Abstract
We performed a meta-analysis of 2 genome-wide association studies of coronary artery disease comprising 1,515 cases with coronary artery disease and 5,019 controls, followed by de novo replication studies in 15,460 cases and 11,472 controls, all of Chinese Han descent. We successfully identified four new loci for coronary artery disease reaching genome-wide significance (P < 5 × 10−8), which mapped in or near TTC32-WDR35, GUCY1A3, C6orf10-BTNL2 and ATP2B1. We also replicated four loci previously identified in European populations (PHACTR1, TCF21, CDKN2A/B and C12orf51). These findings provide new insights into biological pathways for the susceptibility of coronary artery disease in Chinese Han population.
Coronary artery disease (CAD), and its most severe complication myocardial infarction (MI), are leading causes of death and disability worldwide1,2. Recent genome-wide association studies (GWAS) of CAD have discovered multiple chromosomal regions3. However, most of the GWAS have focused on samples of European origin, and the identified loci altogether explain only a small fraction of the risk for CAD. Moreover, the variants identified in populations of European descent might not be applicable in other ethnic groups because of underlying genetic heterogeneity. Therefore, larger scale studies in Chinese and other non-European populations are needed to reveal new susceptibility loci and improve our understanding of causal pathways to CAD.
Herein, we carried out a two-stage GWAS study of CAD in a sample of ~33,000 Han Chinese (Supplementary Fig. 1). In the discovery stage (stage 1), we performed two GWAS studies of CAD in two independent Chinese samples, the Beijing Atherosclerosis Study (BAS) and the China Atherosclerosis Study (CAS), respectively. BAS consisted of 509 cases of MI and 1,034 controls genotyped using the Affymetrix Human Mapping 500K Array Set including 500,568 SNPs. CAS consisted of 1,034 cases of CAD and 4,245 controls genotyped using the Affymetrix Axiom™ Genome-Wide CHB 1 Array Plate including 657,124 SNPs. After a series of quality-control procedures (Online Methods), we retained 367,129 autosomal SNPs in 505 cases and 1,021 controls in BAS study, and 613,724 autosomal SNPs in 1,010 cases and 3,998 controls in CAS (Supplementary Table 1). To facilitate combining results of genome-wide association scans based on the two genotyping platforms, we imputed missing genotypes based on reference haplotypes from the phased CHB+JPT HapMap data release 22 reference dataset4. We performed a meta-analysis of the two GWAS studies with association data for approximately 2.2 million genotyped or imputed autosomal SNPs.
Principal component analysis showed minimal evidence for population stratification in these study populations (Supplementary Fig. 2). Quantile-quantile plot showed that the distribution of observed P values deviated from expected P values only in the extreme tail (Supplementary Fig. 3). The value of genomic inflation factors were 1.04 and 1.04 for BAS and CAS, respectively, and 1.05 for the two studies combined, which indicated population stratification effects were negligible in our study samples.
In this stage-1 discovery analysis, we used a set of criteria to select 96 promising SNPs (Online Methods) and genotyped them in a further replication analysis in a case-control sample including 8,803 CAD cases and 5,183 controls (replication 1) (Supplementary Table 1). We found that two association signals reached genome-wide significance (defined as P < 5 × 10−8) in the replication 1 alone. These included one locus (4 SNPs on 9p21.3) previously identified in populations of European descent and one newly discovered locus (rs9268402 at C6orf10-BTNL2). After meta-analysis of the discovery and replication 1 study, eight additional variants were associated with CAD at a significance level of P < 1 × 10−5. These eight SNPs were further examined in an independent sample (2,408 cases and 2,103 controls) (replication 2). The results of the selected SNPs in the discovery, the replication 1, and the replication 2 were summarized in Supplementary Table 2.
