Abstract
Approximately 80–90% of all retinoids in the body are stored as retinyl esters (REs) in the liver. Adipose tissue also contributes significantly to RE storage. The present studies, employing genetic and nutritional interventions, explored factors that are responsible for regulating RE accumulation in the liver and adipose tissue and how these influence levels of retinoic acid (RA) and RA-responsive gene expression. Our data establish that acyl-CoA:retinol acyltransferase (ARAT) activity is not involved in RE synthesis in the liver, even when mice are nutritionally stressed by feeding a 25-fold excess retinol diet or upon ablation of cellular retinol-binding protein type I (CRBPI), which is proposed to limit retinol availability to ARATs. Unlike the liver, where lecithin:retinol acyltransferase (LRAT) is responsible for all RE synthesis, this is not true for adipose tissue where Lrat-deficient mice display significantly elevated RE concentrations. However, when CrbpI is also absent, RE levels resemble wild-type levels, suggesting a role for CrbpI in RE accumulation in adipose tissue. Although expression of several RA-responsive genes is elevated in Lrat-deficient liver, employing a sensitive liquid chromatography tandem mass spectrometry protocol and contrary to what has been assumed for many years, we did not detect elevated concentrations of all-trans-RA. The elevated RA-responsive gene expression was associated with elevated hepatic triglyceride levels and decreased expression of Pparδ and its downstream Pdk4 target, suggesting a role for RA in these processes in vivo.
Keywords: diacylglycerol acyltransferase 1, cellular retinol-binding protein type I, 9-cis-retinoic acid or 9-cis-RA, retinol-binding protein or RBP4
Retinoids (vitamin A and its analogs) are important transcriptional regulators that act primarily through three nuclear hormone receptors, retinoic acid receptor (RAR)α, RARβ, and RARγ, to modulate the activities of more than 500 genes (1–3). There is also some, albeit controversial, evidence that retinoic acid (RA) is a physiological ligand contributing importantly to the regulation of peroxisome proliferator-activated receptor-δ (PPARδ)-mediated gene expression (4, 5). The great majority of retinoids present in a healthy well-nourished vertebrate are in the form of retinyl esters (REs) (6–8). REs are also found in the postprandial circulation, where they are present in chylomicrons and chylomicron remnants, and in the fasting circulation, where they are present at relatively low levels in very low density lipoproteins (VLDLs) (6–8). Many tissues have some capacity to synthesize REs from retinol, but REs are most abundant in the liver where approximately 80–90% of the body's retinoids are found, primarily in hepatic stellate cells (7–10). REs are also relatively abundant in the eyes, lungs, skin, and adipose tissue (7–10). In times of insufficient dietary vitamin A intake, RE stores undergo enzymatic hydrolysis to retinol which is then secreted into the circulation bound to retinol-binding protein (RBP4)2 (6–10). The accumulation of RE stores within the liver and other tissues relieves the organism from the obligate need to acquire this essential micronutrient regularly from its diet; thus providing an evolutionary advantage to the organism.
The literature, based on in vitro studies, indicates that at least two distinct enzymatic activities present in the liver and other tissues are able to catalyze RE formation; one involves an acyl-CoA-dependent mechanism and the other an acyl-CoA-independent process (11–15). The acyl-Co-A-dependent mechanism is attributed to an acyl-CoA:retinol acyltransferase (ARAT) activity (9–12). The acyl-CoA-independent synthesis of REs was shown early on to involve the transesterification of a fatty acyl group from the sn-1 position of a membrane phosphatidylcholine to retinol, and the enzyme responsible for this transesterification was termed lecithin:retinol acyltransferase (LRAT) (13–15). Studies of Lrat-deficient (Lrat−/−) mice have established that LRAT is responsible for most, but not all, REs synthesized in the body (16–18). Very few REs can be detected in the liver, eyes, or lungs of Lrat−/− mice fed a control chow diet (17). The sole tissue where substantial REs accumulate in Lrat−/− mice is adipose tissue, where concentrations of REs are elevated by 2- to 3-fold over those measured in matched wild-type (WT) mice (17, 18).
