Abstract
The cell interior is a busy and crowded place. A large fraction of the cell volume is taken up by organelles that come in a variety of shapes and sizes. These organelles are surrounded by membrane that not only acts as a diffusion barrier, but also provides each organelle with its unique morphology that contributes to its function, often in ways that are poorly understood. Here we discuss recent discoveries on the relationship between organelle structure and function.
Introduction
Organelles are dynamic, changing size and shape to maintain homeostasis and adjusting to the various needs of the cell. Some changes occur as part of the normal cell cycle, for example during cell division [1–3]. Other changes happen in response to challenges or stress and reflect a modification in organelle function, such as a change in protein folding capacity of the endoplasmic reticulum (ER) or ATP production in mitochondria [4, 5]. It is assumed that alterations to organelle morphology reflect an underlying functional optimization. Yet, this relationship is often poorly understood: for example, does the peripheral endoplasmic reticulum (ER) have to be in the shape of tubules in order to carry out its function? Does mitochondrial size matter? In this review we discuss recent advances in our understanding of the relationship between organelle structure and function, focusing primarily on the ER, nucleus and mitochondria. The reader is referred to excellent reviews that cover earlier work on Golgi [1], peroxosime [6] and endosome [7] structure.
Shaping a membrane-bound organelle
How are organelles shaped? The morphology of most organelles is characterized by a combination of flat and curved membrane, such as in the ER (Figure 1a). Cellular membranes are lipid bilayers made predominantly of phospholipids and proteins, both of which can contribute to membrane curvature. A difference in lipid composition between the two bilayers can itself lead to membrane curvature, and this likely drives the formation of the rims of Golgi cisternae and the tubular structures that connect the Golgi stack to form the ribbon [8]. Recently, a novel ER structure made of a helicoidal surface was shown to connect adjacent ER sheets [9••] (Figure 1b). This configuration, which is akin to the ramps of a parking garage, appears to be an energetically favorable structure that allows the dense packing of ER sheets to accommodate maximum protein synthesis in secretory cells. Thus, inherent properties of membranes contribute to their degree of curvature.
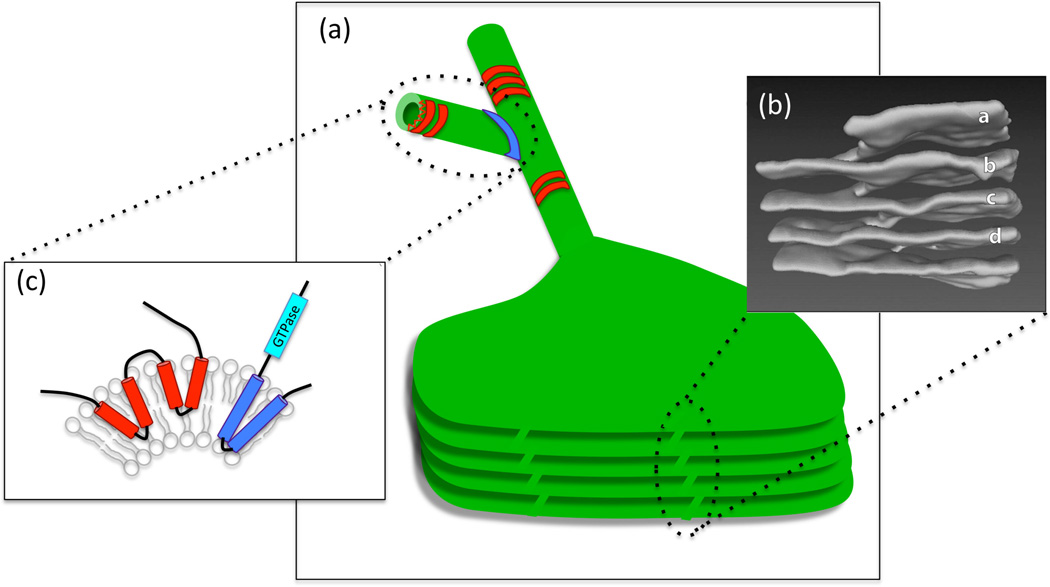
Figure 1.
Diverse membrane structures in the ER. (a) The ER is an interconnected network of composed of branched tubules and sheets, some of which can form stacks, as shown in the illustration. ER tubules are stabilized by the oligomerization of proteins such as reticulons and DP1/Yop1/REEps (in red), while 3-way junctions are mediated by proteins such as atlastins (in blue). (b) The structure of reticulons and atlastin. Membrane curvature is induced by the insertion of protein "wedges" (two in the case of reticulons and one in the case of atlastin) that traverse only one lipid bilayer, forcing the membrane to curve. Atlastin has an in addition GTPase activity that is necessary for fusing membranes and generating 3-way junctions. (c) Helicoidal membrane structure in stacked ER sheets. A 3D reconstruction of a region of stacked ER sheets from an acinar cell of a mouse salivary gland. The letters (a through d) mark the ER sheets that are connected through a helicoidal structure (to the left). From Terasaki et al. Cell 154, 285–296, 2013.
