Abstract
Purpose
To evaluate whether patients with metastatic gastrointestinal (GI) adenocarcinomas refractory to chemotherapy harbor tumor-reactive cytotoxic T cells.
Experimental design
Expansion of CD8+ tumor-infiltrating lymphocytes (TIL) and cancer cell lines was attempted from GI cancer metastases in 16 consecutive patients for the study of anti-tumor immune recognition. Retroviral transduction of genes encoding T-cell receptors (TCR) was used to define HLA-restriction elements and specific reactivity.
Results
TIL were expanded from metastases in all patients, and new tumor cell lines generated in five patients. Autologous tumor-recognition without cross-reactivity against allogeneic HLA-matched GI tumors was found in CD8+ TIL from three of these five patients. In a patient with gastric cancer liver metastases, the repertoire of CD8+ TIL was dominated by cytolytic sister clones reactive to 2 out of 4 autologous cancer cell lines restricted by HLA-C*0701. From the same patient, a rare CD8+ TIL clone with a distinct TCR recognized all four cancer cell lines restricted by HLA-B*4901. In a patient with bile duct cancer, two distinct anti-tumor cytolytic clones were isolated from a highly polyclonal CD8+ TIL repertoire. TCRs isolated from these clones recognized epitopes restricted by HLA-A*0201. In a third patient, CD8+ TIL reactivity was progressively lost against an autologous colon cancer cell line that displayed loss of HLA haplotype.
Conclusions
This study provides a basis for the development of immunotherapy for patients with advanced GI malignancies by first establishing the presence of naturally occurring tumor-reactive CD8+ TIL at the molecular level.
Keywords: Gastrointestinal neoplasms, Metastasis, CD8-positive T-lymphocytes, Tumor-infiltrating lymphocytes, Adoptive cellular immunotherapy
Introduction
Gastrointestinal (GI) adenocarcinomas are among the ten most common malignancies worldwide and the overall mortality associated with their high metastatic potential has not changed significantly over the last decades (1). Current multimodality treatments can slow disease progression but fail to cure patients with metastatic disease. Thus far, immune-based therapies have not shown clinical effectiveness in patients with GI cancers (2–5). A positive association between the density of the tumor-infiltrating lymphocyte (TIL) infiltrate and better outcomes has nonetheless been reported in patients with adenocarcinomas arising in the esophagus, stomach, pancreas, liver, bile ducts, gallbladder, colon, and rectum (6). Multiple immune escape mechanisms have however been proposed that may contribute to the absence, the depletion, and the dysfunction of tumor-reactive TIL in solid tumors (7–9). Thus the specific recognition of human metastatic GI cancers by naturally occurring, autologous, cytotoxic CD8+ T cells harvested from the tumor site has not been defined at the T-cell clonal level with defined HLA restriction elements.
Results from studies in patients with metastatic melanoma have stimulated us to investigate the anti-tumor reactivity of CD8+ TIL in common epithelial malignancies. Indeed, TIL isolated from melanoma metastases can exhibit direct in vitro tumor recognition of defined antigens presented by specific class I HLAs (10–14), and tumor deposits appear to harbor antitumor T cells of sufficient avidity and in sufficient numbers to respond to non-specific systemic modulation of immunity (15–18). Additionally, as now reported by multiple institutions, the adoptive cell transfer of autologous TIL can mediate complete cancer regression in patients with metastatic disease considered incurable with standard therapy, with complete responders reported up to 10 years after treatment (19–23). The curative potential of TIL-based immunotherapy in advanced melanoma represents a paradigmatic shift on how solid cancer treatment is approached, and whether this strategy can be applied for common metastatic epithelial malignancies merits active investigations.
In the current report, an in vitro analysis of TIL was carried in 16 patients with metastatic GI cancer. Detailed CD8+ TIL reactivity to autologous GI cancer metastases was carried out in five patients from whom 13 new cancer cell lines were established. TIL from three of these patients exhibited specific immune reactivity against their autologous metastatic cancer. By defining immune features of metastatic GI cancers cells and TIL, our findings have direct relevance to efforts to develop immunotherapies for patients with these malignancies.
Methods
Patients and tumor processing
Written informed consent was obtained from all patients enrolled under protocols approved by the Institutional Review Board of the National Cancer Institute (NCI) and U.S. Food and Drug Administration. Single cell suspensions were obtained from freshly resected tumors by independent enzymatic digestion and mechanical dispersion as previously described for melanoma specimens (24).
Primary human cancer cell cultures and culture of other cancer cell lines
To develop cancer cell lines, 0.25e6 live nucleated cells were plated in multiple 25 cm2 ultra-low attachment and standard treated canted neck flasks (Corning 3815 and 3056, NY) in RPMI 1640 based medium supplemented with 20% fetal bovine serum (Defined, Hyclone Laboratories, UT), 25 mmol/L HEPES, 2 mmol/L L-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin (all from Life Technologies, Invitrogen, Grand Island, NY), 1.25 μg/ml Amphotericin B (XGen Pharmaceuticals, NY), and 10 μg/ml ciprofloxacin (Bedford laboratories, OH). After 6 to 12 weeks, cell aggregates/tumor spheres (approximately 200 um in diameter) were transferred into standard 25 cm2 flasks for propagation under adherent conditions. For adherent conditions, fibroblasts overgrowth was controlled by differential trypsinization (Trypsin-EDTA 1x, 0,05%, Gibco) and mechanical removal (17 mm blade cell scraper, Sarstedt, Newton, NC), and cultures were fed weekly or according to need, and passaged into larger flasks when reaching confluence. The human cancer cell lines Kato III, NCI N87, NCI H508, Colo205, HCT15, SK-CO-1, KM12, HT29, SW480, SW620, HCC2998, SW1463, Capan1, and Panc 02.03 were purchased from the American Type Culture Collection and grown under the vendor’s suggested conditions. Human melanoma cell lines 3350 and 624, and human pancreatic cancer cell line 2596 and 2742-2, were established in our laboratory.
The authenticity of all cell lines was confirmed by HLA typing and testing expression of melanoma differentiation antigens (MART-1, Melan-A, gp100) and GI cancer antigens (CEA, pan-cytokeratins). Mycoplasma contamination was ruled out on all cell cultures by using MycoAlert (Lonza) according to the manufacturer instructions.
Generation of TIL
TIL establishment and expansion followed techniques used for metastatic melanoma tumor deposits and are presented in supplementary methods (24, 25).
Characteristics of fresh TIL and cultured lines
Staining of paraffin-embedded tissue and cell pellets was done at the NCI Clinical Research Center (NCI/CRC) Pathology and Cytology Laboratory following standard procedures with appropriate positive and isotype controls. Antibodies and staining conditions are presented in supplementary methods. For flow cytometry, the following monoclonal antibodies specific for human antigens and appropriate isotype controls were used: from BD Biosciences, APC-H7-conjugated anti-CD3 (SK7), APC anti-CD137/4-1BB (4B4-1); from Invitrogen: R-PE-Texas Red-conjugated anti-CD8 (3B5). Cell aggregates and dead cells were excluded by forward and side scatter, and with propidium iodide staining. Flow cytometry analysis was carried out with FlowJo F7.5.5 software (Tree Star, OR). HLA typing of cell lines was done by the NCI/CRC HLA lab on genomic DNA following standard procedures (supplementary methods).
Cancer cell-recognition by T cells co-culture assays
T-cell reactivity to cancer cell lines was assessed after 24–36 h co-culture assays in flat-bottom 96 well plates (1e5 T cells, 0.5e5 trypsinized cancer cells, final volume 200 μl). ELISA were carried out for measurement of IFN-γ release in supernatant, and flow cytometry used to quantify CD137 (4-1BB) upregulation on T cells (supplementary methods). Chromium-51 release 4–6 h cytolysis assays were used as described in previous studies (26, 27) and in supplementary methods. The percentage of specific lysis was calculated as: percent lysis = (sample release − spontaneous release)/(maximal − spontaneous release) × 100, average of triplicate samples.
