Summary
A main limitation of therapies that selectively target kinase signaling pathways is the emergence of secondary drug resistance. Cetuximab, a monoclonal antibody that binds the extracellular domain of EGFR, is effective in a subset of KRAS wild type metastatic colorectal cancers1. After an initial response, secondary resistance invariably ensues, thereby limiting the clinical benefit of this drug2. The molecular bases of secondary resistance to cetuximab in colorectal cancer are poorly understood3-8. Here, we show for the first time that molecular alterations (in most instances point mutations) of KRAS are causally associated with the onset of acquired resistance to anti-EGFR treatment in colorectal cancers. Expression of mutant KRAS under the control of its endogenous gene promoter was sufficient to confer cetuximab resistance but resistant cells remained sensitive to combinatorial inhibition of EGFR and MEK. Analysis of metastases from patients who developed resistance to cetuximab or panitumumab showed the emergence of KRAS amplification in one sample and acquisition of secondary KRAS mutations in 60% (6/10) of the cases. KRAS mutant alleles were detectable in the blood of cetuximab treated patients as early as 10 months prior to radiographic documentation of disease progression. In summary, the results identify KRAS mutations as frequent drivers of acquired resistance to cetuximab in colorectal cancers, indicate that the emergence of KRAS mutant clones can be detected non-invasively months prior to radiographic progression and suggest early initiation of a MEK inhibitor as a rational strategy for delaying or reversing drug resistance.
Keywords: KRAS, colorectal cancer, targeted therapy, acquired resistance
Defining the molecular bases of secondary resistance to anti-EGFR therapies is critical to monitor, prevent and/or overcome drug refractoriness. To identify potential mechanisms of cetuximab resistance, we generated cetuximab-resistant variants of two colorectal cancer (CRC) cellular models (DiFi, Lim1215) that are highly sensitive to EGFR inhibition (Fig. S1a). DiFi cells overexpress EGFR as a result of high level amplification of the EGFR gene locus9. In contrast, Lim1215 cells express ‘normal’ levels of EGFR but are similarly sensitive to cetuximab (Figs. S1b, S1c). Both cell lines are wild type for KRAS, BRAF and PIK3CA, paralleling the molecular features of the CRC patients most likely to respond to cetuximab10.
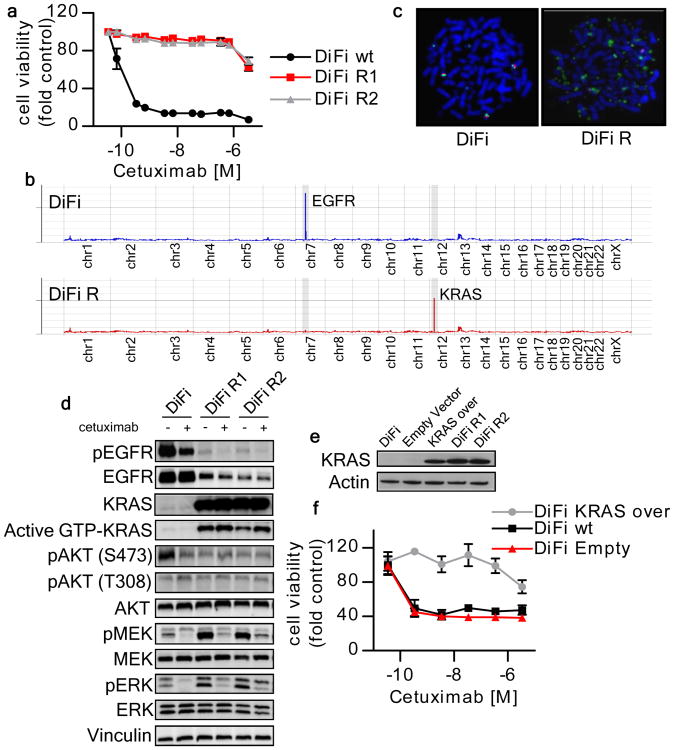
Continuous drug treatment using two different protocols (see methods and Fig. S2) led to the emergence of cetuximab resistant variants (DiFi-R and Lim1215-R, Figs. 1a, 2a). To identify the molecular basis of cetuximab resistance in these cells, we performed gene copy number analysis and mutational profiling of the resistant and parental lines. Cetuximab-resistant DiFi-R cells differed from their sensitive parental counterpart by two focal molecular alterations: EGFR gene copy number was reduced whereas the KRAS gene was amplified (Figs. 1b, c, S3). These genomic changes were accompanied by reduced EGFR and increased KRAS protein expression in the cetuximab resistant cells (Fig. 1d). Sequence analysis confirmed that the EGFR, KRAS, NRAS, HRAS, BRAF and PIK3CA genes were wild type in the cetuximab-resistant clones.
Figure 1. KRAS amplification mediates acquired resistance to cetuximab in DiFi cells.