Several groups have reported in vitro studies demonstrating that recombinant diacylglycerol acyltransferase 1 (DGAT1), an enzyme that catalyzes the formation of triglyceride from diglycerides and fatty acyl-CoAs (19–21), also catalyzes the acyl-CoA-dependent esterification of retinol (17, 22, 23). Thus, DGAT1 possesses ARAT activity. Subsequently, through investigations of Dgat1-deficient (Dgat1−/−), Lrat−/−, and Lrat−/−/Dgat1−/− mice, we established that DGAT1 can act as a physiologically important ARAT in mouse intestine (24). Shih et al. (25), through study of Dgat1−/− mice, demonstrated that DGAT1 acts physiologically in catalyzing RE synthesis in mouse skin. Dgat1 is highly expressed in the liver, where it has been shown to have a role in hepatic triglyceride synthesis (19, 20, 26). Dgat1 is also highly expressed in adipose tissue (26). The literature suggests that ARAT activities may become active only in times of excessive retinol availability (27–29). The literature also proposes that cellular retinol-binding proteins (CRBPs) prevent retinol from being delivered to ARAT activities (13–15, 28, 29). The liver is very much enriched in CRBP type I (CRBPI), and adipose tissue expresses both CRBPI and CRBPIII (28, 30, 31). Moreover, the literature proposes that the ability to esterify retinol contributes to regulation of retinol conversion to RA (32, 33). The present studies are aimed at gaining understanding of the roles of DGAT1 and its ARAT activity and CRBPI in hepatic and adipose RE synthesis, and of how RE formation may influence RA availability and RA-responsive gene expression.
MATERIALS AND METHODS
Animals, animal husbandry, and diets
The mutant mouse lines we employed have all been described in the literature and include Lrat−/− (16, 17), CrbpI−/− (34), Dgat1−/− (35), Rbp4−/− (36), and Lrat−/−/Dgat1−/− (24) mice. The Lrat−/− and CrbpI−/− mice originally described for a mixed C57Bl/6J/129sv genetic background were employed in our studies. Dgat1−/− mice were obtained from Jackson Labs in the C57Bl/6J genetic background. Using conventional breeding protocols we also generated Lrat−/−/CrbpI−/− mice. Genotypes of the mice were determined by protocols already described in the literature (16, 34–36). Routinely, animals were allowed ad libitum access to water and a standard nutritionally complete rodent chow diet (W. F. Fisher and Sons, Inc.). All mice were maintained on a 12 h dark-light cycle, with the period of darkness between 7:00 PM and 7:00 AM, in a conventional barrier facility. For our studies, male and/or female mice at 3 months of age were employed and routinely sacrificed in the morning between 10:00 and 11:00 AM. The animal experiments described in this report were conducted in accordance with the National Research Council's Guide for the Care and Use of Laboratory Animals (37) and were approved by the Columbia University Institutional Animal Care and Use Committee.
Nutritional manipulations
For nutritional studies, mice were maintained on a chow diet (25 IU of retinol/g of diet) until they reached approximately 3 months of age. At 3 months of age, mice were randomized onto one of two different diets: 1) a retinoid-sufficient (basal) diet providing 22 IU of retinol/g diet (Test Diet, 5755, St. Louis, MO); or 2) a retinoid-excess diet containing 25 times the amount of retinol in the basal diet or 550 IU of retinol/g diet. These purified diets were formulated otherwise according to the AIN-93M formulation (38). Each diet was nutritionally complete for all other nutrients and only differed in their retinol content. Depending on the specific experiment, mice were maintained continuously on one of these diets for up to 12 weeks prior to sacrifice. For these nutritional studies, diet and water were provided to all animals on an ad libitum basis until the time of sacrifice.
HPLC analysis of retinol and REs
Tissue and serum retinol and RE levels were determined by HPLC protocols described previously (24). Briefly, serum, liver, and epididymal adipose tissue were flash-frozen in liquid N2 after dissection and stored immediately at −80°C prior to analysis. Tissues were homogenized in 10 vol of PBS [10 mM sodium phosphate (pH 7.2), 150 mM sodium chloride] using a Polytron homogenizer (Brinkmann Instruments, Westbury, NY) set at half-maximal speed for 10 s. An aliquot of serum, the total adipose tissue, or a 200 μl aliquot of the liver homogenate was then treated with an equal volume of absolute ethanol containing a known amount of retinyl acetate as an internal standard. The retinoids present in the homogenates were extracted into hexane. After one backwash against doubly distilled water, the hexane extract was evaporated to dryness under a gentle stream of N2. Immediately upon reaching dryness, the retinoid-containing film was redissolved in 40 μl of benzene for injection onto the HPLC column. The extracted retinoids were separated on a 4.6 × 250 mm Ultrasphere C18 column (Beckmann, Fullerton, CA) preceded by a C18 guard column (Supelco, Bellefonte, PA) using 70% acetonitrile-15% methanol-15% methylene chloride as the running solvent flowing at 1.8 ml/min. Retinol and REs (retinyl palmitate, oleate, linoleate, and stearate) were detected at 325 nm and identified by comparing the retention times and spectral data of experimental compounds with those of authentic standards. Concentrations of retinol and REs in the tissues were quantitated by comparing integrated peak areas of each retinoid against those of known amounts of purified standards. Loss during extraction was accounted for by adjusting for the recovery of internal standard added immediately after homogenization of the samples.