Limiting dilution cloning of 4-1BB+-enriched TIL
After stimulation assays with autologous cancer cell line, CD8+ TIL that exhibited reactivity by high 4-1BB+ expression where sorted by magnetic cell separation (anti-APC IMag, BD Biosciences) or fluorescence-activated cell sorting (FACSAria II, BD Biosciences). Briefly, between 1 to 20 TIL were plated in each well of a 96-well U-bottomed plate in 200 μl of conditioned medium containing 3000 IU/ml rhIL-2, 30 ng/ml of OKT3, with 5 × 104 irradiated (40 Gy) allogeneic PBMCs. On day 5 and every 3 to 4 days thereafter, half of the medium in each well was replaced with fresh medium containing IL-2. About 10 to 14 days after culture initiation, wells in which cell growth was visibly apparent were screened for reactivity to autologous and allogeneic cancer cell line. Clone reactive by IFN-γ secretion and/or 4-1BB expression were subsequently expanded by Rapid Expansion Protocol (REP)(24).
TCR sequencing and cloning, retroviral transduction of PBMC
Clonotypic analysis of the TCR repertoire was done from RNA purified from CD8+ TIL with an RNeasy mini kit (Qiagen) and reverse transcribed into cDNA for 5′Rapid Amplification of cDNA Ends (RACE) by SMARTer RACE cDNA amplification kit per manufacturer instructions (Clontech, Mountain View, CA) and as described with subcloning into a pCR2.1 vector by TA cloning (Life Technologies) (28). Consensus regions were determine using the International Immunogenetics Information System (29) for sequence analysis of 96 colonies picked from each 5′ RACE product of both TCR alpha and beta chains and their variable regions. Murinized human TCRs were cloned into MSGV1 retroviral vectors as described (30) and detailed in supplementary methods. Transduction of PBMC was done as described (31), and efficiency assessed after one week expansion by flow cytometry using an anti-mouse TCR β-chain antibody (H57-597, eBioscience). MSGV1 vector expressing the TCR recognizing the MART-1:27-35 epitope (31) and GFP were used as controls in all experiments, as well as untransduced PBMC.
Statistics
Statistical analyses were performed on GraphPad Prism software version 5.04 (La Jolla, CA). Variances of mean values are presented as standard error of the mean. Two-tailed, non-parametric tests were used and p-values of ≤ 0.05 were considered significant.
Results
Patient and tumor clinicopathological and immunological characteristics
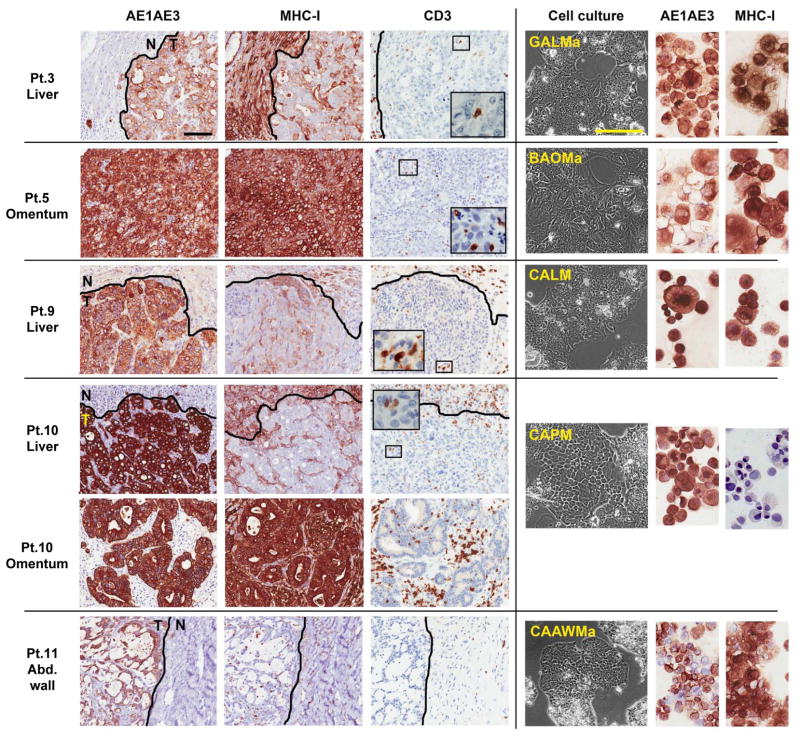
Resected tumors from sixteen patients with widespread, moderately to poorly differentiated metastatic adenocarcinomas originating from the stomach, the bile duct, or the colon were studied (Table 1). All patients had progressive disease after receiving at least one standard first-line chemotherapy regimen (median, 2). The expression of MHC class-I (MHC-I) molecules — required for CD8+ T-cell recognition — was heterogenous across resected metastases and nearly undetectable in 35% of lesions (Table 1, and Fig. 1). All metastases were poorly infiltrated by TIL, which represented less than 5% of the tumor cut surface on histological assessment, with the exception of omental metastases from patients 5 and 10, which were more densely infiltrated by CD3+ T cells. Heterogeneity in antigenic expression by cancer cells within the same patient was exemplified in metastases resected in patient 10, since MHC-I was faintly detected on the liver metastasis but strongly on the omental metastasis (Fig. 1). Patient 11 had a mucinous adenocarcinoma characterized by fine reticulated bands of tumor cells separated by copious pools of mucin and surrounded by a fibrotic stroma containing rare T cells (Fig. 1).
Table 1.
Patient demographics, tumor immune characteristics, and tumor-infiltrating lymphocyte expansion
| Patient characteristics | Adenocarcinoma caracteristics | TIL first outgrowth from tumor | TIL clinical scale expansione | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||||
| No. | Age/Gender | Prior chemotherapy regimens | Primary cancer site | Metastasesa | Tumor cells MHC-I expressionb | Tumor-infiltrating CD3+ T cellsc | New cancer cell lines | Fold expansiond | %CD3+CD8+ of live cells | Fold expansion | %CD3+CD8+ of live cells |
|
|
|
|
|
||||||||
| 1 | 63/M | 2 | Colon | Liver | n/a | n/a | n/a | 12.6 | 10.5 | 990 | 94.0 |
|
|
|
|
|
||||||||
| 2 | 45/M | 1 | Colon | Liver | n/a | n/a | n/a | 19.7 | 29.1 | 522 | 97.0 |
|
|
|
|
|
||||||||
| 3 | 44/F | 3 | Stomach | Liver, ovary | 1+, 5–50% | 1–5% | GALM a, b, c, d | 31.5 | 9.0 | 798 | 98.0 |
|
|
|
|
|
||||||||
| 4 | 45/F | 3 | Colon | Liver, retroperitoneum | 0–1+, <5% | 1–5% | n/a | 12.8 | 10.7 | 788 | 98.4 |
|
|
|
|
|
||||||||
| 5 | 57/M | 3 | Bile duct, intrahepatic | Omentum, liver, ascites | 2–3+, >50% | 5–50% | BAOM a & b, BAAM | 1.1 | n/a | n/a | n/a |
|
|
|
|
|
||||||||
| 6 | 42/M | 1 | Colon | Retroperitoneum, omentum, lung | 2–3+, >50% | 0–1% | n/a | Fg | 51.0 | 904 | 97.4 |
|
|
|
|
|
||||||||
| 7 | 51/M | 4 | Rectum | Lung, bones | 3+, >50% | 1–5% | n/a | 40.0 | 59.4 | 1770 | 98.0 |
|
|
|
|
|
||||||||
| 8 | 51/M | 2 | Colon | Lung, liver, bones, penis | 2–3+, >50% | 1–5% | n/a | 1.8 | 24.9 | n/a | n/a |
|
|
|
|
|
||||||||
| 9 | 57/F | 3 | Colon | Liver | 1+, 5–50% | 1–5% | CALM | Fg | 10.0 | n/a | n/a |
|
|
|
|
|
||||||||
| 10 | 51/F | 1 | Colon | Liver, omentum, retroperitoneum, spleen | (L.) 0–1+, <5% (O.) 3+, >50% |
(L.) 1–5% (O.) 5–50% |
CAPM | Fg | 22.7 | 802 | 98.0 |
|
|
|
|
|
||||||||
| 11 | 53/F | 2 | Colon | Abdominal wall, liver, breast | 0–1+, <5% | 0–1% | CAAWM a, b, c, d | Fg | 3.0 | n/a | n/a |
|
|
|
|
|
||||||||
| 12 | 52/F | 6 | Colon | Lung, liver, omentum, retroperitoneal, pelvis | 3+, >50% | 1–5% | n/a | 60.0 | 50.8 | 1517 | 98.7 |
|
|
|
|
|
||||||||
| 13 | 41/M | 5 | Colon | Axillary lymph node | 1+, <5% | 1–5% | n/a | 11.0 | 41.0 | >743 | 97.9 |
|
|
|
|
|
||||||||
| 14 | 37/F | 2 | Colon | Liver, lung | 3+, >50% | 1–5% | n/a | 47.5 | 20.8 | 1995 | 24.1 |
|
|
|
|
|
||||||||
| 15 | 63/M | 2 | Stomach | Liver, lung | 3+, >50% | 1–5% | n/a | 46.2 | 14.4 | 1454 | 69.6 |
|
|
|
|
|
||||||||
| 16 | 43/F | 2 | Bile duct, intrahepatic | Liver, lung | 2–3+, >50% | 1–5% | n/a | 31.7 | 18.5 | 802 | 40.0 |
NOTE:
Underlined metastases designate the site of TIL harvest, and subsequent columns refer to this tumor.
