SUMMARY
Cardiac dysfunction is the second leading cause of death in myotonic dystrophy type 1 (DM1) primarily due to arrhythmias and cardiac conduction defects. A screen of more than 500 miRNAs in a DM1 mouse model identified 54 that were differentially expressed in heart. More than 80% exhibited down regulation towards the embryonic expression pattern and showed a DM1-specific response. Twenty of 22 miRNAs tested were also significantly down regulated in human DM1 heart tissue. We demonstrate that many of these miRNAs are direct MEF2 transcriptional targets including miRNAs for which depletion is associated with arrhythmias or fibrosis. MEF2 protein is significantly reduced in both DM1 and mouse model heart samples and exogenous MEF2C restores normal levels of MEF2 target miRNAs and mRNAs in a DM1 cardiac cell culture model. We conclude that loss of MEF2 in DM1 heart causes pathogenic features through aberrant expression of both miRNA and mRNA targets.
Keywords: Myotonic dystrophy, microRNA, MEF2, CELF1, MBNL1
INTRODUCTION
Myotonic dystrophy type 1 (DM1) is an autosomal dominant disease caused by an expanded CTG repeat in the last exon of the dystrophia myotonica-protein kinase(DMPK) gene. Pathogenesis is caused primarily by the mRNA containing expanded CUG repeats (CUGexp RNA)that is expressed from the mutated allele(Wheeler and Thornton, 2007). DMPK is expressed in multiple tissues that are subsequently affected in the disease, however, the primary causes of mortality are muscle wasting (60%) and sudden cardiac death (25–30%) (Groh et al., 2008; Heatwole et al., 2012; Phillips and Harper, 1997; Salehi et al., 2007). More than 80% of individuals affected with DM1 have cardiac conduction defects and arrhythmias and a lower percentage are affected by interstitial fibrosis and dilated cardiomyopathy (Groh et al., 2008; Lazarus et al., 2002; Nazarian et al., 2010; Pelargonio et al., 2002; Phillips and Harper, 1997; Sovari et al., 2007). While several molecular mechanisms of DM1 pathogenesis have been defined(Sicot et al., 2011; Udd and Krahe, 2012), the specific mechanisms causing electrophysiological, fibrotic and contractility abnormalities in DM1 heart tissue is unknown.
The best characterized effects of CUGexp RNA are disrupted functions of the RNA binding proteins, muscle blind-like 1 (MBNL1) and CUGBP, Elav-like family member 1(CELF1) which regulate multiple RNA processing events including alternative splicing, translation, mRNA stability, and mRNA intracellular localization (Lee and Cooper, 2009; Timchenko, 2013). Celf1 is down regulated during mouse postnatal heart and skeletal muscle development, while Mbnl1 activity is up regulated driving their target alternative splicing events to the adult patterns (Kalsotra et al., 2008; Lin et al., 2006). Celf1 down regulation is post-transcriptionally mediated by microRNA (miRNA)-repressed translation and protein destabilization by de-phosphorylation (Kalsotra et al., 2010; Kalsotra et al., 2008; Kuyumcu-Martinez et al., 2007). CUGexp RNA reverses normal postnatal regulation of MBNL1 and CELF1 by sequestration of MBNL1 which binds with high affinity to the CUG repeats and stabilization of CELF1 by PKC-activated phosphorylation resulting in a 2–4 fold increase in heart and skeletal muscle (Kuyumcu-Martinez et al., 2007; Savkur et al., 2001; Timchenko et al., 2001; Wang et al., 2007). In addition to disrupted alternative splicing, molecular defects of CUGexp RNA toxicity involve repeat-associated non-ATG (RAN) translation (Zu et al., 2011), abnormal DNA methylation (Lopez Castel et al., 2011), bidirectional transcription (Moseley et al., 2006), and miRNA dysregulation (Fernandez-Costa et al., 2013; Perbellini et al., 2011; Rau et al., 2011).
We previously demonstrated that postnatal downregulation of Celf1 and its paralogue Celf2 in mouse heart results from a dramatic up regulation of miR-23a and miR-23bbetween postnatal day 2 (PN2) and PN21 (Kalsotra et al., 2010; Kalsotra et al., 2008). Therefore, we wanted to determine whether altered miRNA expression in DM1 could be an additional mechanism of CELF1 up regulation. Using an established heart-specific and inducible DM1 mouse model, we found that postnatal upregulation of miR-23a and miR-23b is dramatically reversed upon induction of CUGexp RNA in adult heart. Furthermore, an analysis of >500 miRNAs identified 54 that are mis-regulated within 72 hours of CUGexp RNA induction, >80% of which represent reversal of postnatal up regulation. Twenty of 22 miRNAs affected in the DM1 mouse model were also down regulated in DM1 heart tissues. Pathway analysis of mRNAs and miRNAs mis-regulated in the DM1 mouse heart identified a loss of function of the Mef2 transcriptional network. Loss of MEF2A and MEF2C mRNA and protein expression was demonstrated in heart tissue from the DM1 mouse model and in individuals affected by DM1. In addition, 20 of 20 protein coding genes that are demonstrated targets of MEF2 were down regulated in the DM1 mouse model. Mis regulation of miRNA and mRNA MEF2 targets by CUGexp RNA was rescued by MEF2C. For several of the affected miRNAs, down regulation has previously been shown to produce arrhythmias or fibrotic changes. Our results demonstrate that the MEF2 transcription network is disrupted by CUGexp RNA leading to altered expression of a large number of miRNA and mRNA targets with effects consistent with DM1 heart pathology.
RESULTS
Disrupted expression of postnatally regulated miRNAs in adult DM1 heart
To determine whether CELF1 upregulation in DM1 heart tissue resulted from altered miRNA expression, we quantified miR-23a and miR-23b expression in heart tissue from a heart-specific DM1 mouse model (EpA960; MCM). These mice inducibly express human DMPK exon 15 containing 960 CUG repeats and exhibit Celf1 up regulation (Figure 1A)(Wang et al., 2007). We observed a strong postnatal increase in miR-23a and miR-23b levels in wild type mouse hearts between E14 and adult as described earlier (Figure S1A)(Kalsotra et al., 2010). Importantly, the levels of miR-23a and miR-23bwere significantly reduced (P<0.05) both at 72h and one week following induction of CUGexp RNA expression (Figure S1A). Comparable levels of CUGexp RNA induction are observed at both time points (Figure S1B). This result identified a direct link between induction of CUGexp RNA and mis-regulated expression of miRNAs and suggested that Celf up regulation in DM1 resulted from loss of miRNA expression in addition to the previously described phosphorylation-mediated stabilization(Kuyumcu-Martinez et al., 2007).
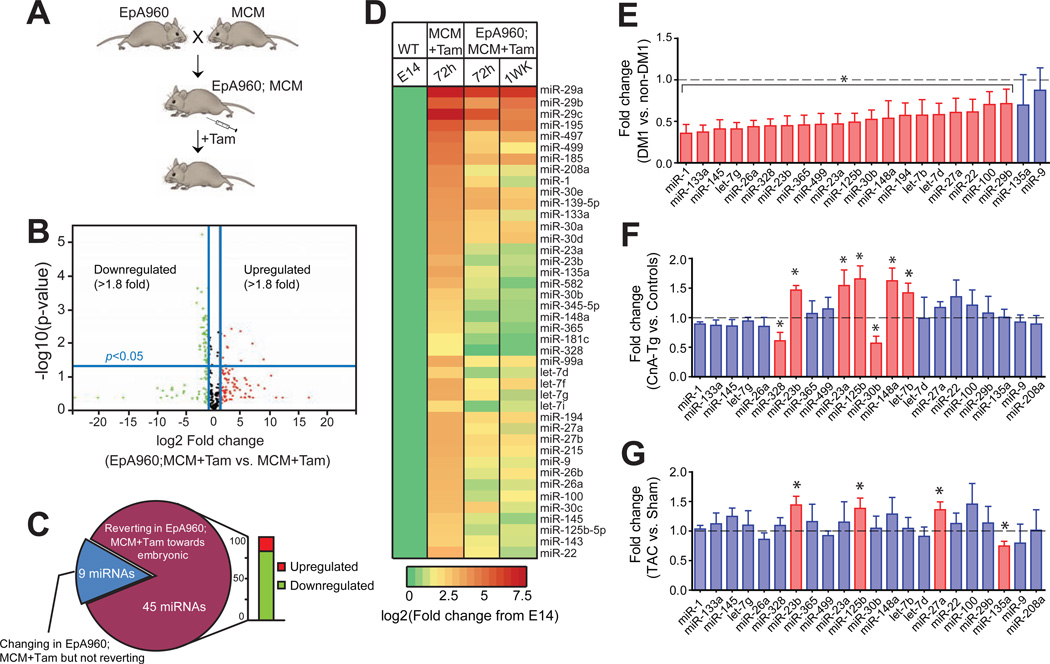
Figure 1. Mis-regulation of a subset of miRNAs in a heart-specific mouse model for DM1.
