Abstract
In the mammalian brain the majority of excitatory synapses are housed in micron-sized dendritic protrusions called spines, which can undergo rapid changes in shape and number in response to increased or decreased synaptic activity. These dynamic alterations in dendritic spines require precise control of the actin cytoskeleton. Within spines, multidomain Rho guanine nucleotide exchange factors (Rho GEFs) coordinate activation of their target Rho GTPases by a variety of pathways. In this review, we focus on the handful of disease-related Rho GEFs (Kalirin; Trio; Tiam1; P-Rex1,2; RasGRF1,2; Collybistin) localized at synapses and known to affect electrophysiology, spine morphology, and animal behavior. The goal is to integrate structure/function studies with measurements of synaptic function and behavioral phenotypes in animal models.
Keywords: protein domain, LTP, receptor localization, knockout mouse
Introduction
Rho family GTPases play essential roles in regulating the complex adjustments to the actin cytoskeleton that underlie structural plasticity in dendritic spines, making the enzymes that regulate them a critical relay system for integrating synaptic activity and intracellular signaling. Rho guanine nucleotide exchange factors (Rho GEFs) catalyze GDP/GTP exchange while Rho GTPase activating proteins (Rho GAPs) accelerate GTP hydrolysis.
The synaptic Rho GEFs are multidomain proteins capable of responding to inputs from several signaling pathways and affecting an array of downstream targets. Proper expression and function of multiple Rho GEFs is necessary for spine formation and synaptic function. While their ability to activate Rho proteins with spatial and temporal precision is critical, the unique ability of the other domains to localize the protein, interact with specific ion channels and respond to intracellular signaling proteins makes the role of each Rho GEF distinct.
A number of recent reviews provide comprehensive summaries of the many Rho GEFs with Dbl homology (DH) domains (Bouquier and others 2009; Kiraly and others 2010a; Mandela and Ma 2012; Penzes and others 2011; Penzes and Jones 2008; Rabiner and others 2005; Rossman and others 2005; Schiller 2006; Tolias and others 2011). We focus here on the handful of Rho GEFs known to affect behavior and synaptic function. Structurally related Rho GEFs are discussed together, with the goal of integrating structure/function studies with measurements of synaptic function and behavior.
Dual Rho GEFs (Kalirin and Trio)
Discovery and expression
Trio was first identified as an interactor with the cytosolic domain of LAR, a receptor protein tyrosine phosphatase localized to focal adhesions (Debant and others 1996). Kalirin was first identified as an interactor with the cytosolic domain of peptidylglycine α-amidating monooxygenase, a secretory granule membrane protein (Alam and others 1996). The proteins are 64% identical (rat Kalirin vs. rat Trio; AF232669.1 vs. XP_003753588.1) (McPherson and others 2002) and, discovered under different circumstances, would likely have been referred to as Trio- or Kalirin-1 and -2. Trio and Kalirin are notable for the presence of two Rho GEF domains of differing specificity along with a Ser/Thr kinase and multiple additional protein-protein interaction domains (Fig.1). A single common ancestor is found in Drosophila (dTrio) and in C.elegans (unc-73). The Drosophila protein is 42–43% identical to both rodent Kalirin and Trio; comparisons to Unc-73 also lead to the conclusion that Kalrn and Trio arose from duplication of a common ancestor.
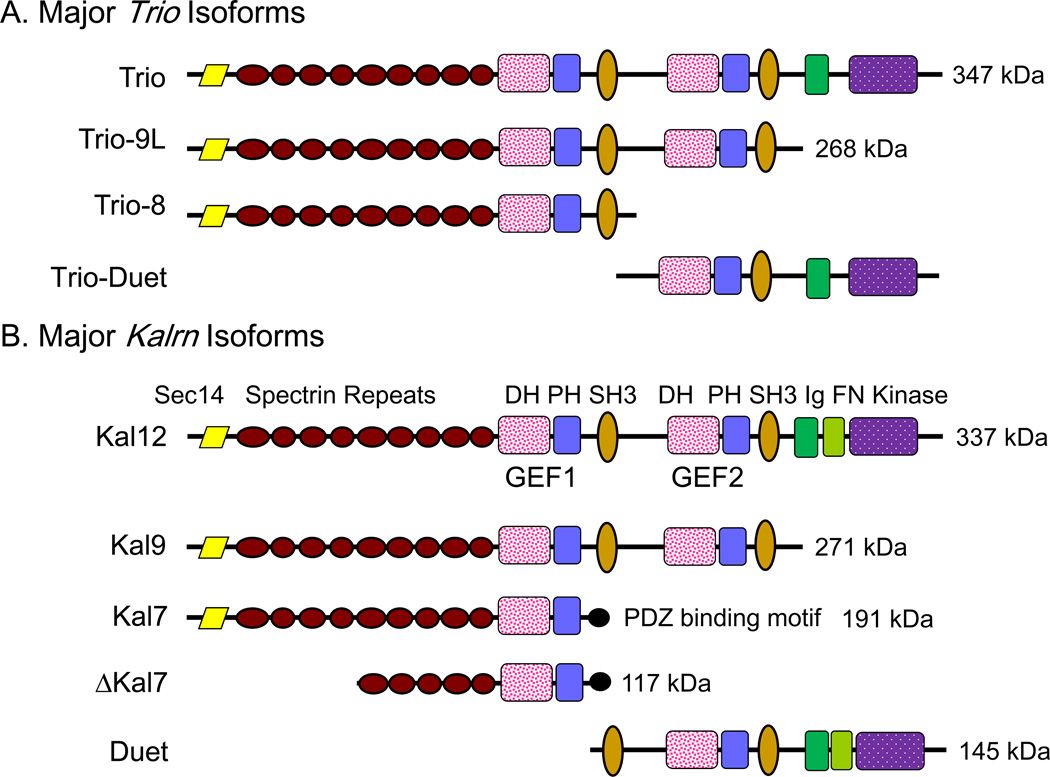
Figure 1. Splice Variants of Kalirin and Trio.
Structural features identified in Kalirin and Trio are drawn to scale; the molecular weights of the intact proteins are indicated. Mouse Kalrn is on chromosome 16 and human KALRN is on chromosome 3. Mouse Trio is on chromosome 15 and human TRIO on chromosome 5. Only the major adult isoforms of Kalirin and Trio, which arise from alternate splicing, are shown. Some of the isoforms are very tissue-specific; Kal7 is expressed exclusively in neurons and Trio-Duet is primarily a cerebellar product, absent from cerebral cortex (McPherson and others 2005). Abbreviations used throughout all figures are: DH, Dbl-homology; PH, pleckstrin homology; SH3, Src3 homology; Ig, immunoglobulin-like; FN, fibronectin-like; PDZ-BD, PDZ binding motif.
Kalirin and Trio are both widely expressed in the brain, primarily localized to neurons at all stages of development (Ma and others 2005; Ma and others 2001; Peng and others 2010). Analysis of dTrio mutants highlighted its role in axon formation and pathfinding (Awasaki and others 2000; Bateman and others 2000; Liebl and others 2000) Trio is most importantly expressed in cerebellar granule cells, while Kalirin is most highly expressed in long projection neurons in the cortex and hippocampus, with less expression in the cerebellum and brainstem. The developmental expression patterns of both genes are isoform- and region-specific. Expression of Kalrn and Trio is regulated independently; for example, compensatory increases in Trio expression have not been observed in Kalrn knockout mice (Ma and others 2008b).
Importantly, both genes are also expressed outside of the nervous system. Kalirin, primarily the Kal9 isoform (Fig.1A), is expressed early in development in a wide variety of tissues throughout the body (Hansel and others 2001), primarily lung, muscle, GI epithelium and pancreas. In the adult, Kal7 is the major form in mature neurons, while Kal9 and Kal12 are found at lower levels in aorta, skeletal muscle and endocrine tissues. Kal7 is primarily expressed in postsynaptic structures such as dendritic spines, while Kal12 is especially elevated in growth cones (Xin and others 2009) (Fig.2A). Expression of Kal7 correlates with the onset of synaptogenesis in the cortex.
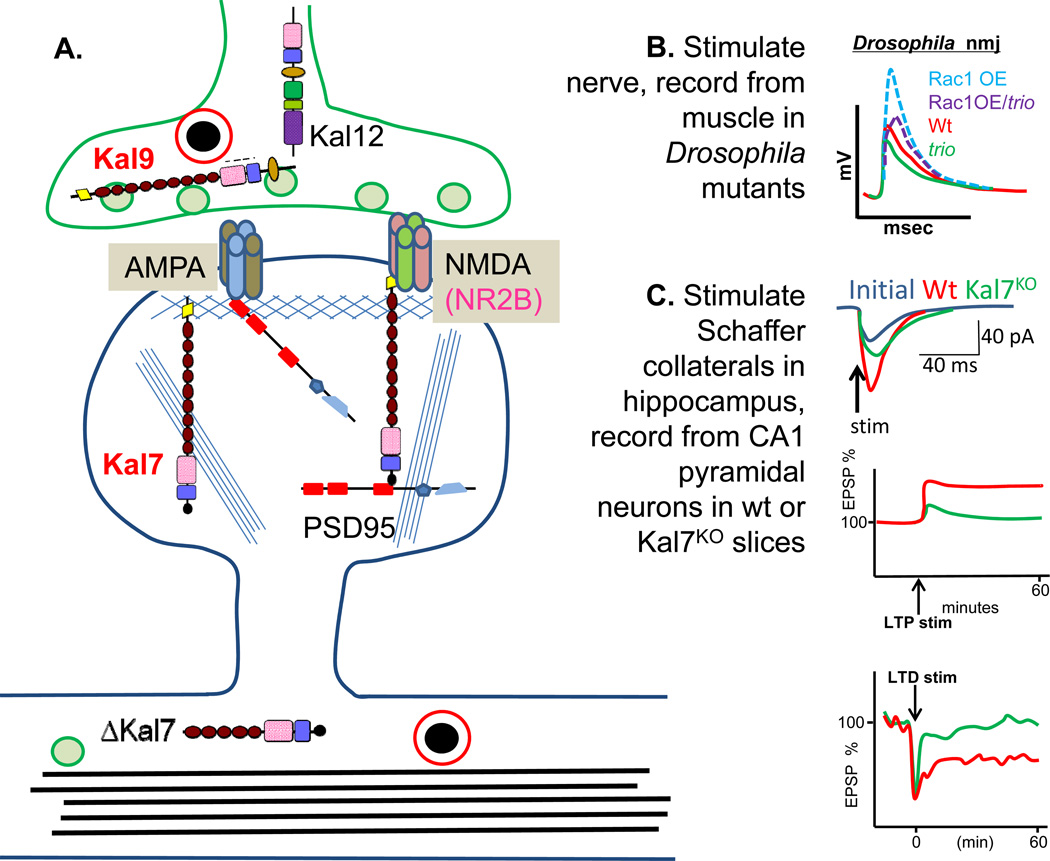
Figure 2. Kalirin and Trio: Synaptic Function.
A. The major sites at which different Kalirin isoforms are localized in neurons are indicated. B. At the Drosophila neuromuscular junction, the dTrioKO (green, trio) shows a much smaller endplate potential than the wildtype (red, Wt) (Ball and others 2010). Over-expression of Rac1 in Wt neurons (dashed red, Rac1/Wt) enhances the endplate potential, and over-expression of Rac1 in dTrioKO neurons (dashed green, Rac1/trio) compensates for the absence of trio. Thus both Kalirin and Trio are likely to enhance synaptic transmission in part by activating Rac1 via their GEF1 domain. C. The Kal7KO mouse shows significantly blunted LTP in hippocampal and cortical neurons compared to wildtype (Wt) mice (green, Kal7KO; red, Wt), and a similar loss of LTP is produced by pharmacological blockade of KalGEF1 (Lemtiri-Chlieh and others 2011; Ma and others 2008b). Kal7KO neurons show little or no LTD.
Protein structure and signaling
Both genes undergo extensive alternative splicing, yielding functionally distinct isoforms ranging from about 100 kDa to nearly 400 kDa in size (Fig.1). For both Kalirin and Trio, GEF1 activates Rac1 and RhoG, while GEF2 activates RhoA; splice variants that include only one or the other GEF domain have been identified. Notably, the kinase domain encoded by Kalrn and Trio is absent from dTrio and unc-73. Both the Sec14 domain of Kalirin, which binds specific phosphoinositides, and its nine spectrin repeats play essential roles in its function (Schiller and others 2008). The spectrin repeat region, SH3 domains and Ig/FN domains of Kalirin and Trio interact with a wide variety of proteins including iNOS, DISC1, Arf6, afadin, PAM, and HAP1.
Recombinant Kal7 is phosphorylated by several of the protein kinases concentrated at the PSD; consistent with this, Kal7 isolated from rat brain is heavily phosphorylated (Kiraly and others 2011b). Several sites in Kalirin are targeted by multiple protein kinases (e.g. protein kinase A, protein kinase C, CaMKII and CKII), leading to the suggestion that Kalirin participates in signaling pathways initiated by a variety of pathways (Kiraly and others 2011b; Penzes and others 2003).