Semi-quantitative measurements of MHC class I performed by immunohistochemistry, with intensity (1+ to 3+) reported in addition to the percent of tumor cells expressing the surface marker (<5%, 5–50%, or >50%).
Semi-quantitative measurement, percent of surface of tumor occupied by tumor-infiltrating CD3+ T cells (0–1%, 1–5% or 5–50%).
Fold expansion cannot be calculated for TIL expanded from tumor fragments (Fg).
Rapid expansion was obtained in all 16 patients, results shown for samples tested with clinical grade reagents. Patient 1 to 13 had CD8+ T cell enrichment prior to clinical scale expansion.
Abbreviations: BAAM, biliary adenocarcinoma metastatic ascites; BAOM, biliary adenocarcinoma omental metastasis; CAAWM, colon adenocarcinoma abdominal wall metastasis; CALM, colon adenocarcinoma liver metastasis; CAPM, colon adenocarcinoma peritoneal metastasis; Fg, tumor fragments; GALM, gastric adenocarcinoma liver metastasis; MHC-I, MHC class I; n/a, not available.
Fig. 1. Cancer cell lines established from gastrointestinal cancer metastases with heterogeneous MHC-I expression and weak T-cell infiltration.
Immunohistochemical staining of paraffin-embedded gastrointestinal cancer metastases (left) in five patients (Pt.) and representative corresponding cancer cell lines (right). Pt. 3, gastric adenocarcinoma liver metastasis, adjacent normal liver, and derived cancer cell line (GALMa). Pt. 5, omental metastasis from an intrahepatic bile duct adenocarcinoma and derived cancer cell line (BAOMa). Pt. 9, colon adenocarcinoma liver metastasis, adjacent liver, and derived cancer cell line (CALM). Patient 10, colon adenocarcinoma liver metastasis, adjacent liver, and omental metastasis, with derived cancer cell line (CAPM). Patient 11, abdominal wall metastasis from a mucinous colon adenocarcinoma, adjacent stroma, and derived cancer cell line (CAAWMa). All epithelial derived-cancer cells are stained by the pan-cytokeratin marker AE1AE3 which delineate tumor (T) from the adjacent normal tissues (N). MHC class I stains all nucleated cells, with weak and heterogeneous expression found in liver metastases in vivo, however most derived cancer cell line express MHC class I in culture except CAPM. Tumor-infiltrating lymphocytes are revealed by CD3 and T-cell marker, and in all cases represent less than 5% of cells in tumor mass.
Representative fields selected from whole slide scan, measure bar = 100 um.
TIL expansion from gastrointestinal metastases
Minced tumor fragments or cell suspensions obtained from GI cancer metastases were cultured in IL-2-containing media. Overgrowth of tumor as well as other adherent cells by TIL was observed between 16 and 29 days from culture initiation (median, 21 days). An average of 25.0±4.6% of the live cells in these cultures were CD3+CD8+ (range 3.0 to 59.4%). TIL were further expanded to large numbers from all samples using soluble anti-CD3 and irradiated allogeneic PBMC feeder cells for 14 days. Separated CD8+ T cells that were positively selected with magnetic beads from the expanded bulk TIL cultures had a median cumulative fold expansion of 853 using clinical-grade reagents (range 522 to 1770) (Table 1). Without CD8+ enrichment, TIL were similarly expanded from bulk cultures in patient 14, 15 and 16 with a median 1454 fold expansion (range 802 to 1995). Overall, TIL from GI cancer metastases in patients heavily pretreated with chemotherapy were found to have good in vitro proliferative potential.
Establishment of new gastrointestinal cancer cell lines and MHC-I expression loss
In parallel to setting up cultures for TIL expansion, as many cancer cell lines as possible were generated for each patient by initial culturing of tumor cell suspensions in ultra-low attachment flasks in addition to standard techniques in adherent flasks. When tumor spheroids were obtained, as in patients 3, 5, and 11, multiple cell lines were initiated from distinct spheroids, and thus 13 cancer cell lines were established in five patients (Table 1, Fig. S1). Complete HLA genotyping of the cancer cell lines and PBMC confirmed the parenthood of the lines for each patient (Table S1). Loss of heterozygosity at the HLA loci (haplotype loss) was found in two out of the 13 newly established cancer cell lines (patients 9 and 10). Total loss of MHC-I protein expression was seen in addition to genomic HLA haplotype loss in the only one colon cancer cell line generated from patient 10 (CAPM) (Fig. 1), rendering this cell line “invisible” for CD8+ T cells.
CD8+ TIL recognition of newly established autologous cancer cell lines
After expansion, CD8+ TIL recognition of autologous GI cancer was evaluated by examining their ability to secrete IFN-γ and to up-regulate expression of the inducible activation marker 4-1BB (CD137) (32) in response to tumor stimulation in the five patients for which new cancer cell lines had been established (Table 2). As expected by lack of MHC-I expression, CAPM failed to stimulate CD8+ TIL in patient 10 (Table S2). The rare fraction of CD8+ TIL expanded from fragments of mucinous abdominal wall colon cancer metastasis in patient 11 were not reactive to 4 autologous cancer cell lines (CAAWM) (Table S2). TIL reactivity to autologous tumors was however detected at different levels in 3 patients with metastatic GI adenocarcinoma originating from 3 different organs: stomach (patient 3), intrahepatic bile ducts (patient 5), and colon (patient 9) (Table 2).
Table 2.
Reactivity of bulk CD8+ tumor infiltrating lymphocytes to autologous cancer cell lines
| Pt. 3 Gastric Ca. CD8+ TIL | Pt. 5 Biliary tract Ca. CD8+ TIL | Pt. 9 Colon Ca. CD8+ TIL | |||||||
|---|---|---|---|---|---|---|---|---|---|
| pg/ml IFN-γ | (% 4-1BB+) | pg/ml IFN-γ | (% 4-1BB+) | pg/ml IFN-γ | (% 4-1BB+) | ||||
|
|
|
|
|
||||||
| pOKT3 | 2302 | (92.3) | 1650 | (38.3) | 607 | - | |||
| Medium | 80 | (0.2) | 71 | (0.3) | 75 | (0.2) | |||
|
|
|
|
|
||||||
| Autologous cancer cell | GALMa | 1236 | (66.2) | BAOMa | 76 | (0.7) | CALM | 200 | (15.7) |
| GALMb | 50 | (1.7) | BAAM | 68 | (1.0) | ||||
| GALMc | 55 | (1.0) | |||||||
| GALMd | 2046 | (83.5) | |||||||
|
|
|
|
|
||||||
| Allogeneic cancer cell | Mel.624 | 37 | (0.3) | GALMa | 79 | (0.1) | GALMd | 62 | - |
| BAOMa | 66 | - | |||||||
NOTE: Mean IFN-γ secretion (pg/ml) in 24 h co-culture supernatants of CD8+ TIL with autologous and allogeneic cancer cell lines in representative independant experiments. After sampling each well for IFN-γ, TIL duplicates were pooled and harvested for 4-1BB staining for flow cytometry analysis (in parenthesis, percent of CD8+ cells, gated on CD3+ cells). Bolded values for IFN-γ reflect secretion of at least 200 pg/ml, whereas bolded 4-1BB+ reflects specific upregulation at least twice as high as the unstimulated condition (media).