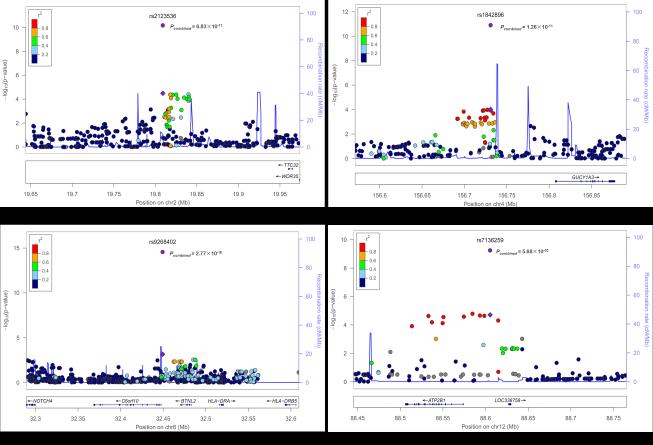
When combining the discovery data with those from replications 1 and 2, we found ten SNPs at seven regions associated with CAD at a pre-specified threshold for genome-wide significance of P < 5 × 10−8. Among these SNPs, the four (rs9632884, rs10757274, rs1333042, rs1333049) on 9p21.3, rs9349379 on 6p24.1 in PHACTR1, and rs11066280 on 12q24.13 near C12orf51 confirmed associations previously reported in Europeans5-11. In addition, rs12524865 on 6q23.2 near TCF21 also showed consistent association evidence in the discovery and replication stages with the same direction of association previously reported in Europeans12, which nearly reached the genome-wide significance threshold in the combined analyses (P = 1.87 × 10−7) (Supplementary Tables 2 and 3). Moreover, we identified four new CAD loci in Chinese (P value ranging from 4.48 × 10−8 to 9.92×10−14) : (i) rs2123536 on 2p24.1 near TTC32-WDR35; (ii) rs1842896 on 4q32.1 near GUCY1A3; (iii) rs9268402 on 6p21.32 near C6orf10-BTNL2; (iv) rs7136259 on 12q21.33 near ATP2B1.
To minimize the chance of false signal for the four new loci with CAD, we performed an additional validation in an independent sample consisting of 4,249 cases and 4,186 controls (replication 3). All four SNPs showed significant association with CAD in this additional sample after adjustment for multiple testing (P < 0.05/4 = 0.0125), and integration of all results for the discovery, replication 1, replication 2, and replication 3 showed unequivocal associations of these loci with CAD (rs2123536, P = 6.83 × 10−11, OR = 1.12; rs1842896, P =1.26 × 10−11, OR =1.14 ; rs9268402, P = 2.77 × 10−15, OR = 1.16 ; rs7136259, P = 5.68 × 10−10, OR = 1.11 ) (Table 1). Figure 1 displays these loci in their genomic coordinate context.
Table 1.
Association results for four CAD susceptibility loci newly identified in the Chinese GWAS
| Discovery meta-analysis
(BAS+CAS) |
Replication1 | Replication2 | Replication3 | Combined Results
(Discovery + Replication) |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||||||||||||
| SNP | Locus | Position | Risk Allele (Freq) |
Nearby gene(s) |
OR (95%CI) |
P | Sample size (case/control) |
OR (95%CI) |
P | Samplesize (case/control) |
OR (95%CI) |
P | Samplesize (case/control) |
OR (95%CI) |
p | Samplesize (case/control) |
OR (95%CI) |
p | Samplesize (case/control) |
| rs2123536 | 2p24.1 | 19809058 | T(0.39) |
TTC32
-WD R35 |
1.2 5 (1.12,1.39) |
4.92×10-5 | 1008/3982 | 1.09 (1.03,1.15) |
1.56×10-3 | 8732/5175 | 1.16 (1.06,1.26) |
9.56×10-4 | 2373/1958 | 1.11 (1.05,1.18) |
8.82×10-4 | 4103/4177 | 1.12 (1.08,1.16) |
6.83×10-11 | 16216/15292 |
| rs1842896 | 4q32.1 | 156730909 | T(0.76) | GUCY1A3 | 1.23 (1.11,1.37) |
9.75×10-5 | 1503/4991 | 1.13 (1.06,1.20) |
5.79×10-5 | 8644/5158 | 1.17 (1.05,1.29) |
2.97×10-3 | 2324/1961 | 1.11 (1.04,1.20) |
3.52×10-3 | 4229/4169 | 1.14 (1.10,1.19) |
1.26×10-11 | 16700/16279 |
| rs9268402 | 6p21.32 | 32449331 | G(0.59) |
C6orf10
-BTNL2 |
1.17 (1.07,1.27) |
6.97×10-4 | 1510/5009 | 1.19 (1.13,1.26) |
332×10-11 | 8708/5120 | 1.11 (1.04,1.18) |
1.52×10-3 | 4208/4071 | 1.16 (1.12,1.20) |
2.77×10-15 | 14426/14200 | |||
| rs7136259 | 12q21.33 | 88605319 | T(0.39) | ATP2B1 | 1.21 (1.11,1.33) |
1.95×10-5 | 1502/4993 | 1.10 (1.04,1.16) |
4.73×104 | 8732/5173 | 1.09 (1.00,1.18) |
5.27 × 10-2 | 2394/2058 | 1.10 (1.03,1.17) |
326× 10-3 | 4218/4126 | 1.11 (1.08,1.15) |
5.68×10-10 | 16846/16350 |
Information for SNP ID and chromosomal position is based on NCBI genome build 36.
