Abstract
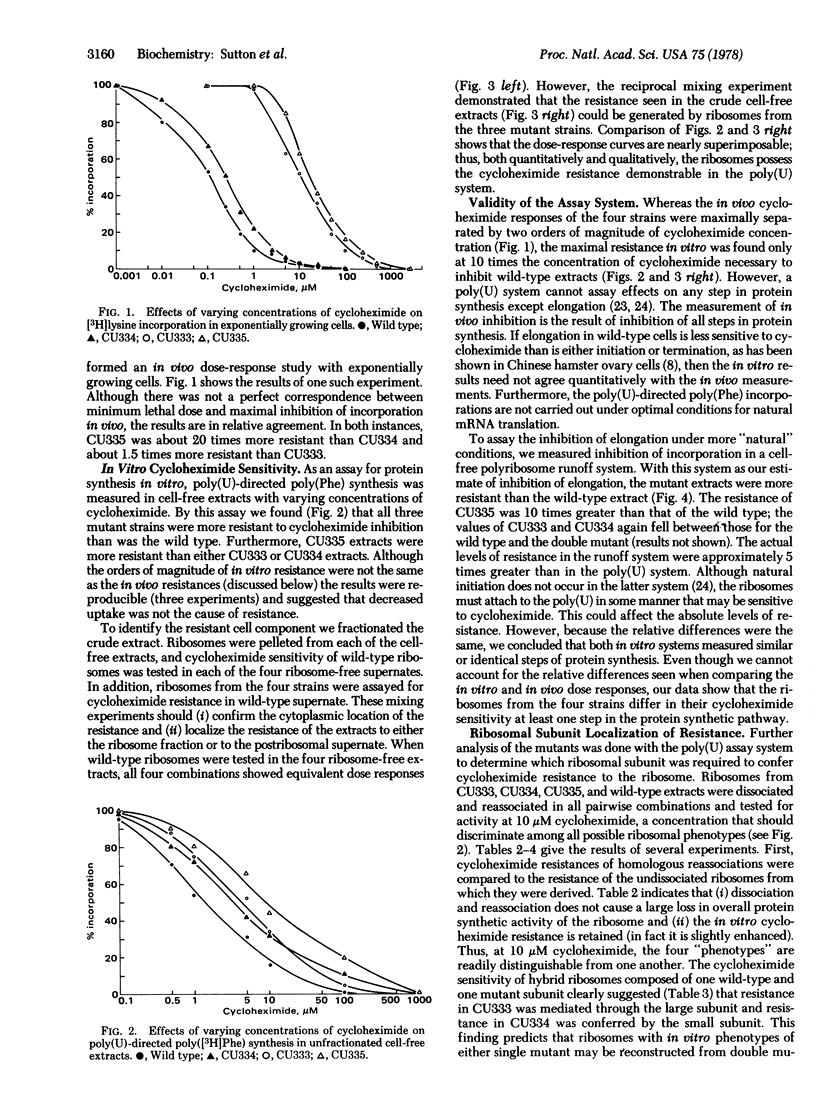
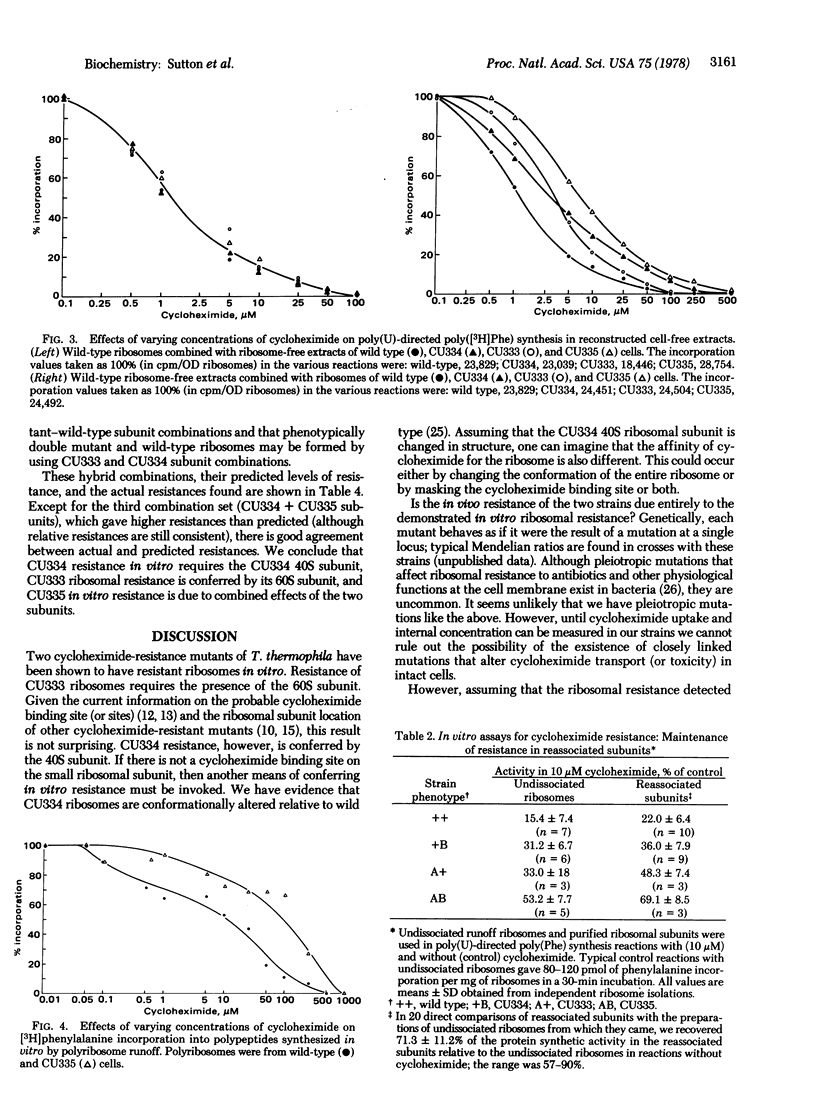
Two cycloheximide-resistant mutants of Tetrahymena thermophila were analyzed to determine the site of their cycloheximide resistance. The mutations in both strains had been previously shown to be genetically dominant and located at separate loci (denoted Chx-A and Chx-B). Strains carrying these mutations were readily distinguished by the extent to which they were resistant to the drug. The homozygous double mutant was more resistant than either single mutant. Cell-free extracts of wild type and of the three mutant strains, assayed for protein synthetic activity by both runoff of natural mRNA and poly(U)-dependent phenylalanine polymerization, demonstrated that in vitro the mutants were all more resistant than the wild type. Further fractionation of the cell-free systems into ribosomes and supernates localized cycloheximide resistance to the ribosome for both Chx-A and Chx-B homozygotes. Ribosome dissociation and pairwise subunit mixing in the in vitro system indicated that ribosome resistance was conferred by the 60S subunit from one strain whereas resistance in the other strain was mediated through the 40S subunit. This was further confirmed by reconstruction of all four cycloheximide-resistance "phenotypes" by mixing ribosomal subunits from appropriate strains. This finding suggests that the mechanisms by which these mutations confer resistance to cycloheximide are different.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baliga B. S., Pronczuk A. W., Munro H. N. Mechanism of cycloheximide inhibition of protein synthesis in a cell-free system prepared from rat liver. J Biol Chem. 1969 Aug 25;244(16):4480–4489. [PubMed] [Google Scholar]
- Bleyman L. K., Bruns P. J. Genetics of cycloheximide resistance in Tetrahymena. Genetics. 1977 Oct;87(2):275–284. doi: 10.1093/genetics/87.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper D., Banthorpe D. V., Wilkie D. Modified ribosomes conferring resistance to cycloheximide in mutants of Saccharomyces cerevisiae. J Mol Biol. 1967 Jun 14;26(2):347–350. doi: 10.1016/0022-2836(67)90302-6. [DOI] [PubMed] [Google Scholar]