(a) Parental and cetuximab resistant DiFi cells were treated for one week with increasing concentrations of cetuximab. Cell viability was assayed by the ATP assay. Data points represent means ± SD of three independent experiments. (b) Whole exome gene copy number analysis of parental and cetuximab resistant DiFi cells. Individual chromosomes are indicated on the x axis. The lines indicate the sequencing depth (y axis) over exome windows of 100,000 bp. (c) FISH analysis confirming KRAS amplification in DiFi-R but not parental DiFi cells. KRAS locus BAC DNA (probe RP11-707G18; green) and chromosome 12 paint (red) were hybridized to the metaphase spreads of DiFi cells. (d) DiFi cells were treated with cetuximab 35 nM for 24 hours, after which whole-cell extracts were subjected to Western blot analysis and compared to untreated cells. DiFi R1 and R2 were plated in the absence of cetuximab for 7 days or maintained in their normal growth medium (with cetuximab 35 nM) before protein analysis. Active KRAS (GTP-KRAS) was assessed by GST-Raf1 pull-down. Whole-cell extracts were blotted with phosphor-EGFR (Tyr 1068), total EGFR, total KRAS, phosphor-AKT (Thr 308), phosphor-AKT (Ser473), total AKT, total MEK1/2 and phospho-MEK1/2, total ERK1/2 and phospho-ERK1/2 antibodies. Vinculin was included as a loading control. (e) Western blot analysis of KRAS protein in DiFi cells infected with a KRAS lentivirus. Actin is shown as a loading control (f) Ectopic expression of wild-type KRAS in parental DiFi cells confers resistance to cetuximab.
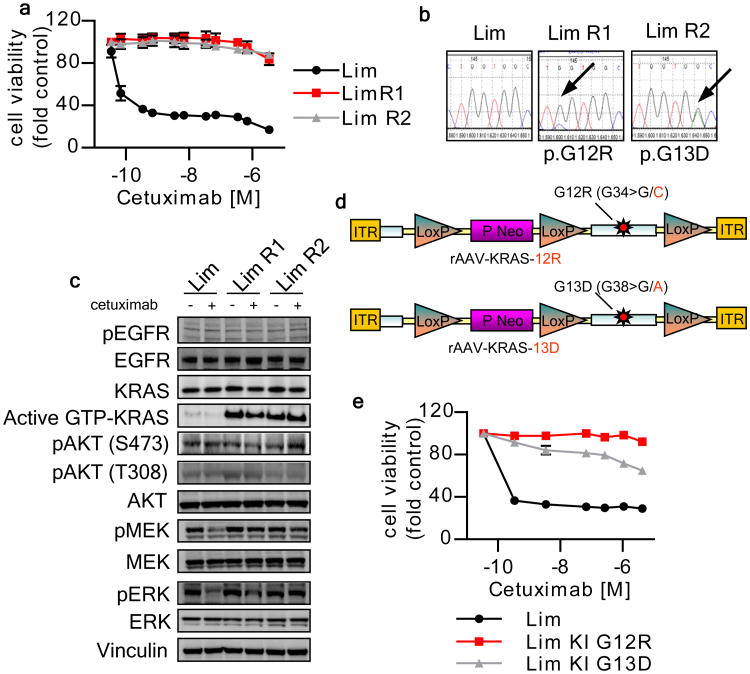
Figure 2. KRAS mutations mediate acquired resistance to cetuximab in Lim1215 cells.
(a) Parental and cetuximab resistant Lim1215 cells were treated for one week with increasing concentrations of cetuximab. Cell viability was assayed by the ATP assay. Data points represent means ± SD of three independent experiments. (b) Sanger sequencing of KRAS exon 2 in parental and two representative cetuximab-resistant Lim1215 cells obtained in independent selection procedures. (c) Western blot analysis of the EGFR signaling pathway in parental and cetuximab resistant Lim1215 cells. (d) Schematic representation of the vectors used to knock-in the G12R and G13D mutations into the genome of Lim1215 parental cell lines by AAV mediated homologous recombination. Targeting was assessed by Sanger sequencing. (e) Parental and isogenic Lim1215 cells carrying the indicated mutations were treated for one week with increasing doses of cetuximab.
Sequence analysis of the Lim1215 cetuximab resistant variants identified acquisition of either G13D or G12R KRAS mutations (Fig. 2b). In both DiFi-R and Lim1215-R cells, KRAS amplification or mutations, respectively, were accompanied by increased KRAS activation relative to their parental counterparts. In the presence of KRAS amplification, cetuximab could partially abrogate phosphorylation of MEK and ERK but, like in KRAS mutant cells, was unable to induce growth arrest (Figs 1a,d, 2a,c).