Proteins also contribute to membrane curvature, as in the case of the ER [10] (Figure 1c). Proteins called reticulons and DP1/Yop1/REEPs contain hydrophobic domains that form wedges on one side of the lipid bilayer, forcing it to bend towards the opposite side. These proteins are essential for maintaining ER tubules and are also involved in the highly curved regions of the NE where nuclear pore complexes are embedded. The tubular ER network is also shaped by the formation of three-way junctions, generated by homotypic membrane fusion between the tip of one ER tubule and the side of another in a process mediated by a conserved family of proteins called atlastin/Sey1 [11•, 12•]. Other proteins that contribute to membrane curvature are the BAR domain proteins, which form a rigid crescent-shaped structure and force membrane bending through electrostatic interactions between the concave surface of the protein dimer and the membrane [13]. Proteins also contribute to the constant luminal width of low curvature double-membrane structures, such as ER cisternae and the nuclear envelope, by acting as spacers within the luminal space [14•, 15]. Thus, organelle morphology is driven, in part, by dedicated proteins that affect membrane curvature and geometry.
Proteins that are not dedicated to altering membrane shape may also contribute to organelle structure. For example, the curvature of the cristae of the mitochondrial inner membrane is stabilized by the presence of ATP synthase [16, 17], and the ER sheets are likely stabilized by attached ribosomes [18]. Finally, membrane shape can be affected by external cytoskeletal forces. One such example is the formation of ER tubules through the attachment of the ER to microtubule associated proteins and the pulling forces exerted by microtubule elongation and microtubule motors [19–21]. The combination of lipid and protein composition, along with external forces, provides each organelle with its unique morphology.
Complex shapes allow for distinct functions within a single organelle
While some organelles, such as the nucleus or the vacuole, are simple in shape, other organelles, such as the Golgi and the ER, have complex shapes made up of a network of cisternae and tubules. This complexity allows for segregation of biochemical processes within the organelle: for example, ribosomes are preferentially associated with the flat ER membrane that forms the rough ER [14, 18]. In contrast, ER tubules are engaged in processes such as lipid synthesis and they are responsible for the majority of contacts between the ER and other organelles. Indeed, when the tubular structure of the ER is disrupted in yeast, the efficiency of lipid transfer between the ER and mitochondria is reduced [22•]. The ER forms membrane contact sites (MCS) with virtually all organelles in the cell; the juxtaposed membranes are typically 20 nm apart and they are held together by protein complexes that are unique to each organelle [23]. Traditionally, MCS were thought of as sites for inter-organelle communication, such as exchange of Ca+2 and the transfer of lipids, which are synthesized predominantly in the ER but are needed by all membrane-bound organelles. More recently, MCS between the ER and the mitochondria were shown to affect mitochondrial fission [24••]: ER tubules wrap around mitochondria at future fission sites and can constrict mitochondria even in the absence of the mitochondrial fission machinery, such as the dynamin related protein Dnm1/Drp1. The mechanism by which this constriction occurs, and whether ER tubules affect the structure of other organelles, remain to be determined.
Unlike the ER, the metazoan nucleus is usually a round or oval structure with limited membrane curvature, except where the inner and outer nuclear membranes meet at nuclear pores. Thus, distinct domains within the nucleus, if they exist, are not defined by nuclear membrane-derived compartments. The budding yeast nucleus, however, changes shape during the cell cycle: while the metazoan nucleus disassembles in mitosis, the budding yeast nucleus remains intact, and in anaphase it forms an hourglass shape with only a narrow opening connecting the nuclear compartments within the mother and daughter cells. This opening is sufficiently small to form a diffusion barrier between the two nuclear halves, allowing the asymmetric accumulation of a transcription factor only in the daughter nuclear compartment [25•]. Widening of the opening connecting the mother and daughter nuclear halves by genetic manipulations allowed diffusion of proteins between the two compartments, indicating that yeast cells take advantage of cell cycle changes in nuclear shape to compartmentalize the nucleus.
Organelle shape and cell function
It is likely that organelle shape, and in particular the membrane configuration, has evolved to suit not only the organelle's biological activity, but also overall cell function. The formation of MCS discussed above is one such example, and recent studies suggest that the reticular nature of the peripheral ER is also important to allow passage of macromolecules from the cytosol to the plasma membrane. In fission yeast, for example, the localization of a plasma membrane protein, Mid1, which usually localizes in a sharp band at the cell midzone, was altered to a more dispersed pattern in a mutant harboring peripheral ER that was in the form of sheets rather than tubules [26]. The subsequent identification of proteins that link the ER to the plasma membrane in budding yeast helped explain this result [27, 28•]: when these ER- plasma membrane tethering proteins were absent, the peripheral ER collapsed to the center of the cell. Interestingly, when these tethering proteins were removed from the fission yeast mutant containing ER sheets described above, the ER sheets detached from the plasma membrane and Mid1 resumed its normal localization pattern [29]. The authors interpreted this result to mean that extensive ER sheets at the cell periphery may obstruct proper protein localization at the plasma membrane, and that the peripheral ER is in the form of tubules in part to allow access of proteins like Mid1 to the plasma membrane. Interestingly, sites of endocytosis do not overlap with plasma membrane regions that are associated with the ER [30], again suggesting that ER presence may limit the accessibility of cytosolic proteins to the plasma membrane. Taken together, these studies suggest that the ER can both facilitate access to the plasma membrane for ER components, through MCSs, but can block plasma membrane accessibility of cytosolic factors.