The risk allele frequency (Freq) in discovery studies is displayed. OR, odds ratio; CI, confidence interval
For each SNP, the odds ratios (with 95% confidence intervals) and P values represent the risk of CAD associated with each copy of the risk allele.
Figure 1.
Regional association plots of four new loci. (a-d) In the top panel of each, SNPs are plotted according to their chromosomal positions (NCBI build36) with their P values (as −log10 P values) of the discovery meta-analysis. The lead SNP is represented by a purple diamond with the discovery meta-analysis result, and represented by a purple circle with the combined analysis result denoted by Pcombined value. The r2 values of LD between the lead SNP and the other SNPs are indicated by different colors. The estimated recombination rates from 1000 Genomes June 2010 CHB+JPT samples are plotted in cyan to reflect the local LD structure. In the bottom panels the genes within the region of interest are annotated and are shown as arrows. Plots were generated using LocusZoom (see URLs).
Of the 4 new CAD loci, 4q32.1 (rs13139571) and 12q21.33 (rs2681492) were recently identified as risk loci for blood pressure in Europeans studies13,14. We observed that rs13139571 and rs1842896 on 4q32.1 show a very weak linkage disequilibrium (LD) (r2 = 0.002, D´ = 0.062 in HapMap CHB; r2 = 0.004, D´ = 0.123 in HapMap CEU), while rs2681492 and rs7136259 on 12q21.33 are in strong LD (r2 = 0.90, D´ = 0.95 in HapMap CHB; r2 = 0.11, D´ = 0.88 in HapMap CEU). To shed some light on the seemingly entangled relationships of the two loci with CAD and hypertension, we examined the associations of the two CAD SNPs (rs1842896 and rs7136259) with blood pressure in all control samples from the discovery and replication samples. Suggestive associations with blood pressure and hypertension were observed (Supplementary Table 4). We further examined whether hypertension could mediate the effect on CAD. After adjustment for hypertension, the associations with CAD remained genome-wide significant (P = 1.31 × 10−9, OR = 1.13 for rs1842896; P = 6.63 ×10−12, OR = 1.13 for rs7136259), indicating the two CAD SNPs might be susceptibility markers independent of hypertension. rs1842896 is located to 76.4kb upstream of the GUCY1A3 locus. The GUCY1A3 gene encodes the α subunit of soluble guanylate cyclase (sGC), a key enzyme of nitric oxide signaling pathway implicated in the pathogenesis CAD and atherosclerosis. Preclinical studies have explored the therapeutic potential of sGC stimulators15. rs7136259 is near ATP2B1, which encodes PMCA1, a plasma membrane calcium ATPase which pumps calcium (Ca2+) ions out of the cytosol into the extracellular milieu16.
The C6orf10-BTNL2 on 6p21.32 is a hotspot associated with immune-related diseases17-19. BTNL2 is a member of the immunoglobulin superfamily that probably functions as a T cell costimulatory molecule. It is noteworthy that rs2076530, a truncating splice site mutation in BTNL2 gene17, is in strong LD with rs9268402 (r2 = 0.59). BTNL2 gene polymorphisms have been found associated with susceptibility to Kawasaki disease (KD)20, a vasculitis of young childhood that particularly affects the coronary arteries, with increased risk of developing ischemic heart disease in the future21. rs2123536 on 2p24.1 is located to ~150kb downstream of TTC32 and WDR35. TTC32 encodes the protein containing the tetratricopeptide repeat motif to bind other peptides22. WDR35 encodes a member of the WD repeat protein family23, which involves cell cycle progression, signal transduction, apoptosis, and gene regulation.