Abbreviations: Pt., patient; pOKT3, plate bound anti-CD3.
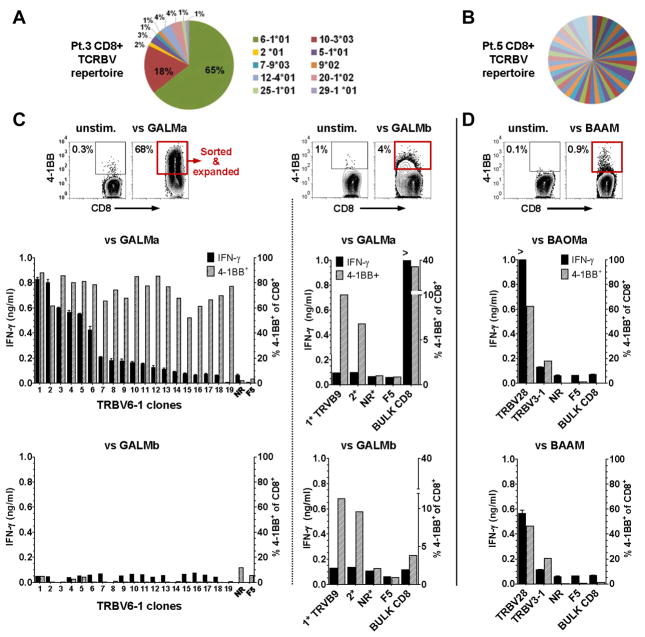
For patient 3, CD8+ T cells expanded from a gastric cancer liver metastasis secreted approximately 1000 pg/ml of IFN-γ in response to stimulation by two of the four autologous tumor cell line (GALMa and d, Table 2 and S2), and at least 65% up-regulated 4-1BB specifically. The other two autologous cancer cell line GALMb and c did not stimulate autologous CD8+ TIL to secrete significant amounts of INF-γ, and only 1 to 2% expressed 4-1BB, compared to 0.3% or less in absence of tumor stimulation or against an allogeneic melanoma cell line (Table 2). Analysis of the diversity of TCR-BV (beta chain gene, variable regions) sequences expressed by the CD8+ TIL from patient 3 revealed a relatively oligoclonal population, where 65% of the TCRs corresponded to a single rearranged TRBV6-1 gene product (Fig. 2A).
Fig. 2. Reactivity CD8+ TIL clones to autologous gastric and bile duct cancers.
(A) Diversity of the TCR repertoire of CD8+ TIL expanded from patient (Pt.) 3 liver metastasis, where distinct TCR variable β chains sequences are labeled by their TCR germline V consensus regions (TRBV). (B) Highly polyclonal TCRBV repertoire of TIL found in malignant ascites from a bile duct cancer in pt. 5. (C) For Pt. 3, FACS plot of 4-1BB expression upregulation in bulk CD8+ TIL after stimulation by autologous GALMa that allowed enrichment of the 4-1BB+ cells for limiting dilution cloning (red rectangle). Inferior to the FACS plot, the bar graphs represent the reactivity of 19 CD8+ TRBV6-1 TIL clones to the autologous GALMa, but not to the autologous GALMb cancer cell line, a non reactive (NR) clone (TRBV12-4), and the melanoma (F5) CD8+ TIL specificity control. FACS plot (middle) of the ~4 % 4-1BB upregulation (red rectangle) seen in bulk CD8+ TIL from pt.3 after stimulation by autologous GALMb. The bar graphs on the right isde of the dotted line represent the low reactivity found in 2 expanded clones (1*:TRBV9, 2*:unsequenced) to GALMa and GALMb, the bulk CD8+ TIL, a non-reactive (NR) clone, and the F5 specificity control by 4-1BB upregulation; none of these clones secreted significant amount of IFN-γ. (D) For Pt.5, FACS plot of the ~1% 4-1BB upregulation seen in the bulk CD8+ TIL. Limiting dilution cloning allowed isolation of 2 clones (TRBV28 and TRBV3-1) reactive to two autologous cancer cell lines established from an omental metastasis BAOMa and the malignant ascites BAAM. For bar graphs, left axis IFN-γ ng/ml; right axis %4-1BB+ of CD3+CD8+.
In contrast to patient 3, studies carried out in patient 5 revealed that the repertoire of CD8+ TIL expanded from the malignant ascites and an omental metastasis was highly polyclonal (Fig. 2B, omental metastasis not shown). Between the ascites and the omental metastasis, only 2% of the TCR-BV sequences overlapped. Approximately 1% of CD8+ TIL expanded from the malignant ascites nonetheless up-regulated 4-1BB after stimulation by the autologous cancer cell lines derived from the malignant ascites (BAAM) and derived from the omental metastasis (BAOMa) (Table 2). CD8+ TIL expanded from the omental nodule showed slightly lower expression of 4-1BB after stimulation with autologous tumor cell lines (data not shown). These percentages of 4-1BB expression upregulation, although low, were reproducible in sequential experiments, increased with prolonged autologous tumor stimulations, and were consistently higher than either the percentage of unstimulated TIL expressing 4-1BB (0.3%), or the percentage of TIL stimulated with allogeneic cancer cell lines, such as GALMa (0.1%) (Table 2).
In patient 9, the CD8+ TIL expanded from a colon cancer liver metastasis were highly oligoclonal, as 93% (62 out of 67 sequences) shared the same rearranged 7-9*03 TCR-BV sequence (Fig. S2). Co-cultures carried out with a low passage of the autologous colon cancer cell line CALM lead to the specific release of approximately 200 pg/ml of IFN-γ as well as up-regulation of 4-1BB expression on 16% of the CD8+ TIL (Table 2). Although recognition of the autologous cancer cell line by CD8+ TIL in this patient was seen in three independent assays, it decreased and was lost by the 6th passage of the cancer line (Fig. S2A). After documenting loss of HLA haplotype in this cancer cell line and that the initial recognition was likely restricted by the A allele (Fig. S2B), we attempted to restore antigen recognition by treating the cell line with IFN-γ to induce gene expression, or by retrovirally transducing it with the HLA-A*0201 gene. Neither attempt restored recognition by TIL.
Isolation of CD8+ T cell clones reactive to autologous cancers
We took advantage of the ability of CD8+ TIL to specifically up-regulate 4-1BB following tumor stimulation in patients 3 and 5 to enrich and isolate reactive T cells. For patient 3, CD8+ TIL expressing 4-1BB after stimulation with GALMa were enriched by magnetic beads prior to limiting dilution cloning (Fig. 2C). Expansion occurred in sixteen percent of the plated microcultures, allowing evaluation of the reactivity of 154 clonal T-cells. Specific up-regulation of 4-1BB expression was observed in 19 microcultures (12.3%). After re-expansion with soluble anti-CD3 and irradiated allogeneic PBMC feeders, each of the 19 clones still up-regulated 4-1BB expression after stimulation by GALMa, but only six of the 19 clones secreted significant amounts of IFN-γ. The TCR of eighteen reactive clones were sequenced and all shared the same dominant TRBV6-1*01 beta chain rearrangement sequence (Table S3 for complete TCR alpha and beta chains). As expected, the TRBV6-1 clones were only stimulated by GALMa and d, but not by GALMb and c, reflecting the reactivity pattern initially found from the bulk CD8+ (Fig. 2C left bar graphs; TRBV6-1 clone reactivity against GALMd, non-reactivity against GALMc, and allogeneic specificity controls are not shown).