Detailed examination of mutations in unc-73 established a role for its first GEF domain in neuronal development (Williams and others 2007) and a role for its second GEF domain in adult neuronal function (Chan and others 2012; McMullan and others 2012; Williams and others 2007). In D.melanogaster, a similar picture emerges; the N-terminal half of dTrio, which includes GEF1, is crucial to neurite outgrowth (Awasaki and others 2000; Bateman and others 2000; Luo 2000), while neuromuscular junction establishment and function require the GEF2 region (Astigarraga and others 2010; Ball and others 2010). Liprin, a cytoplasmic binding partner of LAR, is thought to act through dTrio to promote correct targeting of axons, while retrograde BMP signaling from muscle to the nerve terminal acts through dTrio to regulate synapse development (Astigarraga and others 2010; Ball and others 2010). Expression of BMP in muscle increases dTrio expression in motorneurons quite dramatically (Ball and others 2010).
The GEF2 domain of Trio and Kalirin shares with p63RhoGEF the ability to bind to and be activated by Gαq, placing these RhoGEFs downstream of selected G protein coupled receptors (Lutz and others 2007; Rojas and others 2007). Trio and Gαq co-immunoprecipitate when co-expressed in hEK293 cells (Williams and others 2007). Acetylcholine acting through a muscarinic receptor to activate Gαq results in increased sphingosine kinase activation and increased levels of sphingosine-1-P in the presynaptic terminal, potentially activating Trio or Kalirin GEF2 at the same time and increasing neurotransmitter release (Chan and others 2012; McMullan and others 2012; Williams and others 2007).
Behavior
The Trio−/− mouse dies in late gestation (O'Brien and others 2000) with spherical myofibers and major defects in secondary myogenesis. Subsequent work (Briancon-Marjollet and others 2008) revealed defective neuronal outgrowth, complete lack of the anterior commissure and defects in the internal capsule and corpus callosum. These defects reflect loss of the interaction of Trio with the netrin-1 receptor, Deleted in Colon Cancer (DCC), which is essential for normal Rac1 activation in axonal projections. Because of the lethality of the global Trio−/− knockout, cell-type specific knockouts using Cre recombinase expression driven by the nestin or GFAP promoter were created (Peng and others 2010). Only 10% of Trionestin-KO mice survived for a day; they lacked a cerebellum, were severely ataxic and all died within 3 weeks without ever gaining significant weight. In contrast, TrioGFAP-KO mice appeared unaffected. Southern blot analysis demonstrated complete excision of the floxed allele in brain tissue from Trionestin-KO mice (Peng and others 2010), consistent with early expression of nestin-Cre and elimination of Trio expression in glial, as well as neuronal, lineages.
Knockout of one copy of dTrio results in >50% decrease in Rac1-dependent synaptic bouton numbers (Ball and others 2010). Rac1 overexpression increases the quantal content of neuromuscular junction excitatory junction potentials (EJP), with no change in the unitary size of the quanta (Ball and others 2010) (Fig.2B). Likewise dTrio knockout decreased the quantal content of EJPs, and the EJP enhancement seen with Rac1 overexpression was largely blocked in the dTrio knockout. Interestingly, bone morphogenic protein (BMP) from the muscle is required for normal dTrio expression in the innervating neuron, and decreasing dTrio expression directly (as in trio/+ flies) or indirectly (BMP lowered) results in reduced neuromuscular junction growth; expression of dTrio in motorneurons, but not in the muscle, reverses the deficit. The interpretation is that the level of expression of neuronal dTrio, and consequent activation of Rac1, directly correlates with the size of the presynaptic terminal and thus the quantal content of the EJPs (Ball and others 2010).
Two approaches were used to generate mouse models lacking all of the major Kalrn isoforms (KalGEF1KO and KalSRKO) (Mandela and others 2012; Xie and others 2011) and Kal7 was specifically targeted in another model (Kal7KO) (Ma and others 2008b); each yielded viable homozygous knockout mice. In KalGEF1KO and Kal7KO mice, the density of hippocampal dendritic spines is decreased compared to normal (Ma and others 2008b; Xie and others 2011). Cortical cultures prepared from Kal7KO mice showed almost a 50% decrease in synaptic density; in addition, Vglut1-positive puncta that were not apposed to a PSD-95 positive postsynaptic ending were observed commonly in Kal7KO cultures but only rarely in wildtype neurons, suggesting a role for Kal7 in synaptic maturation (Ma and others 2008b).
Although lack of Rac1 and RhoG activation in the Kal7KO neurons plays a major role (May and others 2002), loss of other protein-protein interactions is also of importance. The first PH domain of Kalirin interacts with the juxtamembrane region connecting the final transmembrane domain of the NR2B subunit of the NMDA receptor with its cytosolic domain (Kiraly and others 2011a). Loss of this interaction presumably contributes to the decrease in NR2B-containing NMDA receptors observed in PSDs isolated from Kal7KO mice (Kiraly and others 2011a; Lemtiri-Chlieh and others 2011). Genotypic differences in several of the behavioral deficits observed in Kal7KO mice (conditioned place preference for cocaine and passive avoidance fear conditioning) were largely abrogated by pretreatment with ifenprodil, a selective blocker of NR2B-containing NMDA receptors, confirming the importance of the Kalirin/NR2B interaction (Kiraly and others 2011a). Kal7KO mice show enhanced locomotor sensitization to cocaine administration (Kiraly and others 2010b).
A striking attribute of both the Kal7KO and the KalSRKO mice is the decrease in anxiety-like behavior on the elevated zero maze, along with an inability to acquire a fear-based passive avoidance task normally (Ma and others 2008b; Mandela and others 2012). Similarly, context and cued fear-based conditioning was impaired in the KalGEF1KO mouse (Xie and others 2011). Deficits in rotarod performance and in the neuromuscular junction were observed in KalSRKO mice, but not in Kal7KO mice; a role for Kalirin/Trio at the neuromuscular junction is consistent with observations made in Drosophila. Analysis of KalSRKO/+ mice revealed a role for Kal9 in the activation of Rac1 in smooth muscle cells and in smooth muscle cell migration and proliferation (Wu and others 2012).
Electrophysiology and synaptic function
When hippocampal LTP was recorded using a theta burst pattern paired with a depolarizing pulse in the clamped cell, pyramidal neurons in acute slices from Kal7KO animals showed less than half as much potentiation as seen using wildtype slices (Fig.2C) (Ma and others 2008b). Further studies established that the NMDA/AMPA current ratio in Kal7KO mice was depressed to half in layer 2/3 cortical neurons, compared to the value in wildtype mice, which almost guarantees the LTP deficit seen in the Kal7KO mice (Kiraly and others 2011a). The decreased amount of NR2B subunit present at the PSD and on the cell surface is presumably a direct result of the loss of Kalirin PH domain binding to the juxtamembrane domain of the NR2B subunit (Kiraly and others 2011a). A similar diminution in LTP was observed in KalGEFKO mice vs. wildtype mice, using a high frequency stimulation paradigm to induce LTP (Xie and others 2011). Additional electrophysiological studies demonstrated that non-NMDA receptor-dependent LTP was normal in Kal7KO mice, while NMDA receptor-dependent LTD was abolished (Fig.2C) (Lemtiri-Chlieh and others 2011), focusing attention on NMDA receptor related deficiencies. Bath application of NPPD, a Kalirin/Trio GEF1-selective blocker, greatly decreased LTP as did the loss of the Kal7 protein, implicating the GEF1 domain in the electrophysiological changes seen in Kal7KO mice (Lemtiri-Chlieh and others 2011). The decreased paired pulse ratio (2nd evoked EPSC not as much larger than the closely paired 1st evoked EPSC as in wildtype) observed in KalGEF1KO mice (Xie and others 2011) may reflect decreased levels of Kal9 and Kal12. Kal9 and Kal12 are known to occur in presynaptic terminals and growth cones (Xin and others 2009), and their loss may underlie some of the hormone secretion abnormalities observed in the KalSRKO mice (Mandela and others 2012).
The Kal7KO mouse was studied extensively, using both hippocampal and cortical slices. Correlating with a 15% decrease in dendritic spine density, recordings from hippocampal slices showed a 50% decrease in sEPSC frequency with no change in sEPSC amplitude, as might be predicted if not all of the remaining dendritic spines are fully functional (Ma and others 2008b). Similarly, the mEPSP frequency was significantly lower in frontal cortex slices from KalGEF1KO mice, again with normal amplitude (Cahill and others 2009).
Disease relevance
Mutations at the KALRN locus have been strongly associated with schizophrenia (Kushima and others 2012) and early onset coronary artery disease (Wang and others 2007). Loss of one copy of Kalrn produces mice with arterial smooth muscle cells which show reduced proliferation, migration and reduced Rac1 activation after carotid endothelial denudation (Wu 2012). Alzheimer’s patients have significantly less Kal7 in hippocampal extracts than non-demented patients (Youn and others 2007). Kal7 mRNA and protein are increased in both rats and mice in response to cocaine (Kiraly and others 2010b; Ma and others 2012; Mains and others 2011); since Kal7 over-expression leads to increased dendritic spines in a number of neuronal systems (Ma and others 2008c), it is not surprising that Kal7KO mice do not exhibit the usual increase in nucleus accumbens dendritic spine numbers in response to chronic cocaine (Kiraly and others 2010b). Chronic prolonged restraint stress has the opposite effect, decreasing the level of Kal7 in the hippocampus to half the normal level (Li and others 2010).
T-cell lymphoma invasion and metastasis family (Tiam1 and Tiam2)
Discovery and expression
Tiam1 was identified as a T-lymphoma invasion and metastasis-inducing gene by retroviral insertional mutagenesis and encodes a GEF specific for Rac (Habets and others 1994; Michiels and others 1995). Tiam1 is widely expressed in the developing central nervous system, and expression continues at postnatal stages (E14.5 to P180), with an established role in neuronal migration and axonal extension both in vitro and in vivo (Ehler and others 1997; Kawauchi and others 2003). Subcellularly, Tiam1 is localized to dendrites and dendritic spines, where it is enriched at the PSD (Tolias and others 2005). Tiam2, a closely related gene, is widely expressed in both in mouse and human brain (Chiu and others 1999).
Due to its roles outside of the CNS, Tiam1 has been extensively studied in cancer research and T-cell migration (Gerard and others 2009; Malliri and others 2002). A recent study demonstrated that Tiam1-Rac1 signaling antagonizes centrosome separation during prophase and is required to balance Kinesin-5-induced forces during bipolar spindle assembly (Woodcock and others 2010). These results suggest that Tiam1 controls cell division, and the disruption of Tiam1 in neuroepithelial cellssupports the observation of severe brain abnormalities in one Tiam−/− mouse line (Yoo and others 2012).
Protein structure and function
Tiam1 is a multi-domain protein with a myristoylation site at its N-terminus; two PEST sequences, a PH-CC-Ex domain (a PH domain, a putative coiled-coil region, and a conserved extra region), a Ras-binding domain and a PDZ domain precede the catalytic DH domain, which is followed immediately by a PH domain and a PDZ binding motif at the C-terminus (Fig.3A). Tiam2, also referred to as STEF (SIF and Tiam1 like-exchange factor), lacks a myristoylation signal and PEST sequences but otherwise shares significant homology to Tiam1; portions of Tiam1 and Tiam2 are 70% identical (Chiu and others 1999; Hoshino and others 1999). Knocking down Tiam1 and Tiam2 using antisense oligonucleotides or shRNAs affects neurite length and morphology (Goto and others 2011; Kunda and others 2001), suggesting that both proteins contribute to neurite outgrowth; we focus on Tiam1 because it is necessary for dendritic arbor and spine formation.
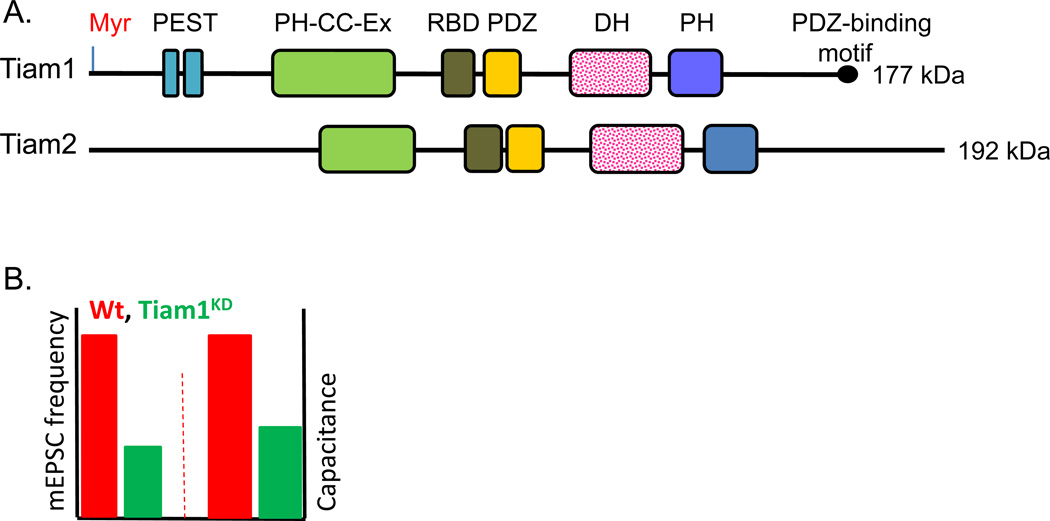
Figure 3. Tiam1 and Tiam2.
A. Structure of Tiam1 (TIAM1, human 21q22.11) (177 kDa) and Tiam2 (TIAM2, human 6q25.2) (192 kDa). B. Tiam1 RNAi in rat hippocampal neurons results in a ~62% decrease in mEPSC frequency and a decrease in membrane capacitance compared to the control neurons, suggesting that Tiam1 knockdown results in fewer synapses and smaller cell size (Tolias and others 2005).