To determine whether resistance was due to selection of pre-existing drug resistant cells, we analyzed in depth the parental cell lines for the presence of a minority population of KRAS amplified or mutant cells. In the parental DiFi cells, we identified a sub-population with high level KRAS amplification at a prevalence of approximately 1:40,000 (Fig. S4). Similarly, deep sequencing and BEAMing (Bead Emulsion Amplification and Magnetics)11 indicated that approximately 0.2% of the parental Lim1215 cells harbored the KRAS G13D mutation (Table S1). Notably, the G12R mutation was not detectable in the earliest available passage of parental cells, even when the analysis was performed at high coverage (>50,000 fold). These results suggest that the emergence of a cetuximab resistant population could derive from selection of a pre-existing KRAS amplified or mutant clone, or as the result of ‘de novo’ acquisition of a KRAS mutation under the pressure of cetuximab treatment. To formally assess this latter possibility, we performed dilution cloning of the earliest available passage of Lim1215 cells in order to generate a homogenous, KRAS wild-type Lim1215 subline. As schematized in Fig. S5, two successive dilution cloning experiments were performed and the derivative cells (hereafter referred to as E4.1) were confirmed as KRAS wild-type by both mass spectrometry (MS) based genotyping and by 454 analysis. We then cultured the E4.1 cells in increasing concentrations of cetuximab, analogous to the experiment performed with the original Lim1215 parental line. Cells were collected during intermediate passages and subjected to MS based genotyping and/or 454 analysis (Fig. S5a). MS genotyping identified a KRAS A146T mutation following four passages in increasing concentrations of cetuximab (20 nM and higher, Fig.S5b). These cells were indeed resistant to the drug (Fig S5c, d) and displayed biochemical activation of KRAS (Fig. S5e). In parallel, genetic analysis of the E4.1 cells grown in medium without cetuximab found them to be KRAS wild-type. In sum, these data suggest that resistance to cetuximab in Lim1215 cells may emerge not only from the selection of pre-existing KRAS mutant clones but also as a result of ongoing mutagenesis.
To formally prove that amplification or mutations of KRAS were causally responsible for cetuximab resistance in our in vitro models, we performed two sets of forward genetic experiments. First, ectopic overexpression of wild type KRAS in DiFi conferred resistance to cetuximab (Fig. 1e, f). Second, AAV-mediated targeted homologous recombination was employed to introduce (knock-in) the G13D and G12R alleles into the endogenous KRAS locus of Lim1215 cells12. Knock-in of the G13D or G12R mutant alleles rendered Lim1215 cells resistant to cetuximab (Figs. 2 d, e).
Chemotherapy-refractory CRC patients who initially respond and then become resistant to cetuximab have no further therapeutic options. We reasoned that cetuximab resistance resulting from constitutive KRAS activation could be prevented or reversed by pharmacologic inhibition of KRAS signaling. We thus co-treated the resistant clones with cetuximab and selective inhibitors of the MEK and PI3 kinases, two key downstream effectors of oncogenic KRAS. While inhibitors of PI3 kinase were ineffective in the cetuximab resistant cells, both the Lim1215-R and DiFi-R cells were sensitive to combinatorial targeting of MEK and EGFR (Fig. S6).
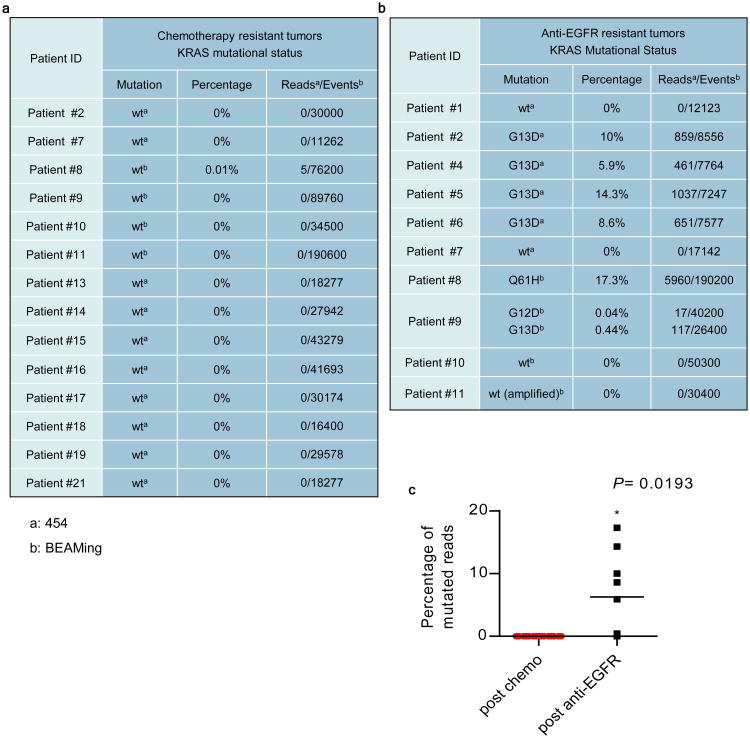
To determine whether KRAS mutation and/or amplification are clinically relevant mechanisms of acquired cetuximab-resistance, we examined tumor biopsies from 10 CRC patients who had become refractory to either cetuximab or the anti-EGFR antibody panitumumab (Table S2). We identified one individual (Table S3a, Patient 11) whose tumor at progression displayed KRAS amplification that was not present in a matched pre-cetuximab biopsy (Fig. S7). In a different patient, Sanger sequencing identified a KRAS Q61H mutation in a biopsy obtained following disease progression on cetuximab (Table S3a, Patient 8); whereas the remaining eight tumor samples obtained in patients with acquired resistance to anti-EGFR therapy were KRAS wild-type by this technique (Table S3a). To determine whether the Sanger technology may have been underpowered to detect the presence of KRAS mutations in the biopsies obtained following cetuximab or panitumumab progression, these remaining cases were analyzed using either 454 deep sequencing or BEAMing. These techniques identified KRAS G13D mutation in four samples and the simultaneous presence of G12D and G13D mutations in one case (Fig. 3b). In the six patients for whom sufficient pre-treatment tumor samples were available for high coverage 454 sequence analysis or BEAMing, KRAS mutations were absent pre-treatment (Table S3b). Tumors from an additional eight patients treated with cytotoxic chemotherapy but not previously exposed to anti-EGFR therapies were also analyzed by 454 deep sequencing. In all eight cases (Patients #13 – 21), 454 analyses identified no evidence of KRAS mutation (Fig. 3a). These results indicate that treatment with anti-EGFR antibodies but not cytotoxic chemotherapy is associated with acquisition of KRAS mutations (P = 0.0193) (Fig.3c). Our data support the initiation of clinical trials to define the prevalence of KRAS alterations as mechanism of acquired resistance to anti-EGFR therapies through systematic collection of biopsies.