In an attempt to isolate rare CD8+ TIL clones reactive to GALMb, FACS sorting was carried out to enrich a population corresponding to ~4% of the bulk TIL that up-regulated 4-1BB expression in response to stimulation by this autologous cancer cell line (Fig. 2C, right FACS plot). Clonal growth efficiency was 2.1% of the sorted TIL that were plated at 1 and 2 cells per well and lead to expansion of 32 clones. Five clones could successfully be re-expanded, 2 of which demonstrating approximately 10% 4-1BB up-regulation following co-culture with all GALM cell lines, without significant IFN-γ secretion capacity (Fig. 2C, right bar graphs; clones 1* and 2* reactivity to GALMc and GALMd are not shown). The TCR of the CD8+ T cell clones most reactive to GALM cell lines was sequenced and found to be derived from the TRBV9 germline gene (Table S3). We estimated that the frequency of the TRBV9 TCR was of less than 1.3% of CD8+ TIL expanded from the metastasis, since this specific TCR sequence did not match any of the one initially analyzed (n=76) from the bulk CD8+ TIL (Fig. 2A).
Thus from the gastric cancer liver metastasis of patient 3, multiple TRBV6-1 sister clones constituting a dominant T-cell clonotype in CD8+ TIL recognized 2 out of 4 autologous cancer cell lines (GALMa and d), whereas a genetically distinct TIL clone found at low frequency recognized all 4 autologous cell with lower reactivity. Of note, the TRBV6-1 sister clones, with genetically identical TCRs, up-regulated 4-1BB to similar levels upon tumor recognition but differed in their capacity to secrete IFN-γ, implying distinct functional attributes regulated at the epigenetic level. Our findings also supported distinct antigen expression profiles found in 4 autologous cancer cell lines derived from a single liver metastasis constituted by heterogeneous cancer cells.
For patient 5 with bile duct cancer, approximately 4000 4-1BB+CD8+ TIL from the malignant ascites and the omental metastasis were FACS-sorted after stimulation with autologous cell lines (Fig. 2D). Overall 604 clones grew at a clonal efficiency of 9, 17, and 32% when plated at 1, 2, and 5 cells per well. A preliminary assessment of the T-cell clone reactivity by stimulation with autologous tumor indicated that 3 clones (0.5%) were reactive by standard criteria for IFN-γ release and 10 (1.6%) possessed relatively low reactivity with IFN-γ secretion at least two time higher than the unstimulated condition, however not reaching the 200 pg/ml cut-off. Among 32 microcultures assessed for 4-1BB up-regulation, 4 clones (12.5%) up-regulated the surface marker by at least 20%. After re-expansion, two out of the 17 clones showed signs of specific TCR engagement with autologous BAOMa and BAAM cancer cell lines by 4-1BB up-regulation, but only one clone secreted significant amounts of IFN-γ (Fig. 2D bar graphs; non-reactivity to allogeneic targets not shown). The two clones did not share the same TCR (TRBV28 and TRBV3-1, see Table S3) and were found at a frequency of less than 1.8% of the expanded CD8+ TIL. Again, the differential reactivity of these clones to autologous cancer cell lines here established from distinct body compartments (omental metastases and malignant ascites), suggested a heterogenous expression of immune epitopes by these cell lines.
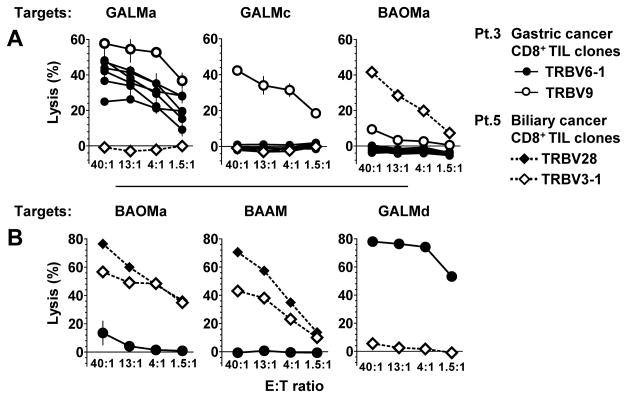
Importantly, all of the CD8+ TIL clones isolated from the Patient 3 and 5 lysed autologous tumors with specificity, irrespective of their ability to specifically secrete IFN-γ (Fig 3). For patient 3, the TRBV6-1 sister clones maintained the same pattern of reactivity noted by 4-1BB up-regulation by not lysing the autologous GALMc line, whereas the TRBV9 clone could lyse both cell line GALMa and c (Fig. 3A). For patient 2 similarly, the 4-1BB up-regulation seen by the TRBV28 and TRBV3-1 clones after stimulation by BAOMa and BAAM was associated with cytolysis of tumor lines from patient 5 but not from patient 3. Thus CD8+ TIL clones with specific lytic capacity toward autologous cancer cells were isolated from a metastatic gastric adenocarcinoma and a cholangiocarcinoma.
Fig. 3. Specific CD8+ TIL clone lysis of autologous gastric and a bile duct cancer cell lines.
Chromium 51-release assays testing cancer cell line lysis by the TRBV6-1 and TRBV9 CD8+ TIL clones from patient 3, and TRBV28 and TRBV3-1 clones from patient 5 in different experiments. (A) Patient 3 TRBV9 CD8+ TIL clone lysed autologous GALMa and GALMc, whereas the TRBV6-1 sister CD8+ clones only lysed autologous GALMa and GALMd (in B). (B) From patient 5, TRBV28 and TRBV3-1 CD8+ TIL clones lysed two autologous cancer cell lines (BAOMa and BAAM). For all clones, reactivity was specific to autologous tumor cell lines.
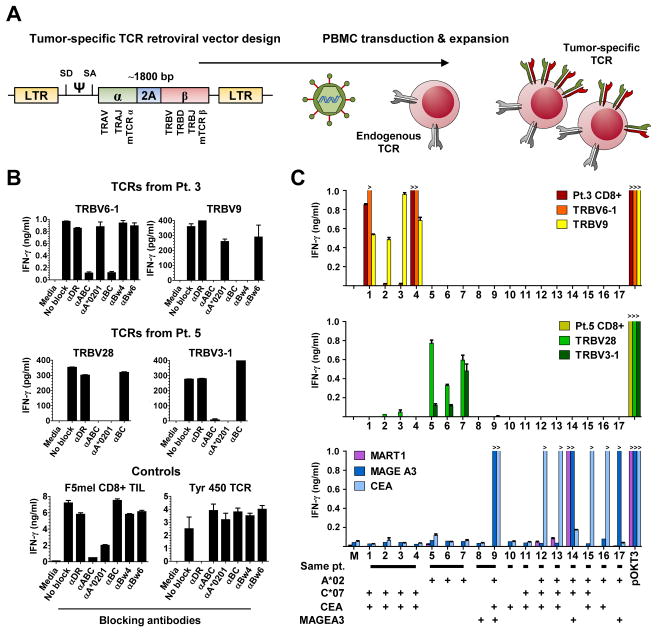
TCR-transduced lymphocytes redirected to recognize gastric and bile duct cancers restricted to the autologous context by common HLA
We next synthesized the genes encoding the TCRs derived from reactive TIL clones isolated from patients 3 and 5, as well as the dominant TCR found in patient 9 (Fig. 4A) and transduced these genes into PBMC. TCR-transduced PBMC from patient 3 and 5 were co-cultured with autologous cell lines in the presence of absence of HLA blocking antibodies to define the HLA restriction elements mediating tumor recognition (Fig. 4B). For patient 3, the TRBV6-1 and TRBV9 TCRs appeared to recognize tumors in the context of the HLA-C*0701 and HLA-B*4901 class I alleles, respectively. For patient 5, TRBV28 and TRBV3-1 TCRs both appeared to recognize autologous tumor cells in the context of HLA-A*0201.
Fig. 4. Retroviral transduction of lymphocytes with four TCRs and assessment of their HLA restriction elements and specificity.