(A) Schematic of an inducible heart-specific DM1 mouse model. EpA960 mice contain a transgene containing DMPK exon 15 with 960 CTG interrupted repeats that were crossed with Mer Cre Mer (MCM) (Sohal et al., 2001)mice to generate heart-specific and tamoxifen-inducible expression of CUGexp RNA. (B) Expression profiling using quantitative real-time qRT-PCR based TaqMan arrays of >500 miRNAs in EpA960; MCM vs. MCM control mouse heart 1 wk after tamoxifen injection. The volcano plot shows up or downregulated miRNAs in DM1 mice compared to MCM controls. Data is normalized relative to U6 small nuclear (sn) RNA (n=3). (C) Adult-to-embryonic shift in miRNA expression in DM1 mice upon CUGexp RNA expression. Pie chart summarizing miRNAs that are differentially expressed and exhibit a developmental shift towards the embryonic pattern. (D) Heat map showing developmental up regulation of 42 miRNAs during normal mouse heart development, which are down regulated at 72h and 1 wk after repeat RNA expression. (E) Reduced miRNA expression in DM1 heart tissue. Each bar represents fold change in individual miRNA expression (mean ± SD) from heart samples of adults with DM1 (n=8) relative to heart samples from unaffected individuals (n=4).(F) Developmentally regulated miRNAs do not show a coordinate reduction in expression in two distinct models of heart disease: calcineurin transgenic nA-Tg) mice and (G) wild type mice 8 wks after Transverse Aortic Constriction (TAC). Each bar represents fold change in individual miRNA expression (mean ± SD) from heart samples of CnA-Tg mice relative to littermate controls (n=3) or from mice that underwent TAC surgery relative to shams (n=3). *P< 0.05.
To determine if additional miRNAs are mis-regulated in DM1, we used quantitative real time PCR (qRT-PCR) based TaqMan arrays to profile expression of >500 miRNAs from the hearts of wild type embryonic day (E)14 mice, adult EpA960; MCMDM1 mice and MCM control mice. We identified 54 miRNAs that were differentially expressed (fold change >1.8; P<0.05) between MCM controls and DM1 mice 1 wk following induction of CUGexp RNA (Figure 1B and Table S1). Eighty-three percent (45/54) of the miRNAs are regulated during normal postnatal heart development and exhibit an adult-to-embryonic shift in expression in DM1 mice, while 17 percent (9/54)exhibit a change but are not regulated postnatally (Figure 1C). A heat map representation of 42 miRNAs that are up regulated during normal heart development shows as triking decrease in their expression both at 72h and 1 wk following CUGexp RNA induction (Figure 1D), indicating cardiac expression of CUGexp RNA results in developmental reprogramming of a subset of miRNAs.
Next, we assayed miRNA expression in heart tissues of eight DM1 and four unaffected individuals. Twenty out of the top 22 miRNAs found to be mis-regulated in the DM1 mouse model were significantly reduced (P<0.05) in DM1 heart tissue(Figure 1E). We also determined that reduced levels of developmentally regulated miRNAs in DM1 are not a general response secondary to cardiomyopathy, as the same miRNA subset is not coordinately affected in two separate models of heart disease (Figure 1F and G) or among a subset of the miRNAs tested in human heart failure samples (Figure S1E). Based on these data we conclude that reduced expression of developmentally regulated miRNAs is specific to DM1 rather than a general response to cardiac injury. In addition, the DM1 mouse model does not show activation of hypertrophy markers suggesting that the response of the heart to induced CUGexp RNA is distinct from a hypertrophic response (Figure S1B and C).
Altered miRNA expression induced by CUGexp RNA is not reproduced by loss of Mbnl1 or gain of Celf1
CUGexp RNA disrupts the functions of the RNA binding proteins MBNL1 and CELF1 resulting in mis-splicing of their pre-mRNA targets(Lee and Cooper, 2009; Wheeler and Thornton, 2007). Altered expression of one miRNA, miR-1, in DM1 heart was proposed to result from disrupted pre-miRNA processing due to loss of MBNL1 activity(Rau et al., 2011). To determine whether the adult-to-embryonic shift in miRNA expression observed in DM1 is driven by mis-regulation of Mbnl1 or Celf1, we quantified expression of 23 miRNAs most affected in the DM1 mouse model in heart tissue from Mbnl1••3/••3 mice(Kanadia et al., 2003) and a previously described heart-specific and tetracycline (tet)-inducible human CELF1 transgenic mouse line (Kalsotra et al., 2008; Koshelev et al., 2010). None of the miRNAs were mis regulated in CELF1 inducible transgenic animals (Figure 2A) and only the three let-7 family members were reduced significantly (P<0.05) in Mbnl1••3/••3 mice (Figure 2C). Loss of Mbnl1 and gain of CELF1 activities were confirmed in the same RNA samples by showing altered splicing of Mbnl1 or CELF1 targets(Figure 2B and D).
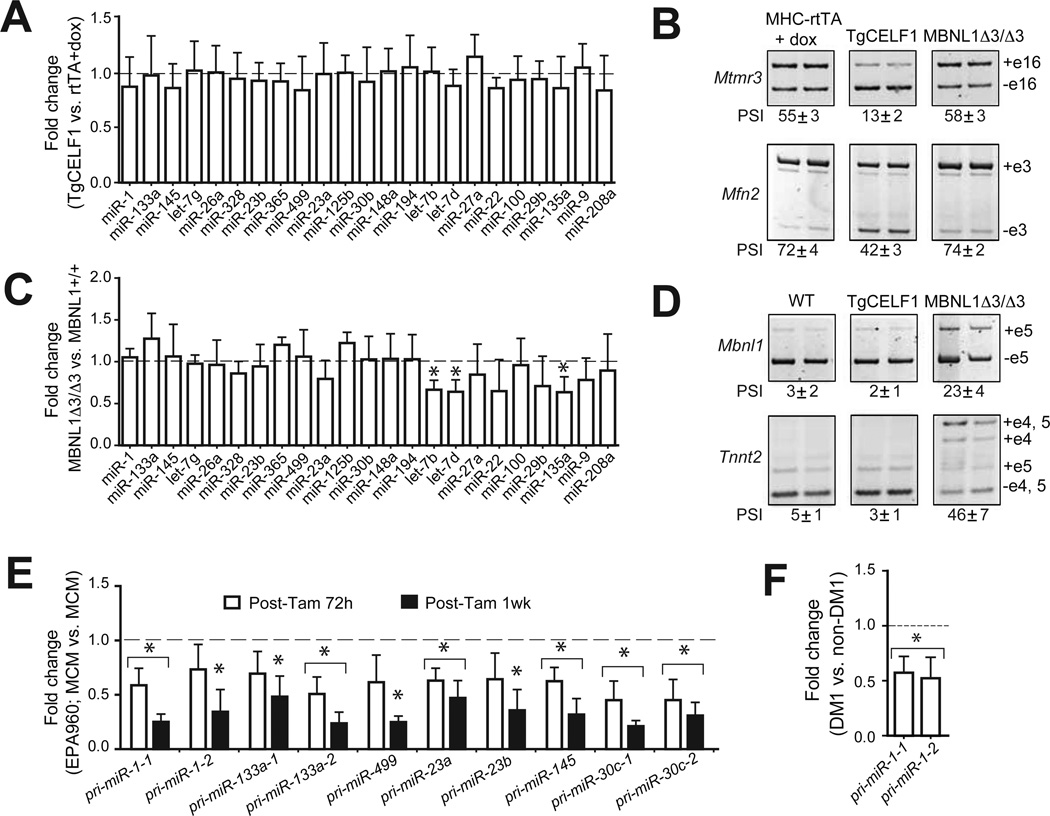
Figure 2. Altered miRNA expression identified in DM1 is not reproduced by loss of Mbnl1 or gain of CELF1.