The chromosome 12q24 region is of particular interest. The association signal on 12q24 spans ~0.7 Mb and rs11066280 is in almost perfect LD (r2 = 0.95~0.97) with rs3782886, rs4646776, rs671, rs2074356, and rs77768175 (Supplementary Fig. 4). Previous studies showed significant evidence supporting signatures of natural selection11,24 and pleiotropic effects (CAD10,11,25, plasma lipids24,26,27 and blood pressure14,24,28). All variants on 12q24 associated with CAD10,11 in Europeans are not polymorphic in Chinese, whereas all CAD-associated variants on 12q24 in Chinese are monomorphic in Europeans (Supplementary Table 5). In the present study, rs11066280 also showed significant or suggestive evidence of association with high-density lipoprotein cholesterol (HDL-C), triglycerides (TG), total cholesterol (TC), and blood pressure (Supplementary Table 4). Because there is no substantial evidence for functional variants in this locus, further in-depth analysis is to explain the long-range LD and uncover causal mechanisms for CAD in this region.
While the association between the CDKN2A/B on 9p21 locus and CAD was replicated in our study, we note that the LD structures of the 9p21 region are different between populations of European and Asian descent (Supplementary Fig. 5). The 4 SNPs showing significant CAD association in 9p21.3 region were in almost perfect LD in European descent (pairwise r2 from 0.84 to 0.90). In Chinese, however, two of the SNPs (rs10757274 and rs1333049) are in strong LD with each other (r2 = 0.78), but only in moderate LD with the other two SNPs (rs9632884, rs1333042) (pairwise r2 from 0.27 to 0.43, Supplementary Table 6). Conditional logistic analyses showed the association evidence for rs1333042 or rs10757274 remained significant after control for the genetic effect at any of the other three SNPs (P value ranging from 3.71×10−8 to 7.96×10−9, and from 1.44×10−6 to 1.18×10−18, respectively) (Supplementary Table 7). Therefore, the two SNPs appear to represent independent signals.
We evaluated whether the CAD variants identified in Chinese were associated with CAD in Europeans using the results of the CARDIoGRAM consortium12, a meta-analysis for 22,233 cases and 64,762 controls (Supplementary Table 8). Of the four SNPs, rs2123536 (P = 0.0038, OR = 1.10) and rs7136259 (P = 0.035, OR = 1.03) showed nominal association with CAD in the population of European ancestry, and the direction of the effect was consistent with our findings. The associations for the other two SNPs were not detected in CARDIoGRAM consortium.
Conversely, we also investigated whether the 35 CAD-associated SNPs (in 29 loci) identified by previous GWAS in European populations were associated with CAD in our sample (Supplementary Table 9). In addition to the 4 loci that were confirmed by our discovery and replication studies (Supplementary Table 3), 7 loci within 1p32.2, 1q41, 10q23.31, 10q24.32, 11q22.3, 15q25.1 and 17p13.3 showed directionally consistent and nominally significant associations in the discovery study (P < 0.05) (Supplementary Table 9, Group 1). We observed 11 SNPs in 10 loci were monomorphic or had low minor allele frequencies (MAF ≤ 0.1) in Chinese Han population, which were quite different in European populations (Supplementary Table 9, Group 2). We examined the associations of other correlated SNPs in strong LD with these 11 SNPs in HapMap CEU data with CAD in Chinese. Of interest, the associations of the proxy SNPs in 3 loci, 3q22.3, 6q26 and 17p11.2, were also supported by our discovery analysis albeit with only suggestive evidence (P < 0.05). These data suggest that the difference in the LD structure may partially explain the discrepancy of the association between the European and Chinese populations. No associations were observed for the remaining 8 SNPs with common minor allele frequencies (MAF > 0.1) in the Chinese Han population (Supplementary Table 9, Group 3). The disparities between Chinese and European populations might be due to differences in genetic architecture and environmental factors, or insufficient power in the present study. In addition, a recent GWAS on CAD in the Chinese Han population29 identified rs6903956 on 6p24.1 (C6orf105) as a susceptibility locus. We could not replicate the association of rs6903956 in our data, though our present discovery analyses have >90% power to detect a SNP with an OR = 1.51 even using a P value threshold of 1.0 × 10−5.