Figure 3. Mutational analysis of the KRAS gene in patients.
(a) Mutational analysis of KRAS in chemorefractory patients. (b) Mutational analysis of the KRAS gene in patients who progressed on anti-EGFR antibodies. The results are based on assays performed by Deep sequencing technologies a: 454 pyrosequencing; b: BEAMing. (c) Dot plot of percentage of mutated KRAS alleles in chemorefractory and anti-EGFR resistant patients: p-value was calculated by two-tailed unpaired Mann-Whitney test.
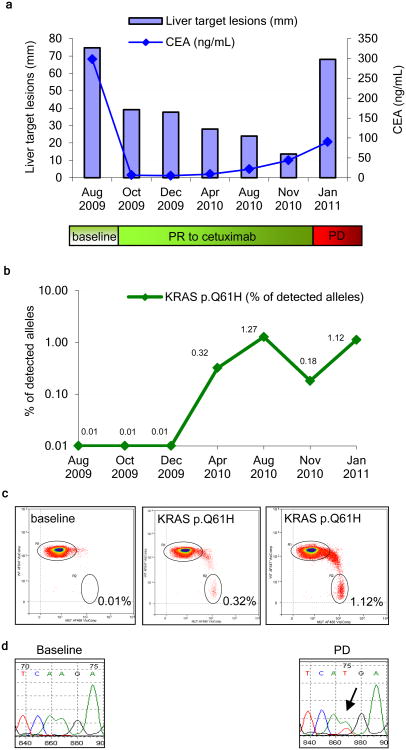
Emergence of secondary resistance to cetuximab (disease progression) is presently established by radiological evaluation and typically occurs within 9-18 months. We reasoned that the detection of KRAS mutant alleles in the plasma of patients treated with cetuximab or panitumumab may allow the early identification of individuals at risk for this mechanism of drug resistance prior to radiographic documentation of disease progression. We thus performed BEAMing analysis of serial plasma samples from patients treated with cetuximab (Table S4a, b). This analysis confirmed that the same KRAS variants that were ultimately identified in the post-treatment (disease progression) biopsies were detectable in plasma as early as 10 months prior to the documentation of disease progression by radiological assessment (Fig. 4).
Figure 4. Detection of circulating KRAS mutant DNA in a patient with acquired resistance to cetuximab therapy.
(a) Size of liver metastasis (blue bars) and CEA levels in blood (blue line) at the indicated time points showing an initial response to cetuximab followed by progression (Patient 8). (b) Quantitative analysis of Q61H mutant DNA in plasma as assessed by BEAMing (green line). (c) Two dimensional dot plot showing quantitative analysis of the KRAS Q61H mutation in plasma using BEAMing at individual time points (d) Mutational analysis of KRAS on tumor samples collected pre-cetuximab treatment and at the time of disease progression.
Drugs that target activated kinase pathways have profound but often temporary anti-tumor effects in subsets of patients with advanced solid tumors. In patients with advanced CRC, antibodies that bind to the extracellular domain of EGFR induce tumor regressions in 10-15% of patients when used alone and enhance the effects of cytotoxic chemotherapies when used in combination13,14. Whereas several prior studies have identified somatic mutations in the KRAS gene as a biomarker of intrinsic resistance to EGFR targeted agents in patients with CRC2,15, the molecular basis for acquired resistance to these agents has remained obscure. We now report for the first time that a substantial fraction of CRC patients who exhibit an initial response to anti-EGFR therapies have, at the time of disease progression, tumors with focal amplification or somatic mutations in KRAS which were not detectable prior to initiation of therapy. Our data indicate that drug resistance resulting from alterations in KRAS can be attributed not only the selection of pre-existent KRAS mutant and amplified clones but also to new mutations that arise as the result of ongoing mutagenesis. The percentage of KRAS mutant alleles detected in the resistant tumors ranged from 0.4 to 17% (figure 3). At least three (not mutually exclusive) possibilities could account for this low allele frequency. First, despite our efforts to maximize tumor content by macrodissecting each sample, the individual tumor biopsies consisted of variable proportions of tumor and intermixed KRAS wild-type stromal cells. Second, only a fraction of the tumor cells in the disease progression samples may have harbored the ‘resistance’ mutation. The latter scenario has been observed in lung cancer patients with secondary resistance to the EGFR inhibitor erlotinib where only a fraction of the tumor cells collected at the time of radiographic disease progression harbor the EGFR T790M ‘resistant’ allele16-18. Analogously, a recent study indicates that a subset of colorectal cancers found to be KRAS wild type by conventional Sanger sequencing but KRAS mutated with more sensitive techniques, do not respond to anti-EGFR treatment19. These data suggest that clinical drug resistance may results from the acquisition of a drug ‘resistant’ allele in sub-population of tumor cells. Finally, it is plausible that independent cell populations harbouring different ‘resistant’ mechanisms may evolve in parallel within the same metastatic lesion. Nevertheless, our functional analysis in cell models show that KRAS mutations are causally responsible for acquired resistance to cetuximab
Finally, we find that the KRAS mutant alleles found in the tumors of patients collected following radiographic disease progression can be detected in plasma using highly sensitive DNA analysis methods. As such tumors may be sensitive to combined treatment with a MEK inhibitor, our results suggest that blood based non-invasive monitoring of patients undergoing treatment with anti-EGFR therapies for the emergence of KRAS mutant clones could allow for the early initiation of combination therapies that may delay or prevent disease progression.