LC/MS/MS analysis of RA
Serum and tissue levels of all-trans-RA were determined by ultra high-performance liquid chromatography tandem mass spectrometry (LC/MS/MS) using a Waters Xevo TQ MS ACQUITY UPLC system (Waters, Milford, MA). For this analysis, we only employed LC/MS grade acetonitrile and LC/MS grade water purchased from Thermo Fisher (Pittsburgh, PA). All-trans- and 9-cis-RA were purchased from Sigma-Aldrich. Penta-deuterated all-trans-RA was employed as an internal standard and was purchased from Toronto Research Chemicals (North York, Ontario, Canada). Retinoid concentrations were verified spectrophotometrically using published ε values (39). Tissue homogenates were extracted using the two-step acid-base extraction described by Kane et al. (40). All-trans-RA was detected and quantified using the multiple reaction monitoring mode employing the following transitions: all-trans-RA, m/z 301.16→123.00; penta-deuterated all-trans-RA, m/z 306.15→127.03; and 9-cis-RA, m/z 301.16→123.00.
Triglyceride analysis
Triglyceride concentrations were determined enzymatically using a commercial colorimetric triglyceride kit (Wako), according to the manufacturer's instructions.
RNA isolation, reverse transcription, and qualitative real-time PCR
Total RNA from the liver was isolated using the RNA-Bee (Tel-Test) reagent according to the manufacturer's instructions. Potential contaminating genomic DNA present in the liver RNA isolates was removed by DNase treatment and chromatography on RNeasy columns (Qiagen). Reverse transcription was performed using random hexamer primers to generate cDNAs according to the supplier's instructions (Invitrogen). Quantitative polymerase chain reaction (qPCR) was performed for 40 cycles for 15 s at 95°C and 60 s at 60°C using an ABI 7000 sequence detection system (Applied Biosystems). TaqMan probes and primers for Pparα, Pparγ, Pparδ, Pdk4, Chrebp, Fas, Scd1, Acc, Cpt1, Dgat1, Dgat2, Lrat, Rarβ isoform 2 (Rarβ2), cytochrome 26A1 (Cyp26A1), cytochrome 26B1 (Cyp26B1), cellular-retinoic acid-binding protein type I (CrabpI), CrabpII, and 18S transcripts were designed by and obtained from ABI (Applied Biosystems). Quantification of mRNA levels was performed by comparing the Ct value of each sample to a standard curve generated by serial dilution of the appropriate tissue cDNA. For each of these standard curves, the correlation coefficients were 0.99 or greater. Values are normalized to 18S rRNA levels.
Hepatic VLDL production and triglyceride analysis
To assess the roles or effects of LRAT, DGAT1, and RBP4 in facilitating RE incorporation into nascent VLDLs, mice were fasted for 4 h and then injected with the total lipase inhibitor P-407, at 1 mg/g body weight by ip injection (41, 42). Immediately prior to injection (0 h) and 6 h after injection (a time previously shown to assure a linear rate of triglyceride accumulation in P-407-treated mice (43), serum was obtained and processed for retinoid analysis by HPLC and triglyceride analysis as described above.
Statistical analyses
All data were analyzed for statistically significant differences using standard procedures consisting of an unpaired t-test for comparisons of two groups or an ANOVA followed by post hoc analysis if more than two groups of mice were being compared.
RESULTS
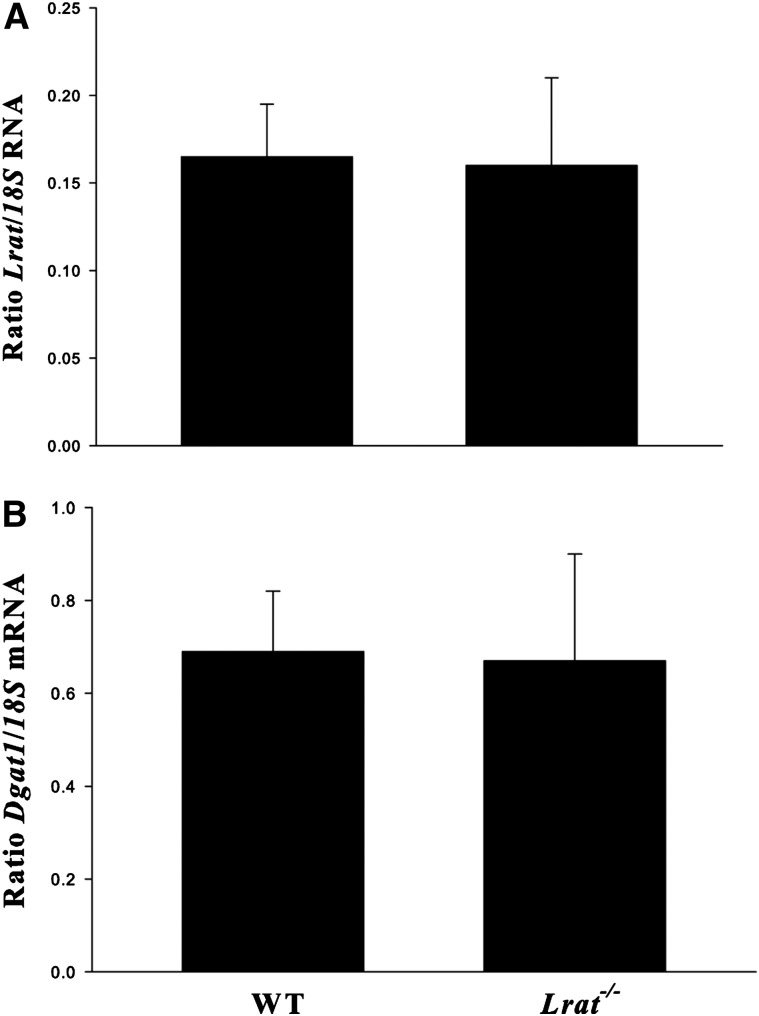