(A) Design of gammaretroviral TCR construct used for transduction of peripheral blood mononuclear cells (PBMC) with four TCRs derived from patient 3 and 5 cytolytic TIL clones. The human constant chains were replaced with mouse constant chains (mTCRα and mTCRβ) to reduce mispairing of transduced alpha and beta chains with endogenous TCR chains. A ribosomal skipping motif (2A) is inserted between the alpha and beta chains for separate transcription and expression of the two TCR chains.
(B) Determination of the restriction elements by which TCR react to autologous cancers (See Table S1 for HLA listing). For TCRs in patient 3, TRBV6-1 TCR recognizes its epitope by HLA-C*0701, whereas TRBV9 is restricted by HLA-B*4901, blocked by Bw4 and not by Bw6, the latter having the potential to block HLA-B*0801. The reactivity of both TCRs in patient 5 are not blocked by the anti-BC antibody, and are restricted by HLA-A*0201. The specificity of the Class I and Class II blocking antibodies was demonstrated in the same experiment by the selective block of known Class I (F5 mel) and Class II (Tyr 450) lymphocyte lines.
(C) Reactivity of native TIL and PBMC transduced with TCRs from patient 3 (upper), TCRs from patient 5 (middle), TCR that recognize MART1, MAGE A3 and CEA presented by HLA-A*020101, to 17 targets (see Table S2 for all target tested): 1. GALMa, 2. GALMb, 3. GALMc, 4. GALMd, 5. BAOMa, 6. BAOMb, 7. BAAM, 8. CALM, 9. CALM transduced to express HLA-A*020101, 10. CAPM, 11. CAAWMa, 12. NCI H508 colon cancer (Ca.), 13. SK-CO-1 colon Ca., 14. 624 melanoma TC, 15. SW1463 rectal Ca, 16. KATO III gastric Ca, 17. HCT15 colon Ca. Patients 3 and 5 TCRs reactivity are restricted to the autologous setting.
To define the specificity of the TCRs derived from the reactive clones and to investigate shared reactivities across cancer cell lines, large co-culture assays were carried out to assess IFN-γ responses against a variety of targets including autologous cancer cell lines, normal autologous or HLA-matched B cells, and a panel of 17 commercially available GI cancer cell lines that were HLA genotyped (Table S2). The reactivity observed from patient 3 bulk CD8+ TIL was recapitulated with the TRBV6-1 TCR-engineered T cells, specifically recognizing the autologous GALMa and d, but not b and c, and T cells transduced with the TRVB9 TCR recognizing all four GALM tumor lines (Fig. 4C, top panel, orange bar for TRBV6-1, yellow bar for TRBV9, red for bulk CD8+). The TRBV6-1 lymphocyte reactivity was entirely specific to the autologous tumor, despite the fact that the restriction element HLA-C*0701 allele was potentially expressed by 13 other allogeneic tumor targets (Table S1). For patient 5, the TRBV28 TCR-transduced PBMC recognized the autologous cell lines established from this patient’s omental nodule (BAOMa and b) and malignant ascites (BAAM) (Fig. 4C, middle, lighter green bar). The reactivity of the TRBV3-1 was weaker but specific to the autologous tumor (Fig. 4C, dark green bar). Again, although 13 cancer cell lines potentially expressed HLA-A*0201, there was no reactivity suggestive of shared antigen recognition by T cells transduced with TRBV28 and TRBV3-1.
In addition, other CD8+ TIL derived from HLA-A*0201 patients or with known HLA-A*0201-restricted epitopes (Pt.9, Pt.10, F5 mel with MART1 reactivity (31), CEA (33) and MAGE-A3 (27) transduced T cells) failed to recognize BAOM and BAAM tumor cell lines. PBMC engineered to expressed the dominant TCR from patient 9 (TRBV7) failed to recognize CALM, even when the cancer cell line was transduced to expressed HLA-A*0201 which expression it had lost. Finally, native CD8+ TIL from patient 3, 5, 9, 10, and 11 failed to recognize allogeneic tumor cell lines.
Altogether, these results showed that it was possible to redirect PBMC specificity toward GI cancer antigens presented by distinct HLA class I alleles, and that the antigens recognized in the autologous setting were likely unique to each patient.
Discussion
Whether common human epithelial malignancies such as GI cancers harbor tumor-reactive T cells has been debated for decades, with arguments mainly relying on associative findings (6) rather than direct demonstration. Suggestive evidence has supported the idea that CD8+ TIL reactive to tumor cell suspensions could be expanded from metastatic GI cancers (34, 35), but the lack of well characterized cancer cell lines precluded assays necessary to define tumor reactivity and specificity at the TCR-HLA molecular level. Ten years ago, a colon cancer cell line established from a liver metastasis was used to identify CD4+ TIL clones reactive to a self-epitope restricted by HLA-DRβ1, but required engineering of the cancer cell line for expression of MHC Class II molecules (36). Here, we establish the presence of naturally occurring CD8+ TIL able to specifically recognize autologous metastatic gastric, bile duct, and colon adenocarcinomas by generating new cancer cell lines and TIL cultures. Three main observations can be made from our findings: First, tumor-reactive CD8+ TIL were found at low frequency. Second, metastatic GI cancer recognition by CD8+ TIL was seen only in the autologous setting, in absence of shared recognition across allogeneic cancer cell lines. Third, cancer cell lines generated from given patients were not equally recognized by T cells, and in other patients were deficient in MHC-I expression.
Unlike melanoma, only anecdotal responses to systemic immunomodulation have been reported in patients with metastatic GI cancers (2–5), implying that these tumors lacked tumor-reactive T cells in sufficient numbers or with the quality necessary to mediate tumor regression. One of the factors that may contribute to the low frequency of tumor-reactive TIL in GI cancer is the relatively low number of exomic mutations those tumors generally carry (14). Whole exome sequencing studies have reported an average of approximately 200 mutations per melanoma compared to approximately 55 in GI cancers, representing less potential opportunities for anti-tumor CD8+ T-cell recognition (37). In silico-based epitope prediction analysis has suggested that 2 to 17 HLA-A*0201 mutated epitopes may be generated per colon cancer (38). Additionally, even if a productive anti-tumor immune response occurred against mutated epitopes, it is possible that over time, a natural selection of the least immunogenic cancer cells occurs, as suggested in animal models (39, 40). Here, our ability to establish new cancer cell lines in a limited number of patients with advanced GI cancers consistently supported a low frequency of tumor-reactive CD8+ TIL, but more samples should be tested to further examine this phenomenon. Higher frequencies of GI tumor-reactive TIL may be found in earlier stages of disease, or in GI cancer subsets, such as those with a high mutation rate due to mismatch repair gene deficiencies.
Our study thus demonstrates that tumor-reactive T cells with proliferative potential may be isolated from advanced GI cancer, provided that suitable tumor targets are available for testing. Thus far, CD137 (4-1BB), a cell surface marker of recent TCR engagement, had been used for isolation of precursor T cells derived from PBMC, using peptides from known tumor or viral antigens as stimulators (32, 41, 42). Without knowing the antigens recognized by polyclonal TIL, the use of 4-1BB expression here allowed to isolate polyclonal cytolytic CD8+ T cells, independent of their capacity to secrete IFN-γ (Fig. 2 and 3). Since the difficulty in generating cancer cell lines has limited the study of the immune recognition of epithelial cancers in vitro, it appears critical that new methods be developed to increase the yield of new cell line establishment, as shown recently by the use of a ROCK inhibitor with stromal cells (43, 44). As clinical-grade flow cytometry cell sorting becomes available for enriching cell products in tumor-reactive T cells (45), it may be possible to design adoptive cell transfer immunotherapy for patients with tumors that harbor a small fraction of tumor-reactive T cells that can be expanded in vitro.