(A) qRT-PCR analysis of miRNA expression in hearts of tet-inducible and heart-specific CELF1 transgenic (TgCELF1) mice. Each bar represents fold change in individual miRNA expression (mean ± SD) in TgCELF1 mice relative to MHC-rtTA controls given doxycycline (dox). Data is normalized relative to U6 snRNA (n=3). (B) RT-PCR analysis monitoring percent spliced in (PSI) of two CELF1-regulated alternative splicing events, Mtmr3 exon 16 and Mfn2 exon 3 in MHCrtTA or TgCELF1 mice given dox or Mbnl1 3/ 3 mice. (C) miRNA expression in hearts of Mbnl1 3/ 3 relative to Mbnl1+/+ mice showing fold change in individual miRNA expression (mean ± SD). Data is normalized relative to U6 snRNA (n=3). (D) RT-PCR analysis monitoring PSI of two Mbnl1-regulated alternative splicing events, Mbnl1 exon 5 and Tnnt2 exon 5 in Mbnl1+/+, TgCELF1 + dox, or Mbnl1••3/••3 mice. (E) Reduced expression of ten primary (pri-) miRNA transcripts at 72h and 1 wk after CUGexp RNA induction in DM1 mice. Each bar represents fold change in individual pri-miRNAs in DM1 mice relative to MCM controls at 72h or 1 wk after tamoxifen injection. (F) Reduced steady state levels of pri-miR-1-1 and pri-miR-1-2 transcripts in human heart samples from DM1 patients relative to unaffected individuals (n=3). *P< 0.05.
To determine whether the expression of miRNA primary transcripts (pri-miRNA)were affected in the DM1 mouse model, we performed qRT-PCR analysis using TaqMan probes specific for the pri-miRNAs of ten down regulated miRNAs. We found that ten out of ten primary miRNA transcripts examined showed decreased expression that paralleled the reduced expression of their mature miRNAs when assayed at 72h and 1 wk after CUGexp RNA induction (Figure 2E). Moreover, we found that expression ofpri-miR-1-1 and pri-miR-1-2 are significantly reduced in the hearts of DM1 patients relative to unaffected controls (Figure 2F). Overall these data demonstrate that gain of Celf1 or loss of Mbnl1 activity is not responsible for the altered miRNA expression in DM1. Our data are consistent with an upstream defect in transcription rather than a downstream RNA processing defect.
Large-scale shift in gene expression in DM1 is due in part to loss of miRNAs and inactivation of the Mef2 transcriptional program
To assess if in addition to splicing and miRNA defects, CUGexp RNA also perturbed mRNA steady state levels, we carried out a microarray study on heart RNA from wild type E14 and adult mice as well as adult MCM control and DM1 mice. As anticipated we noted a large number of genes to be developmentally regulated in wild type hearts, however, within 72h of CUGexp RNA induction many genes showed a coordinated adult-to-embryonic shift in mRNA expression(Figure 3A). Strikingly, 1 wk of CUGexp RNA expression resulted in a pervasive shift in transcript levels of a large number of genes towards the embryonic pattern(Figure 3A).
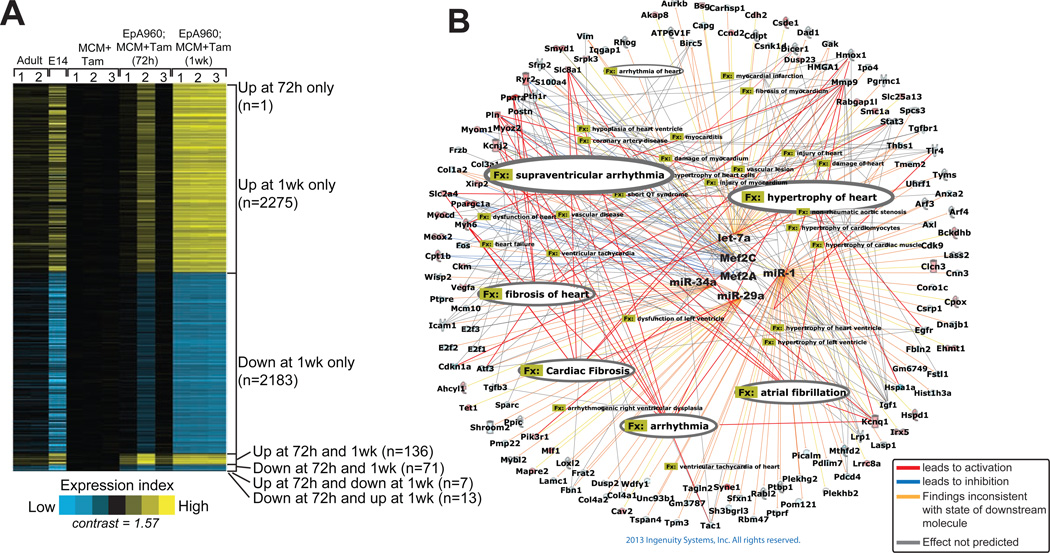
Figure 3. DM1 heart mouse model shows large-scale shift in gene expression that identifies disrupted Mef2 network.
(A) Gene expression profiling in mouse heart development and adult DM1 mice shows a developmental reversion in mRNA expression. Heat map representation of transcripts over expressed (yellow) and under expressed (blue) in hearts of wild type adult mice, wild type embryonic day 14 (E14), and DM1 mice induced to express CUGexp RNA for 72 h and 1 wk, when compared to MCM controls (P< 0.01, fold change >1.5). Rows, transcripts (values centered on MCM group); columns, profiled samples.(B) Ingenuity pathway analysis identified Mef2 as a key regulator of both miRNA and mRNA with altered expression in heart tissue expressing CUGexp RNA. Cardiovascular gene function categories with P<1E-05 are highlighted in the figure.
Gene Ontology analysis using the Ingenuity Pathway Analysis (IPA) showed the mitochondrial pathway as the most significantly affected pathway (P=5.36E-08 and threshold ratio=0.259) (Figure S2). To identify the transcription factors and miRNAs that are potentially responsible for these gene expression changes and the ensuing phenotype in DM1 mice we performed an upstream regulator analysis by IPA. This analysis examined the enrichment of known targets of each transcriptional regulator present in our gene list to that in the database, resulting in an estimation of an overlap P-value. Based on activation or suppression of target genes (for a transcriptional regulator) compared with observed changes in gene expression an activation Z-score was assigned. Z-score >2 illustrates activation and Z-score <-2 illustrates inhibition of activity. Using this approach we discovered the cardiac transcription factorsMef2a and Mef2c as most significantly inhibited (Mef2a Z-score of −2.941, P=1.70E-04;Mef2c Z-score of −3.017, P=1.20E-07) (Figure 3B and Table S2).
IPA also predicted inhibition of miRNA families miR-29, let-7, miR-1 and miR-34a corroborating the reciprocal up regulation of their corresponding targets (Table S2). Finally, to understand how Mef2 and the predicted miRNAs interact with one another and their targets we made an interaction network and overlaid it with the cardiac disease function from the function and disease tools of IPA. These analyses showed genes involved in cardiac arrhythmia, hypertrophy and fibrosis as over represented in the network (Figure 3B). Importantly, these categories correlate strongly with the phenotypic changes observed in DM1 patients and mice including prolonged PR intervals and QRS duration, decreased contractility, dilated 2007).
qRT-PCR assays confirmed significant down regulation (P<0.05) of Mef2a and Mef2 cmRNAs in our DM1 mouse model (top panel, Figure 4A) as well as in DM1 patient samples (bottom panel, Figure 4A). As a control we assessed Gata4 transcript levels, which were unchanged in both the DM1 mouse model and DM1 heart tissues. MEF2A and MEF2C mRNA levels were not affected in human heart failure samples (Figure S3D) indicating that reduced expression is not a general response to heart disease. We also found that the alternative exons in Mef2a (α&β••••••) and Mef2c (γ••••) did not exhibit a significant difference in PSI (percent spliced in) values following 1 wk of CUGexp RNA expression when compared to MCM controls (Figure S3A). Similarly, while alternative splicing of the MEF2A β• and MEF2C γ• were affected in some DM1 heart samples, differences are not statistically significant (Figure S3E) indicating that down regulation rather than altered splicing is likely to have the larger impact on MEF2 activity.