Methods Summary
DiFi and Lim1215 were exposed to different doses of cetuximab as described in figure S2 to obtain the resistant variants. Cell viability was assessed by ATP content. Cells were seeded in 100μl medium in 96-well plastic culture plates. The experimental procedures for knock in of cancer mutations, the vectors, AAV production, cell infection and screening for recombinants have already been described elsewhere12. Tumor specimens were obtained through protocols approved by the Institutional Review Board of Memorial Sloan-Kettering Cancer Center (protocol 10-029) and Ospedale Niguarda Ca' Granda, Milano, Italy (protocols 1014/09 and 194/2010). Details about the clinical characteristics of patients are provided in Supplementary Table 2. Identification of cancer mutations in the KRAS, HRAS, NRAS, BRAF, PIK3CA and EGFR genes was performed with different sequencing platforms (Sanger, 454 pyrosequencing and Mass Spectrometry) as described in details in the supplementary methods. For immunoblot analysis, total cellular proteins were extracted by solubilizing the cells in boiling SDS buffer. Western blot detection was done by enhanced chemiluminescence. The analysis of KRAS activation was performed by immunoprecipitation assay with GST-Raf1-RBD. Real time PCR was performed using an ABI PRISM® 7900HT apparatus (Applied Biosytems). KRAS protein expression was evaluated by immunohistochemistry performed on 3μm thick tissue sections using a specific KRAS (F234) antibody (SC-30, mouse monoclonal IgG2a Santa Cruz Biotechnology). BEAMing was performed essentially as described previously10, deviation from the original protocol are outlined in the supplementary methods. FISH experiments were conducted according with the histology FISH accessory kit (Dako, Glostrup, Danmark). Data are presented as the mean ± SD and n = 3. Statistical significance was determined by paired Student's t test. P < 0.05 was considered statistically significant.
Methods
Cell culture and generation of resistant cells
DiFi cells were cultured in F12 medium (Invitrogen) supplemented with 5% FBS and Lim1215 cells were cultured in RPMI-1640 medium (Invitrogen) supplemented with 5% FBS and Insulin (1 μg/ml). DiFi parental cells were plated in 100 mm Petri dishes with 2.5% FBS and exposed to a constant dose of cetuximab (350 nM), for one year in order to obtain the resistant counterpart DiFi R1. The DiFi R2 derivative was obtained by increasing cetuximab dosage stepwise starting from 3.5 nM, to 35 nM and finally to 350 nM during a time of one year. The same protocols were applied to Lim1215 with variations regarding cetuximab concentrations: for Lim R1 cetuximab was used at 1400 nM and for Lim R2 drug concentration started from 350 nM, to 700 nM and finally 1400 nM. For Lim1215 both protocols required at least 3 months' drug treatment. The Lim1215 parental cell line had been described previously1 and was obtained from Prof Robert Whitehead, Vanderbilt University, Nashville, with permission from the Ludwig Institute for Cancer Research, Zurich, Switzerland. The genetic identity of the cell lines used in this study was confirmed by STR profiling (Cell ID, Promega)
Drug viability assays
Cetuximab was obtained from Pharmacy at Niguarda Ca' Granda Hospital, Milan, Italy. AZD6244 and GSK1059615 were purchased from Sequoia Chemicals (Pangbourne, UK) and Selleck Chemicals (Houston, USA), respectively. Cell lines were seeded in 100μl medium at appropriate density (5×104, 1,5×104 for DiFi and Lim1215 cells, respectively) in 96-well plastic culture plates. After serial dilutions, drugs in serum free medium were added to cells and medium-only containing wells were added as controls. Plates were incubated at 37°C in 5% CO2 for 72-168 h, after which cell viability was assessed by ATP content using the CellTiter-Glo® Luminescent Assay (Promega Madison, WI, USA).