- Cooper T. G., Bossinger J. Selective inhibition of protein synthesis initiation in Saccharomyces cerevisiae by low concentrations of cycloheximide. J Biol Chem. 1976 Nov 25;251(22):7278–7280. [PubMed] [Google Scholar]
- Falvey A. K., Staehelin T. Structure and function of mammalian ribosomes. I. Isolation and characterization of active liver ribosomal subunits. J Mol Biol. 1970 Oct 14;53(1):1–19. doi: 10.1016/0022-2836(70)90042-2. [DOI] [PubMed] [Google Scholar]
- Godchaux W., 3rd, Adamson S. D., Herbert E. Effects of cycloheximide on polyribosome function in reticulocytes. J Mol Biol. 1967 Jul 14;27(1):57–72. doi: 10.1016/0022-2836(67)90351-8. [DOI] [PubMed] [Google Scholar]
- Grant P., Sánchez L., Jiménez A. Cryptopleurine resistance: genetic locus for a 40S ribosomal component in Saccharomyces cerevisiae. J Bacteriol. 1974 Dec;120(3):1308–1314. doi: 10.1128/jb.120.3.1308-1314.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. S., Siminovitch L. The molecular basis of emetine resistance in Chinese hamster ovary cells: alteration in the 40S ribosomal subunit. Cell. 1977 Jan;10(1):61–66. doi: 10.1016/0092-8674(77)90140-4. [DOI] [PubMed] [Google Scholar]
- Haugli F. B., Dove W. F., Jimenez A. Genetics and biochemistry of cycloheximide resistance in Physarum polycephalum. Mol Gen Genet. 1972;118(2):97–107. doi: 10.1007/BF00267081. [DOI] [PubMed] [Google Scholar]
- Kisilevsky R. The regulatory parameter of protein synthesis most affected by ethionine, and cycloheximide. A comparison of computer and in vivo studies. Biochim Biophys Acta. 1972 Jul 20;272(3):463–472. doi: 10.1016/0005-2787(72)90398-x. [DOI] [PubMed] [Google Scholar]
- LEDERBERG J. Streptomycin resistance; a genetically recessive mutation. J Bacteriol. 1951 May;61(5):549–550. doi: 10.1128/jb.61.5.549-550.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish H. F. Alpha and beta globin messenger ribonucleic acid. Different amounts and rates of initiation of translation. J Biol Chem. 1971 Dec 10;246(23):7131–7138. [PubMed] [Google Scholar]
- Mizuno T., Yamada H., Yamagata H., Mizushima S. Coordinated alterations in ribosomes and cytoplasmic membrane in sucrose-dependent, spectinomycin-resistant mutants of Escherichia coli. J Bacteriol. 1976 Feb;125(2):524–530. doi: 10.1128/jb.125.2.524-530.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obrig T. G., Culp W. J., McKeehan W. L., Hardesty B. The mechanism by which cycloheximide and related glutarimide antibiotics inhibit peptide synthesis on reticulocyte ribosomes. J Biol Chem. 1971 Jan 10;246(1):174–181. [PubMed] [Google Scholar]