The literature has long indicated that an acyl-CoA-dependent enzymatic activity, an ARAT, present in liver homogenates, can catalyze synthesis of REs (9–12). DGAT1, which is expressed in the liver, has been shown to be a physiologically significant ARAT in the intestine and skin (24, 25). It also has been proposed in the literature that ARAT activities can contribute to RE synthesis when retinol is present in excess of normal amounts (27–29). We investigated these possibilities in matched male WT, Lrat−/−, Dgat1−/−, and Lrat−/−/Dgat1−/− mice fed a diet containing a 25-fold excess of retinol compared with standard dietary levels for 4 weeks. However, we were unable to detect substantial RE concentrations in the livers of Lrat−/− or Lrat−/−/Dgat1−/− mice (Table 1). This is contrary to what has been reported in the literature by Yamaguchi et al., who proposed, based on cell culture studies, that DGAT1 is the major contributor to the ARAT activity contributing to RE formation in hepatic stellate cells (44), the cellular site for RE storage in the liver (7, 8, 10). These investigators also reported that ablation of Dgat1 expression in cultured cells using antisense oligonucleotides results in increased expression of Lrat (44). We were unable to confirm this published finding in our studies of Dgat1−/− mice. Lrat mRNA levels assessed by qPCR for matched WT and Dgat1−/− livers were identical (Fig. 1A). Similarly, Dgat1 mRNA levels were not different for WT and Lrat−/− livers (Fig. 1B). We also attempted to confirm the published studies of Yamaguchi et al. (44) in vivo, using adenovirus constructs to rescue RE synthesis in Lrat−/− or Lrat−/−/Dgat1−/− mice. However, adenovirus rescue vectors injected into the circulation of these mice were cleared predominantly by hepatocytes with very little being taken up by hepatic stellate cells, the cellular site of retinoid storage in the liver. Consequently, it was not possible to use this standard approach for rescuing hepatic Lrat expression to further validate our findings from nutritional and genetic studies.
TABLE 1.
Hepatic RE concentrations for 3-month-old male WT, Lrat−/−, Lrat−/−/Dgat1−/−, CrbpI−/−, and Lrat−/−/CrbpI−/− mixed C57Bl/6J/129sv genetic background mice
| Strain | n | Hepatic RE (nmole/g tissue) |
| WT | 5 | 4272.0 ± 828.0 |
| Lrat−/− | 4 | 0.1 ± 0.1ab |
| Lrat−/−/Dgat−/− | 4 | 0.1 ± 0.1ab |
| CrbpI−/− | 5 | 679.5 ± 265.8ac |
| Lrat−/−/CrbpI−/− | 5 | 5.0 ± 3.1a |
Mice were maintained for 4 weeks on a diet providing 25 times more retinol than a standard vitamin A-sufficient basal diet. Prior to being placed on the excess-retinol diet, all mice were maintained from weaning on a standard vitamin A-sufficient chow diet. All values are given as mean ± SD.
P < 0.01 different from WT mice.
P < 0.05 different from CrbpI−/− mice.
P < 0.05 different from Lrat−/− mice.
Fig. 1.
Ablation of either the Lrat or the Dgat1 gene does not change the expression level of the other gene, as assessed in the livers of Dgat1−/− or Lrat−/− mice. mRNA levels of Lrat and Dgat1 were determined by qPCR for 3-month-old male chow-fed WT (n = 6) and Dgat1−/− (n = 6) mice (A) or WT (n = 8) and Lrat−/− (n = 8) mice (B). Expression levels are normalized for hepatic expression of 18S mRNA. All values are given as means ± SD. No statistically significant differences were observed.