Another example of organelle shape adapting to the cell's needs is seen in mitosis. Most organelles must change shape and/or size (e.g. undergo fragmentation) during cell division to ensure their own proper segregation to the daughter cells. Less appreciated, until recently, was the need to change shape in order to get out of the way of the mitotic apparatus. Under certain pathological conditions that lead to abnormal cellular structure, chromosome segregation or nuclear division can be obstructed. For example, the FAB1 gene, which is required to regulate vacuole morphology, was initially identified in a screen for chromosome segregation mutants in budding yeast because the grossly enlarged vacuole in fab1 mutants prevented nuclear elongation [31]. But do organelles remodel to avoid obstructing the mitotic apparatus under non-pathological conditions?
Following NE break down in vertebrate cells, many NE components are resorbed into the ER. The ER itself is conspicuously absent from the area of the mitotic spindle and becomes enriched at spindle poles [18]. Two recent papers describe microtubule-dependent mechanisms that serve to keep the ER clear of the mitotic spindle, and in one case this is essential for proper NE architecture (Figure 2). Smyth et al. [32••] showed that phosphorylation of Stromal interaction molecule 1 (STIM1) keeps ER off of spindle microtubules by dissociating it from the microtubule plus end binding protein 1 (EB1). Schlaitz et al. [33••] identified REEP4 in a proteomic screen for membrane proteins that bind microtubules in Xenopus egg extracts, and showed that depletion of REEP4 and its close homolog REEP3 caused ER membrane to accumulate on mitotic chromosomes and become trapped inside reforming daughter nuclei. Whereas expression of wild-type REEP4 rescued the phenotype, a REEP4 mutant defective in microtubule binding could not. Thus, while phosphorylation of STIM1 prevents association of ER membranes with microtubule plus ends, REEP3/4 function to concentrate these membranes near microtubule minus ends at spindle poles, away from the chromosomes, through an uncharacterized mechanism.
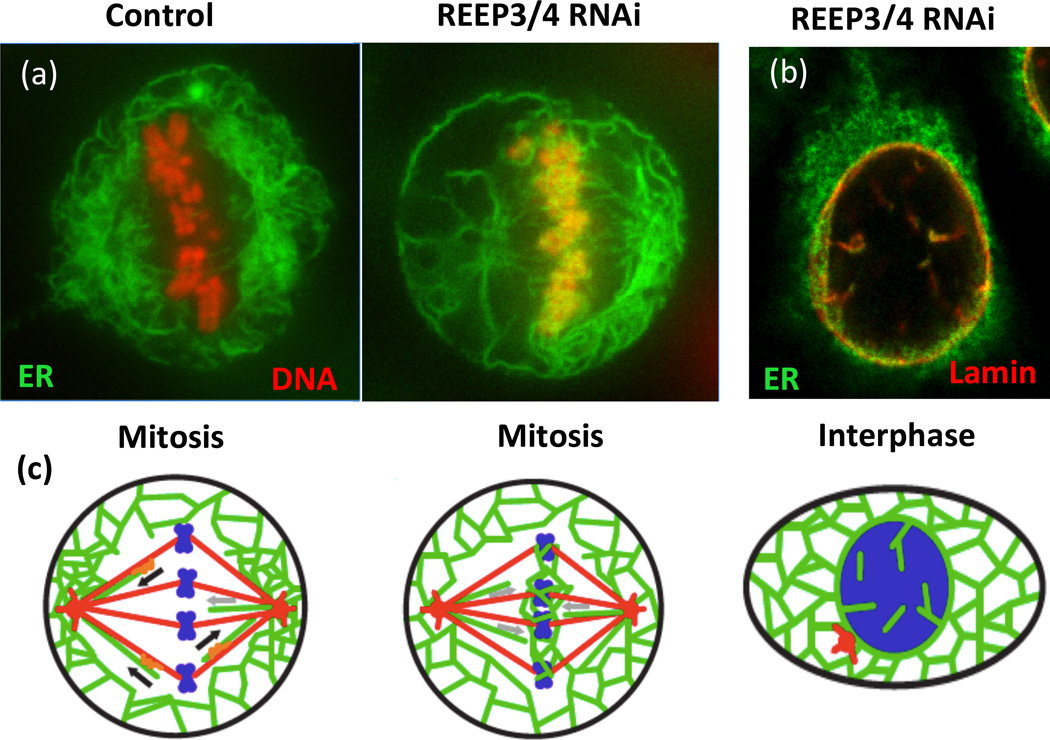
Figure 2.
Depletion of REEP3/4 causes accumulation of ER on mitotic chromosomes and leads to intranuclear membranes and lamina. A. HeLa cells expressing RFP-histone (red) to label the DNA and GFP-Sec61 (green) to mark the ER were subjected to control or REEP3/4 RNAi and imaged during mitosis. Note the colocalization of ER and mitotic chromosomes in the absence of REEP3/4. (b) An interphase REEP3/4 RNAi-treated HeLa cell expressing GFP-Sec61 (green) was fixed and stained for nuclear lamin B1 (red). Both NE markers are aberrantly localized to structures inside the nucleus. (c) Schematic of phenotypes with microtubules in red, DNA in blue and ER in green. Adapted from Schlaitz et al. Dev. Cell 26, 316–323, 2013. Images courtesy of Anne-Lore Schlaitz and Rebecca Heald.