Behavior
Tiam1 knockout mouse lines were created by Malliri et al. (Malliri and others 2002) and by Yoo et al.(Yoo and others 2012). Tiam1−/− mice generated by ablating exon 2 grow and reproduce normally (Malliri and others 2002); these mice are primarily used in cancer biology and immunology research. The other Tiam1 knockout mouse line was generated using a Tiam1 gene-trapped embryonic stem cell line, which targets the intron separating exons 5 and 6, potentially creating a truncated Tiam-β-galactosidase fusion protein (Yoo and others 2012). Although no other genes are known to lie in the targeted region, these E9.5 and E10.5 Tiam−/− embryos exhibit anencephaly and exencephaly and do not survive beyond E13.5, indicating that Tiam1 is important for early forebrain development (Yoo and others 2012). Despite high expression levels in the brain and an obvious role in CNS function, to our knowledge there have been no behavioral or electrophysiological studies performed with homozygous or heterozygous Tiam1−/− animals.
Electrophysiology and synaptic function
RNAi-mediated knockdown of Tiam1 in rat hippocampal neurons dramatically reduces dendritic arborization and spine density (Tolias and others 2005; Tolias and others 2007; Zhang and Macara 2006) as well as frequency of spontaneous miniature excitatory postsynaptic currents (mEPSC) (Fig.3B). Decreasing Tiam1 inhibits neuronal migration and prevents axon formation, with most neurons exhibiting smaller growth cone size and abnormal axon cytoskeletal organization (Kunda and others 2001; Matsuo and others 2003) as well as neuronal migration (Kawauchi and others 2003). MAP1B interacts directly with Tiam1 (Fig.4), and over-expression of Tiam1 rescues the axonal growth defects observed in MAP1B-deficient neurons. These data suggest that the MAP1-Tiam1 interaction plays an important role in Rac1 regulation and is required for axonal elongation (Fig.4) (Montenegro-Venegas and others 2010).
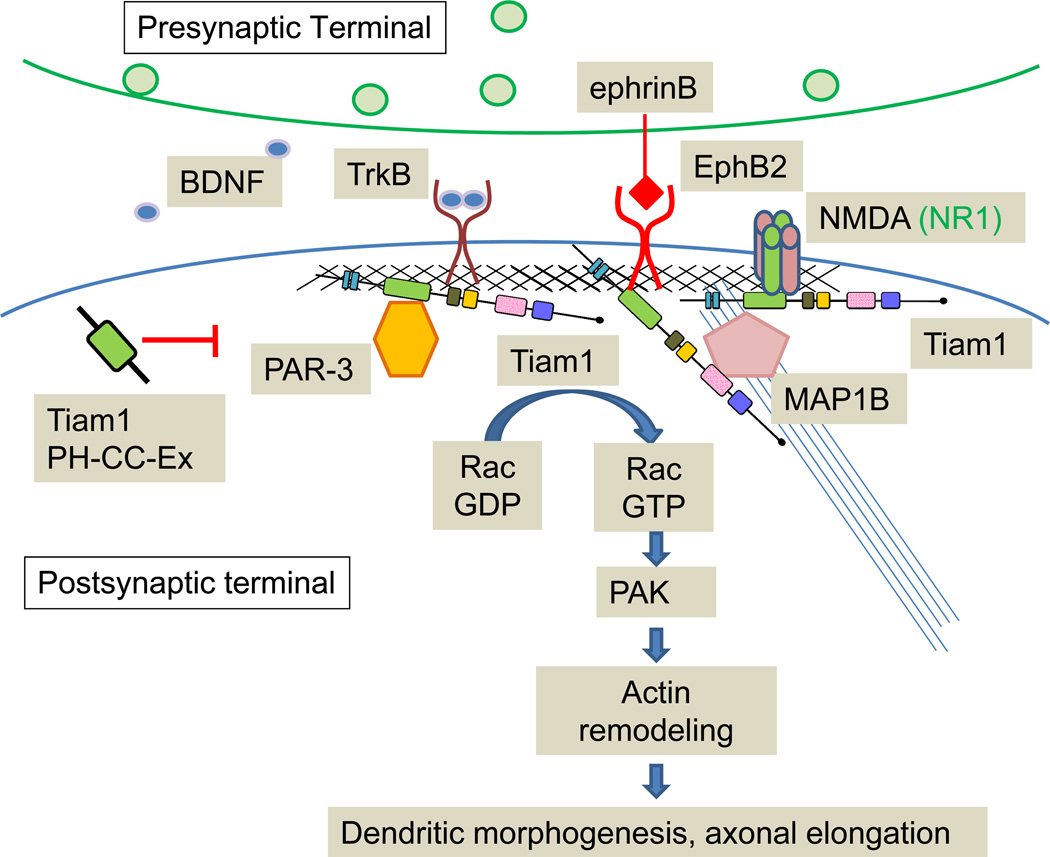
Figure 4. Tiam1: Synaptic Functions.
Schematic illustrates proposed signaling mechanisms of Tiam1 at the synapse. Tiam1 interacts with many postsynaptic proteins including the NMDA receptor, EphB, MAP1B, PAR-3 and TrkB, which controls dendritic morphogenesis and axon elongation.
Tiam1 over-expression induces extension of multiple axon-like neurites (Kunda and others 2001; Matsuo and others 2003), and also results in increased dendritic spine density (Tolias and others 2005), further suggesting a roles in spine and axon formation. Tiam1 interacts directly with the NR1, but not any of the NR2 subunits of the NMDA receptor, and is phosphorylated in a Ca2+-dependent manner following NMDA receptor stimulation. Knocking down Tiam1 expression in rat cortical neurons blocks NMDA receptor-induced increases in dendritic spine density, indicating that Tiam1-dependent Rac1 activation is required for NMDA receptor-mediated spine formation (Tolias and others 2005).
The PH-CC-Ex domain of Tiam1 interacts with the EphB2 receptor tyrosine kinase in a kinase-dependent manner. RNAi-mediated knockdown of Tiam1 expression blocks the ability of ephrin B1 to increase dendritic spine density in rat hippocampal neurons (Tolias and others 2007). Interestingly, the Tiam1 PH-CC-Ex region acts in a dominant-negative manner, probably by binding to Tiam1 interacting proteins and preventing recruitment of endogenous Tiam1 to the membrane (Fig.4) (Tolias and others 2005; Tolias and others 2007). The crystal structure of PH-CC-Ex reveals that this region is a novel protein- and membrane-binding module, strongly binding the NMDA receptor loop I peptide (residues 591–610) and not the C-terminal tail peptide (residues 921–938) (Fig. 4) (Terawaki and others 2010). This type of interaction is reminiscent of the binding of Kalirin-PH1 to the juxtamembrane domain of the cytoplasmic tail of NR2B (Kiraly and others 2011a).
Tiam1 also binds to partitioning-defective gene 3 (PAR-3), a member of the PAR polarity complex, which contributes to spine morphogenesis (Zhang and Macara 2006). PAR-3 spatially restricts Tiam1 to dendritic spines, contributing to proper spine formation (Fig. 4) (Zhang and Macara 2006). Finally, Tiam1 is involved in the BDNF-TrkB signaling pathway. Tiam1 is required for BDNF-induced spine morphogenesis and neurite extension; BDNF-activated TrkB directly binds to and activates Tiam1 by phosphorylating Tyr829, and a point mutation at this residue blocks BDNF-induced morphological changes (Lai and others 2012; Miyamoto and others 2006). In maturing postnatal cerebellar granule cells, BDNF up-regulated expression of Tiam1 and the NR2C NMDA receptor via the TrkB-Erk cascade. This up-regulation is blocked not only by inhibition of signaling pathways, but also by suppression of the Etv1/Er81 transcription factor using Etv1 siRNA (Fig. 4) (Abe and others 2012).
Disease relevance
In hippocampal neurons, Tiam1 is activated and recruited to membranes following exposure to amyloid beta peptide (Aβ), and thus may be involved in Alzheimer’s disease pathology (Ma and others 2008a; Mendoza-Naranjo and others 2007). In addition to the roles of Tiam1 in the nervous system, Tiam1−/− mice are resistant to Ras-induced skin tumors, suggesting that Tiam1 is a critical regulator of tumor formation and invasion (Malliri and others 2002; Vigil and others 2010).
Phosphatidylinositol (3,4,5)-trisphosphate-dependent Rac Exchange Factor Family (P-Rex1 and P-Rex2)
Discovery and expression
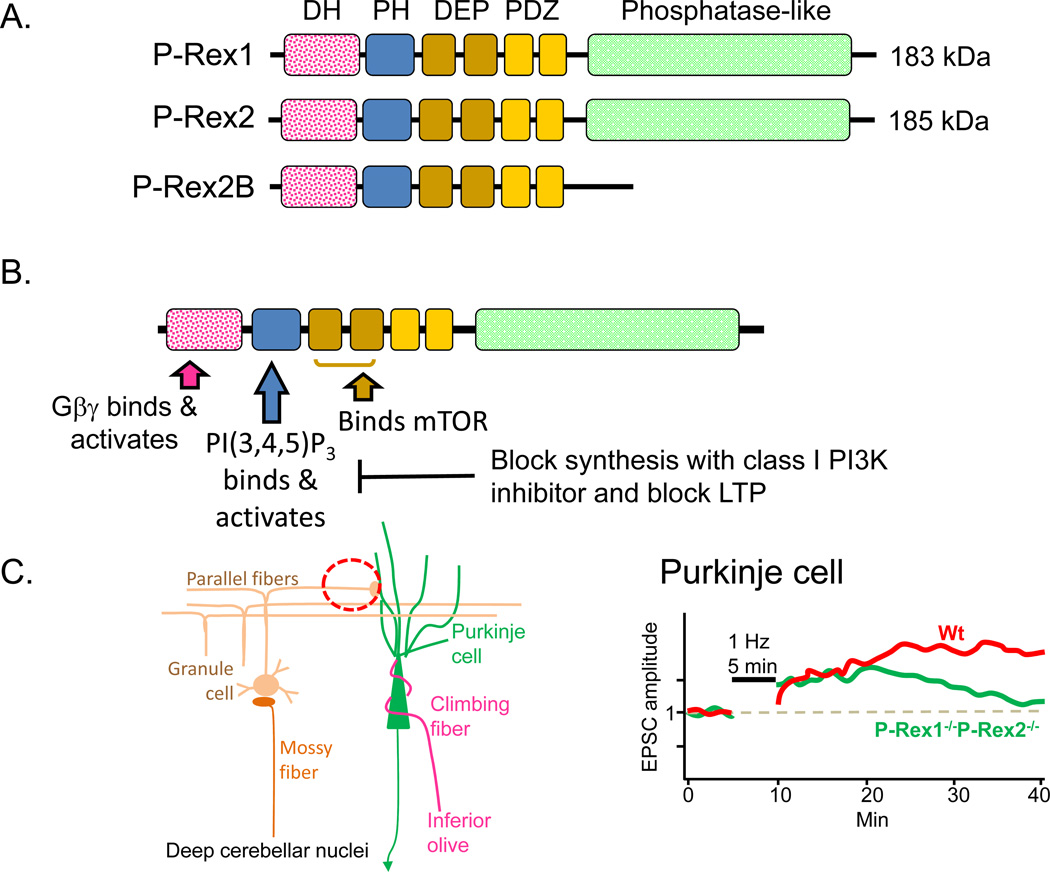
Two gene family members have been characterized in human (PREX1, PREX2) and mouse (Prex1, Prex2) (Fig.5A). P-Rex1 was purified from neutrophil cytosol on the basis of its phosphatidylinositol(3,4,5)trisphosphate [PI(3,4,5)P3]-sensitive Rac GEF activity (Welch and others 2002); Rac activation in these cells plays an essential role in chemotaxis, phagocytosis and reactive oxygen species (ROS) formation. The Rac GEF activity of purified P-Rex1 was also shown to be stimulated by the addition of recombinant Gβ1γ2 (Welch and others 2002; Hill and others 2005). The effects of PI(3,4,5)P3 and Gβγ on Rac activation are synergistic, leading to the concept that P-Rex1 could serve as a coincidence detector for the simultaneous activation of both signaling pathways. P-Rex2, which was identified by homology, is 59% identical to P-Rex1. As for P-Rex1, PI(3,4,5)P3 and Gβγ exert a synergistic effect on the ability of P-Rex2 to activate Rac1 (Donald and others 2004).
Figure 5. P-Rex1 and P-Rex2.
A. Human P-Rex1 (PREX1, 20q13.13) (185 kDa) accounts for 0.1% of the cytosolic protein in neutrophils and is also highly expressed in brain (Welch and others 2002). Human P-Rex2 (PREX2, 8q13.2) (183 kDa) is not expressed in neutrophils, but is highly expressed in skeletal muscle, heart and placenta. Mouse P-Rex1 and P-Rex2 are on Chr2 and 1, respectively. In addition to a DH and adjacent PH domain, both full-length proteins include two DEP (Disheveled, EGL-10, Pleckstrin) domains, two PDZ domains and an inositol polyphosphate 4-phosphatase-like domain; the phosphatase domain is absent from P-Rex2B, a 980 amino acid splice variant expressed only in the heart (Donald and others 2008). B. Key signaling pathways known to affect P-Rex1 function are identified, along with some of its major downstream targets in the nervous system. The DEP and PDZ domains play a role in protein-protein interactions and membrane targeting; phosphorylation of P-Rex1 by PKA disrupts these interactions and diminishes the ability of Gβγ to bind to and activate P-Rex1 (Balamatsias and others 2011). C. The cell types involved in the parallel fiber Purkinje cell synapses examined electrophysiologically are illustrated (red dashed circle). Parallel fiber stimulation causes LTP at wildtype synapses; while P-Rex1−/−P-Rex2−/− mice produce a similar response immediately after stimulation, the response is not sustained (Jackson and others 2010).