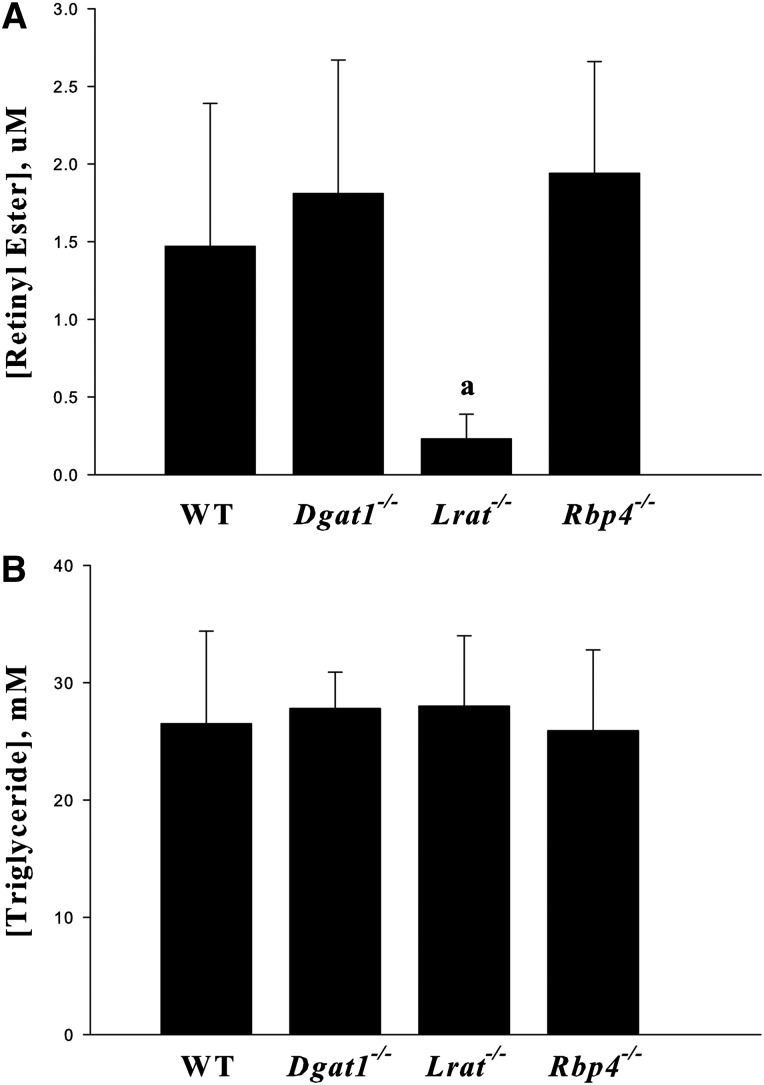
The literature indicates that DGAT1 contributes to triglyceride-rich lipoprotein (VLDL) secretion from hepatocytes (45, 46). Because REs are present in VLDLs, we asked whether DGAT1 might act to facilitate RE incorporation into VLDLs. Figure 2 provides evidence that LRAT is responsible for the synthesis of most REs that are incorporated into VLDLs and secreted from the liver. When RE concentrations were normalized for VLDL triglyceride levels, these concentrations were not different for WT or Dgat1−/− mice. Very little RE was detected in VLDLs obtained from Lrat−/− mice. Thus, LRAT-catalyzed RE formation appears to be primarily responsible for most of the REs that are incorporated into VLDLs. Interestingly, mice totally lacking expression of Rbp4, and hence unable to mobilize hepatic retinol (36), are able to mobilize REs from the liver bound to VLDL at levels that are identical to those of WT mice (Fig. 2).
Fig. 2.
LRAT but not DGAT1 accounts for synthesis of REs that is present in circulating VLDLs and the absence of RBP4 does not affect RE secretion. Serum concentrations of REs (A) and triglycerides (B) 6 h after administration of a dose of P-407 (1 g/kg body weight) for 3-month-old male WT, Lrat−/−, Dgat1−/−, and Rbp4−/− mice that had been fasted 4 h prior to P-407 administration by ip injection. All values are given as means ± SD for six mice per group. Statistical significance: a, P < 0.01 compared with WT, Dgat1−/−, or Rbp4−/− mice.