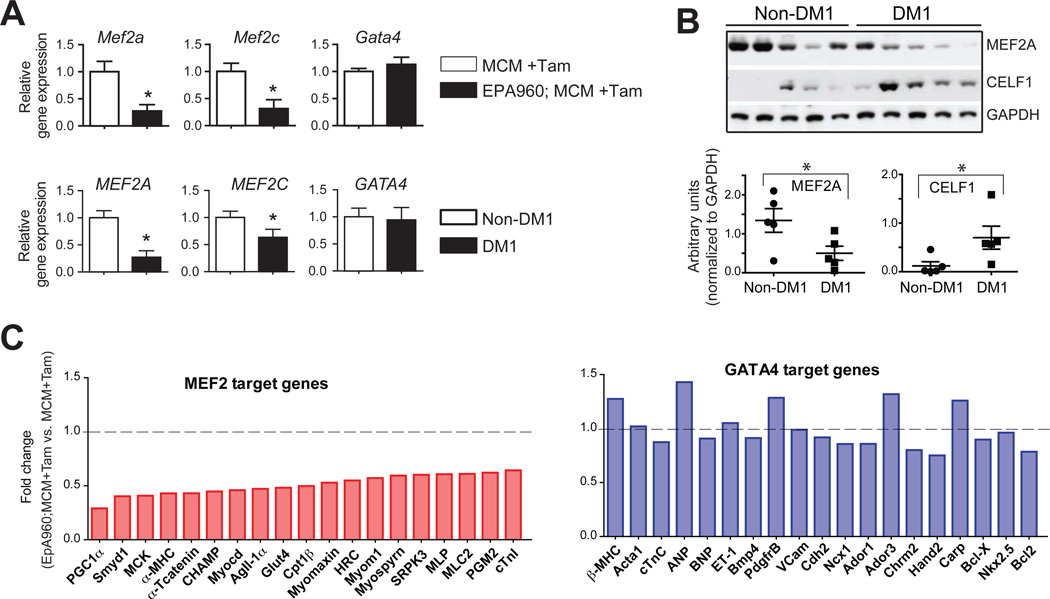
Figure 4. Disruption of Mef2 transcription program in DM1.
(A) Reduced Mef2a and Mef2c expression in heart tissue from the EpA960; MCM DM1 mouse model (n=3) and individuals with DM1 (n=8) or normal controls (n=4). Gata4 mRNA levels are not affected. Data is normalized to ribosomal protein L30 (Rpl30).(B) Western blot showing reduction in steady state MEF2A protein levels in human DM1 heart samples. CELF1 protein levels are up regulated in these samples, as previously described(Savkur et al., 2001; Timchenko et al., 2001). Quantification of relative band intensities, normalized to GAPDH levels are shown below. (C) Decreased Mef2a and Mef2c expression affects mRNA steady-state levels of Mef2 target genes in mouse DM1 heart tissue. Representative Mef2 target genes show a significant reduction in expression (light red bars) whereas Gata4 target genes are unaffected (light blue bars). *P< 0.05.
Western blot analysis showed a 2-fold decrease in MEF2A protein levels in DM1 compared to unaffected hearts (Figure 4B). As expected CELF1 protein levels were induced >2-fold in DM1 heart tissue. Importantly, Mef2a and Mef2ctranscripts were unaltered in hearts of either Celf1 transgenic or Mbnl1 3/3 mice compared to their littermate controls (Figure S3B);suggesting reduced Mef2expression in DM1 is unrelated to the mis-regulation of either RNA binding protein.
Furthermore, we tested whether high confidence Mef2 target genes expressed in heart were altered in the DM1 mouse model. A high confidence Mef2 target was defined as a gene that has published chromatin immunoprecipitation (ChIP) evidence of Mef2 occupancy, for which promoter analysis has implicated Mef2 as a direct transcriptional activator and/or which displays decreased transcript levels upon loss of Mef2 activity. Twenty such targets were chosen(see Supplemental references). Remarkably, 100% (20/20) of the Mef2 target genes examined were down regulated in DM1 mouse heart tissue(left panel, Figure 4C). As a control data set we tested a target gene set for Gata4 (see Supplemental references), which is unaffected in DM1 heart tissue (Figure 4A). We observed no significant change in expression of Gata4 targets in DM1 mice (right panel, Figure 4C). These results indicate that the vast majority of experimentally supported Mef2 target genes expressed in heart are down regulated in DM1.
Identification of Mef2 regulated miRNAs in cardiac cells
In addition to many muscle-specific genes, Mef2 directly activates transcription of bicistronic primary transcripts encoding miR-1-2/-133a-1 and miR-1-1/-133a-2(Liu et al., 2007), which we found to be down regulated in DM1 patient and mouse model heart tissues. However, it is not known what other miRNAs are regulated by Mef2 in heart. We searched 10 kb genomic regions spanning each of the 54 differentially expressed miRNAs in DM1 mice for Mef2 binding sites [CTA(A/T)4TAG] and found 65 putative sites in 34 different miRNA genes.
To determine the fraction of miRNA genes physically bound by Mef2 proteins in heart, we performed ChIP assays on wild type adult mouse hearts using a pan-Mef2 antibody. Non immune IgGs and RNA Pol II antibodies served as negative and positive controls respectively. The genomic regions harboring the ChIP-ed Mef2 consensus sites were amplified using specific primer sets. PCR analyses of the precipitated chromatin showed strong Mef2 binding along the genomic regions of previously characterized Mef2 targets(Smyd1, Pgc1a, Myom1 and cTnI3) while no significant binding was detected to an intergenic negative control region (Figure S4A). Using the ChIP assay we confirmed 20 of 65 sites in 15 different pri-miRNAs to be occupied by Mef2 (Figure S4B). let-7d upstream region does not contain a Mef2 site and showed no binding while a previously characterized pri-miR-1-2 binding site showed positive binding as expected. RNA Pol II showed preferential association with all miRNAs and mRNA genomic regions tested confirming these regions are actively transcribed in wild type mouse hearts (Figure S4B).