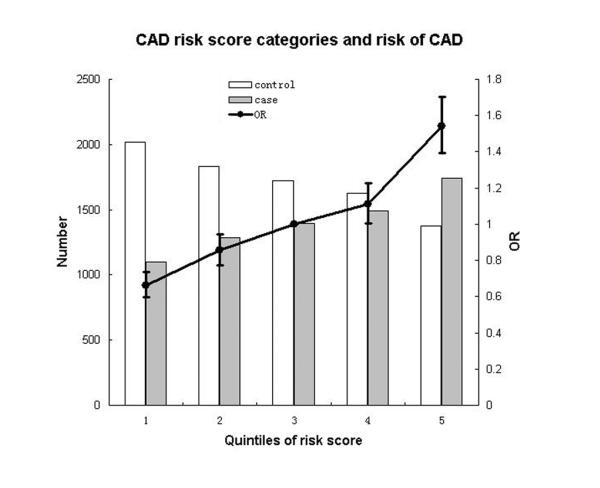
To examine the effect of nine SNPs in aggregate on the risk of CAD, a CAD risk score was calculated by using weighted sum across the SNPs combining effect size and doses of risk alleles. The CAD risk score can explain ~1.92% of the variance in risk for CAD. The mean CAD risk score of cases was significantly higher than that of controls (P < 1 × 10−74). Logistic regression model was applied to test the association of risk score categories with CAD. Compared with bottom quintile, individuals in the top quintile of CAD risk score had greater than twofold increased risk for CAD (OR=2.34, 95%CI: 2.11-2.59). Risk of CAD across quintiles of CAD risk score is shown in Figure 2.
Figure 2.
CAD risk score categories and risk of CAD. This figure shows odds ratios for CAD compared with quintile 3 as solid dot with whiskers extending to OR±95%CI. The white and gray bars represent number of controls and cases in each quintile. This analysis was performed in 15,591 individuals (7,012 cases and 8,579 controls) with all phenotype information for nine SNPs in Table 1 and Supplementary Table 3, including rs2123536, rs1842896, rs9349379, rs9268402, rs12524865, rs10757274, rs1333042, rs7136259, and rs11066280.
In conclusion, we identified four new loci for CAD (TTC32-WDR35, GUCY1A3, C6orf10-BTNL2 and ATP2B1) in Chinese and confirmed four previously reported loci (PHACTR1, TCF21, CDKN2A/B and C12orf51). These results suggest that both shared and unique genetic backgrounds of CAD are present between different ethnic groups and highlight the importance of fine-mapping efforts to pinpoint causal variants and mechanisms. Further study and integration of multiethnic GWAS findings will surely promote a fuller and better understanding of the global genetic architecture of CAD.
ONLINE METHODS
Study populations
We performed a two-stage case-control analysis in participants of Chinese Han ancestry. The general characteristics of the study participants are summarized in Supplementary Table 1. In the discovery stage, we performed a meta-analysis of two independent GWAS studies, BAS and CAS study. BAS study30 consisted of 505 cases of MI and 1,021 controls. All participants were from Beijing, China. All cases had a validated history of MI and were verified by hospital records and by cardiologists according to standard protocol31. Controls were randomly selected from subjects participating in a community-based survey of cardiovascular risk factors in Beijing. The control subjects were judged to be free of CAD by history, clinical examination, electrocardiography, and Rose questionnaire32. Detailed data were collected through in-person interviews with each case and control. Subjects with congenital heart disease, cardiomyopathy, valvular disease, and renal or hepatic disease were excluded. CAS consisted of 1,010 cases of CAD and 3,998 controls. 1,010 cases from the Northern provinces in China were enrolled from Fuwai Hospital, National Center of Cardiovascular Diseases. 83.8% of cases have a family history of CAD. Diagnoses of MI cases follow strict diagnostic rules based on signs, symptoms, electrocardiograms and cardiac enzymes31. The CAD subjects without known history of MI had > 70% stenosis in at least one of a major epicardial vessel with the exception of the left main coronary artery where a >50% stenosis was sufficient to meet the diagnosis of CAD. Controls of CAS study were recruited from the International Collaborative Study of Cardiovascular Disease in Asia (InterASIA in China)33. InterASIA used a four-stage stratified sampling method to select a nationally representative sample of the general population aged 35 to 74 years in China. A total of 15,838 persons completed the survey and examination in 2001, and a follow-up of this survey in 2008 was conducted. 3,998 controls were individuals who did not develop incident CAD and had no family history of CAD during the 8 yr follow-up period of the study from four northern field centers of InterASIA.
