004;11:835–844. doi: 10.1016/j.chembiol.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 32.Olson CA, Liao HI, Sun R, Roberts RW. mRNA display selection of a high-affinity, modification-specific phospho-IkappaBalpha-binding fibronectin. ACS Chem. Biol. 2008;3:480–485. doi: 10.1021/cb800069c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parthasarathy R, Subramanian S, Boder ET. Sortase A as a novel molecular “stapler” for sequence-specific protein conjugation. Bioconjug. Chem. 2007;18:469–476. doi: 10.1021/bc060339w. [DOI] [PubMed] [Google Scholar]
- 34.Kim Y, Bhandari R, Cochran J, Kuriyan J, Wittrup K. Directed evolution of the epidermal growth factor receptor extracellular domain for expression in yeast. Proteins. 2006;62:1026–1035. doi: 10.1002/prot.20618. [DOI] [PubMed] [Google Scholar]
- 35.Ackerman M, Levary D, Tobon G, Hackel B, Orcutt KD, Wittrup K. Highly avid magnetic bead capture: an efficient selection method for de novo protein engineering utilizing yeast surface display. Biotechnol. Prog. 2009;25:774–783. doi: 10.1002/btpr.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chao G, Lau W, Hackel B, Sazinsky S, Lippow S, Wittrup KD. Isolating and engineering human antibodies using yeast surface display. Nat. Protocols. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.