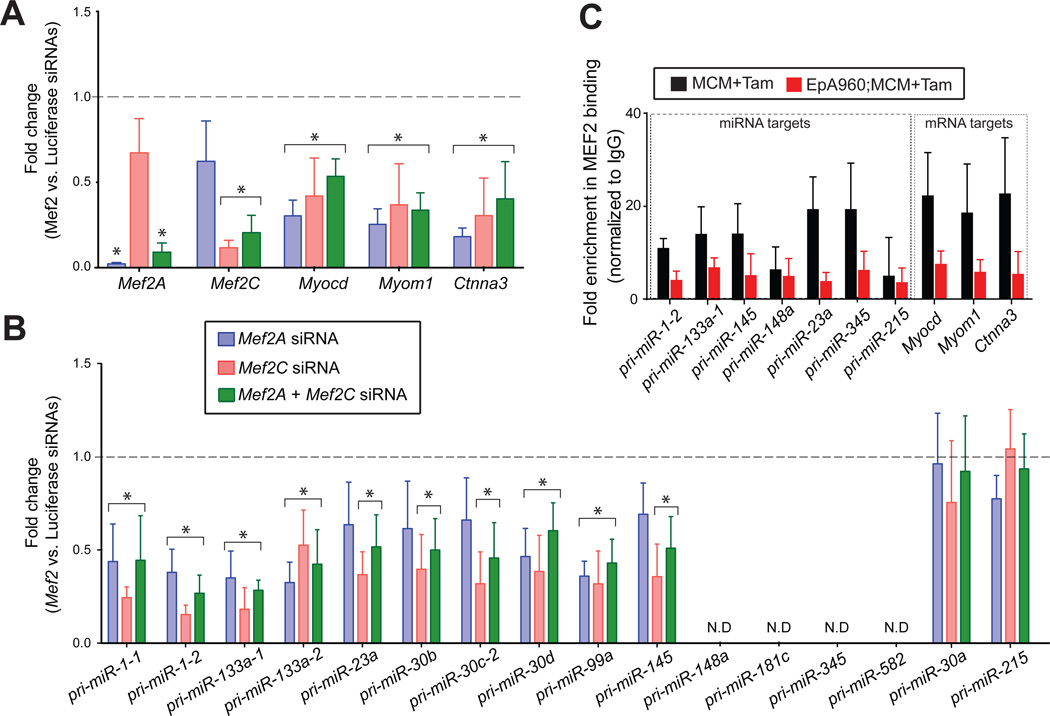
To determine whether expression of the miRNAs downregulated in DM1 heart require Mef2, we used short interfering RNAs (siRNAs) to knock down Mef2a and Mef2c genes individually or in combination in mouse a trial cardiac HL-1 cells. We consistently achieved over 80% knockdown efficiency for both Mef2a and Mef2cfrom their endogenous levels (Figure 5A). We tested expression of three known Mef2 target genes (Myocd, Myom1 and Ctnna3) in Mef2 knock down cultures and observed an expected reduction in their steady state levels compared to the control knock downs (Figure 5A). Importantly, both individual and combined Mef2 knockdowns resulted in a significantly lower expression (P<0.05)of 10 out of 15pri-miR transcripts that showed Mef2 binding in the ChIP assays (Figure 5B&Figure S4B). Four out of 15 were not expressed in HL-1 cells while pri-miR-30a was expressed but was unaffected by the knock downs (Figure 5B). These results indicate that in addition to pri-miR-1/-133a clusters, Mef2 proteins drive expression of several other pri-miRNAs in cardiac cells. We next used a pan Mef2 antibody to perform ChIP analysis of DM1 mouse heart tissue. Consistent with reduced Mef2a and Mef2c levels we noted their association with the response elements was decreased on both miRNA and mRNA targets in DM1 mouse heart tissues in comparison to the MCM controls (Figure 5C). Collectively, these data demonstrate that loss of Mef2 expression in DM1 is likely to have a significant impact on expression of downstream targets directly due to their reduced occupancy on the target genes.
Figure 5. Identification of Mef2-regulated miRNAs in cardiac cells.
RNAi-based Mef2 knockdowns coupled with chromatin immunoprecipitation (ChIP) assays identify miRNAs directly regulated by Mef2. (A)Knockdown efficiency of Mef2a and Mef2c siRNAs in HL-1 cardiac cells was determined by qRT-PCR in three independent experiments. Reduced steady state levels of Mef2 mRNA targets Myocardin (Myocd), Myomesin (Myom1) and α-T-catenin (Ctnna3) in response to Mef2A, Mef2C individual or double knockdowns (mean ± SD; n=3). (B) Reduced steady state levels of pri-miRNAs in Mef2 knockdowns. All data is plotted relative to a luciferase control siRNA and expression is normalized to Rpl30. *P< 0.05, N.D, not detected. (C) Reduced interaction of Mef2 with its primary miRNA and mRNA gene targets in DM1 mice. Quantification of genomic DNA in chromatin immunoprecipitates using Mef2 antibody in heart tissue of MCM controls and DM1 mice. The primers used for qRT-PCR assays span the Mef2 binding sites in target primary miRNAs or mRNAs. Each bar represents mean ± SD of the fraction of input detected in the Mef2 precipitates normalized to IgG precipitates (n=3).
Mis-regulation of miRNAs and mRNAs in a DM1 cardiac cell model is rescued by exogenous Mef2c
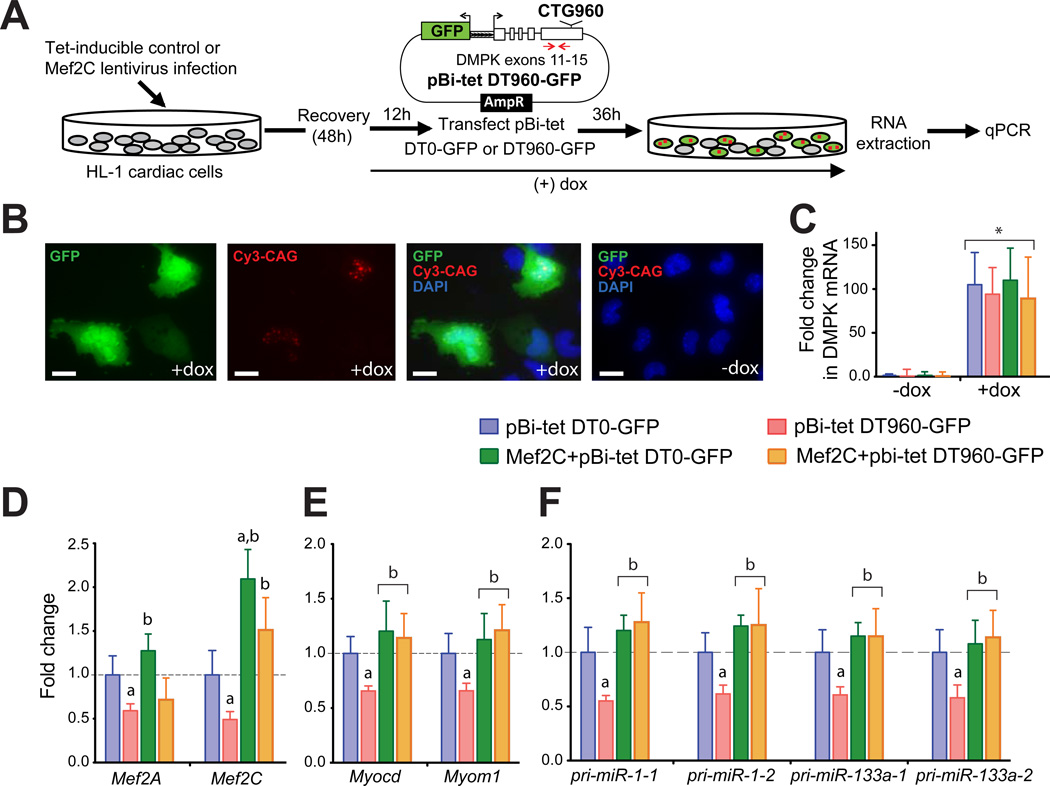
To test whether re-expression of Mef2 could rescue the loss of expression of select miRNA and mRNA targets in DM1, we infected HL-1 cells with control or tet-inducible Mef2c lentivirus that co-expresses an rtTA trans activator(Figure 6A). After a 12h induction of Mef2c expression with doxycycline, cells were transfected with pBi-tet-DT0-GFP or -DT960-GFP plasmids. These plasmids express GFP and the DMPK 3’-UTR with or without 960 CUG repeats through a tet-inducible bidirectional promoter(Figure 6A)(Lee et al., 2012). Thirty-six hours later, cells were either fixed for combined FISH/immunofluorescence analysis or lysed to extract total RNA. As shown previously(Lee et al., 2012)we found that most transfected cells that expressed GFP also formed CUG repeat-containing RNA foci(Figure 6B). GFP expression or foci formation was not observed in the absence of doxycycline (Figure 6B and C), and no significant differences in DMPK mRNA levels were noted upon exogenous Mef2c expression (Figure 6C).
Figure 6. Rescued expression of Mef2 miRNA and mRNAs targets in a cardiac cell DM1 model by exogenous Mef2c.