By defining the reactivity of CD8+ T cells against GI cancers at the TCR and the HLA molecular level and by testing allogeneic reactivity against a comprehensive panel of HLA-genotyped GI cancer cell lines, our study highlighted an additional difference with melanoma, which is the absence of shared recognition across allogeneic tumors (Table S2, Fig. 4). TIL in melanoma not only recognize mutated epitopes, but many self-epitopes, such as cancer testis antigens (MAGE, NY-ESO1, etc.) and melanocyte-differentiation antigens (gp100, MART-1, etc.) (10–13). While ongoing studies are aimed at identifying the genes that encode the antigens recognized by GI TIL, conceivably these may consist of mutation-generated epitopes unique to each tumor, or self-antigens overexpressed by the tumor. Mining exomic mutation expression in this context could represent a powerful tool for defining new tumor antigens recognized by T cells (14). Thus for advanced GI cancers, T-cell based immunotherapy may have to rely on high through put screening of unique reactive TIL, especially considering the infrequent expression of cancer-testis antigens (46) by GI cancers and the potential toxicity seen in trial targeting shared self-differentiation antigens such as CEA (47). Further studies should also aim at clarifying whether tumor-reactive CD8+ TIL represents dominant clonotypes in freshly resected tumors, and if current in vitro TIL expansion protocols lead to overgrowth of non-tumor reactive bystander rather than tumor-reactive T cells.
Although the genetic heterogeneity found in a given tumor and across distinct metastases in a given patient is now well established using second generation genomic sequencing (37, 48, 49), our data support that this heterogeneity can translate into the generation of various cell lines with the distinct potential of being recognized by autologous CD8+ TIL. For example, in the case of a gastric cancer metastatic to the liver (patient 3), 2 of 4 cancer cell lines appeared to express the gene encoding an epitope recognized by a TCR (TRBV6-1) that dominated the CD8+ TIL repertoire and was restricted by the HLA-C*0701 (Fig. 4B–C). A second epitope, restricted by HLA-B*4901, was present on the 4 autologous cancer cell lines and recognized by a distinct TCR (TRBV9) expressed by less than 1.3% of the CD8+ bulk TIL expanded from the tumor. Adding to the complexity of in vitro assessment of tumor-recognition by T cells, the loss of HLA expression by cancer cells — a well-known mechanism of tumor immune-escape (9, 50) — was seen in two of the five patients in which new cancer cell lines were established, and in 4 of the 17 commercially available GI cancer cell lines tested. MHC expression on paraffin-embedded GI cancer metastases further suggested frequent deficient expression in vivo (Fig. 1 and Table 1). Implications of these findings for advanced GI cancer immunotherapy are twofold: First, a non-polyclonal immunologic approach that only targets one tumor antigen is unlikely to mediate sustained tumor regression. Second, the antigen presentation capacity of cancer cells should be evaluated as a potential biomarker of response to immunotherapy, while strategies that aim at restoring antigen presentation have to be developed to broaden the use of T-cell based immunotherapy.
In conclusion, this study defines at the clonal and molecular level the existence of naturally-occurring cytolytic CD8+ TIL specifically reactive to autologous metastatic GI cancers. These findings propose avenues for the development of T-cell based immunotherapies for GI cancers by pointing to main challenges to be addressed, such as the low frequency of tumor-reactive TIL, the absence of shared antigen recognition across allogeneic tumors, the difficulty in establishing suitable tumor targets for the selection of reactive TIL, the heterogeneity of antigen expression by tumors, and the loss of MHC-I expression by immune-escape tumor variants.
Supplementary Material
Translational Relevance.
Little is known about T-cell immune response of patients to advanced gastrointestinal (GI) adenocarcinomas. How immunotherapy could be a relevant alternative to current therapeutics for these common malignancies is unclear. Conversely, adoptive transfer of tumor-infiltrating lymphocytes (TIL) can mediate durable complete regression of metastatic melanoma in patients with refractory disease. By overcoming the difficulty of establishing new cancer cell lines and by expanding TIL from the same patients, this study first provides evidence at the molecular level for the presence of cytolytic tumor reactive CD8+ T cells in growing GI cancer metastases. The difficulty in establishing tumor targets to test for T-cell reactivity, the heterogeneity and defects in antigen expression by cancer cell lines, the low frequency of tumor-reactive CD8+ TIL, and the lack of shared reactivity across HLA-matched allogeneic tumors appear as major challenges for the development of effective T-cell based immunotherapy for patients with advanced GI cancer.
Acknowledgments
Funding: This work was supported by the Intramural Research Program of the NIH, NCI, Center for Cancer Research.
The authors are grateful to P. Fetsch and A.C. Filie for cytoimmunohistochemisty staining and interpretation, L.T. Ngo for technical support, K. Hogan for expertise in TIL expansion, Q.J. Wang and Y.F. Li for TCR repertoire assessment and cloning, A. Mixon and S. Farid for cell sorting by flow cytometry, S. Adams for MHC genotyping, and to S.A. Williams for high resolution whole slide scanning, and to N.P. Restifo for thoughtful discussions on T-cell biology and editorial review.
Footnotes
Conflicts of interests: none
Reference List
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Brahmer JR, Drake CG, Wollner I, Powderly JD, Picus J, Sharfman WH, et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J Clin Oncol. 2010;28:3167–75. doi: 10.1200/JCO.2009.26.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung KY, Gore I, Fong L, Venook A, Beck SB, Dorazio P, et al. Phase II study of the anti-cytotoxic T-lymphocyte-associated antigen 4 monoclonal antibody, tremelimumab, in patients with refractory metastatic colorectal cancer. J Clin Oncol. 2010;28:3485–90. doi: 10.1200/JCO.2010.28.3994. [DOI] [PubMed] [Google Scholar]
- 4.Rosenberg SA, Lotze MT, Yang JC, Aebersold PM, Linehan WM, Seipp CA, et al. Experience with the use of high-dose interleukin-2 in the treatment of 652 cancer patients. Ann Surg. 1989;210:474–84. doi: 10.1097/00000658-198910000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Royal RE, Levy C, Turner K, Mathur A, Hughes M, Kammula US, et al. Phase 2 trial of single agent Ipilimumab (anti-CTLA-4) for locally advanced or metastatic pancreatic adenocarcinoma. J Immunother. 2010;33:828–33. doi: 10.1097/CJI.0b013e3181eec14c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 7.Mantovani A, Romero P, Palucka AK, Marincola FM. Tumour immunity: effector response to tumour and role of the microenvironment. Lancet. 2008;371:771–83. doi: 10.1016/S0140-6736(08)60241-X. [DOI] [PubMed] [Google Scholar]
- 8.Baitsch L, Fuertes-Marraco SA, Legat A, Meyer C, Speiser DE. The three main stumbling blocks for anticancer T cells. Trends Immunol. 2012;33:364–72. doi: 10.1016/j.it.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Khong HT, Restifo NP. Natural selection of tumor variants in the generation of “tumor escape” phenotypes. Nat Immunol. 2002;3:999–1005. doi: 10.1038/ni1102-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Bruggen P, Traversari C, Chomez P, Lurquin C, De PE, Van den Eynde B, et al. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254:1643–7. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 11.Kawakami Y, Eliyahu S, Delgado CH, Robbins PF, Rivoltini L, Topalian SL, et al. Cloning of the gene coding for a shared human melanoma antigen recognized by autologous T cells infiltrating into tumor. Proc Natl Acad Sci U S A. 1994;91:3515–9. doi: 10.1073/pnas.91.9.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang RF, Appella E, Kawakami Y, Kang X, Rosenberg SA. Identification of TRP-2 as a human tumor antigen recognized by cytotoxic T lymphocytes. J Exp Med. 1996;184:2207–16. doi: 10.1084/jem.184.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guillaume B, Stroobant V, Bousquet-Dubouch MP, Colau D, Chapiro J, Parmentier N, et al. Analysis of the processing of seven human tumor antigens by intermediate proteasomes. J Immunol. 2012;189:3538–47. doi: 10.4049/jimmunol.1103213. [DOI] [PubMed] [Google Scholar]