In the stage 2, replication analyses were conducted in three independent samples with a total of 15,460 cases and 11,472 controls (8,803 cases and 5,183 controls for replication 1; 2,408 cases and 2,103 controls for replication 2; 4,249 cases and 4,186 controls for replication 3). All the cases in the replication stage were recruited using uniform criteria, and the clinical information were collected using the same questionnaire as that used in CAS study. For replication 1, CAD cases were recruited using standardized protocol through collaboration among multiple hospitals in China; and controls were selected from samples of the China Collaborative Study of Cardiovascular Epidemiology. For replication 2, cases were enrolled from Fuwai Hospital, Beijing, and controls were selected from urban and suburban communities in Beijing. For replication 3, cases from the northern China were enrolled from Fuwai Hospital, Beijing and other medical centers, and controls were selected from northern field centers of China Cardiovascular Health Study (CCHS) project. CCHS has been a population-based investigation of risk factors for cardiovascular diseases in China since 2006. The controls for the stage 2 validation were selected using the identical criteria as for the discovery populations.
Each study obtained approval from institutional review boards of Fuwai Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, and other medical institutions, and all participants gave written informed consents.
Genotyping and quality control
For BAS in the discovery stage, a total of 509 cases of MI and 1,034 controls were genotyped with the Affymetrix GeneChip® Human Mapping 500K Array Set, including 500,568 SNPs. Principal component analysis using EIGENSOFT34,35 was used to compare all samples with reference samples from the HapMap YRI, CHB, JPT and CEU panels. We excluded SNPs with minor allele frequency (MAF) < 0.01 in cases or controls (n = 100,865, including 46,048 monomorphic SNPs); genotype call rates below 95% in cases or controls (n = 20,030); or deviations from Hardy-Weinberg equilibrium (P value < 10−4, n = 12,544). We also excluded 17 samples because of gender discordance, high genotype missing rate (>3.0%), cryptic relatedness (IBD > 0.1875) or population outliers. After quality control, 1,526 samples and 367,129 autosomal SNPs remained for the subsequent analyses. For CAS in the discovery stage, a total of 1,034 cases of CAD and 4,245 controls were genotyped with the Axiom™ Genome-Wide CHB 1 Array Plate, which was designed for Chinese population and includes 657,124 SNPs. After these quality control procedures, 5,008 samples and 613,724 autosomal SNPs were retained for subsequent analyses.
In stage 2, 96 SNPs were selected and genotyped using TaqMan SNP Genotyping Assays in Fludigm EP1 platform in replication 1 sample. Of the 96 SNPs genotyped, 5 SNPs with Hardy-Weinberg equilibrium P-value <0.001 were removed from further association analysis. To assess genotyping reproducibility, 48 duplicate samples were genotyped, and concordance rate is over 99.4%. 9 non-9p21 loci SNPs with P < 1 × 10−5 in the combined discovery and replication 1 analysis were selected and genotyped using the iPLEX MassARRAY platform (Sequenom) in replication 2 sample, but rs9268402 was not taken forward into replication 2 because of difficulty in the design of the replication array, leaving 8 SNPs for replication. The concordance rate for 96 replicate samples was 99.7%. To evaluate the quality of the genotype data between different genotyping platform, 48 random replication samples genotyped by Fludigm EP1 platform were re-genotyped for 8 SNPs on the iPLEX Sequenom MassARRAY platforms, and the concordance rate between the genotypes from the two platforms was 99.5%. The replication 3 sample for 4 new SNPs was genotyped using a TaqMan genotyping platform (ABI 7900HT Real Time PCR system, Applied Biosystems). The cluster patterns of the genotyping data from the Fludigm EP1, Sequenom and TaqMan analyses were checked to confirm their good quality.
Genotype imputation
In the discovery stage, imputation of allele dosage of ungenotyped SNPs using the HapMap Phase 2 (JPT+CHB) data was carried out using MACH36,37. After excluding imputed SNPs with imputation quality scores below a threshold (R2 < 0.30), call rate < 0.90 in either cases or controls, MAF < 0.01 in either cases or controls, Hardy-Weinberg equilibrium P < 1 × 10−5 in controls, and significantly different missing genotype rates between cases and controls (P < 1 × 10−5), a total of 1,532,051 genotyped and imputed autosomal SNPs from the BAS, 2,042,781 from the CAS, and 2,228,999 SNPs from the combined two GWAS samples were retained for subsequent association analysis.