Northern blot analysis of human tissue reveals high levels of P-Rex1 mRNA in neutrophils, with slightly lower levels in brain (Donald and others 2008; Welch and others 2002). P-Rex1 mRNA expression in brain is readily detected from E13 through adulthood, with levels highest from E17 through P7 (Yoshizawa and others 2005). Based on Western blot analysis, P-Rex1 expression is widespread, but highest in the cerebellum (Donald and others 2008). In situ hybridization reveals a complex and rapidly changing pattern for P-Rex1, with expression in cerebral cortex, trigeminal ganglion and dorsal root ganglia readily apparent at E12 (Donald and others 2008). In the adult, expression of P-Rex1 is most notable in layers II/III of the cortex and in the Purkinje cell layer of the cerebellum. In tissue sections and cultured neurons, endogenous P-Rex1 protein localizes to the shafts of neurites (Yoshizawa and others 2005). As hippocampal neurons differentiate in culture, P-Rex1 localizes to the distal part of the axon and axonal growth cones; expression of exogenous P-Rex1 enlarges axonal, but not dendritic, growth cones (Waters and others 2008).
P-Rex2 mRNA is most highly expressed in heart, skeletal muscle and placenta, but is also present in adult brain (Donald and others 2004). Western blots reveal similar levels of P-Rex2 in brain and lung (Donald and others 2008); in situ hybridization and immunostaining identify cerebellar Purkinje cells as the major site of P-Rex2 expression in adult brain, with lower levels in frontal cortex, striatum, amygdala and hippocampus.
Protein structure and function
The catalytic Dbl homology domain of P-Rex occurs near its N-terminus, followed by a PH domain, two DEP (Disheveled, EGL-10, Pleckstrin) domains, two PDZ domains and an inositol polyphosphate 4-phosphatase-like domain (Fig.5A). PI(3,4,5)P3 binds to the PH domain while Gβγ subunits interact with the DH domain (Fig.5B); binding of either PI(3,4,5)P3 or Gβγ stimulates the ability of P-Rex to activate Rac, with a synergistic effect of the two activators. P-Rex1 activates Rac1 and Rac3 (Rac1B), a neuron-specific Rho GTPase (Waters and others 2008).
Intact P-Rex1 is 60-fold less active than its isolated DH/PH domain; the DEP and PDZ domains play a role in keeping P-Rex1 inactive. PI(3,4,5)P3 increases the activity of the isolated DH/PH domain of P-Rex1, but has a far greater effect on the activity of full-length P-Rex1. Gβγ seems to require only the DH domain to activate P-Rex1; as for PI(3,4,5)P3, the effect of Gβγ on Rac activation is substantial (Hill and others 2005). The ability of the Gβγ dimer released upon GPCR stimulation to activate P-Rex1 depends on its Gγ subunit (Mayeenuddin and others 2006). A yeast two-hybrid screen using the tandem DEP domains of P-Rex1 and a human brain cDNA library identified the C-terminal region of mammalian target of rapamycin (mTOR), including its serine/threonine kinase domain, as an interactor (Fig.5B) and both P-Rex1 and P-Rex2 have been identified as components of the mTORC2 complex (Hernandez-Negrete and others 2007). Phosphorylation of P-Rex1 by PKA alters its intramolecular interactions and prevents its binding and activation by Gβγ (Urano and others 2008).
The activation of Rac1 and Rac2 in neutrophils triggers a variety of responses including chemotaxis, generation of ROS and secretion of azurophilic granules. It is striking that the absence of P-Rex1 has quite distinct effects on these responses (Welch and others 2005). The ability of unprimed or tumor necrosis factor α-primed P-Rex1−/− neutrophils to produce ROS in response to formyl-Met-Leu-Phe (fMLP) is only slightly diminished; in contrast, the ability fMLP to stimulate ROS production by lipopolysaccharide-primed neutrophils is almost entirely eliminated (Welch and others 2005).
The GEF activity of P-Rex1, which is expressed endogenously in PC12 pheochromocytoma cells, is activated by NGF, resulting in increased membrane ruffling and cell motility (Yoshizawa and others 2005). Along with the DH domain, the PDZ and phosphatase-like domains of P-Rex1 play essential roles in its ability to stimulate membrane ruffling. PC12 cell migration in response to NGF is greatly increased upon expression of exogenous P-Rex1; the process is dependent on activation of Rac1. Deletion of the DH domain of P-Rex1 (ΔDH-P-Rex1) yielded a dominant negative variant of P-Rex1, which blocks NGF-induced PC12 cell migration.
Behavior
P-Rex1, P-Rex2 and P-Rex1/P-Rex2 knockout mice have been characterized. P-Rex1−/− mice are viable and healthy, perform normally in tests of motor behavior, and are slightly smaller than wildtype mice (Donald and others 2008; Welch and others 2005) P-Rex2−/− mice are viable and fertile although females have lower than normal body weight (Donald and others 2008). The brains of P-Rex2−/− mice show no major anatomical defects, but Purkinje cell dendritic structure is abnormal. Rotarod performance in adult female P-Rex2−/− mice is somewhat compromised, while male performance is less affected (Donald and others 2008). Basic sensory motor functions and gait are normal. P-Rex1−/−/P-Rex2−/− mice are viable and fertile, with sex-specific changes in body weight. Brain anatomy is grossly normal, with disordered Purkinje cell dendrites, as observed in the P-Rex2−/− mouse brain. The motor behavior of male and female P-Rex1−/−/P-Rex2−/− mice is more impaired than that of P-Rex2−/− mice, with substantial deficits in rotarod performance, open field behavior, beam walking and gait (Donald and others 2008). Deficits worsen with age and are more pronounced in females than in males.
Electrophysiology and synaptic function
The prevalence of P-Rex1 and P-Rex2 in cerebellar Purkinje cells, the altered Purkinje cell dendritic morphology and altered motor function observed in mice lacking both P-Rex proteins identified parallel fiber/Purkinje cell synapses as a likely site at which to observe alterations in synaptic transmission (Fig.5C). LTD at these synapses, which can be evoked by co-stimulation of parallel fiber and climbing fiber inputs, involves increased phosphorylation of AMPA receptor GluR2 subunits, which promotes their internalization and decreases the response to transmitter release (Jackson and others 2010). The parallel fiber-Purkinje synapse is essential for the eye-blink reflex.
LTP at these same synapses can be evoked by stimulating the parallel fibers alone; although less well understood than LTD, it is clear that LTP involves phosphatase-mediated reversal of GluR2 phosphorylation and decreased AMPA receptor internalization. Consistent with a role for P-Rex, inhibition of PI3K abolishes LTP entirely. Synthesis of NO is required for LTP at these synapses and NO donors can evoke LTP. P-Rex1 and P-Rex2 are activated by PI(3,4,5)P3, the product of Class I PI3Ks, which are essential for the induction of hippocampal LTP and NMDA receptor evoked increased surface expression of GluR1-containing AMPA receptors in hippocampal neurons.
Sagittal slices from the cerebellar vermis of P21-P28 mice, were used (Jackson and others 2010); despite the altered dendritic morphology observed in P-Rex1−/−/P-Rex2−/− mice, synaptic transmission, short-term plasticity and passive membrane properties were unaltered. LTP was induced at parallel fiber Purkinje cell synapses by stimulating at 1 Hz for 5 min. In wildtype mice, this triggers an increase in parallel fiber EPSC amplitude that lasts over 30 min; with no change in paired-pulse ratio, the effect is thought to be post-synaptic (Fig.5C, red). In the P-Rex1−/−/P-Rex2−/− mice, EPSC amplitude shows an initial increase but this increase is not sustained (Fig.5C, green). Parallel fiber LTP can also be evoked by exogenous NO donors; the initial response of P-Rex-deficient slices to an exogenous NO donor resembles the wildtype response, but potentiation is not sustained. Hence, P-Rex family enzymes are required for the maintenance of cerebellar LTP.
Disease relevance
PREX1 maps to a Type 2 diabetes mellitus susceptibility locus. Insulin is known to activate PI3K, yielding PI(3,4,5)P3 and muscle cells that lack Rac1 are known to exhibit impaired insulin-stimulated GLUT4-translocation. Although Rac1 does not promote GLUT4 translocation when insulin levels are saturating (100 nM), it does at physiological levels of insulin (1–10 nM). In the absence of insulin, expression of exogenous P-Rex1 in adipocytes has no effect on GLUT4 trafficking; however, the effect of insulin on GLUT4 trafficking is enhanced. Pre-treating adipocytes with a PI3K inhibitor decreases the ability of P-Rex1 to enhance the actions of insulin (Balamatsias and others 2011).
Ras-guanine nucleotide-releasing factor family (RasGRF1 and RasGRF2)
Discovery and expression
Although initially identified due to their sequence homology to yeast CDC25, a potent Ras GEF, the mammalian Ras-guanine nucleotide-releasing factors (RasGRFs) are large, multidomain proteins containing both Ras GEF and Rac GEF domains. RASGRF1 (mouse chromosome 9, human 15) is paternally imprinted and expressed only after birth. RASGRF2 (mouse chromosome 13, human 5) is not imprinted, but its expression is developmentally regulated; low early in development, expression increases around postnatal day 10 in the rat and remains high in adulthood. Both RasGRF1 and RasGRF2 are predominantly expressed in neurons of the adult brain, although RasGRF2 is also highly expressed in other tissues (Chen and others 1993; Feig 2011). In addition to the two full-length proteins, smaller alternatively spliced transcripts have been identified; some have unique developmental expression patterns and differential effects on downstream signaling, suggesting diverging physiological roles (see (Fernandez-Medarde and Santos 2011) for review). Our discussion here will focus on the full-length gene products, which have been studied in more detail.
Protein structure and function
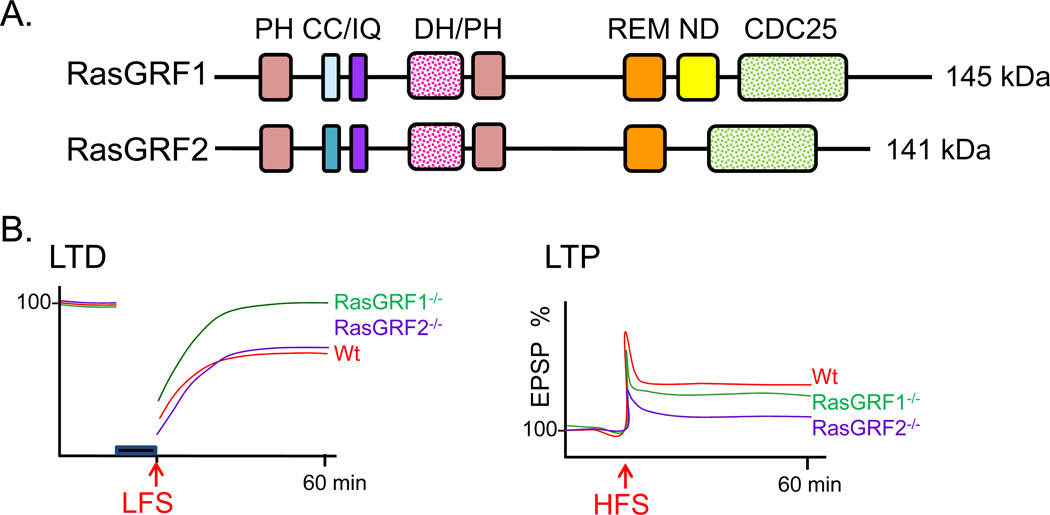
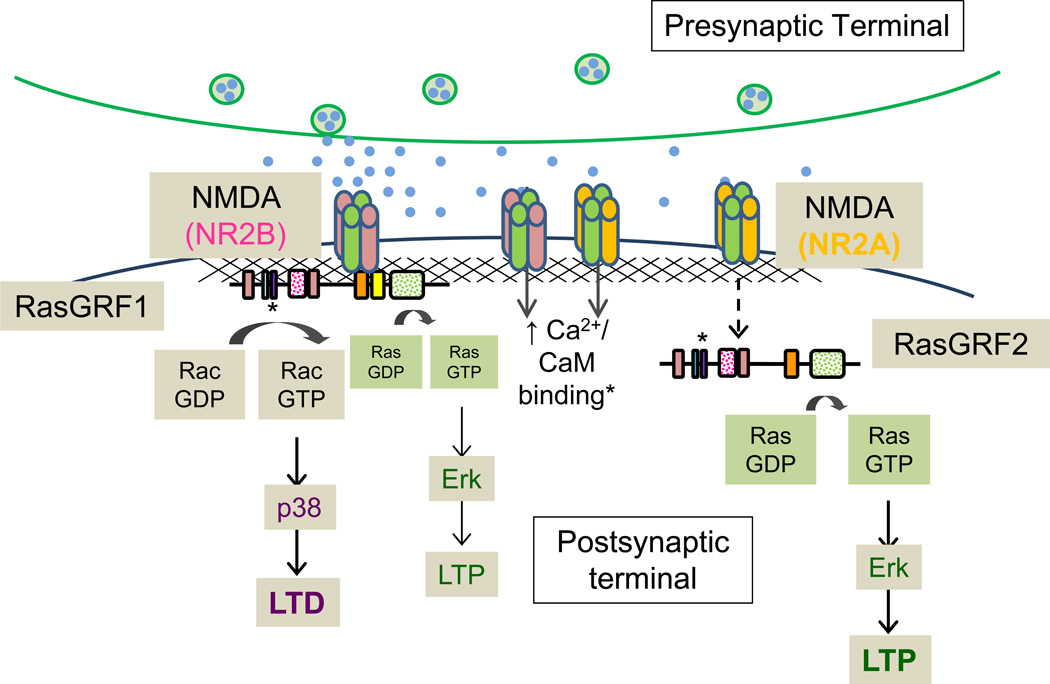
Full length RasGRF proteins contain an N-terminal PH domain followed by a coiled coil domain, a Ca2+/calmodulin-binding IQ domain, the tandem DH/PH domains common to Dbl family Rho GEFs, a Ras exchange motif (REM), and a Ras GEF domain (CDC25) (Fig.6A). In vitro and in vivo studies have shown that both full-length RasGRFs are capable of activating Ras family members as well as the Rho GTPase Rac1. The IQ domain of RasGRF1 binds calmodulin in a Ca2+-dependent manner, leading to increased GEF activity in cells (Farnsworth and others 1995), but not in vitro (Baouz and others 1997). A RasGRF2 mutant lacking the IQ domain altogether is still able to activate Ras, but does not result in activation of its downstream effector, Erk (de Hoog and others 2000). These results have not been entirely reconciled, but the hypothesis that RasGRF activity requires proper protein localization and assembly of downstream networks prevails. The N-terminal PH and CC domains may also be involved in protein localization. RasGRF1, but not RasGRF2, contains a “Neuronal domain (ND)”, which lets RasGRF1 associate with NMDA-type glutamate receptors through a direct interaction (Fig.6A); the ND interacts directly with the cytosolic tail of the NR2B subunit (Krapivinsky and others 2003) (Fig.7). Disruption of this interaction results in reduced Erk activation in cultured hippocampal neurons, while leaving NMDA receptor-mediated current intact following bath application of NMDA receptor agonists (Krapivinsky and others 2003).