Changing shape in adaptation to stress
In addition to changes associated with the cell cycle, cells may experience a need to increase the functional capacity of an organelle, either due to specialization following differentiation or under stress conditions, when the activity of a certain cellular compartment must be increased in order achieve homeostasis. A well-documented case is the unfolded protein response (UPR), which is activated due to the accumulation of unfolded proteins in the ER and leads to increased phospholipid synthesis that drives ER expansion [34]. In a more recent example, a pathway linking mitochondria shape changes to stress in the form of nutrient deprivation has been identified [35••, 36••]. A catabolic process termed autophagy is induced by starvation and proceeds through the formation of a double-membrane vesicle, the autophagosome. Organelle and cytosolic components engulfed by the autophagosome are recycled following fusion with lysosomes, thereby prolonging cell survival when nutrients are scarce. In separate studies, Gomes et al. and Rambold et al. observed that mitochondria elongate during autophagy, which spares their degradation (Figure 3). Starvation-induced mitochondrial elongation is mediated by down regulation of dynamin-related protein 1 (Drp1), which prevents mitochondrial fission, leading to unopposed fusion. It is unclear whether elongated mitochondria cannot be targeted to autophagosomes because of their size or because they are not recognized in their fused form. Why mitochondria are spared is another question. Elongated mitochondria have a higher density of cristae and increased ATP production, which could spare amino acids for protein synthesis rather than catabolism thereby promoting cell survival under starvation conditions. In addition, mitochondria may also provide membrane to autophagosomes during starvation [37••].
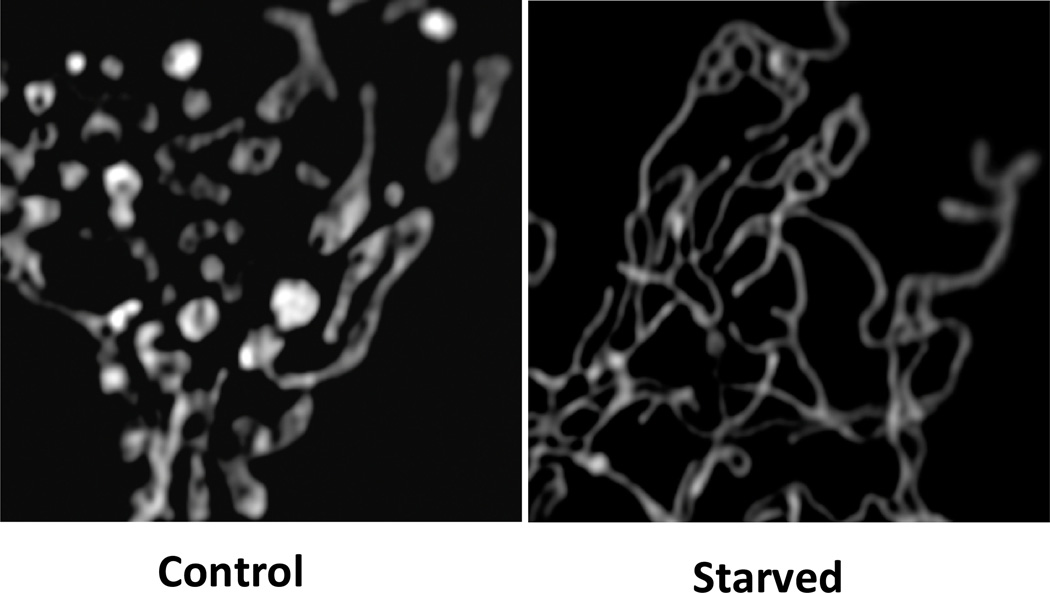
Figure 3.
Starvation leads to unopposed mitochondrial fusion. Mouse embryo fibroblasts transfected with mitoRFP were incubated in full nutrient medium (Control), or starvation medium (Starved) for 6 hours. Starved cells show a continuous network of mitochondria. Cells were fixed and images acquired by Structured Illumination Microscopy. Images courtesy of Angelika Rambold and Jennifer Lippencott-Schwartz.
The importance of organelle size
In addition to having a distinct shape, organelles also have a specific size. Organelle size may scale with cell size as was shown in yeast for the nucleus [38, 39] and more recently for mitochondria [40•]. Organelle size may also expand to accommodate the cell's changing needs, as in the case of ER expansion during the UPR or when mitochondria fuse to evade autophagy, described above. Thus, the size of an organelle undoubtedly affects its function, but in only very few cases has organelle size been manipulated to evaluate the consequences.