Mutational analysis
RAS genotyping was performed using the iPLEX assay (Sequenom, Inc.), which is based on a single-base primer extension assay. Briefly, multiplexed PCR and extension primers are designed for a panel of known mutations. After PCR and extension reactions, the resulting extension products are analyzed using a matrix-assisted laser desorption/ionization–time-of-flight (MALDI-TOF) mass spectrometer. For 454 picotiter plate Pyrosequencing (Roche Inc.), PCR products were generated using primers designed to span exons 2, 3 and 4 in KRAS and adapted with 5′ overhangs to facilitate emulsion polymerase chain reaction (emPCR) and sequencing. After amplification by emPCR, beads containing DNA were isolated. 34,000 beads were sequenced in both directions, yielding 1000 - 5000 sequencing reads on average per sample (∼1000 reads per amplicon per sample) using GSFLX. To detect variants in 454 sequencing data, reads were mapped with the BWA aligner using the bwasw mode for aligning long reads. The generated SAM file was then run through the Picard MarkDuplicate program to remove duplicated reads (reads with the same initial starting point). The file was then processed with the GATK BaseQ recalibrator. Finally, we generated pileups using Samtools and called variants using VarScan. For Sanger Sequencing all samples were subjected to automated sequencing by ABI PRISM 3730 (Applied Biosystems, Foster City, CA, USA). All mutations were confirmed twice, starting from independent PCR reactions.
All primer sequences are available upon request. Exome sequencing was carried out by exome capture using the SeqCap EZ Human Exome Library v1.0 (Nimblegen Inc.) and subsequent pyrosequencing of the captured fragments by means of 454Flx sequencer (Roche Inc.), according to manufacturer's protocols. A total of 1.2 Million reads were sequenced for an average exome depth of 4×. The reads were mapped using the manufacturer's mapping tools and the reads' depth used as an estimator of the copy number value in the two Difi parental and Difi resistant samples. Average reads' depths were calculated within overlapping 100,000 bp wide windows for Fig1b, while average reads' depths were calculated for exons and genes and respectively plotted as dots and segments in Supplementary Fig3ab.
Tissue procurement
Tumor specimens were obtained through protocols approved by the Institutional Review Board of Memorial Sloan-Kettering Cancer Center (protocol 10-029) and Ospedale Niguarda Ca' Granda, Milano, Italy (protocols 1014/09 and 194/2010). All tumor specimens were formalin fixed paraffin embedded (FFPE). All patients provided informed consent and samples were procured and the study was conducted under the approval of the Review Boards and Ethical Committees of the Institutions. Details about the clinical characteristic of the patients are provided in Table S2.
BEAMing procedure
DNA was extracted from plasma using the QIAamp Circulating Nucleic Acid Kit (QIAGEN) according to manufacturer's instructions. BEAMing was performed as describedpreviously12. The first amplification was performed in 50-μL PCR reaction, containing DNAisolated from 1 ml of plasma, 1× Phusion high-fidelity buffer, 1.5 U of Hotstart Phusionpolymerase (NEB, BioLabs), 0.5 μM of each primer with tag sequence, 0.2 mM of each deoxynucleoside triphosphate, and 0.5 mmol/L MgCl2. Amplification was carried out using the following cycling conditions: 98°C for 45 sec; 2 cycles of 98°C for 10 sec, 67°C for 10sec, 72°C for 10 sec; 2 cycles of 98°C for 10 sec, 64°C for 10 sec, 72°C for 10 sec; 2cycles of 98°C for 10 sec, 61°C for 10 sec, 72°C for 10 sec; 31 cycles of 98°C for 10 sec,58°C for 10 sec, 72°C for 10 sec. PCR products were diluted, and quantified using the PicoGreen double-stranded DNA assay (Invitrogen, Carlsbad, CA). A clonal bead population is generated performing an emulsion PCR (emPCR). 150 ul PCR mixture was prepared containing 18 pg template DNA, 40 U of Platinum Taq DNA polymerase (Invitrogen), 1× Platinum buffer, 0.2 mM dNTPs, 5 mM MgCl2, 0.05 μM Tag1 (tcccgcgaaattaatacgac), 8 μM Tag2 (gctggagctctgcagcta) and 6×107 magnetic streptavidin beads (MyOne, Invitrogen) coated with Tag1 oligonucleotide (dual biotin-TSpacer18-tcccgcgaaattaatacgac). The 150 μl PCR reactions were distributed into the wells of a 96-well PCR plate together with 70 μl of the Emusifire oil. The water-in-oil emulsion was obtained by pipetting. The PCR cycling conditions were: 94°C for 2 min; 50 cycles of 94°C for 10 sec, 58°C for 15 sec, 70°C for 15 sec. All primer sequences are available upon request.
Immunoblot analysis
Prior to biochemical analysis, all cells were grown in their specific media supplemented with 5% FBS. Total cellular proteins were extracted by solubilizing the cells in boiling SDS buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 1% SDS). Western blot detection was done by enhanced chemiluminescence (GE Healthcare). The following antibodies were used for western blotting (all from Cell Signaling Technology, except where indicated): anti-phospho-AKT S473; anti-phospho-AKT T308; anti-AKT; anti-phospho- p44/42 ERK (thr202/tyr204); anti-p44/42 ERK; anti-P-MEK1/2 (Ser217/221), anti-MEK1/2; anti-KRAS (Santa Cruz); anti-EGFR (clone13G8, Enzo Life Sciences); anti-phospho EGFR (tyr1068); anti–actin and anti-vinculin (Sigma-Aldrich, St. Louis, MO);
KRAS activation assay (RAS-GTP)
The analysis of KRAS activation was performed by an immunoprecipitation assay with GST-Raf1-RBD (ras binding domain), as previously described2. The KRAS protein was detected with Anti-KRAS (F234) mAb (Santa Cruz, Santa Cruz, CA).