Figure 6. RasGRF1 and RasGRF2.
A. RasGRF1 and RasGRF2 are encoded by separate genes (RASGRF1 and RASGRF2), respectively located on chromosomes 15 and 5 in human (Chromosomes 9 and 13 in mouse). Key features of RasGRF1 and RasGRF2 are identified: CC, coiled coil; IQ, Ca2+/calmodulin-binding domain; REM, Ras exchange motif; ND, neuronal domain; CDC25, Ras GEF domain. B. Electrophysiological recordings revealed significantly impaired LTP but normal LTD in slices from the hippocampus of P25 and older RasGRF2−/− mice, while LTD but not LTP was disrupted in hippocampal slices from RasGRF1−/− mice (Li and others 2006).
Figure 7. Distinct roles for RasGRF family members in NMDA-mediated long term plasticity.
Schematic illustrates proposed signaling mechanisms of the major RasGRF family members, RasGRF1 and RasGRF2. While both proteins are known to contribute to NMDA receptor-mediated Erk activity in vivo, data from biochemical and electrophysiological experiments suggest that RasGRF1 acts downstream of NR2B-containing NMDA receptors and signals predominantly through Rac and p38 to promote LTD in hippocampal slices. RasGRF2, however, signals almost exclusively through Ras and Erk following activation of NR2A-containing NMDA receptors to promote LTP.
A role for RasGRF1 has also been identified in Ca2+-dependent signaling downstream of various GPCRs and receptor tyrosine kinases. PKA-mediated phosphorylation and activation of RasGRF1 is also necessary for Erk activation following stimulation of exogenous Gαs-coupled 5-HT7 serotonin receptors in HEK-293 cells (Norum and others 2005). Stimulation of TrkA receptors, dopamine receptors and muscarinic acetylcholine receptors also results in RasGRF1 activation. The mechanisms and consequences of these interactions have not been fully elucidated, but all appear to require RasGRF1 phosphorylation, most at Ser916, as well as its Ca2+-dependent association with calmodulin (Mattingly 1999; Mattingly and Macara 1996; Norum and others 2005; Yang and others 2003).
Behavior
The availability of several transgenic mouse lines has established the physiological significance of the RasGRF family. Phenotypically, mice lacking RasGRF1 (RasGRF1−/−) are smaller than their wildtype littermates, and behavioral tests have revealed impaired fear conditioning in RasGRF1−/− mice. Normal performance on hippocampal memory tasks was seen in mice lacking both the full-length RasGRF1 and a less studied splice variant of RasGRF1, p55-GRF1; however numerous studies using mice lacking only the full-length RasGRF1 isoform have shown impaired hippocampal-dependent memory (Brambilla and others 1997; d'Isa and others 2011; Giese and others 2001; Li and others 2006). Locomotor sensitization and conditioned place preference for cocaine are significantly reduced in RasGRF1−/− mice, while both are exacerbated in animals over-expressing RasGRF1 (Fasano and others 2009). RasGRF1 may also be involved in CB1 (endocannabinoid) receptor sensitization and tolerance to Δ9-THC, since RasGRF1−/− mice have altered gene expression profiles following chronic Δ9-THC exposure (Rubino and others 2006). Visual phenotypes, including deficient photoreception, have also been observed and likely arise from RasGRF functions in the retina (Fernandez-Medarde and others 2009). RasGRF2−/− mice grow and reproduce normally (Fernandez-Medarde and others 2002) and most reported phenotypes of single and combined knockout strains have indicated RasGRF2 involvement in immune mechanisms.
Electrophysiology and Synaptic Function
Using theta burst stimulation, Brambilla et al. showed impaired LTP in the amygdala of RasGRF1−/− mice, though hippocampal LTP appeared normal (Brambilla and others 1997). They also observed elevated basal field activity in both amygdala and hippocampal slices (Brambilla and others 1997). Furthermore, recordings from cultured neurons and acute brain slices revealed hippocampal hyperexcitability in RasGRF1−/− mice, and convulsant drug administration studies have shown increased seizure susceptibility in these animals (Tonini and others 2001).
Although the RasGRF proteins are structurally similar (Fig.6A), diverging roles in synaptic function are apparent. Both RasGRF1 and RasGRF2 act downstream of NMDA receptors. While both RasGRFs are known to contribute to neuronal Ras/Erk signaling in vivo, evaluation of Erk activity following NMDA stimulation in acute hippocampal brain slices from knockout animals revealed larger deficiencies in NMDA receptor-mediated Erk activation in RasGRF2−/− mice than in RasGRF1−/− mice (Li and others 2006). Conversely, hippocampal slices from RasGRF1−/− mice show reduced NMDA receptor-mediated activation of the Rac effector p38 MAP kinase, while recordings from RasGRF2−/− slices are indistinguishable from wildtype (Fig.7).
Although basal synaptic transmission appears to be normal in hippocampal slices from mice (P25–36) lacking both RasGRF1 and RasGRF2, expression of both LTP and LTD is blunted in hippocampal slices from adult animals; this deficit is not seen in P14–18 brains (Li and others 2006). Electrophysiological analysis of single knockout brains revealed that hippocampal LTD, but not LTP, is disrupted in RasGRF1−/− mice, while LTP, but not LTD, is impaired in RasGRF2−/− mice (Li and others 2006) (Fig.6B). Further manipulation suggests that these effects are mediated by different NMDA receptor subtypes. APV blockade of NMDA receptors blocks NMDA-mediated activation of Erk, but not p38, while selective blockade of NR2B-containing NMDA receptors with ifenprodil prevents NMDA-mediated p38 activation with no effect on Erk activation. Thus, electrophysiological and biochemical studies using knockout animals suggest that RasGRF2 signals predominantly through the Ras/Erk pathway in response to NR2A-containing NMDA receptor activation and contributes to LTP induction. Conversely, RasGRF1 contributes mostly to NMDA receptor-dependent LTD via Rac/p38 activation downstream of NR2B-containing NMDA receptor activation (Krapivinsky and others 2003; Li and others 2006) (Fig.7).
Furthermore, cortical Ca2+-permeable AMPA receptor currents are reduced in RasGRF1−/− and 2−/− knockout animals and are abolished in double knockout animals. Both RasGRF proteins co-immunoprecipitate with the GluR1 subunit, but not the GluR2 subunit, of the AMPA receptor in lysates from cortical brain slices following bath application of AMPA (Tian and Feig 2006). Tian et al. also showed that RasGRF expression is required for Ras/Erk signaling following activation of Ca2+-permeable AMPA receptors in the adult brain (Tian and Feig 2006).
Disease relevance
Increased RasGRF1 expression is seen in the striatum, cortex and cerebellum following chronic cocaine, amphetamine or THC administration (Fasano and others 2009; Parelkar and others 2009; Rubino and others 2005; Rubino and others 2006; Zhang and others 2007). Furthermore, RasGRF1−/− mice display decreased locomotor sensitization and conditioned place preference for cocaine (Fasano and others 2009). RasGRF1 may also be involved in CB1 cannabinoid receptor adaptation and drug tolerance following THC administration (Rubino and others 2005; Rubino and others 2006). RasGRF1−/− mice and non-human primates expressing dominant negative forms of RasGRF1 are significantly less susceptible to L-dopa-induced dyskinesias associated with dopamine replacement therapies (Fasano and others 2010), and mice lacking both full-length isoforms have higher susceptibility to cerebral ischemia.
Collybistin
Discovery and expression
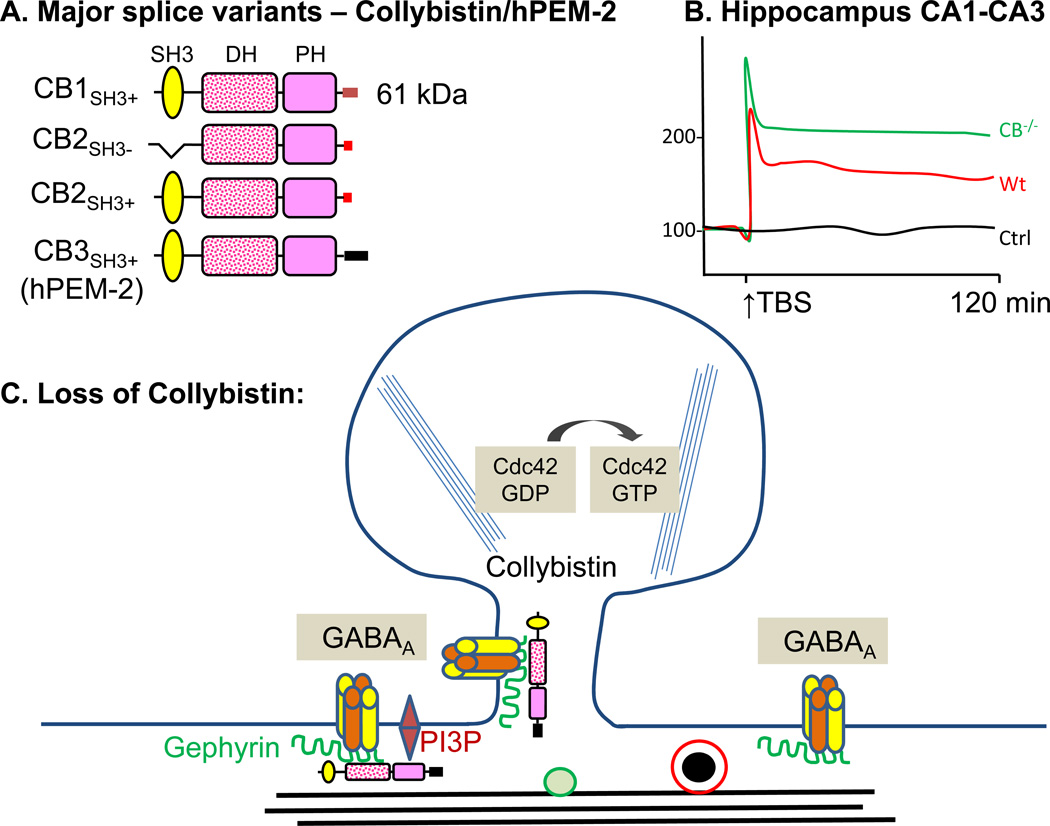
Collybistin [Arhgef9; human hPEM2 (Posterior End Mark-2), ARHGEF9] was discovered in a yeast two hybrid screen for gephyrin interactors (Kins and others 2000); gephyrin, a scaffolding protein, plays an essential role in the postsynaptic clustering of GABAA and glycine receptors, both of which are ligand gated inhibitory ion channels. The collybistin gene is located on the X chromosome. In the rat, alternative splicing generates collybistin transcripts that encode functionally distinct proteins (Fig.8A). Some lack the SH3 domain that precedes the catalytic DH/PH domain and three different C-termini (CB1, CB2, CB3) have been identified; CB2SH3+ and CB3SH3+ are the major splice variants in the rat (Harvey and others 2004). Two promoters generate hPEM2, producing slightly different sequences that precede the SH3 domain (Harvey and others 2004).
Figure 8. Collybistin. Collybistin structure and synaptic function.
A. The major splice variants of rat collybistin (CB; Arhgef9) are shown; the human homologue, hPEM-2 (ARHGEF9) has alternate promoters, but does not have similar splice variants (Harvey and others 2004). B. Slices prepared from collybistin−/− mice show exaggerated hippocampal LTP; theta-burst stimulation was applied to fibers from CA3 neurons and fEPSPs were recorded in the stratum radiatum of the CA1 region (Papadopoulos and others 2007). The genotypic difference was abrogated in the presence of picrotoxin, a GABAA receptor blocker, which increased potentiation only in wildtype slices. C. Collybistin localizes the scaffolding protein gephyrin, which in turn clusters GABAA and the group of GlyR receptors (as depicted).