(A) Experimental schematic of Mef2c rescue in CUGexp RNA expressing cardiac cells. (B) GFP expression was detected by indirect fluorescence using anti-GFP antibody. RNA foci containing DT960 RNA were detected by FISH using Cy3-labeled probes. Nuclei were counterstained with DAPI. GFP expression or RNA foci formation was not detected in the absence of dox. All images were taken at the same exposure time. Scale bars: 20 m. (C) Induction of DT0 and DT960 containing DMPK mRNA after dox treatment. Each bar represents fold change in expression relative to the DT0 control without dox treatment. Positions of qRT-PCR primers used to quantitate mRNA expression are indicated with red arrows. Data is normalized relative to Rpl30. *P< 0.05. Reduced (D) Mef2A and Mef2C; (E) Myocd and Myom1; and (F) pri-miR-1-1, 1-2, 133a-1 and 133a-2 steady state levels in response to CUGexp RNA expression. Exogenous Mef2c restores the mRNA and miRNA expression in CUGexp RNA expressing cells. Each bar represents fold change in expression relative to DT0 control. a, significantly different from DT0 (infected with control virus). b, significantly different from DT960 (infected with control virus). (G) Model of Mef2-miRNA circuitry in normal and DM1 cardiac cells. In healthy adult cardiomyocytes, Mef2 proteins coordinate expression of cardiac-enriched mRNA and miRNA genes. miRNAs post-transcriptionally down regulate expression of multiple gene targets to maintain adult cardiac gene expression program. This transcriptional circuit becomes defective in DM1 (black panel); Mef2 levels are reduced in response to CUGexp RNA expression, which results in loss of expression of Mef2-driven cardiac genes and miRNAs resulting in developmental reprogramming of gene expression.
Similar to the DM1 mouse model and the human patient samples, transient transfection of HL-1 cells with the DT960 plasmid led to a significant decrease (P<0.05) in endogenousMef2a and Mef2ctranscript levels when compared to the DT0 plasmid (Figure 6D). Forced expression of exogenous Mef2c in the DM1 cardiac cell model not only increased the Mef2a and Mef2ctranscript levels (Figure 6D) but also rescued the expression of its mRNA and miRNA targets (Figure 6E and F). These results thus provide direct evidence that mis-regulation of the cardiac Mef2 regulatory network plays a fundamental role in the pathological response to CUGexp RNA in DM1 hearts.
DISCUSSION
We demonstrated a hierarchical relationship between expression of CUGexp RNA and loss of Mef2 activity using two independent experimental systems (inducible heart-specific DM1 mouse model and a DM1 cardiac cell culture model) and validated the results in DM1 heart tissue. CUGexp RNA leads to an overall decrease in MEF2 expression and decreased expression of MEF2 miRNAs and mRNA targets resulting in global reprogramming of the cardiac transcriptome. Our results identify several miRNA families that are deregulated in DM1 heart tissues. This is predicted to have a cascade effect as individual miRNAs can target multiple mRNAs (Bartel, 2009) and therefore, modulate DM1 phenotype by regulating functionally related networks. For instance, miR-1 is known to regulate gap junction proteins and cardiac channels including Gja1, Cacna1c and Kcnd2 and a greater than 50% reduction in its expression may directly contribute to the conduction defects seen in DM1 (Rau et al., 2011; Zhao et al., 2007). This is consistent with a previous report where genetic loss of one of the two miR-1 family members, resulted in a range of cardiac abnormalities, including postnatal electrophysiological defects with a spectrum of cardiac arrhythmias (Zhao et al., 2007).
Interstitial fibrosis is another important feature of DM1 heart tissue(Nazarian et al., 2010). Our study identified dysregulation of a network of four miRNA families that may be directly responsible for this phenotype. CUGexp RNA expression leads to an up regulation of miR-21 and down regulation of miR-29, miR-30 and miR-133 family members. miR-21 is known to repress the Sprouty homolog 1 (3-fold down regulated in our study), a negative regulator of ERK-MAP kinase signaling, thereby leading to fibrosis (Thum et al., 2008). miR-29 represses expression of collagens (van Rooij et al., 2008), many of which are up regulated in our microarray study, and miR-30 and −133 repress expression of the connective tissue growth factor (Duisters et al., 2009), a positive regulator of fibrosis (4.8 fold up regulated in our study). Thus, the results from this study show tight reciprocal relationships between gain and loss of these four miRNAs and their target genes that support the critical role of this core network in DM1 cardiac fibrosis.
One of the clear down stream implications of miRNA dysfunction in DM1 is that reduced miR-23a/b levels lead to increased expression of its target, CELF1 protein. The miR-23a/b family regulates post-transcriptional loss of Celf1 protein during mouse postnatal heart development (Kalsotra et al., 2010). Reduced levels of both miR-23a and −23b in DM1 heart tissue, therefore is expected to result in an overall increase in CELF1 protein levels thus contributing to mis-regulation of CELF1 splicing targets. Together, these data indicate that dysregulation of specific miRNAs are likely to contribute to specific cardiac phenotypes observed in DM1.
MBNL1 and CELF1 are RNA binding proteins that are required for alternative splicing regulation during normal skeletal muscle and heart development (Kalsotra et al., 2008; Lin et al., 2006). Disruption of their functions by CUGexp RNA results in mis-splicing of their pre-mRNA targets such that adult tissues express embryonic splice forms. It was recently described that reduced expression of miR-1 in DM1 patients is due in part to mis-processing of pre-miR-1(Rau et al., 2011). It was proposed that MBNL1 binding within the loop of pre-miR-1 disrupts LIN28 binding to this region and thereby promotes its processing by dicer (Rau et al., 2011). Our data argue against this model as we show that: (i) primary and mature miRNAs exhibit a parallel decrease in expression in heart tissues from DM1 mouse model and patient samples; and (ii) Mbnl1 knock out mice do not show a significant change in miR-1 and many other miRNAs in heart. Instead, we provide evidence that a select set of miRNAs in DM1, including miR-1, is down regulated due to a reduced MEF2 transcriptional program.
The MEF2 paralogues are a conserved family of proteins that bind to a consensus DNA sequence CTA(A/T)4TAG in the promoter region of target genes (Molkentin and Olson, 1996). Although MEF2 proteins are expressed in various tissues, the expression of the Mef2 target genes in mouse is highest in skeletal muscle, heart, and brain (Potthoff and Olson, 2007). In addition to Gata4 and Tbx5, Mef2c is a key transcription factor required for direct reprogramming of cardiac fibroblasts into induced cardiomyocytes (Ieda et al., 2010; Qian et al., 2012). This study identifies loss of Mef2 activity as causal to deregulation of many miRNAs and mRNAs in a DM1 cardiac cell culture model and heart tissue from DM1 mouse model. Reduced levels of Mef2a and Mef2c in response to transient expression of repeats in cultured cardiac cells argues for a direct effect of CUGexp RNA in reduction of Mef2 levels. The results we obtained in DM1 heart tissue are in contrast to results from microarray studies showing increased expression of MEF2A and MEF2C in skeletal muscle from DM1 as well as other neuromuscular disorders (Bachinski et al., 2010) and suggest different pathogenic effects of CUGexp RNA in heart and skeletal muscle. In summary, our data supports a model in which nuclear accumulation of CUGexp RNA in DM1 affects a MEF2-miRNA regulatory circuit such that reduced MEF2 activity results in loss of expression of its miRNA and mRNA targets in cardiac cells. The specific mechanism by which CUGexp RNA affects mRNA and protein levels of MEF2 paralogs remains to be determined.
EXPERIMENTAL PROCEDURES
Animal models and human tissue samples
Tamoxifen (Tam)-inducible and heart-specific EpA960; MCM DM1 mouse model was described previously (Wang et al., 2007). CUGexp RNA was induced in 2–4 month old EpA960; MCM bitransgenic animals with a single or five consecutive daily intra-peritoneal injections of 20 mg/kg Tam (Sigma-Aldrich). Tet-inducible CELF1 bitransgenic mice (TRECUGBP1/Myh6-rtTA) were previously described (Kalsotra et al., 2008). All experiments were conducted in accordance with the NIH Guide for the Use and Care of Laboratory Animals and approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine. Human tissue or RNA samples were provided by Drs. C. Thornton and T. Ashizawa, X. Wehrens, NDRI and the University of Miami Tissue bank. DM1 samples: 50 yr male (RF), 48 yr female (1500 repeats, RF), 55 yr male (PN), 52 yr female (>1000 repeats, RF), 46 yr male (PN), 50 yr female (RF), 53 yr male (unknown cause), 26 yr male (glioma), 55 yr male (pulmonary embolism); RF-respiratory failure, PN-pneumonia. Normal heart samples: range from 21–55 yr, pooled samples. Heart failure samples: 44 yr old male, 18 yr old male, and 51 yr old female.