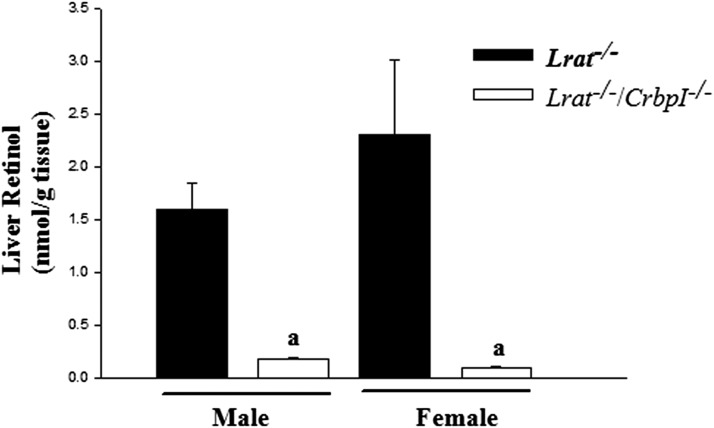
Cellular retinol-binding proteins, like CRBPI, which is highly expressed in the liver, have been proposed to sequester retinol and prevent it from being acted upon by ARAT activities (27–29). To address whether this might account for our inability to demonstrate the existence of a hepatic ARAT in vivo, we conventionally bred Lrat−/− with CrbpI−/− mice to generate mice deficient in both genes, Lrat−/−/CrbpI−/− mice. Very low levels of REs, approximately 0.12% those of littermate controls, were detected in the livers of Lrat−/−/CrbpI−/− mice fed the 25-fold excess retinol diet (Table 1). In agreement with reports by others (34), hepatic RE levels for the CrbpI−/− mice were also low, approximately 15% those of WT mice fed the 25-fold excess retinol diet. Although hepatic REs are absent in the livers of Lrat−/− mice (Table 1), retinol is still present in these livers. Interestingly, as seen in Fig. 3, hepatic retinol concentrations for male and female Lrat−/−/CrbpI−/− mice fed a control diet were markedly diminished, by 10- to 20-fold, compared with matched Lrat−/− mice. Moreover, for age- and diet-matched male and female WT mice, the hepatic retinol levels were much greater, by approximately 50-fold, than those of Lrat−/− mice; 81.5 ± 46.7 nmol/g for males and 49.3 ± 14.4 nmol/g for females. We examined both male and female mice because of known gender differences in hepatic total retinol accumulation (17).
Fig. 3.
Hepatic unesterified retinol levels are lower in Lrat−/−/CrbpI−/− mice than Lrat−/− mice. Hepatic unesterified retinol levels were measured for both 3-month-old chow-fed male and female Lrat−/− mice (n = 10 males and 4 females) and Lrat−/−/CrbpI−/− mice (n = 7 males and 5 females). All values are given as means ± SD. Statistical significance: a, P < 0.01 compared with Lrat−/− mice of the same gender.
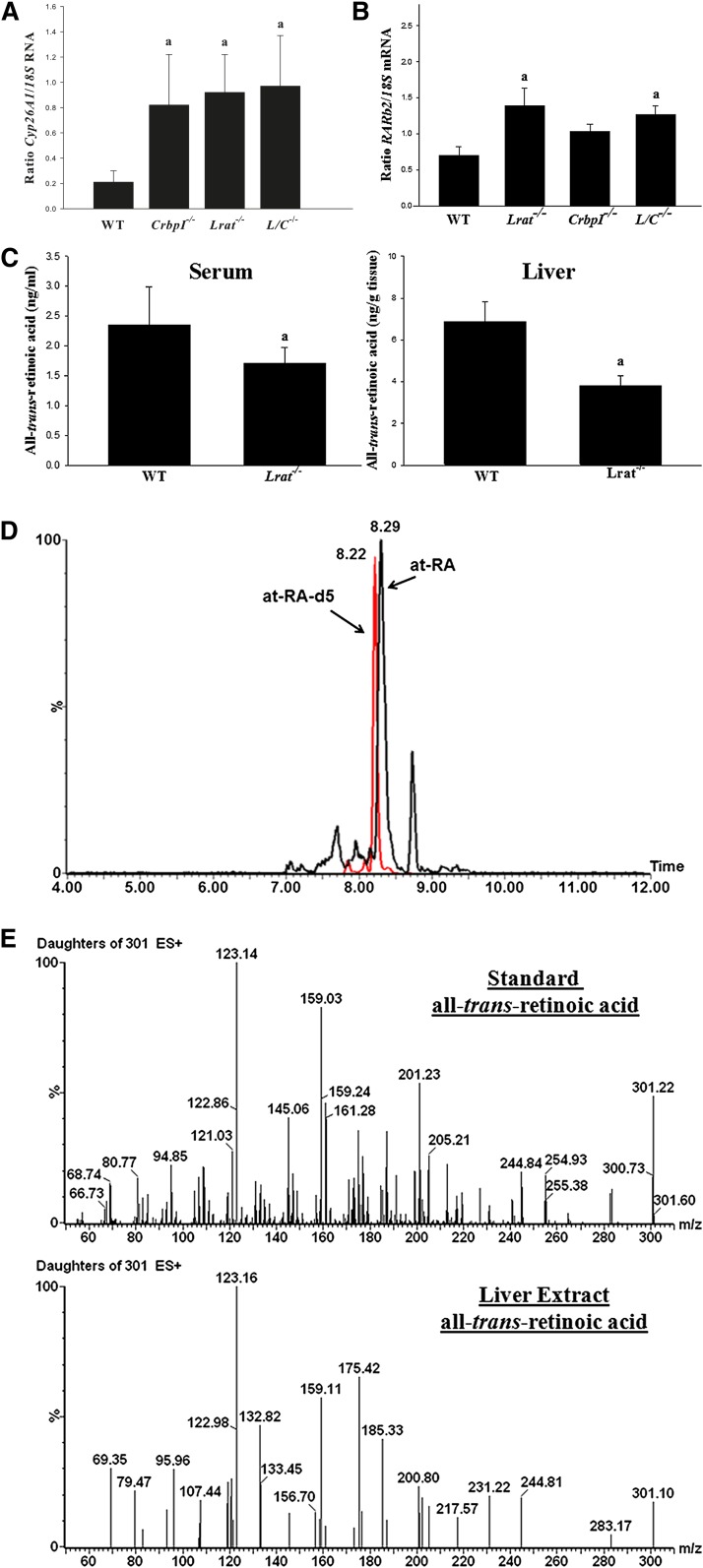
Liu and Gudas (18) reported that the expression level of Cyp26A1, an enzyme that is induced by RA and that catalyzes retinoid degradation, is upregulated in Lrat−/− mice. We were able to confirm this finding and able to extend it to CrbpI−/− and Lrat−/−/CrbpI−/− mice, which also showed elevated levels of Cyp26A1 mRNA (Fig. 4A). In addition to elevated expression of Cyp26A1, we observed statistically significant elevations in hepatic expression of another RA-inducible transcript, Rarβ2, for Lrat−/− and Lrat−/−/CrbpI−/− mice (Fig. 4B). However, we did not detect differences in hepatic mRNA expression levels of CrabpI or CrabpII. Thus, expression levels for a number of RA-inducible genes are likely elevated in the livers of these mutant mice. It is generally assumed that elevated expression levels of Cyp26A1 and Rarβ2 reflect elevated cellular all-trans-RA concentrations but, as far as we are aware, this has not been directly established. Consequently, we assessed serum and hepatic all-trans-RA concentrations for Lrat−/− and matched WT mice using very sensitive LC/MS/MS methodologies (Fig. 4C–E). Our LC/MS/MS methods