Gene copy number analysis (qPCR and NGSeq)
Parental and resistant cell lines were trypsinized, washed with PBS and centrifuged; pellets were lysed and DNA was extracted using Wizard SV Genomic kit (Promega) according to the manufacturer's directions. Real time PCR was performed with 150 ng of DNA per single reaction using GoTaq QPCR Master Mix (Promega) and determined by real time PCR using an ABI PRISM® 7900HT apparatus (Applied Biosytems). All primer sequences are available upon request. Exome sequencing was carried out by exome capture using the SeqCap EZ Human Exome Library v1.0 (Nimblegen Inc.) and subsequent pyrosequencing of the captured fragments by means of 454Flx sequencer (Roche Inc.), according to manufacturer's protocols. A total of 1.2 Million reads were sequenced for an average exome depth of 4×. The reads were mapped using the manufacturer's mapping tools and the reads' depth was determined and used as an estimator of the copy number value in the two DiFi parental and DiFi resistant samples. Average reads' depths within overlapping 100,000 bp wide windows were calculated and plotted in Fig 1c; average reads' depths within exons and genes were calculated and respectively plotted as dots and segments in Supplementary Fig 4 a,b.
Immunohistochemistry Assay
KRAS protein expression was evaluated by immunohistochemistry performed on 3um thick tissue sections using a specific KRAS (F234) antibody (SC-30, mouse monoclonal IgG2a Santa Cruz Biotechnology; dilution 1:100) and the automated system BenchMark Ultra (Ventana Medical System, Inc., Roche) according to the manufactures instructions, with minimum modifications. KRAS protein expression was detected at cytoplasmatic and membrane level. Samples were considered positive when the expression of protein was present in at least 10 % of cells. Healthy tissue, i.e, normal colon mucosa, was used as internal negative control; slide with DiFi R2 cell line was used as external positive control. Images were captured with the AxiovisionLe software (Zeiss, Gottingen, Germany) using a Axio Zeiss Imager 2 microscope (Zeiss, Gottingen, Germany).
Fluorescent in situ hybridisation (FISH) analysis
All analyses were performed on 3μm thick sections of formalin-fixed paraffin-embedded tumour tissue, provided by the department of anatomy pathology of Niguarda Hospital, and on metaphase chromosomes and interphase nuclei, obtained from DiFi cell line culture following the standard procedures. Tissue sections for FISH experiment were prepared according to the manufacturer's instructions of Histology FISH Accessory kit (Dako, Glostrup, Danmark). For both types of samples the last steps before hybridization were: dehydration in ethanol series (70%, 90%, 100%), 3 washes (5′ each) and air drying. Dual colour FISH analysis was performed using a 10 μl mix- probe made up by 1 μl CEP12 alpha satellite probe (12p11-q11) labelled in SpectrumOrange (Vysis, Downers Grove, IL. USA), 1 μl BAC (Bacterial Artificial Chromosome) genomic probe RP11-707G18 (12p12.1) spanning an approximately 176 kb region encompassing the KRAS gene, labelled in SpectrumGreen (Bluegnome;3) and 8 μl LSI-WCP hybridisation buffer (Vysis, Downers Grove, IL. USA) for each slide. Probes and target DNA of specimens were co-denatured in HYBRite System (Dako Glostrup, Danmark) for 5 min at 75°C and then hybridized overnight at 37°C. Slides were washed with post-hybridisation buffer (Dako Glostrup, Danmark) at 73°C for 2 min and counterstained with 4,6-diamino-2phenylindole (DAPI II; Vysis, Downers Grove, IL. USA). Fluorescent in situ hybridisation signals were evaluated with a Zeiss Axioscope Imager. Z1 (Zeiss, Gottingen, Germany) equipped with single and triple band pass filters. Images for documentation were captured with CCD camera and processed using the MetaSystems Isis software. Samples with a ratio greater than 3 between KRAS gene and chromosome 12 centromere signals, in at least 10% of 100 cells analysed in 10 different fields, were scored as positive for KRAS gene amplification. Healthy tissue, i.e, normal colon mucosa, was used as internal negative control.
Plasmids and viral vectors
All experimental procedures for targeting vector construction, AAV production, cell infection and screening for recombinants have already been described elsewhere12.
Statistical analysis
Data are presented as the mean ± SD and n = 3. Statistical significance was determined by paired Student's t test or two-tailed unpaired Mann-Whitney test (Fig 3c). P < 0.05 was considered statistically significant
Supplementary Material
Acknowledgments
We are particularly indebted to Simona Lamba for generating the KRAS G12R KI in Lim1215 cells. We thank Carlotta Cancelliere and Simona Destefanis for technical assistance. We thank Sabrina Arena, Mariangela Russo and Davide Zecchin for critically reading the manuscript. We also thank Adriana Heguy of the MSKCC Beene Core, Agnes Viale of the MSKC Genomic Core and Nicolas Socci and Mono Pirun for assistance with analysis of next generation sequencing data. This work was supported by European Union Seventh Framework Programme, grant agreement 259015 COLTHERES (A.B. and S.S.), AIRC, Associazione Italiana per la Ricerca sul Cancro – 2010 Special Program Molecular Clinical Oncology 5 × 1000, project 9970 (A.B. and S.S.); Regione Piemonte (A.B, F.D.N.); FPRC, Intramural Grant – 5×mille 2008 – Fondazione Piemontese per la Ricerca sul Cancro –ONLUS (A.B, F.D.N.); AIRC MFAG 11349 (F.D.N.); OCGO, Oncologia Ca’ Granda ONLUS (S.S.); Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research; the Experimental Therapeutics Center of Memorial Sloan-Kettering Cancer Center (D.S.) the Society of MSKCC (M.W.) and the National Institute of Health (D.S.) and the Beene Foundation (D.S.).