- Oleinick N. L. Initiation and elongation of protein synthesis in growing cells: differential inhibition by cycloheximide and emetine. Arch Biochem Biophys. 1977 Jul;182(1):171–180. doi: 10.1016/0003-9861(77)90296-x. [DOI] [PubMed] [Google Scholar]
- Pongratz M., Klingmüller W. Role of ribosomes in cycloheximide resistance of Neurospora mutants. Mol Gen Genet. 1973 Aug 28;124(4):359–363. doi: 10.1007/BF00267664. [DOI] [PubMed] [Google Scholar]
- Pöche H., Junghahn I., Geissler E., Bielka H. Cycloheximide resistance in Chinese hamster cells. III. Characterization of cell-free protein synthesis by polysomes. Mol Gen Genet. 1975;138(2):173–177. [PubMed] [Google Scholar]
- Rajalakshmi S., Liang H., Sarma D. S., Kisilevsky R., Farber E. Cycloheximide, an inhibitor of peptide chain termination or release in liver in vivo and in vitro. Biochem Biophys Res Commun. 1971 Jan 22;42(2):259–265. doi: 10.1016/0006-291x(71)90096-9. [DOI] [PubMed] [Google Scholar]
- Rao S. S., Grollman A. P. Cycloheximide resistance in yeast: a property of the 60s ribosomal subunit. Biochem Biophys Res Commun. 1967 Dec 15;29(5):696–704. doi: 10.1016/0006-291x(67)90273-2. [DOI] [PubMed] [Google Scholar]
- Roberts C. T., Jr, Orias E. A cycloheximide-resistant mutant of Tetrahymena pyriformis. Exp Cell Res. 1973 Oct;81(2):312–316. doi: 10.1016/0014-4827(73)90520-x. [DOI] [PubMed] [Google Scholar]
- Skogerson L., Wakatama E. A ribosome-dependent GTPase from yeast distinct from elongation factor 2. Proc Natl Acad Sci U S A. 1976 Jan;73(1):73–76. doi: 10.1073/pnas.73.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasundaran U., Skogerson L. A cycloheximide sensitivity factor from yeast required for N-acetylphenylalanylpuromycin formation. Biochemistry. 1976 Nov 2;15(22):4760–4764. doi: 10.1021/bi00667a002. [DOI] [PubMed] [Google Scholar]
- Stanners C. P. The effect of cycloheximide on polyribosomes from hamster cells. Biochem Biophys Res Commun. 1966 Sep 8;24(5):758–764. doi: 10.1016/0006-291x(66)90390-1. [DOI] [PubMed] [Google Scholar]