A recent study addressed the effects of assembling a very small nucleus [41••]. Micronuclei form around an individual or broken chromosome when it segregates improperly during mitosis and becomes separated from the main chromatin mass. Hetzer and colleagues showed that although micronuclei appear to be structurally normal, their nuclear envelopes frequently collapse due to defects in assembly of the underlying network of intermediate filaments, the nuclear lamina. Nuclear envelope collapse is accompanied by chromatin compaction, invasion of the ER, and loss of nuclear functions including transcription and DNA replication, and can trigger massive DNA damage, termed chromothripsis (Figure 4). Indeed, Pellman and colleagues [42] showed that DNA subjected to fragmentation within micronuclei can be re-integrated into the genome in subsequent cell divisions. Although it is not currently known that size, per se, caused micronuclei-dependent damage, the underlying causes of micronucleus instability are likely to shed light on nuclear function.
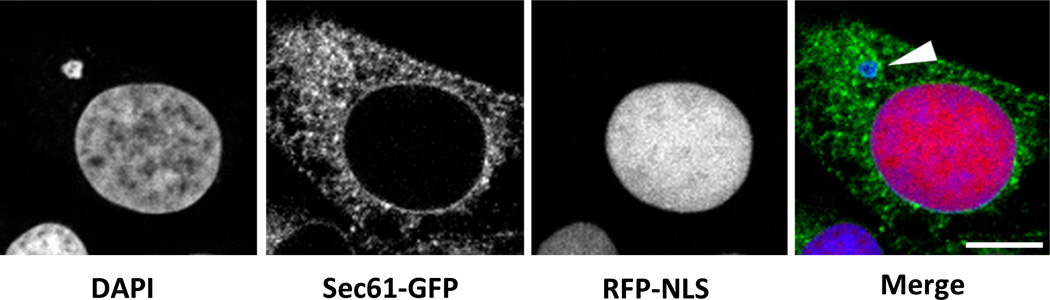
Figure 4.
Micronuclei are unstable in somatic cells. The images show a U2OS cell containing an intact nucleus and a disrupted micronucleus (arrowhead). The micronucleus fails to accumulate the fluorescent nuclear protein RFP-NLS, and has been invaded by ER as indicated by the presence of Sec61-GFP. Scale bar = 10 microns. Images courtesy of Emily Hatch and Martin Hetzer.
It is also the case that not all small nuclei are defective. In certain large and rapidly dividing embryos, such as fish [43] and frog [44], nuclear envelopes form around individual or groups of chromosomes, rather than around the entire DNA mass, likely so that DNA replication can commence quickly. These micronuclei, termed karyomeres, fuse to form a mononucleus prior to the next division. Abrams et al. identified a protein in zebrafish, brambleberry, which is required for karyomere fusion [45•]. Brambleberry mutants fail to fuse their karyomeres but still develop normally, indicating that unlike micronuclei in differentiated cells, these karyomeres are functional. One possible explanation is that the embryonic nuclear lamina stabilizes micronuclei during embryogenesis, since expression of a B-type lamin largely rescued collapse of micronuclei in somatic cells. Or, perhaps embryonic karyomere nuclear envelopes avoid collapse because they need only persist for 30 minutes prior to the next division, compared to the many hours of a typical somatic cell interphase. The potential instability of micronuclei highlights how nuclear morphology can dramatically affect nuclear function. The many diseases associated with defects in nuclear structure, such as laminopathies, further illustrate that less dramatic structural defects than nuclear envelope collapse nevertheless have strong negative consequences.
Conclusions
In this review we presented examples of the relationship between organelles, their surrounding membranes and morphology, and their function. In describing recent studies, we highlighted some of the possible mechanisms determining organelle shape, as well as the functional consequences of altering their structure. Organelles differ in shape because the lipid and protein building blocks involved in determining membrane shape are distinct, resulting in the prototypical organelle shape we see by light and electron microscopy. While textbooks typically present a canonical set of organelle shapes, it is important to remember that most organelles are dynamic, displaying a rather wide range of possible shapes in different cell types, under different conditions and among different organisms.
There is still a lot that we don't understand about organelle morphology, and in particular how organelle size is determined and how morphology contributes to organelle function. For example, at membrane contact sites, the curved nature of ER tubules along with their specific lipid composition [46] may provide an energetically favorable conformation for the detachment of proteins or lipids that move between adjacent membranes. Another example is organelle fragmentation, which may have dual roles: it likely increases the odds of equitable partitioning to daughter cells during mitosis, but it may also serve to increase the surface area to volume ratio under conditions where surface-associated processes need to be up-regulated. Finally, a fascinating question is the reason for a constant nuclear/cell volume ratio: it could simply be a byproduct of protein synthesis rate, which may provide a proportional amount of building blocks for the cell and the nucleus (i.e. both are controlled by the same upstream machinery). Alternatively, this ratio could be kept constant by a dedicated, yet unknown, regulatory mechanism that scales nuclear size to cell size in order to regulate the intra-nuclear concentration of signaling and chromatin associated factors, thereby matching the transcriptional response to the cell's needs. Future studies on organelle structure-function relationships will benefit from identifying the entire repertoire of building blocks that determine organelle morphology, and elucidating how they contribute to organelle and cell function.