Quantitative miRNA profiling
TaqMan stem-loop RT-PCR MicroRNA Arrays (Applied Biosystems) were used to quantify mature miRNA expression. Briefly, 500 ng of total RNA from each sample was reverse-transcribed using Megaplex RT Primers and the TaqMan miRNA reverse transcription kit. Quantitative real-time PCR reactions were performed in triplicate in a 384-well plate on a 7900HT Real-Time PCR System using ABI TaqMan Universal PCR Master Mix. An initial denaturation step of 10 min at 95°C was followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Cycle threshold (Ct) values were calculated using the SDS software v.2.3 using automatic baseline settings and a threshold of 0.2. Only the miRNAs with a Ct • 35 were included in the analyses. The Ct value of an endogenous control gene (MammU6) was subtracted from the corresponding Ct value for the target gene resulting in the Ct value, which was used for relative quantification. Fold change of miRNA expression was calculated by the equation 2− Ct.
Microarrays
Total RNA was prepared using RN easy Kit (Qiagen) and the RNA quality was tested with Agilent Bioanalyzer 2100. Total RNA was amplified and labeled using the Illumina Total Prep RNA Amplification Kit (catalog AMIL1791, Ambion) and 500ng of cRNA were applied to Illumina Mouse WG-6 v2 Whole-Genome Expression Bead chips. Following hybridization, washing and detection, chips were scanned using the Illumina 500GX scanner at the Genomics and Proteomics Core Laboratory at Texas Children’s hospital. Expression values were quantile normalized. Genes were selected that showed differential expression between 1 wk and MCM (t test p<0.01 and fold change >1.5) or between 72h and MCM; expression values were clustered as previously described (Creighton et al., 2008)
Protein and mRNA expression analysis
Normal (non DM1) and DM1 human heart tissue lysates were prepared and protein concentrations were determined by BCA assays (Pierce). 50 µg of lysate from each sample were separated on 10% SDS-PAGE followed by western blot. MEF2A (54 kDa; Cell signaling) CELF1 (50 kDa; 3B1), and GAPDH (36 kDa; Abcam) antibodies were used at 0.5-2 mg/mL dilution as previously described (Kalsotra et al., 2008). Anti-mouse IgG-HRP (Invitrogen, 1:5000) or Anti-rabbit IgG-HRP (Calbiochem; 1:10000) were used as a secondary antibodies. After appropriate washing in PBST (0.1% Tween 20), immunoreactivity was detected by using HRP-chemiluminescence system (Pierce). Total RNA was prepared from human heart samples using Trizol. Steady state mRNA expression was measured by qRT-PCR as previously described (Kalsotra et al., 2008). All individual pri-miRNA, miRNA and mRNA qRT-PCR assays were performed using pre-designed TaqMan primers and probes (Applied Biosystems) according to the manufacturer’s instructions.
Alternative splicing assays
Total RNA (0.3–0.5 µg) was used for RT-PCR as described (Kalsotra et al., 2010). Primer sequences for detecting alternative splicing of Mef2a (α & β•••••) and Mef2c (γ••••)are provided in Table S3. Percent spliced in (PSI) values for the variable region were calculated using Kodak Gel logic 2200 and Molecular Imaging Software as: [(inclusion band)/ (inclusion band + exclusion band) × 100].
Cell Culture and transfections
HL-1 cells were cultured on gelatin (0.02%, w/v)/fibronectin (10µg/ml) coated plates and maintained in Claycomb medium (JRH Biosciences) as previously described (Kalsotra et al., 2010). The HL-1 cells were infected in T25 flasks with tet-inducible control or Mef2c expressing virus in presence of 5µg/ml polybrene. After 48h of recovery, the cells were switched to 1µg/ml doxycycline containing media for 12 hrs. Next, the cells were transiently transfected with pBi-tet DT0-GFP or pBi-tet DT960-GFP plasmids with Lipofectamine 2000 using manufacturer’s instructions. Thirty-six hours later cells were harvested to isolate total RNA or were fixed in 4% paraformaldehyde and permeabilized with 0.02% Triton X-100 in PBS. CUG transcripts were detected using (CAG)5-Cy3-labeled LNA probes (Exiqon) as described(Wang et al., 2007). Nuclei were stained with DAPI using Vectashield (Vector).
Chromatin immunoprecipitation (ChIP) assays
Mef2-ChIP was performed using Imprint® Chromatin Immunoprecipitation Kit (Sigma) according to the manufacturer’s instructions with minor modifications. Three mouse hearts each from wild type adults, MCM mice given tamoxifen for a week, and DM1 mice induced to express CUG repeat RNA for a week were collected in cold PBS, chopped into smaller on ice and then incubated in 1% formaldehyde in PBS for 10 minutes at room temperature. Formaldehyde cross-linking was stopped by adding 10X Glycine to a final concentration of 1X and incubating at room temperature for 5 minutes. Tissue was spun at 4°C at 220g for 5 minutes and the remaining tissue pellet was rinsed twice in ice-cold PBS. Tissues were harvested and lysed to isolate nuclei in a hypotonic buffer, then re-suspended, lysed in lysis buffer, and sonicated in 15 ml tubes with Bioruptor UCD-200 Diagenode (ultrasonic wave output power 250W, 14 × 30 seconds) to yield chromatin size of 200–400 bp. ChIP was performed two times with 2µg of anti-RNA Pol II rabbit polyclonal antibody (Santa Cruz, sc-900) and anti-pan Mef2A and Mef2C goat polyclonal antibody (Santa Cruz, sc-313), anti-Mef2c specific rabbit polyclonal antibody (Cell signaling, 5030) and 2µg of normal rabbit IgG (Santa Cruz, sc-2027) or goat IgG (Santa Cruz, sc-2028) as isotype controls. Co-precipitated DNA was then analyzed by qRT-PCR performed with Sybr Green mix (Applied Biosystems). The primers used are listed in Table S3.
Statistics
Data are presented as mean ± SD. Statistical significance was determined using a two-tailed Student’s t-test or one-way ANOVA followed by post-hoc Tukey’s multiple range tests. P value of less than 0.05 was considered significant.
Supplementary Material
HIGHLIGHTS.
The MEF2 regulatory network is disrupted in DM1 cardiac tissue
Altered expression is specific to DM1 and not a general response to cardiac injury
Multiple new miRNA genes identified as MEF2 targets in postnatal heart development
miRNA expression is rescued in a DM1 cardiomyocyte model by MEF2C expression
ACKNOWLEDGEMENTS
We thank Donnie Bundman, Marissa A Ruddy, Yiqun Zhang, and Chaitali Chakraborty (BCM) for technical assistance. We thank Dr. J. Molkentin for providing MCM mice, Dr. C. Thornton for providing Mbnl1 3/ 3 mice and tissue samples, Dr. Tetsuo Ashizawa for tissue samples, Dr. A. Rodriguez and Dr. X. Wehrens for providing heart tissue from CnA-Tg mice and TAC operated mice respectively. AK was supported by a Myotonic Dystrophy Foundation postdoctoral fellowship and a Scientist Development Grant (11SDG4980011) from the American Heart Association. RKS is supported by the postdoctoral fellowship from American Heart Association. CJC is supported in part by National Institutes of Health grant P30 CA125123. This project is funded by the National Institutes of Health (R01HL045565, R01AR060733, and R01AR045653) and Muscular Dystrophy Association grants to TAC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
All raw microarray data files are available for download from NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/), accession number GSE48991.
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures, three tables, and Supplemental References.
Authors’ contributions. AK designed research, performed the experiments, analyzed the data and wrote the manuscript. RKS performed experiments, analyzed data and contributed to the manuscript. PG performed experiments and contributed to the manuscript. AJW isolated heart tissues of EpA960; MCM animals and its controls and performed alternative splicing analysis. CJC analyzed the microarray data and contributed to the manuscript. TAC supervised and designed research, analyzed the data and wrote the manuscript.