Collybistin transcripts are highly expressed in neurons throughout the adult brain and spinal cord, with little expression outside of the nervous system. Collybistin is expressed in all cerebellar neurons, with highest levels in Purkinje cells (Kneussel and others 2001). Expression of collybistin is upregulated during the time of major neuronal differentiation and synaptogenesis and it is thought to play a role in post-mitotic neurons (Kneussel and others 2001; Tolias and others 2011). Collybistin immunostaining is strong in dendrites, with very little in cell body (Tyagarajan and others 2011); Collybistin is detected at only a subset of gephyrin-positive inhibitory synapses containing all major GABAAR subunits (Patrizi and others 2011).
Protein structure and signaling
The DH domain of collybistin activates Cdc42, but does not activate Rac1 or RhoA; this domain is involved in binding gephyrin (Kiraly and others 2010a; Tolias and others 2011; Tyagarajan and others 2011). The SH3 domain of collybistin interacts with neuroligin-2, a transmembrane protein that interacts with presynaptic neurexins localized at GABAergic synapses (Chiou and others 2011), and the PH domain binds selectively with the phosphoinositide PI3P (Patrizi and others 2011). Initial comparisons of collybistin variants with and without the SH3 domain suggested an inhibitory role for this region: CB2SH3- forms a ternary complex with gephyrin and Cdc42 while CB2SH3+ does not (Tyagarajan and others 2011). Other work argues that all four major isoforms of Collybistin concentrate at GABAergic synapses (Chiou and others 2011).
Collybistin plays a role in localizing gephyrin, which is thought to form a scaffold beneath the plasma membrane that contributes to the anchoring of receptors. Gephyrin, which interacts with microtubules, actin filaments and many additional proteins, plays an essential role in the formation of postsynaptic GABAA and glycine receptor clusters. The PH domain of collybistin is essential for gephyrin clustering and for interaction with PI3P, but Cdc42 activation by collybistin does not appear to be essential (Tyagarajan and others 2011).
In collybistin−/− mice, a subset of the GABAergic synapses fail to form normally, but glycinergic synapses are unaffected. The effect on GABAA receptors is both subtype and region-specific: γ2-containing GABAA receptor levels drop in the basolateral amygdala and hippocampus, but not in the brainstem or cerebellum (Papadopoulos and others 2007). In affected areas, the absence of collybistin results in a decrease in the density of gephyrin clusters at postsynaptic densities and in decreased staining for the γ2 and α2 subunits of the GABAA receptor. Why the effects are so regionally specific is not yet clear.
Behavior
Given its role in inhibitory receptor clustering, it is not surprising that collybistin−/− mice have significantly fewer GABAA receptor clusters in the amygdala and hippocampus than wt mice (Papadopoulos and others 2007). Surprisingly, spinal cord and brainstem Gly receptors are properly assembled (Tyagarajan and others 2011). The number of postsynaptic gephyrin puncta was dramatically decreased in the CA1 region of the hippocampus, while the decreases were evident but much less striking in other regions such as the amygdala; paradoxically, the number of VIAAT puncta (presynaptic marker for GABA terminals) was normal (Papadopoulos and others 2007; Jedlicka and others 2009).
Consistent with loss of GABAA receptor clusters in the amygdala and the hippocampus, Collybistin−/− mice have impaired spatial learning, reduced exploratory behavior, and increased anxiety-like behavior (Papadopoulos and others 2007). Time spent in the open arm of the elevated plus maze was dramatically reduced. The Barnes maze is learned well by wildtype mice, but not by Collybistin−/− mice. Motor performance was normal in Collybistin−/− mice, consistent with normal glycinergic inhibition (Papadopoulos and others 2007).
Electrophysiology and neuronal function
Electrophysiological recordings from collybistin−/− mice have been carried out in vitro and in vivo (Papadopoulos and others 2007). The frequency and amplitude of GABAergic miniature inhibitory postsynaptic potentials (mIPSPs) in CA1 pyramidal neurons were both decreased in Collybistin−/− mice. As expected, the field EPSP slope in CA1 is dramatically increased in wildtype mice in the presence of picrotoxin and only a blunted increase is seen in CB−/− mice (consistent with loss of necessary clustering of GABAAR) (Jedlicka and others 2009). LTP induction using theta-burst stimulation of the CA3-CA1 hippocampal collateral pathway produces an enhanced response in Collybistin−/− mice (Fig.8B); the difference between genotypes is eliminated by picrotoxin, again consistent with diminished GABA function in CB−/− mice. A low frequency stimulation paradigm that produces LTD in wildtype mice does not do so in Collybistin−/− mice.
Disease relevance
Harvey et al. (Harvey and others 2004) identified a mutation in the SH3 domain of collybistin when examining 32 hereditary hyperekplexia (excessive startle response) patients; the child with the mutation survived until age 4 but suffered from tonic seizures provoked by tactile stimulation and was severely mentally retarded. Expression of CB3SH3+ bearing this mutation results in loss of endogenous gephyrin clusters and mislocalization of GABAA receptors (Papadopoulos and others 2007). Expression of CB2SH3+ mutated at the same site does not support gephyrin localization to the cell surface (Tyagarajan and others 2011). Loss of function mutations in the ARHGEF9 gene lead to mental retardation and refractory epilepsy in both boys and heterozygous girls (Harvey and others 2004; Shimojima and others 2011).
Acknowledgments
Support: National Institutes of Health Grants DK-32948, DK-32949, DA-23082
References
- Abe H, Okazawa M, Nakanishi S. Gene regulation via excitation and BDNF is mediated by induction and phosphorylation of the Etv1 transcription factor in cerebellar granule cells. Proc Natl Acad Sci U S A. 2012;109:8734–8739. doi: 10.1073/pnas.1206418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam MR, Caldwell BD, Johnson RC, Darlington DN, Mains RE, Eipper BA. Novel proteins that interact with the COOH-terminal cytosolic routing determinants of an integral membrane peptide-processing enzyme. J Biol Chem. 1996;271:28636–28640. doi: 10.1074/jbc.271.45.28636. [DOI] [PubMed] [Google Scholar]
- Astigarraga S, Hofmeyer K, Farajian R, Treisman JE. Three Drosophila liprins interact to control synapse formation. J Neurosci. 2010;30:15358–15368. doi: 10.1523/JNEUROSCI.1862-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasaki T, Saito M, Sone M, Suzuki E, Sakai R, Ito K, et al. The Drosophila trio plays an essential role in patterning of axons by regulating their directional extension. Neuron. 2000;26:119–131. doi: 10.1016/s0896-6273(00)81143-5. [DOI] [PubMed] [Google Scholar]
- Balamatsias D, Kong AM, Waters JE, Sriratana A, Gurung R, Bailey CG, et al. Identification of P-Rex1 as a novel Rac1-guanine nucleotide exchange factor (GEF) that promotes actin remodeling and GLUT4 protein trafficking in adipocytes. J Biol Chem. 2011;286:43229–43240. doi: 10.1074/jbc.M111.306621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball RW, Warren-Paquin M, Tsurudome K, Liao EH, Elazzouzi F, Cavanagh C, et al. Retrograde BMP signaling controls synaptic growth at the NMJ by regulating trio expression in motor neurons. Neuron. 2010;66:536–549. doi: 10.1016/j.neuron.2010.04.011. [DOI] [PubMed] [Google Scholar]
- Baouz S, Jacquet E, Bernardi A, Parmeggiani A. The N-terminal moiety of CDC25(Mm), a GDP/GTP exchange factor of Ras proteins, controls the activity of the catalytic domain. Modulation by calmodulin and calpain. J Biol Chem. 1997;272:6671–6676. doi: 10.1074/jbc.272.10.6671. [DOI] [PubMed] [Google Scholar]
- Bateman J, Shu H, Van Vactor D. The guanine nucleotide exchange factor trio mediates axonal development in the Drosophila embryo. Neuron. 2000;26:93–106. doi: 10.1016/s0896-6273(00)81141-1. [DOI] [PubMed] [Google Scholar]
- Bouquier N, Vignal E, Charrasse S, Weill M, Schmidt S, Leonetti JP, et al. A cell active chemical GEF inhibitor selectively targets the Trio/RhoG/Rac1 signaling pathway. Chem Biol. 2009;16:657–666. doi: 10.1016/j.chembiol.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Brambilla R, Gnesutta N, Minichiello L, White G, Roylance AJ, Herron CE, et al. A role for the Ras signalling pathway in synaptic transmission and long-term memory. Nature. 1997;390:281–286. doi: 10.1038/36849. [DOI] [PubMed] [Google Scholar]
- Briancon-Marjollet A, Ghogha A, Nawabi H, Triki I, Auziol C, Fromont S, et al. Trio mediates netrin-1-induced Rac1 activation in axon outgrowth and guidance. Mol Cell Biol. 2008;28:2314–2323. doi: 10.1128/MCB.00998-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill ME, Xie Z, Day M, Photowala H, Barbolina MV, Miller CA, Penzes P. Kalirin regulates cortical spine morphogenesis and disease-related behavioral phenotypes. Proc Natl Acad Sci U S A. 2009;106:13058–13063. doi: 10.1073/pnas.0904636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JP, Hu Z, Sieburth D. Recruitment of sphingosine kinase to presynaptic terminals by a conserved muscarinic signaling pathway promotes neurotransmitter release. Genes Dev. 2012;26:1070–1085. doi: 10.1101/gad.188003.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhang LJ, Greer P, Tung PS, Moran MF. A murine CDC25/ras-GRF-related protein implicated in Ras regulation. Dev Genet. 1993;14:339–346. doi: 10.1002/dvg.1020140503. [DOI] [PubMed] [Google Scholar]
- Chiou TT, Bonhomme B, Jin H, Miralles CP, Xiao H, Fu Z, et al. Differential regulation of the postsynaptic clustering of gamma-aminobutyric acid type A (GABAA) receptors by collybistin isoforms. J Biol Chem. 2011;286:22456–22468. doi: 10.1074/jbc.M111.236190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CY, Leng S, Martin KA, Kim E, Gorman S, Duhl DM. Cloning and characterization of T-cell lymphoma invasion and metastasis 2 (TIAM2), a novel guanine nucleotide exchange factor related to TIAM1. Genomics. 1999;61:66–73. doi: 10.1006/geno.1999.5936. [DOI] [PubMed] [Google Scholar]
- d'Isa R, Clapcote SJ, Voikar V, Wolfer DP, Giese KP, Brambilla R, et al. Mice Lacking Ras-GRF1 Show Contextual Fear Conditioning but not Spatial Memory Impairments: Convergent Evidence from Two Independently Generated Mouse Mutant Lines. Front Behav Neurosci. 2011;5:78. doi: 10.3389/fnbeh.2011.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoog CL, Fan WT, Goldstein MD, Moran MF, Koch CA. Calmodulin-independent coordination of Ras and extracellular signal-regulated kinase activation by Ras-GRF2. Mol Cell Biol. 2000;20:2727–2733. doi: 10.1128/mcb.20.8.2727-2733.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debant A, Serra-Pages C, Seipel K, O'Brien S, Tang M, Park SH, Streuli M. The multidomain protein Trio binds the LAR transmembrane tyrosine phosphatase, contains a protein kinase domain, and has separate rac-specific and rho-specific guanine nucleotide exchange factor domains. Proc Natl Acad Sci U S A. 1996;93:5466–5471. doi: 10.1073/pnas.93.11.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald S, Hill K, Lecureuil C, Barnouin R, Krugmann S, Coadwell JW, et al. P-Rex2, a new guanine-nucleotide exchange factor for Rac. FEBS Lett. 2004;572:172–176. doi: 10.1016/j.febslet.2004.06.096. [DOI] [PubMed] [Google Scholar]
- Donald S, Humby T, Fyfe I, Segonds-Pichon A, Walker SA, Andrews SR, et al. P-Rex2 regulates Purkinje cell dendrite morphology and motor coordination. Proc Natl Acad Sci U S A. 2008;105:4483–4488. doi: 10.1073/pnas.0712324105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehler E, van Leeuwen F, Collard JG, Salinas PC. Expression of Tiam-1 in the developing brain suggests a role for the Tiam-1-Rac signaling pathway in cell migration and neurite outgrowth. Mol Cell Neurosci. 1997;9:1–12. doi: 10.1006/mcne.1997.0602. [DOI] [PubMed] [Google Scholar]
- Farnsworth CL, Freshney NW, Rosen LB, Ghosh A, Greenberg ME, Feig LA. Calcium activation of Ras mediated by neuronal exchange factor Ras-GRF. Nature. 1995;376:524–527. doi: 10.1038/376524a0. [DOI] [PubMed] [Google Scholar]