Acknowledgements
We thank Will Prinz, Rebecca Meseroll and Alison Walters for excellent comments on the manuscript, and apologize for all those whose work could not be cited due to space limitations. RH is supported by NIH: R01 GM057839 and GM098766. OCF is supported by intramural funding from NIDDK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nakamura N, Wei JH, Seemann J. Modular organization of the mammalian Golgi apparatus. Curr Opin Cell Biol. 2012;24:467–474. doi: 10.1016/j.ceb.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnone JT, Walters AD, Cohen-Fix O. The dynamic nature of the nuclear envelope: Lessons from closed mitosis. Nucleus. 2013;4 doi: 10.4161/nucl.25341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Puhka M, Joensuu M, Vihinen H, Belevich I, Jokitalo E. Progressive sheet-to-tubule transformation is a general mechanism for endoplasmic reticulum partitioning in dividing mammalian cells. Mol Biol Cell. 2012;23:2424–2432. doi: 10.1091/mbc.E10-12-0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bereiter-Hahn J, Jendrach M. Mitochondrial dynamics. Int Rev Cell Mol Biol. 2010;284:1–65. doi: 10.1016/S1937-6448(10)84001-8. [DOI] [PubMed] [Google Scholar]
- 5.Schuck S, Prinz WA, Thorn KS, Voss C, Walter P. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J Cell Biol. 2009;187:525–536. doi: 10.1083/jcb.200907074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tabak HF, Hoepfner D, Zand A, Geuze HJ, Braakman I, Huynen MA. Formation of peroxisomes: present and past. Biochim Biophys Acta. 2006;1763:1647–1654. doi: 10.1016/j.bbamcr.2006.08.045. [DOI] [PubMed] [Google Scholar]
- 7.Huotari J, Helenius A. Endosome maturation. Embo J. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graham TR, Kozlov MM. Interplay of proteins and lipids in generating membrane curvature. Curr Opin Cell Biol. 2010;22:430–436. doi: 10.1016/j.ceb.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Terasaki M, Shemesh T, Kasthuri N, Klemm RW, Schalek R, Hayworth KJ, Hand AR, Yankova M, Huber G, Lichtman JW, et al. Stacked endoplasmic reticulum sheets are connected by helicoidal membrane motifs. Cell. 2013;154:285–296. doi: 10.1016/j.cell.2013.06.031. •• Using ultrathin section electron microscopy of secretory cells, this study shows that stacked ER sheets are connects via helicoidal memrbane structures, resembling the ramps between different levels in a parking garage. This not only demonstrates that stacked ER sheets are connected, but it also uncovers a novel memrbane structure.
- 10.Friedman JR, Voeltz GK. The ER in 3D: a multifunctional dynamic membrane network. Trends Cell Biol. 2011;21:709–717. doi: 10.1016/j.tcb.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Orso G, Pendin D, Liu S, Tosetto J, Moss TJ, Faust JE, Micaroni M, Egorova A, Martinuzzi A, McNew JA, et al. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 2009;460:978–983. doi: 10.1038/nature08280. • Mutations in atlastin, coding for a protein of the dynamin superfamily, were previouly known to be associated with heredetary spastic paraplagia. This study shows that loss of atlasin in flies leads to ER fragmentation and that atlastin can promote memrbane fusion in vitro and in vivo by trans-oligomerization in a GTPasedependent manner. This study established atlastin as a key compenent in homotypic ER memrbane fusion.
- 12. Anwar K, Klemm RW, Condon A, Severin KN, Zhang M, Ghirlando R, Hu J, Rapoport TA, Prinz WA. The dynamin-like GTPase Sey1p mediates homotypic ER fusion in S. cerevisiae. J Cell Biol. 2012;197:209–217. doi: 10.1083/jcb.201111115. • By mating yeast cells expressing differentially labeled ER porteins (fused to two different fluorescent proteins), the authors demontrate that the budding yeast atlastin homolog, Sey1, is involved in homotypic ER fusion. They also showed that in the absence of Sey1 there is residual ER fusion that is dependent on the ER SNARE Ufe1.
- 13.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 14. Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA. Mechanisms determining the morphology of the peripheral ER. Cell. 2010;143:774–788. doi: 10.1016/j.cell.2010.11.007. • By examining the distribution of different ER proteins between ER sheets and ER tubules, the authors show that in animal cells rough ER proteins, namely ribosomes and traslocation proteins, segregate into ER sheets. Using biochemical analysis of ER from secretory cells the authors identified Climp63, which is involved in dictating the constant spacing between the two mambranes of ER cisternae. Finally, the authors show that membrane curving proteins (e.g. reticulons) are present at the edge of ER sheets, perhaps to stablize the high membrane curvature in this region.