- 14.Robbins PF, Lu YC, El-Gamil M, Li YF, Gross C, Gartner J, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19:747–52. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atkins MB, Lotze MT, Dutcher JP, Fisher RI, Weiss G, Margolin K, et al. High-dose recombinant interleukin 2 therapy for patients with metastatic melanoma: analysis of 270 patients treated between 1985 and 1993. J Clin Oncol. 1999;17:2105–16. doi: 10.1200/JCO.1999.17.7.2105. [DOI] [PubMed] [Google Scholar]
- 16.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–23. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–54. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, et al. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369:122–33. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenberg SA, Yang JC, Sherry RM, Kammula US, Hughes MS, Phan GQ, et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin Cancer Res. 2011;17:4550–7. doi: 10.1158/1078-0432.CCR-11-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Besser MJ, Shapira-Frommer R, Treves AJ, Zippel D, Itzhaki O, Hershkovitz L, et al. Clinical responses in a phase II study using adoptive transfer of short-term cultured tumor infiltration lymphocytes in metastatic melanoma patients. Clin Cancer Res. 2010;16:2646–55. doi: 10.1158/1078-0432.CCR-10-0041. [DOI] [PubMed] [Google Scholar]
- 21.Ellebaek E, Iversen TZ, Junker N, Donia M, Engell-Noerregaard L, Met O, et al. Adoptive cell therapy with autologous tumor infiltrating lymphocytes and low-dose Interleukin-2 in metastatic melanoma patients. J Transl Med. 2012;10:169–80. doi: 10.1186/1479-5876-10-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radvanyi LG, Bernatchez C, Zhang M, Fox P, Miller P, Chacon J, et al. Specific lymphocyte subsets predict response to adoptive cell therapy using expanded autologous tumor-infiltrating lymphocytes in metastatic melanoma patients. Clin Cancer Res. 2012;18:6758–70. doi: 10.1158/1078-0432.CCR-12-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pilon-Thomas S, Kuhn L, Ellwanger S, Janssen W, Royster E, Marzban S, et al. Efficacy of adoptive cell transfer of tumor-infiltrating lymphocytes after lymphopenia induction for metastatic melanoma. J Immunother. 2012;35:615–20. doi: 10.1097/CJI.0b013e31826e8f5f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dudley ME, Wunderlich JR, Shelton TE, Even J, Rosenberg SA. Generation of tumor-infiltrating lymphocyte cultures for use in adoptive transfer therapy for melanoma patients. J Immunother. 2003;26:332–42. doi: 10.1097/00002371-200307000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dudley ME, Gross CA, Somerville RP, Hong Y, Schaub NP, Rosati SF, et al. Randomized Selection Design Trial Evaluating CD8+-Enriched Versus Unselected Tumor-Infiltrating Lymphocytes for Adoptive Cell Therapy for Patients With Melanoma. J Clin Oncol. 2013 doi: 10.1200/JCO.2012.46.6441. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Topalian SL, Solomon D, Rosenberg SA. Tumor-specific cytolysis by lymphocytes infiltrating human melanomas. J Immunol. 1989;142:3714–25. [PubMed] [Google Scholar]
- 27.Chinnasamy N, Wargo JA, Yu Z, Rao M, Frankel TL, Riley JP, et al. A TCR Targeting the HLA-A*0201-Restricted Epitope of MAGE-A3 Recognizes Multiple Epitopes of the MAGE-A Antigen Superfamily in Several Types of Cancer. J Immunol. 2011;186:685–96. doi: 10.4049/jimmunol.1001775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang QJ, Hanada K, Robbins PF, Li YF, Yang JC. Distinctive features of the differentiated phenotype and infiltration of tumor-reactive lymphocytes in clear cell renal cell carcinoma. Cancer Res. 2012;72:6119–29. doi: 10.1158/0008-5472.CAN-12-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cohen CJ, Zhao Y, Zheng Z, Rosenberg SA, Morgan RA. Enhanced antitumor activity of murine-human hybrid T-cell receptor (TCR) in human lymphocytes is associated with improved pairing and TCR/CD3 stability. Cancer Res. 2006;66:8878–86. doi: 10.1158/0008-5472.CAN-06-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson LA, Heemskerk B, Powell DJ, Jr, Cohen CJ, Morgan RA, Dudley ME, et al. Gene transfer of tumor-reactive TCR confers both high avidity and tumor reactivity to nonreactive peripheral blood mononuclear cells and tumor-infiltrating lymphocytes. J Immunol. 2006;177:6548–59. doi: 10.4049/jimmunol.177.9.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolfl M, Kuball J, Ho WY, Nguyen H, Manley TJ, Bleakley M, et al. Activation-induced expression of CD137 permits detection, isolation, and expansion of the full repertoire of CD8+ T cells responding to antigen without requiring knowledge of epitope specificities. Blood. 2007;110:201–10. doi: 10.1182/blood-2006-11-056168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parkhurst MR, Joo J, Riley JP, Yu Z, Li Y, Robbins PF, et al. Characterization of genetically modified T-cell receptors that recognize the CEA:691–699 peptide in the context of HLA-A2. 1 on human colorectal cancer cells. Clin Cancer Res. 2009;15:169–80. doi: 10.1158/1078-0432.CCR-08-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hom SS, Rosenberg SA, Topalian SL. Specific immune recognition of autologous tumor by lymphocytes infiltrating colon carcinomas: analysis by cytokine secretion. Cancer Immunol Immunother. 1993;36:1–8. doi: 10.1007/BF01789124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turcotte S, Gros A, Hogan K, Tran E, Hinrichs CS, Wunderlich JR, et al. Phenotype and function of T cells infiltrating visceral metastases from gastrointestinal cancers and melanoma: implications for adoptive cell transfer therapy. J Immunol. 2013;191:2217–25. doi: 10.4049/jimmunol.1300538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maccalli C, Li YF, El-Gamil M, Rosenberg SA, Robbins PF. Identification of a colorectal tumor-associated antigen (COA-1) recognized by CD4(+) T lymphocytes. Cancer Res. 2003;63:6735–43. [PMC free article] [PubMed] [Google Scholar]
- 37.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segal NH, Parsons DW, Peggs KS, Velculescu V, Kinzler KW, Vogelstein B, et al. Epitope landscape in breast and colorectal cancer. Cancer Res. 2008;68:889–92. doi: 10.1158/0008-5472.CAN-07-3095. [DOI] [PubMed] [Google Scholar]
- 39.DuPage M, Mazumdar C, Schmidt LM, Cheung AF, Jacks T. Expression of tumour-specific antigens underlies cancer immunoediting. Nature. 2012;482:405–9. doi: 10.1038/nature10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–4. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watanabe K, Suzuki S, Kamei M, Toji S, Kawase T, Takahashi T, et al. CD137-guided isolation and expansion of antigen-specific CD8 cells for potential use in adoptive immunotherapy. Int J Hematol. 2008;88:311–20. doi: 10.1007/s12185-008-0134-z. [DOI] [PubMed] [Google Scholar]
- 42.Han S, Huang Y, Liang Y, Ho Y, Wang Y, Chang LJ. Phenotype and functional evaluation of ex vivo generated antigen-specific immune effector cells with potential for therapeutic applications. J Hematol Oncol. 2009;2:34. doi: 10.1186/1756-8722-2-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B, et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am J Pathol. 2012;180:599–607. doi: 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan H, Myers S, Wang J, Zhou D, Woo JA, Kallakury B, et al. Use of reprogrammed cells to identify therapy for respiratory papillomatosis. N Engl J Med. 2012;367:1220–7. doi: 10.1056/NEJMoa1203055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaye DL, Bray RA, Gebel HM, Harris WA, Waller EK. Translational applications of flow cytometry in clinical practice. J Immunol. 2012;188:4715–9. doi: 10.4049/jimmunol.1290017. [DOI] [PubMed] [Google Scholar]
- 46.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–25. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 47.Parkhurst MR, Yang JC, Langan RC, Dudley ME, Nathan DA, Feldman SA, et al. T Cells Targeting Carcinoembryonic Antigen Can Mediate Regression of Metastatic Colorectal Cancer but Induce Severe Transient Colitis. Mol Ther. 2010 doi: 10.1038/mt.2010.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–13. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aptsiauri N, Cabrera T, Garcia-Lora A, Lopez-Nevot MA, Ruiz-Cabello F, Garrido F. MHC class I antigens and immune surveillance in transformed cells. Int Rev Cytol. 2007;256:139–89. doi: 10.1016/S0074-7696(07)56005-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.