Footnotes
Author contributions: A.B., D.S., S.S. and F.D.N. planned the project and supervised all research. A.B., D.S, and F.D.N. wrote the manuscript. S.M., R.Y., S.H., E.S., M.W. and F.D.N. designed the experiments. A.B. conceived the molecular analysis of plasma samples. S.M., R.Y., S.H., E.S., M.J., D.L., E.V., R.S., M.B., G.S., C.C., S.V., M.G., and V.B. performed the experiments. C.Z., A.S-B., M.G., E.M. analyzed data. K.B., A.C., E.V. provided samples for analysis.
References
- 1.Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 2.Karapetis CS, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 3.Wheeler DL, et al. Mechanisms of acquired resistance to cetuximab: role of HER (ErbB) family members. Oncogene. 2008;27:3944–3956. doi: 10.1038/onc.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benavente S, et al. Establishment and characterization of a model of acquired resistance to epidermal growth factor receptor targeting agents in human cancer cells. Clin Cancer Res. 2009;15:1585–1592. doi: 10.1158/1078-0432.CCR-08-2068. 10.1158/1078-0432.CCR-08-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li C, Iida M, Dunn EF, Ghia AJ, Wheeler DL. Nuclear EGFR contributes to acquired resistance to cetuximab. Oncogene. 2009 doi: 10.1038/onc.2009.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatakeyama H, et al. Regulation of heparin-binding EGF-like growth factor by miR-212 and acquired cetuximab-resistance in head and neck squamous cell carcinoma. PLoS One. 2010;5:e12702. doi: 10.1371/journal.pone.0012702. 10.1371/journal.pone.0012702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yonesaka K, et al. Activation of ERBB2 signaling causes resistance to the EGFR-directed therapeutic antibody cetuximab. Science translational medicine. 2011;3:99ra86. doi: 10.1126/scitranslmed.3002442. 10.1126/scitranslmed.3002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montagut C, et al. Identification of a mutation in the extracellular domain of the Epidermal Growth Factor Receptor conferring cetuximab resistance in colorectal cancer. Nature medicine. 2012 doi: 10.1038/nm.2609. 10.1038/nm.2609. [DOI] [PubMed] [Google Scholar]
- 9.Moroni M, et al. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. The lancet oncology. 2005;6:279–286. doi: 10.1016/S1470-2045(05)70102-9. 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 10.De Roock W, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 11.Diehl F, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Nicolantonio F, et al. Replacement of normal with mutant alleles in the genome of normal human cells unveils mutation-specific drug responses. Proc Natl Acad Sci U S A. 2008;105:20864–20869. doi: 10.1073/pnas.0808757105. 10.1073/pnas.0808757105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bardelli A, Siena S. Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J Clin Oncol. 2010;28:1254–1261. doi: 10.1200/JCO.2009.24.6116. [DOI] [PubMed] [Google Scholar]
- 14.Van Cutsem E, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 15.Amado RG, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 16.Janne PA. Challenges of detecting EGFR T790M in gefitinib/erlotinib-resistant tumours. Lung Cancer. 2008;60(2):S3–9. doi: 10.1016/S0169-5002(08)70099-0. S0169-5002(08)70099-0 [pii] 10.1016/S0169-5002(08)70099-0. [DOI] [PubMed] [Google Scholar]
- 17.Engelman JA, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest. 2006;116:2695–2706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arcila ME, et al. Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid-based assay. Clin Cancer Res. 2011;17:1169–1180. doi: 10.1158/1078-0432.CCR-10-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molinari F, et al. Increased detection sensitivity for KRAS mutations enhances the prediction of anti-EGFR monoclonal antibody resistance in metastatic colorectal cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:4901–4914. doi: 10.1158/1078-0432.CCR-10-3137. 10.1158/1078-0432.CCR-10-3137. [DOI] [PubMed] [Google Scholar]
- 1.Whitehead RH, Macrae FA, St John DJ, Ma J. A colon cancer cell line (LIM1215) derived from a patient with inherited nonpolyposis colorectal cancer. J Natl Cancer I. 1985;74:759–765. [PubMed] [Google Scholar]
- 2.Di Nicolantonio F, et al. Replacement of normal with mutant alleles in the genome of normal human cells unveils mutation-specific drug responses. Proc Natl Acad Sci U S A. 2008;105:20864–20869. doi: 10.1073/pnas.0808757105. 10.1073/pnas.0808757105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith G, et al. Activating K-Ras mutations outwith ‘hotspot’ codons in sporadic colorectal tumours - implications for personalised cancer medicine. Br J Cancer. 2010;102:693–703. doi: 10.1038/sj.bjc.6605534. 10.1038/sj.bjc.6605534. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.