allowed for a very clean separation of all-trans-RA in tissue extracts. We did not encounter any problems that might be associated with matrix effects for either the LC separations (Fig. 4D) or the fragmentation as assessed from the daughter ion spectrum of the endogeneous all-trans-RA (Fig. 4E). Surprisingly, and contrary to what has been inferred based on gene expression data, serum and hepatic steady-state concentrations of all-trans-RA were not elevated for Lrat−/− compared with WT mice (Fig. 4C). These levels were actually significantly reduced in the serum and livers of the mutant mice. This was also the case for hepatic all-trans-RA levels for CrbpI−/− and Lrat−/−/CrbpI−/− mice as well (data not shown). We take this to indicate that elevated expression of CYP26A1 results in increased catabolism and lower hepatic all-trans-RA concentrations.
Fig. 4.
A: Cyp26A1 mRNA levels are significantly elevated in the livers of 3-month-old male chow-fed CrbpI−/− (n = 5), Lrat−/− (n = 5), and Lrat−/−/CrbpI−/− (L/C−/−) (n = 7) mice compared with age- and gender-matched WT (n = 6) mice. mRNA levels were determined in triplicate for each liver by qPCR. Expression levels are normalized for hepatic expression of 18S rRNA. Statistical significance: a, P < 0.01 compared with WT mice. B: Rarβ2 mRNA levels are significantly elevated in the same livers from Lrat−/− and Lrat−/−/CrbpI−/− (L/C−/−) mice compared with WT mice. mRNA levels were determined in triplicate for each liver by qPCR. Expression levels are normalized for hepatic expression of 18S rRNA. Statistical significance: a, P < 0.05 compared with WT mice. C: Serum and liver all-trans-RA concentrations are significantly lower for Lrat−/− (n = 9) compared with WT (n = 9) mice. Statistical significance: a, P < 0.01 compared with WT mice. D: A representative LC/MS/MS profile for RA for an extract obtained for a 3-month-old male Lrat−/− liver showing the multiple reaction monitoring peaks due to all-trans-RA (at-RA, retention time 8.29 min) and penta-deuterated all-trans-RA (at-RA-d5, retention time 8.22 min) employed as the internal standard. E: Fragmentation spectra for authentic all-trans-RA standard (upper spectrum) and for the endogenous all-trans-RA detected in an Lrat−/− liver extract (lower spectrum).
We were also interested in measuring 9-cis-RA concentrations in addition to all-trans-RA by LC/MS/MS. However, 9-cis-RA was not present in the livers at a level that we felt we could accurately measure. This can be seen in the LC/MS/MS profile provided as Fig. 4D. The peak for all-trans-RA is very substantial for this liver extract, and for all other liver extracts we analyzed. There is a small peak with a retention time of approximately 8.15 min, which is the retention time at which authentic 9-cis-RA elutes. Given the size of this peak, it is possible that the small amount of 9-cis-RA present may have been formed as an artifact during extraction and processing, because it is well known that all-trans-RA can undergo some isomerization to its cis-isomers.