Selection of replication SNPs
After genome-wide association analyses for each of the two discovery studies and meta-analysis in the combined sample, SNPs fulfilling the following criteria were taken forward to replication: (i) SNPs showed potential associations from meta-analysis of the two study (P < 1.0 × 10−4); (ii) SNPs had a consistent association at P ≤ 1.0 × 10−2 in both discovery populations; (iii) SNPs with P < 1.0 × 10−3 in meta-analysis discovery within ±500 kb of the locus previously reported at genome-wide significance or suggestive significance in a prior publication; (iv) the SNP had nearby correlated SNPs (within 25 kb) that also showed a signal (P < 0.01). With the exception of four SNPs at the 9p21.3 locus, SNPs in strong LD (r2 > 0.5) with the most significant SNP at each locus were removed. When a SNP could not be genotyped, alternative tagging SNPs (r2 > 0.8) were considered.
Association analysis
The association of imputed and genotyped SNPs with CAD was tested with multiple logistic regression analysis in an additive genetic model (with 1 degree of freedom) after adjusting for age (onset of the first event for cases or time of recruitment for controls) and sex. We used allele dosages from the imputation to account for uncertainty of imputed genotypes. Association analyses were performed using PLINK38 (see URLs). A fixed-effects inverse variance-weighted meta-analysis as implemented by METAL39 (see URLs) was used to combine the two discovery studies and the results for each SNP across all replication studies. A quantile-quantile plot generated using R was used to evaluate the overall significance of the GWAS results and the potential impact of population stratification. The genomic inflation factor (λ)40 was estimated from the median of the χ2 statistic divided by 0.456.
The association of the loci with established cardiovascular risk factors was examined in all control samples from the discovery and replication samples. For quantitative traits (high-density lipoprotein, low-density lipoprotein, total cholesterol, triglyceride, blood pressure, fasting plasma glucose, body mass index), linear regressions were used whereas for binary trait (hypertension) logistic regression model was applied. We then combined the respective regression estimates from each stage in a meta-analysis using inverse variance weighting.
Conditional analyses were performed to test the independence of significant SNPs in each region conditioning on the genotype of the SNP chosen for replication. These analyses were carried out using PLINK with the --logistic and --condition options.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by the National Basic Research Program of China (973 Plan) (2011CB503901,2006CB503805) from the Ministry of Science and Technology of China, the National Science Foundation of China (30930047), the High-Tech Research and Development Program of China (863 Plan) (2009AA022703, 2012AA02A516, 2006AA02A406), and a grand (2006BAI01A01) from the Ministry of Science and Technology of China. This study was also supported by Biomedical Project from the Council of Science and Technology, Beijing (H020220030130).
Footnotes
URLs
PLINK v1.07, http://pngu.mgh.harvard.edu/~purcell/plink/;
METAL, http://www.sph.umich.edu/csg/abecasis/metal/;
The International HapMap Project, http://hapmap.ncbi.nlm.nih.gov/;
LocusZoom, http://csg.sph.umich.edu/locuszoom/.