- 15.Tzur YB, Wilson KL, Gruenbaum Y. SUN-domain proteins: 'Velcro' that links the nucleoskeleton to the cytoskeleton. Nat Rev Mol Cell Biol. 2006;7:782–788. doi: 10.1038/nrm2003. [DOI] [PubMed] [Google Scholar]
- 16.Rafelski SM. Mitochondrial network morphology: building an integrative, geometrical view. BMC Biol. 2013;11:71. doi: 10.1186/1741-7007-11-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zick M, Rabl R, Reichert AS. Cristae formation-linking ultrastructure and function of mitochondria. Biochim Biophys Acta. 2009;1793:5–19. doi: 10.1016/j.bbamcr.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 18.Puhka M, Vihinen H, Joensuu M, Jokitalo E. Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J Cell Biol. 2007;179:895–909. doi: 10.1083/jcb.200705112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waterman-Storer CM, Salmon ED. Endoplasmic reticulum membrane tubules are distributed by microtubules in living cells using three distinct mechanisms. Curr Biol. 1998;8:798–806. doi: 10.1016/s0960-9822(98)70321-5. [DOI] [PubMed] [Google Scholar]
- 20.Grigoriev I, Gouveia SM, van der Vaart B, Demmers J, Smyth JT, Honnappa S, Splinter D, Steinmetz MO, Putney JW, Jr., Hoogenraad CC, et al. STIM1 is a MT-plus-end-tracking protein involved in remodeling of the ER. Curr Biol. 2008;18:177–182. doi: 10.1016/j.cub.2007.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wozniak MJ, Bola B, Brownhill K, Yang YC, Levakova V, Allan VJ. Role of kinesin-1 and cytoplasmic dynein in endoplasmic reticulum movement in VERO cells. J Cell Sci. 2009;122:1979–1989. doi: 10.1242/jcs.041962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Voss C, Lahiri S, Young BP, Loewen CJ, Prinz WA. ER-shaping proteins facilitate lipid exchange between the ER and mitochondria in S. cerevisiae. J Cell Sci. 2012;125:4791–4799. doi: 10.1242/jcs.105635. • To identify processes that require ER tubulation in budding yeast, the authors screened for mutants in which ER tubulating proteins, reticulons and Sey1, become essential for proper cell growth. They identified mutations in the ERMES complex that was previouly shown to be inovlved in membrane contact sites between the ER and mitochondria. When both ER tubulating proteins and ERMES complex proteins were absent, lipid transfer between the ER and mitochondria was compromized. These findings suggest that ER structure, and in particular ER tubules, is important for the formation of functional ER-mitochondria memrbane contact sites.
- 23.Helle SC, Kanfer G, Kolar K, Lang A, Michel AH, Kornmann B. Organization and function of membrane contact sites. Biochim Biophys Acta. 2013;1833:2526–2541. doi: 10.1016/j.bbamcr.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 24. Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. •• The contact sites between mitochondria and ER in budding yeast were analyzed using electron microscopy and tomography. In many cases, the ER wrapped around the mitochondria, leading to a constriction of the mitochondria that was independent of the mitochondrial fission machinery. Live cell imaging revealed that mitochondrial fission was associated with contact sites between the mitochondrai and ER in both yeast and mammlain cells. The authors proposed that ER-mediated mitochondrial constriction facilitates the subsequent recruitment of the mitochondrial fission machinery.
- 25. Boettcher B, Marquez-Lago TT, Bayer M, Weiss EL, Barral Y. Nuclear envelope morphology constrains diffusion and promotes asymmetric protein segregation in closed mitosis. J Cell Biol. 2012;197:921–937. doi: 10.1083/jcb.201112117. • During anaphase in budding yeast, the nucleus elongates and forms an hourglass shape, with the DNA masses destined for the two daughter cells at the two poles of the nucleus. In this study, the authors show that the narrow neck of this hourglass nucleus provides a diffusion barrier between the two halves of the nucleus. This diffusion barrier allows the selective accumulation of proteins in only one half of the nucleus.
- 26.Zhang D, Vjestica A, Oliferenko S. The cortical ER network limits the permissive zone for actomyosin ring assembly. Curr Biol. 2010;20:1029–1034. doi: 10.1016/j.cub.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 27.Toulmay A, Prinz WA. A conserved membrane-binding domain targets proteins to organelle contact sites. J Cell Sci. 2012;125:49–58. doi: 10.1242/jcs.085118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Manford AG, Stefan CJ, Yuan HL, Macgurn JA, Emr SD. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev Cell. 2012;23:1129–1140. doi: 10.1016/j.devcel.2012.11.004. • In this study, the authors identified fission yeast proteins that participate in tethering the ER to the plasma memrbane. In the absence of these proteins, the ER detaches from the cell periphery. Taking advantage of this phenomenon, the authors used mutants in these tethering proteins to detach peripheral ER that was mostly in the form of sheets from the plasma membrane. They previously showed that when the peripheral ER looses its tubular conformation, a plasma memrbane protein, Mid1, fails to localize properly. Here they show that detachment of these ER sheets from the plasma memrbane restored Mid1 proper localization. The authors suggest that the formation of ER tubules is important to allow proper access of cytosolic proteins to the plasma membrane.
- 29.Zhang D, Vjestica A, Oliferenko S. Plasma membrane tethering of the cortical ER necessitates its finely reticulated architecture. Curr Biol. 2012;22:2048–2052. doi: 10.1016/j.cub.2012.08.047. [DOI] [PubMed] [Google Scholar]
- 30.Stradalova V, Blazikova M, Grossmann G, Opekarova M, Tanner W, Malinsky J. Distribution of cortical endoplasmic reticulum determines positioning of endocytic events in yeast plasma membrane. PLoS One. 2012;7:e35132. doi: 10.1371/journal.pone.0035132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamoto A, DeWald DB, Boronenkov IV, Anderson RA, Emr SD, Koshland D. Novel PI(4)P 5-kinase homologue, Fab1p, essential for normal vacuole function and morphology in yeast. Mol Biol Cell. 1995;6:525–539. doi: 10.1091/mbc.6.5.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smyth JT, Beg AM, Wu S, Putney JW, Jr., Rusan NM. Phosphoregulation of STIM1 leads to exclusion of the endoplasmic reticulum from the mitotic spindle. Curr Biol. 2012;22:1487–1493. doi: 10.1016/j.cub.2012.05.057. •• The authors show that mitotic phosporylation of the ER protein STIM1 dissociates it from the microtubule plus end binding protein EB1. A non-phosphorylatable STM1 mutant causedd ER mislocalization by pulling ER tubules into the mitotic spindle.