REFERENCES
- Bachinski LL, Sirito M, Böhme M, Baggerly KA, Udd B, Krahe R. Altered MEF2 isoforms in myotonic dystrophy and other neuromuscular disorders. Muscle Nerve. 2010;42:856–863. doi: 10.1002/mus.21789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton CJ, Casa A, Lazard Z, Huang S, Tsimelzon A, Hilsenbeck SG, Osborne CK, Lee AV. Insulin-like growth factor-I activates gene transcription programs strongly associated with poor breast cancer prognosis. J Clin Oncol. 2008;26:4078–4085. doi: 10.1200/JCO.2007.13.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duisters RF, Tijsen AJ, Schroen B, Leenders JJ, Lentink V, van der Made I, Herias V, van Leeuwen RE, Schellings MW, Barenbrug P, et al. miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res. 2009;104:170–178. doi: 10.1161/CIRCRESAHA.108.182535. [DOI] [PubMed] [Google Scholar]
- Fernandez-Costa JM, Garcia-Lopez A, Zuniga S, Fernandez-Pedrosa V, Felipo-Benavent A, Mata M, Jaka O, Aiastui A, Hernandez-Torres F, Aguado B, et al. Expanded CTG repeats trigger miRNA alterations in Drosophila that are conserved in myotonic dystrophy type 1 patients. Hum Mol Genet. 2013;22:704–716. doi: 10.1093/hmg/dds478. [DOI] [PubMed] [Google Scholar]
- Groh WJ, Groh MR, Saha C, Kincaid JC, Simmons Z, Ciafaloni E, Pourmand R, Otten RF, Bhakta D, Nair GV, et al. Electrocardiographic abnormalities and sudden death in myotonic dystrophy type 1. N Engl J Med. 2008;358:2688–2697. doi: 10.1056/NEJMoa062800. [DOI] [PubMed] [Google Scholar]
- Heatwole C, Bode R, Johnson N, Quinn C, Martens W, McDermott MP, Rothrock N, Thornton C, Vickrey B, Victorson D, et al. Patient-reported impact of symptoms in myotonic dystrophy type 1 (PRISM-1) Neurology. 2012;79:348–357. doi: 10.1212/WNL.0b013e318260cbe6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra A, Wang K, Li PF, Cooper TA. MicroRNAs coordinate an alternative splicing network during mouse postnatal heart development. Genes Dev. 2010;24:653–658. doi: 10.1101/gad.1894310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, Cooper TA. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci U S A. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. A muscle blind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- Koshelev M, Sarma S, Price RE, Wehrens XH, Cooper TA. Heart-specific overexpression of CUGBP1 reproduces functional and molecular abnormalities of myotonic dystrophy type 1. Hum Mol Genet. 2010;19:1066–1075. doi: 10.1093/hmg/ddp570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased Steady-State Levels of CUGBP1 in Myotonic Dystrophy 1 Are Due to PKC-Mediated Hyperphosphorylation. Mol Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus A, Varin J, Babuty D, Anselme F, Coste J, Duboc D. Long-term follow-up of arrhythmias in patients with myotonic dystrophy treated by pacing: a multicenter diagnostic pacemaker study. J Am Coll Cardiol. 2002;40:1645–1652. doi: 10.1016/s0735-1097(02)02339-2. [DOI] [PubMed] [Google Scholar]
- Lee JE, Bennett CF, Cooper TA. RNase H-mediated degradation of toxic RNA in myotonic dystrophy type 1. Proc Natl Acad Sci U S A. 2012;109:4221–4226. doi: 10.1073/pnas.1117019109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Cooper TA. Pathogenic mechanisms of myotonic dystrophy. Biochem Soc Trans. 2009;37:1281–1286. doi: 10.1042/BST0371281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Miller JW, Mankodi A, Kanadia RN, Yuan Y, Moxley RT, Swanson MS, Thornton CA. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum Mol Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, Richardson JA, Bassel-Duby R, Olson EN. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci U S A. 2007;104:20844–20849. doi: 10.1073/pnas.0710558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Castel A, Nakamori M, Tome S, Chitayat D, Gourdon G, Thornton CA, Pearson CE. Expanded CTG repeat demarcates a boundary for abnormal CpG methylation in myotonic dystrophy patient tissues. Hum Mol Genet. 2011;20:1–15. doi: 10.1093/hmg/ddq427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin JD, Olson EN. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:9366–9373. doi: 10.1073/pnas.93.18.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley ML, Zu T, Ikeda Y, Gao W, Mosemiller AK, Daughters RS, Chen G, Weatherspoon MR, Clark HB, Ebner TJ, et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat Genet. 2006;38:758–769. doi: 10.1038/ng1827. [DOI] [PubMed] [Google Scholar]
- Nazarian S, Bluemke DA, Wagner KR, Zviman MM, Turkbey E, Caffo BS, Shehata M, Edwards D, Butcher B, Calkins H, et al. QRS prolongation in myotonic muscular dystrophy and diffuse fibrosis on cardiac magnetic resonance. Magn Reson Med. 2010;64:107–114. doi: 10.1002/mrm.22417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelargonio G, Dello Russo A, Sanna T, De Martino G, Bellocci F. Myotonic dystrophy and the heart. Heart. 2002;88:665–670. doi: 10.1136/heart.88.6.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbellini R, Greco S, Sarra-Ferraris G, Cardani R, Capogrossi MC, Meola G, Martelli F. Dysregulation and cellular mislocalization of specific miRNAs in myotonic dystrophy type 1. Neuromus Disorders. 2011;21:81–88. doi: 10.1016/j.nmd.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Phillips MF, Harper PS. Cardiac disease in myotonic dystrophy. Cardiovasc Res. 1997;33:13–22. doi: 10.1016/s0008-6363(96)00163-0. [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu JD, Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau F, Freyermuth F, Fugier C, Villemin JP, Fischer MC, Jost B, Dembele D, Gourdon G, Nicole A, Duboc D, et al. Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nat Struct Mol Biol. 2011;18:840–845. doi: 10.1038/nsmb.2067. [DOI] [PubMed] [Google Scholar]
- Salehi LB, Bonifazi E, Stasio ED, Gennarelli M, Botta A, Vallo L, Iraci R, Massa R, Antonini G, Angelini C, et al. Risk prediction for clinical phenotype in myotonic dystrophy type 1: data from 2,650 patients. Genet Test. 2007;11:84–90. doi: 10.1089/gte.2006.0511. [DOI] [PubMed] [Google Scholar]
- Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- Sicot G, Gourdon G, Gomes-Pereira M. Myotonic dystrophy, when simple repeats reveal complex pathogenic entities: new findings and future challenges. Hum Mol Gen. 2011;20:R116–R123. doi: 10.1093/hmg/ddr343. [DOI] [PubMed] [Google Scholar]
- Sohal DS, Nghiem M, Crackower MA, Witt SA, Kimball TR, Tymitz KM, Penninger JM, Molkentin JD. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ Res. 2001;89:20–25. doi: 10.1161/hh1301.092687. [DOI] [PubMed] [Google Scholar]
- Sovari AA, Bodine CK, Farokhi F. Cardiovascular manifestations of myotonic dystrophy-1. Cardiol Rev. 2007;15:191–194. doi: 10.1097/CRD.0b013e318070d1a7. [DOI] [PubMed] [Google Scholar]
- Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- Timchenko L. Molecular mechanisms of muscle atrophy in myotonic dystrophies. The Internat. J Biochem Cell Biol. 2013 doi: 10.1016/j.biocel.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko NA, Cai ZJ, Welm AL, Reddy S, Ashizawa T, Timchenko LT. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J Biol Chem. 2001;276:7820–7826. doi: 10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- Udd B, Krahe R. The myotonic dystrophies: molecular, clinical, and therapeutic challenges. Lancet Neurology. 2012;11:891–905. doi: 10.1016/S1474-4422(12)70204-1. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GS, Kearney DL, De Biasi M, Taffet G, Cooper TA. Elevation of RNA-binding protein CUGBP1 is an early event in an inducible heart-specific mouse model of myotonic dystrophy. J Clin Invest. 2007;117:2802–2811. doi: 10.1172/JCI32308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TM, Thornton CA. Myotonic dystrophy: RNA-mediated muscle disease. Curr Opin Neurol. 2007;20:572–576. doi: 10.1097/WCO.0b013e3282ef6064. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, Margolis J, Peterson M, Markowski TW, Ingram MA, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.