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
Reference
- 1.Roger VL, et al. Heart disease and stroke statistics--2011 update: a report from the American Heart Association. Circulation. 2011;123:e18–e209. doi: 10.1161/CIR.0b013e3182009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He J, et al. Major causes of death among men and women in China. N. Engl. J. Med. 2005;353:1124–1134. doi: 10.1056/NEJMsa050467. [DOI] [PubMed] [Google Scholar]
- 3.Peden JF, Farrall M. Thirty-five common variants for coronary artery disease: the fruits of much collaborative labour. Hum. Mol. Genet. 2011;20:R198–205. doi: 10.1093/hmg/ddr384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 5.Kathiresan S, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McPherson R, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samani NJ, et al. Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helgadottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 10.Gudbjartsson DF, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat. Genet. 2009;41:342–347. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 11.Soranzo N, et al. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat. Genet. 2009;41:1182–1190. doi: 10.1038/ng.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schunkert H, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehret GB, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy D, et al. Genome-wide association study of blood pressure and hypertension. Nat. Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stasch JP. Soluble Guanylate Cyclase as an Emerging Therapeutic Target in Cardiopulmonary Disease. Circulation. 2011;123:2263–2273. doi: 10.1161/CIRCULATIONAHA.110.981738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tempel BL, Shilling DJ. The plasma membrane calcium ATPase and disease. Subcell. Biochem. 2007;45:365–383. doi: 10.1007/978-1-4020-6191-2_13. [DOI] [PubMed] [Google Scholar]
- 17.Valentonyte R, et al. Sarcoidosis is associated with a truncating splice site mutation in BTNL2. Nat. Genet. 2005;37:357–364. doi: 10.1038/ng1519. [DOI] [PubMed] [Google Scholar]
- 18.Clancy RM, et al. Identification of candidate loci at 6p21 and 21q22 in a genome-wide association study of cardiac manifestations of neonatal lupus. Arthritis. Rheum. 2010;62:3415–3424. doi: 10.1002/art.27658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin Y, et al. Genome-wide analysis identifies a quantitative trait locus in the MHC class II region associated with generalized vitiligo age of onset. J. Invest. Dermatol. 2011;131:1308–1312. doi: 10.1038/jid.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsueh KC, Lin YJ, Chang JS, Wan L, Tsai FJ. BTNL2 gene polymorphisms may be associated with susceptibility to Kawasaki disease and formation of coronary artery lesions in Taiwanese children. Eur. J. Pediatr. 2010;169:713–719. doi: 10.1007/s00431-009-1099-5. [DOI] [PubMed] [Google Scholar]
- 21.Kato H, et al. Long-term consequences of Kawasaki disease. A 10- to 21-year follow-up study of 594 patients. Circulation. 1996;94:1379–1385. doi: 10.1161/01.cir.94.6.1379. [DOI] [PubMed] [Google Scholar]
- 22.Smith DF. Tetratricopeptide repeat cochaperones in steroid receptor complexes. Cell. Stress. Chaperones. 2004;9:109–121. doi: 10.1379/CSC-31.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakajima D, et al. Construction of expression-ready cDNA clones for KIAA genes: manual curation of 330 KIAA cDNA clones. DNA. Res. 2002;9:99–106. doi: 10.1093/dnares/9.3.99. [DOI] [PubMed] [Google Scholar]
- 24.Kato N, et al. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat. Genet. 2011;43:531–538. doi: 10.1038/ng.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeuchi F, et al. Genome-wide association study of coronary artery disease in the Japanese. Eur. J. Hum. Genet. 2011;184:333–340. doi: 10.1038/ejhg.2011.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim YJ, et al. Large-scale genome-wide association studies in East Asians identify new genetic loci influencing metabolic traits. Nat. Genet. 2011;43:990–995. doi: 10.1038/ng.939. [DOI] [PubMed] [Google Scholar]
- 27.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newton-Cheh C, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat. Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang F, et al. Genome-wide association identifies a susceptibility locus for coronary artery disease in the Chinese Han population. Nat. Genet. 2011;43:345–349. doi: 10.1038/ng.783. [DOI] [PubMed] [Google Scholar]
- 30.Hou L, et al. Associations of PLA2G7 gene polymorphisms with plasma lipoprotein-associated phospholipase A2 activity and coronary heart disease in a Chinese Han population: the Beijing atherosclerosis study. Hum. Genet. 2009;125:11–20. doi: 10.1007/s00439-008-0587-4. [DOI] [PubMed] [Google Scholar]
- 31.Alpert JS, Thygesen K, Antman E, Bassand JP. Myocardial infarction redefined--a consensus document of The Joint European Society of Cardiology/American College of Cardiology Committee for the redefinition of myocardial infarction. J. Am. Coll. Cardiol. 2000;36:959–969. doi: 10.1016/s0735-1097(00)00804-4. [DOI] [PubMed] [Google Scholar]
- 32.Rose GA. The diagnosis of ischaemic heart pain and intermittent claudication in field surveys. Bull. World. Health. Organ. 1962;27:645–658. [PMC free article] [PubMed] [Google Scholar]
- 33.Gu D, et al. Prevalence of the metabolic syndrome and overweight among adults in China. Lancet. 2005;365:1398–1405. doi: 10.1016/S0140-6736(05)66375-1. [DOI] [PubMed] [Google Scholar]
- 34.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS. Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu. Rev. Genomics. Hum. Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.