- 33. Schlaitz AL, Thompson J, Wong CC, Yates JR, 3rd, Heald R. REEP3/4 Ensure Endoplasmic Reticulum Clearance from Metaphase Chromatin and Proper Nuclear Envelope Architecture. Dev Cell. 2013;26:315–323. doi: 10.1016/j.devcel.2013.06.016. •• Through a biochemical screen, the authors identified REEP4 as an ER membrane and microtubule binding protein. Depletion of REEP4 and its close homologue REEP3 from HeLa cells caused proliferation of intranuclear membranes derived from the nuclear envelope. This defect originated in mitosis as ER membranes accumulated on metaphase chromosomes.
- 34.Brewer JW, Jackowski S. UPR-Mediated Membrane Biogenesis in B Cells. Biochem Res Int. 2012;2012:738471. doi: 10.1155/2012/738471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. •• The authors examined the role of mitochondria and their morphology during autophagy, a cellular recycling process triggered by starvation. They show that mitochondria elongate as cyclic AMP levels increase and activate protein kinase A, leading to phosphorylation and inhibition of the mitochondrial fission protein Drp1 (dynamin related protein 1) and unopposed mitochondrial fusion. If mitochondrial elongation was inhibited in nutrient-deprived cells, mitochondria consumed ATP, which led to starvation-induced death.
- 36. Rambold AS, Kostelecky B, Elia N, Lippincott-Schwartz J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci U S A. 2011;108:10190–10195. doi: 10.1073/pnas.1107402108. •• The authors looked at the effects of starvation on mitochondria in mouse embryo fibroblasts and show that mitochondrial elements become elongated and interconnected soon after nutrient depletion due to phosphorylation and down regulation of the mitochondrial fission protein Drp1 (dynamin related protein 1). Furthermore, mitochondrial tubulation was shown to protect mitochondria from autophagosomal degradation.
- 37.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jorgensen P, Edgington NP, Schneider BL, Rupes I, Tyers M, Futcher B. The size of the nucleus increases as yeast cells grow. Mol Biol Cell. 2007;18:3523–3532. doi: 10.1091/mbc.E06-10-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neumann FR, Nurse P. Nuclear size control in fission yeast. J Cell Biol. 2007;179:593–600. doi: 10.1083/jcb.200708054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rafelski SM, Viana MP, Zhang Y, Chan YH, Thorn KS, Yam P, Fung JC, Li H, Costa Lda F, Marshall WF. Mitochondrial network size scaling in budding yeast. Science. 2012;338:822–824. doi: 10.1126/science.1225720. • The authors measured the physical size of mitochonrdial networks in budding yeast cells and showed that mitochondria size scales with growing cell size primarily in the bud.
- 41. Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell. 2013;154:47–60. doi: 10.1016/j.cell.2013.06.007. •• In this study, the authors show that micronuclei have reduced function compared to primary nuclei in the same cell. Disruption of micronuclei occured as the ER invaded the chromatin. NE collapse was induced by defects in nuclear lamina assembly, and could trigger massive DNA damage.
- 42.Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schoft VK, Beauvais AJ, Lang C, Gajewski A, Prufert K, Winkler C, Akimenko MA, Paulin-Levasseur M, Krohne G. The lamina-associated polypeptide 2 (LAP2) isoforms beta, gamma and omega of zebrafish: developmental expression and behavior during the cell cycle. J Cell Sci. 2003;116:2505–2517. doi: 10.1242/jcs.00450. [DOI] [PubMed] [Google Scholar]
- 44.Montag M, Spring H, Trendelenburg MF. Structural analysis of the mitotic cycle in pre-gastrula Xenopus embryos. Chromosoma. 1988;96:187–196. doi: 10.1007/BF00302357. [DOI] [PubMed] [Google Scholar]
- 45. Abrams EW, Zhang H, Marlow FL, Kapp L, Lu S, Mullins MC. Dynamic assembly of brambleberry mediates nuclear envelope fusion during early development. Cell. 2012;150:521–532. doi: 10.1016/j.cell.2012.05.048. • The authors identified a zebrafish mutant called brambleberry that failed to fuse karyomeres, the micronuclei formed around individual chromosomes, during the rapid embryo cleavage divisions. Brambleberry shares homology with Kar5p, which functions in pronuclear fusion during yeast mating.
- 46.Bigay J, Antonny B. Curvature, lipid packing, and electrostatics of membrane organelles: defining cellular territories in determining specificity. Dev Cell. 2012;23:886–895. doi: 10.1016/j.devcel.2012.10.009. [DOI] [PubMed] [Google Scholar]