- Fasano S, Bezard E, D'Antoni A, Francardo V, Indrigo M, Qin L, et al. Inhibition of Ras-guanine nucleotide-releasing factor 1 (Ras-GRF1) signaling in the striatum reverts motor symptoms associated with L-dopa-induced dyskinesia. Proc Natl Acad Sci U S A. 2010;107:21824–21829. doi: 10.1073/pnas.1012071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano S, D'Antoni A, Orban PC, Valjent E, Putignano E, Vara H, et al. Ras-guanine nucleotide-releasing factor 1 (Ras-GRF1) controls activation of extracellular signal-regulated kinase (ERK) signaling in the striatum and long-term behavioral responses to cocaine. Biol Psychiatry. 2009;66:758–768. doi: 10.1016/j.biopsych.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig LA. Regulation of Neuronal Function by Ras-GRF Exchange Factors. Genes Cancer. 2011;2:306–319. doi: 10.1177/1947601911408077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Medarde A, Barhoum R, Riquelme R, Porteros A, Nunez A, de Luis A, et al. RasGRF1 disruption causes retinal photoreception defects and associated transcriptomic alterations. J Neurochem. 2009;110:641–652. doi: 10.1111/j.1471-4159.2009.06162.x. [DOI] [PubMed] [Google Scholar]
- Fernandez-Medarde A, Esteban LM, Nunez A, Porteros A, Tessarollo L, Santos E. Targeted disruption of Ras-Grf2 shows its dispensability for mouse growth and development. Mol Cell Biol. 2002;22:2498–2504. doi: 10.1128/MCB.22.8.2498-2504.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Medarde A, Santos E. The RasGrf family of mammalian guanine nucleotide exchange factors. Biochim Biophys Acta. 2011;1815:170–188. doi: 10.1016/j.bbcan.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Gerard A, van der Kammen RA, Janssen H, Ellenbroek SI, Collard JG. The Rac activator Tiam1 controls efficient T-cell trafficking and route of transendothelial migration. Blood. 2009;113:6138–6147. doi: 10.1182/blood-2008-07-167668. [DOI] [PubMed] [Google Scholar]
- Giese KP, Friedman E, Telliez JB, Fedorov NB, Wines M, Feig LA, et al. Hippocampus-dependent learning and memory is impaired in mice lacking the Ras-guanine-nucleotide releasing factor 1 (Ras-GRF1) Neuropharmacology. 2001;41:791–800. doi: 10.1016/s0028-3908(01)00096-x. [DOI] [PubMed] [Google Scholar]
- Goto A, Hoshino M, Matsuda M, Nakamura T. Phosphorylation of STEF/Tiam2 by protein kinase A is critical for Rac1 activation and neurite outgrowth in dibutyryl cAMP-treated PC12D cells. Mol Biol Cell. 2011;22:1780–1790. doi: 10.1091/mbc.E10-09-0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habets GG, Scholtes EH, Zuydgeest D, van der Kammen RA, Stam JC, Berns A, et al. Identification of an invasion-inducing gene, Tiam-1, that encodes a protein with homology to GDP-GTP exchangers for Rho-like proteins. Cell. 1994;77:537–549. doi: 10.1016/0092-8674(94)90216-x. [DOI] [PubMed] [Google Scholar]
- Hansel DE, Quinones ME, Ronnett GV, Eipper BA. Kalirin, a GDP/GTP exchange factor of the Dbl family, is localized to nerve, muscle, and endocrine tissue during embryonic rat development. J Histochem Cytochem. 2001;49:833–844. doi: 10.1177/002215540104900704. [DOI] [PubMed] [Google Scholar]
- Harvey K, Duguid IC, Alldred MJ, Beatty SE, Ward H, Keep NH, et al. The GDP-GTP exchange factor collybistin: an essential determinant of neuronal gephyrin clustering. J Neurosci. 2004;24:5816–5826. doi: 10.1523/JNEUROSCI.1184-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Negrete I, Carretero-Ortega J, Rosenfeldt H, Hernandez-Garcia R, Calderon-Salinas JV, Reyes-Cruz G, et al. P-Rex1 links mammalian target of rapamycin signaling to Rac activation and cell migration. J Biol Chem. 2007;282:23708–23715. doi: 10.1074/jbc.M703771200. [DOI] [PubMed] [Google Scholar]
- Hill K, Krugmann S, Andrews SR, Coadwell WJ, Finan P, Welch HC, et al. Regulation of P-Rex1 by phosphatidylinositol (3,4,5)-trisphosphate and Gbetagamma subunits. J Biol Chem. 2005;280:4166–4173. doi: 10.1074/jbc.M411262200. [DOI] [PubMed] [Google Scholar]
- Hoshino M, Sone M, Fukata M, Kuroda S, Kaibuchi K, Nabeshima Y, et al. Identification of the stef gene that encodes a novel guanine nucleotide exchange factor specific for Rac1. J Biol Chem. 1999;274:17837–17844. doi: 10.1074/jbc.274.25.17837. [DOI] [PubMed] [Google Scholar]
- Jackson C, Welch HC, Bellamy TC. Control of cerebellar long-term potentiation by P-Rex-family guanine-nucleotide exchange factors and phosphoinositide 3-kinase. PLoS One. 2010;5:e11962. doi: 10.1371/journal.pone.0011962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedlicka P, Papadopoulos T, Deller T, Betz H, Schwarzacher SW. Increased network excitability and impaired induction of long-term potentiation in the dentate gyrus of collybistin-deficient mice in vivo. Mol Cell Neurosci. 2009;41:94–100. doi: 10.1016/j.mcn.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Kawauchi T, Chihama K, Nabeshima Y, Hoshino M. The in vivo roles of STEF/Tiam1, Rac1 and JNK in cortical neuronal migration. EMBO J. 2003;22:4190–4201. doi: 10.1093/emboj/cdg413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kins S, Betz H, Kirsch J. Collybistin, a newly identified brain-specific GEF, induces submembrane clustering of gephyrin. Nat Neurosci. 2000;3:22–29. doi: 10.1038/71096. [DOI] [PubMed] [Google Scholar]
- Kiraly DD, Eipper-Mains JE, Mains RE, Eipper BA. Synaptic plasticity, a symphony in GEF. ACS Chem Neurosci. 2010a;1:348–365. doi: 10.1021/cn100012x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiraly DD, Lemtiri-Chlieh F, Levine ES, Mains RE, Eipper BA. Kalirin binds the NR2B subunit of the NMDA receptor, altering its synaptic localization and function. J Neurosci. 2011a;31:12554–12565. doi: 10.1523/JNEUROSCI.3143-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiraly DD, Ma XM, Mazzone CM, Xin X, Mains RE, Eipper BA. Behavioral and morphological responses to cocaine require kalirin7. Biol Psychiatry. 2010b;68:249–255. doi: 10.1016/j.biopsych.2010.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiraly DD, Stone KL, Colangelo CM, Abbott T, Wang Y, Mains RE, Eipper BA. Identification of Kalirin-7 as a potential post-synaptic density signaling hub. J Proteome Res. 2011b;10:2828–2841. doi: 10.1021/pr200088w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneussel M, Engelkamp D, Betz H. Distribution of transcripts for the brain-specific GDP/GTP exchange factor collybistin in the developing mouse brain. Eur J Neurosci. 2001;13:487–492. doi: 10.1046/j.0953-816x.2000.01411.x. [DOI] [PubMed] [Google Scholar]
- Krapivinsky G, Krapivinsky L, Manasian Y, Ivanov A, Tyzio R, Pellegrino C, et al. The NMDA receptor is coupled to the ERK pathway by a direct interaction between NR2B and RasGRF1. Neuron. 2003;40:775–784. doi: 10.1016/s0896-6273(03)00645-7. [DOI] [PubMed] [Google Scholar]
- Kunda P, Paglini G, Quiroga S, Kosik K, Caceres A. Evidence for the involvement of Tiam1 in axon formation. J Neurosci. 2001;21:2361–2372. doi: 10.1523/JNEUROSCI.21-07-02361.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushima I, Nakamura Y, Aleksic B, Ikeda M, Ito Y, Shiino T, et al. Resequencing and association analysis of the KALRN and EPHB1 genes and their contribution to schizophrenia susceptibility. Schizophr Bull. 2012;38:552–560. doi: 10.1093/schbul/sbq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai KO, Wong AS, Cheung MC, Xu P, Liang Z, Lok KC, et al. TrkB phosphorylation by Cdk5 is required for activity-dependent structural plasticity and spatial memory. Nat Neurosci. 2012;15:1506–1515. doi: 10.1038/nn.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemtiri-Chlieh F, Zhao L, Kiraly DD, Eipper BA, Mains RE, Levine ES. Kalirin-7 is necessary for normal NMDA receptor-dependent synaptic plasticity. BMC Neurosci. 2011;12:126. doi: 10.1186/1471-2202-12-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Tian X, Hartley DM, Feig LA. Distinct roles for Ras-guanine nucleotide-releasing factor 1 (Ras-GRF1) and Ras-GRF2 in the induction of long-term potentiation and long-term depression. J Neurosci. 2006;26:1721–1729. doi: 10.1523/JNEUROSCI.3990-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Li QJ, An SC. Preventive effect of estrogen on depression-like behavior induced by chronic restraint stress. Neurosci Bull. 2010;26:140–146. doi: 10.1007/s12264-010-0609-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebl EC, Forsthoefel DJ, Franco LS, Sample SH, Hess JE, Cowger JA, et al. Dosage-sensitive, reciprocal genetic interactions between the Abl tyrosine kinase and the putative GEF trio reveal trio's role in axon pathfinding. Neuron. 2000;26:107–118. doi: 10.1016/s0896-6273(00)81142-3. [DOI] [PubMed] [Google Scholar]
- Luo L. Trio quartet in D. (melanogaster) Neuron. 2000;26:1–2. doi: 10.1016/s0896-6273(00)81129-0. [DOI] [PubMed] [Google Scholar]
- Lutz S, Shankaranarayanan A, Coco C, Ridilla M, Nance MR, Vettel C, et al. Structure of Galphaq-p63RhoGEF-RhoA complex reveals a pathway for the activation of RhoA by GPCRs. Science. 2007;318:1923–1927. doi: 10.1126/science.1147554. [DOI] [PubMed] [Google Scholar]
- Ma QL, Yang F, Calon F, Ubeda OJ, Hansen JE, Weisbart RH, et al. p21-activated kinase-aberrant activation and translocation in Alzheimer disease pathogenesis. J Biol Chem. 2008a;283:14132–14143. doi: 10.1074/jbc.M708034200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Huang JP, Eipper BA, Mains RE. Expression of Trio, a member of the Dbl family of Rho GEFs in the developing rat brain. J Comp Neurol. 2005;482:333–348. doi: 10.1002/cne.20404. [DOI] [PubMed] [Google Scholar]
- Ma XM, Huang JP, Xin X, Yan Y, Mains RE, Eipper BA. A role for kalirin in the response of rat medium spiny neurons to cocaine. Mol Pharmacol. 2012;82:738–745. doi: 10.1124/mol.112.080044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Johnson RC, Mains RE, Eipper BA. Expression of kalirin, a neuronal GDP/GTP exchange factor of the trio family, in the central nervous system of the adult rat. J Comp Neurol. 2001;429:388–402. doi: 10.1002/1096-9861(20010115)429:3<388::aid-cne3>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Ma XM, Kiraly DD, Gaier ED, Wang Y, Kim EJ, Levine ES, Eipper BA, Mains RE. Kalirin-7 is required for synaptic structure and function. J Neurosci. 2008b;28:12368–12382. doi: 10.1523/JNEUROSCI.4269-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Wang Y, Ferraro F, Mains RE, Eipper BA. Kalirin-7 is an essential component of both shaft and spine excitatory synapses in hippocampal interneurons. J Neurosci. 2008c;28:711–724. doi: 10.1523/JNEUROSCI.5283-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mains RE, Kiraly DD, Eipper-Mains JE, Ma XM, Eipper BA. Kalrn promoter usage and isoform expression respond to chronic cocaine exposure. BMC Neurosci. 2011;12:20. doi: 10.1186/1471-2202-12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malliri A, van der Kammen RA, Clark K, van der Valk M, Michiels F, Collard JG. Mice deficient in the Rac activator Tiam1 are resistant to Ras-induced skin tumours. Nature. 2002;417:867–871. doi: 10.1038/nature00848. [DOI] [PubMed] [Google Scholar]
- Mandela P, Ma XM. Kalirin, a key player in synapse formation, is implicated in human diseases. Neural Plast. 2012;2012:728161. doi: 10.1155/2012/728161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandela P, Yankova M, Conti LH, Ma X, Grady J, Eipper BA, Mains RE. Kalrn plays key roles within and outside of the nervous system. BMC Neurosci. 2012;13:136. doi: 10.1186/1471-2202-13-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo N, Terao M, Nabeshima Y, Hoshino M. Roles of STEF/Tiam1, guanine nucleotide exchange factors for Rac1, in regulation of growth cone morphology. Mol Cell Neurosci. 2003;24:69–81. doi: 10.1016/s1044-7431(03)00122-2. [DOI] [PubMed] [Google Scholar]
- Mattingly RR. Phosphorylation of serine 916 of Ras-GRF1 contributes to the activation of exchange factor activity by muscarinic receptors. J Biol Chem. 1999;274:37379–37384. doi: 10.1074/jbc.274.52.37379. [DOI] [PubMed] [Google Scholar]
- Mattingly RR, Macara IG. Phosphorylation-dependent activation of the Ras-GRF/CDC25Mm exchange factor by muscarinic receptors and G-protein beta gamma subunits. Nature. 1996;382:268–272. doi: 10.1038/382268a0. [DOI] [PubMed] [Google Scholar]
- May V, Schiller MR, Eipper BA, Mains RE. Kalirin Dbl-homology guanine nucleotide exchange factor 1 domain initiates new axon outgrowths via RhoG-mediated mechanisms. J Neurosci. 2002;22:6980–6990. doi: 10.1523/JNEUROSCI.22-16-06980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeenuddin LH, McIntire WE, Garrison JC. Differential sensitivity of P-Rex1 to isoforms of G protein betagamma dimers. J Biol Chem. 2006;281:1913–1920. doi: 10.1074/jbc.M506034200. [DOI] [PubMed] [Google Scholar]
- McMullan R, Anderson A, Nurrish S. Behavioral and immune responses to infection require Galphaq-RhoA signaling in C. elegans. PLoS Pathog. 2012;8:e1002530. doi: 10.1371/journal.ppat.1002530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson CE, Eipper BA, Mains RE. Genomic organization and differential expression of Kalirin isoforms. Gene. 2002;284:41–51. doi: 10.1016/s0378-1119(02)00386-4. [DOI] [PubMed] [Google Scholar]
- McPherson CE, Eipper BA, Mains RE. Multiple novel isoforms of Trio are expressed in the developing rat brain. Gene. 2005;347:125–135. doi: 10.1016/j.gene.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Mendoza-Naranjo A, Gonzalez-Billault C, Maccioni RB. Abeta1–42 stimulates actin polymerization in hippocampal neurons through Rac1 and Cdc42 Rho GTPases. J Cell Sci. 2007;120:279–288. doi: 10.1242/jcs.03323. [DOI] [PubMed] [Google Scholar]
- Michiels F, Habets GG, Stam JC, van der Kammen RA, Collard JG. A role for Rac in Tiam1-induced membrane ruffling and invasion. Nature. 1995;375:338–340. doi: 10.1038/375338a0. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y, Yamauchi J, Tanoue A, Wu C, Mobley WC. TrkB binds and tyrosine-phosphorylates Tiam1, leading to activation of Rac1 and induction of changes in cellular morphology. Proc Natl Acad Sci U S A. 2006;103:10444–10449. doi: 10.1073/pnas.0603914103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montenegro-Venegas C, Tortosa E, Rosso S, Peretti D, Bollati F, Bisbal M, et al. MAP1B regulates axonal development by modulating Rho-GTPase Rac1 activity. Mol Biol Cell. 2010;21:3518–3528. doi: 10.1091/mbc.E09-08-0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norum JH, Methi T, Mattingly RR, Levy FO. Endogenous expression and protein kinase A-dependent phosphorylation of the guanine nucleotide exchange factor Ras-GRF1 in human embryonic kidney 293 cells. FEBS J. 2005;272:2304–2316. doi: 10.1111/j.1742-4658.2005.04658.x. [DOI] [PubMed] [Google Scholar]
- O'Brien SP, Seipel K, Medley QG, Bronson R, Segal R, Streuli M. Skeletal muscle deformity and neuronal disorder in Trio exchange factor-deficient mouse embryos. Proc Natl Acad Sci U S A. 2000;97:12074–12078. doi: 10.1073/pnas.97.22.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos T, Korte M, Eulenburg V, Kubota H, Retiounskaia M, Harvey RJ, et al. Impaired GABAergic transmission and altered hippocampal synaptic plasticity in collybistin-deficient mice. EMBO J. 2007;26:3888–3899. doi: 10.1038/sj.emboj.7601819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parelkar NK, Jiang Q, Chu XP, Guo ML, Mao LM, Wang JQ. Amphetamine alters Ras-guanine nucleotide-releasing factor expression in the rat striatum in vivo. Eur J Pharmacol. 2009;619:50–56. doi: 10.1016/j.ejphar.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrizi A, Viltono L, Frola E, Harvey K, Harvey RJ, Sassoe-Pognetto M. Selective localization of Collybistin at a subset of inhibitory synapses in brain circuits. J Comp Neurol. 2012;520:130–141. doi: 10.1002/cne.22702. [DOI] [PubMed] [Google Scholar]
- Peng YJ, He WQ, Tang J, Tao T, Chen C, Gao YQ, et al. Trio is a key guanine nucleotide exchange factor coordinating regulation of the migration and morphogenesis of granule cells in the developing cerebellum. J Biol Chem. 2010;285:24834–24844. doi: 10.1074/jbc.M109.096537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Beeser A, Chernoff J, Schiller MR, Eipper BA, Mains RE, et al. Rapid induction of dendritic spine morphogenesis by trans-synaptic ephrinB-EphB receptor activation of the Rho-GEF kalirin. Neuron. 2003;37:263–274. doi: 10.1016/s0896-6273(02)01168-6. [DOI] [PubMed] [Google Scholar]
- Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14:285–293. doi: 10.1038/nn.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Jones KA. Dendritic spine dynamics--a key role for kalirin-7. Trends Neurosci. 2008;31:419–427. doi: 10.1016/j.tins.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabiner CA, Mains RE, Eipper BA. Kalirin: a dual Rho guanine nucleotide exchange factor that is so much more than the sum of its many parts. Neuroscientist. 2005;11:148–160. doi: 10.1177/1073858404271250. [DOI] [PubMed] [Google Scholar]
- Rojas RJ, Yohe ME, Gershburg S, Kawano T, Kozasa T, Sondek J. Galphaq directly activates p63RhoGEF and Trio via a conserved extension of the Dbl homology-associated pleckstrin homology domain. J Biol Chem. 2007;282:29201–29210. doi: 10.1074/jbc.M703458200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- Rubino T, Forlani G, Vigano D, Zippel R, Parolaro D. Ras/ERK signalling in cannabinoid tolerance: from behaviour to cellular aspects. J Neurochem. 2005;93:984–991. doi: 10.1111/j.1471-4159.2005.03101.x. [DOI] [PubMed] [Google Scholar]
- Rubino T, Vigano D, Premoli F, Castiglioni C, Bianchessi S, Zippel R, et al. Changes in the expression of G protein-coupled receptor kinases and beta-arrestins in mouse brain during cannabinoid tolerance: a role for RAS-ERK cascade. Mol Neurobiol. 2006;33:199–213. doi: 10.1385/MN:33:3:199. [DOI] [PubMed] [Google Scholar]
- Schiller MR. Coupling receptor tyrosine kinases to Rho GTPases--GEFs what's the link. Cell Signal. 2006;18:1834–1843. doi: 10.1016/j.cellsig.2006.01.022. [DOI] [PubMed] [Google Scholar]
- Schiller MR, Ferraro F, Wang Y, Ma XM, McPherson CE, Sobota JA, Mains RE, Eipper BA. Autonomous functions for the Sec14p/spectrin-repeat region of Kalirin. Exp Cell Res. 2008;314:2674–2691. doi: 10.1016/j.yexcr.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojima K, Sugawara M, Shichiji M, Mukaida S, Takayama R, Imai K, Yamamoto T. Loss of function mutation of collybistin is responsible for X-linked mental retardation associated with epilepsy. J Hum Genet. 2011;56:561–565. doi: 10.1038/jhg.2011.58. [DOI] [PubMed] [Google Scholar]
- Terawaki S, Kitano K, Mori T, Zhai Y, Higuchi Y, Itoh N, et al. The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module. EMBO J. 2010;29:236–250. doi: 10.1038/emboj.2009.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Feig LA. Age-dependent participation of Ras-GRF proteins in coupling calcium-permeable AMPA glutamate receptors to Ras/Erk signaling in cortical neurons. J Biol Chem. 2006;281:7578–7582. doi: 10.1074/jbc.M512060200. [DOI] [PubMed] [Google Scholar]
- Tolias KF, Bikoff JB, Burette A, Paradis S, Harrar D, Tavazoie S, et al. The Rac1-GEF Tiam1 couples the NMDA receptor to the activity-dependent development of dendritic arbors and spines. Neuron. 2005;45:525–538. doi: 10.1016/j.neuron.2005.01.024. [DOI] [PubMed] [Google Scholar]
- Tolias KF, Bikoff JB, Kane CG, Tolias CS, Hu L, Greenberg ME. The Rac1 guanine nucleotide exchange factor Tiam1 mediates EphB receptor-dependent dendritic spine development. Proc Natl Acad Sci U S A. 2007;104:7265–7270. doi: 10.1073/pnas.0702044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolias KF, Duman JG, Um K. Control of synapse development and plasticity by Rho GTPase regulatory proteins. Prog Neurobiol. 2011;94:133–148. doi: 10.1016/j.pneurobio.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonini R, Franceschetti S, Parolaro D, Sala M, Mancinelli E, Tininini S, et al. Involvement of CDC25Mm/Ras-GRF1-dependent signaling in the control of neuronal excitability. Mol Cell Neurosci. 2001;18:691–701. doi: 10.1006/mcne.2001.1050. [DOI] [PubMed] [Google Scholar]
- Tyagarajan SK, Ghosh H, Harvey K, Fritschy JM. Collybistin splice variants differentially interact with gephyrin and Cdc42 to regulate gephyrin clustering at GABAergic synapses. J Cell Sci. 2011;124:2786–2796. doi: 10.1242/jcs.086199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Nakata A, Mizuno N, Tago K, Itoh H. Domain-domain interaction of P-Rex1 is essential for the activation and inhibition by G protein betagamma subunits and PKA. Cell Signal. 2008;20:1545–1554. doi: 10.1016/j.cellsig.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10:842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Hauser ER, Shah SH, Pericak-Vance MA, Haynes C, Crosslin D, et al. Peakwide mapping on chromosome 3q13 identifies the kalirin gene as a novel candidate gene for coronary artery disease. Am J Hum Genet. 2007;80:650–663. doi: 10.1086/512981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JE, Astle MV, Ooms LM, Balamatsias D, Gurung R, Mitchell CA. P-Rex1 - a multidomain protein that regulates neurite differentiation. J Cell Sci. 2008;121:2892–2903. doi: 10.1242/jcs.030353. [DOI] [PubMed] [Google Scholar]
- Welch HC, Coadwell WJ, Ellson CD, Ferguson GJ, Andrews SR, Erdjument-Bromage H, et al. P-Rex1, a PtdIns(3,4,5)P3- and Gbetagamma-regulated guanine-nucleotide exchange factor for Rac. Cell. 2002;108:809–821. doi: 10.1016/s0092-8674(02)00663-3. [DOI] [PubMed] [Google Scholar]
- Welch HC, Condliffe AM, Milne LJ, Ferguson GJ, Hill K, Webb LM, et al. P-Rex1 regulates neutrophil function. Curr Biol. 2005;15:1867–1873. doi: 10.1016/j.cub.2005.09.050. [DOI] [PubMed] [Google Scholar]
- Williams SL, Lutz S, Charlie NK, Vettel C, Ailion M, Coco C, et al. Trio's Rho-specific GEF domain is the missing Galpha q effector in C. elegans. Genes Dev. 2007;21:2731–2746. doi: 10.1101/gad.1592007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock SA, Rushton HJ, Castaneda-Saucedo E, Myant K, White GR, Blyth K, et al. Tiam1-Rac signaling counteracts Eg5 during bipolar spindle assembly to facilitate chromosome congression. Curr Biol. 2010;20:669–675. doi: 10.1016/j.cub.2010.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JH, Fanaroff AC, Sharma KC, Zhang L, Smith LS, Brian L, Eipper BA, Mains RE, Freedman NJ. Kalirin Promotes Neointimal Hyperplasia by Activating Rac in Smooth Muscle Cells. Arterioscler Thromb Vasc Biol. 2012 doi: 10.1161/ATVBAHA.112.300234. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Cahill ME, Radulovic J, Wang J, Campbell SL, Miller CA, et al. Hippocampal phenotypes in kalirin-deficient mice. Mol Cell Neurosci. 2011;46:45–54. doi: 10.1016/j.mcn.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin X, Rabiner CA, Mains RE, Eipper BA. Kalirin12 interacts with dynamin. BMC Neurosci. 2009;10:61. doi: 10.1186/1471-2202-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Cooley D, Legakis JE, Ge Q, Andrade R, Mattingly RR. Phosphorylation of the Ras-GRF1 exchange factor at Ser916/898 reveals activation of Ras signaling in the cerebral cortex. J Biol Chem. 2003;278:13278–13285. doi: 10.1074/jbc.M209805200. [DOI] [PubMed] [Google Scholar]
- Yoo S, Kim Y, Lee H, Park S, Park S. A gene trap knockout of the Tiam-1 protein results in malformation of the early embryonic brain. Mol Cells. 2012;34:103–108. doi: 10.1007/s10059-012-0119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa M, Kawauchi T, Sone M, Nishimura YV, Terao M, Chihama K, et al. Involvement of a Rac activator,P-Rex1, in neurotrophin-derived signaling and neuronal migration. J Neurosci. 2005;25:4406–4419. doi: 10.1523/JNEUROSCI.4955-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn H, Jeoung M, Koo Y, Ji H, Markesbery WR, Ji I, et al. Kalirin is under-expressed in Alzheimer's disease hippocampus. J Alzheimers Dis. 2007;11:385–397. doi: 10.3233/jad-2007-11314. [DOI] [PubMed] [Google Scholar]
- Zhang GC, Hoffmann J, Parelkar NK, Liu XY, Mao LM, Fibuch EE, et al. Cocaine increases Ras-guanine nucleotide-releasing factor 1 protein expression in the rat striatum in vivo. Neurosci Lett. 2007;427:117–121. doi: 10.1016/j.neulet.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Zhang H, Macara IG. The polarity protein PAR-3 and TIAM1 cooperate in dendritic spine morphogenesis. Nat Cell Biol. 2006;8:227–237. doi: 10.1038/ncb1368. [DOI] [PubMed] [Google Scholar]