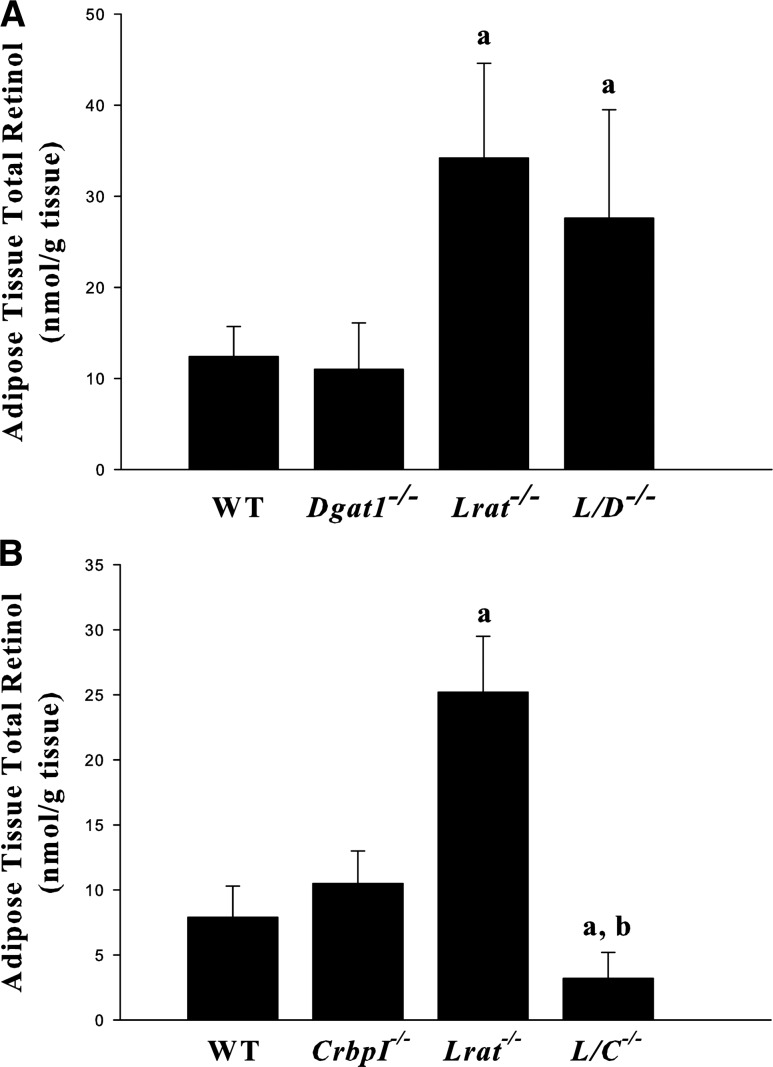
To understand whether DGAT1 is responsible for the REs present in Lrat-deficient adipose tissue, we measured total retinol levels (retinol + REs) for epididymal fat pads obtained from mice lacking both Lrat−/− and Dgat1−/−, Lrat−/−/Dgat1−/− mice. These levels were not statistically different for Lrat−/− or Lrat−/−/Dgat1−/− mice (Fig. 5A). Because CrbpI is expressed in adipose tissue, in a separate study we asked whether the absence of CrbpI affects adipose retinol levels as it does in the liver. Indeed, adipose tissue total retinol levels, which are elevated by approximately 3-fold for Lrat−/− compared with WT mice, were diminished in adipose tissue from matched Lrat−/−/CrbpI−/− mice to levels identical to WT mice (Fig. 5B). We also undertook studies to identify whether there might be differences in expression of known RA-responsive genes in adipose tissue obtained from these mice. However, unlike the liver, we did not detect statistically significant differences in mRNA expression levels for Rarβ2, Cyp26A1, or Cyp26B1 for the different mouse lines (data not shown). We also did not observe differences in Rbp4, CrabpI, or CrabpII mRNA levels between the different lines.
Fig. 5.
Epididymal adipose tissue total retinol (retinol + REs) levels. A: Total retinol levels are significantly elevated for 3-month-old male chow-fed Lrat−/− (n = 12) and Lrat−/−/Dgat1−/− (L/D−/−) (n = 13) mice compared with WT (n = 8) or Dgat1−/− (n = 4) mice. All values are given as means ± SD. Statistical significance: a, P < 0.01 compared with WT mice or Dgat1−/− mice. B: Total retinol levels are significantly lower in Lrat−/−/CrbpI−/− (L/C−/−) mice compared with WT, CrbpI−/−, or Lrat−/− mice. Epididymal adipose tissue retinol and RE levels were assessed for 3-month-old male chow-fed WT (n = 5), CrbpI−/− (n = 10), Lrat−/− (n = 8), and Lrat−/−/CrbpI−/− (n = 22) mice. All values are given as means ± SD. Statistical significance: a, P < 0.01 compared with WT mice or CrbpI−/− mice; b, P < 0.01 compared with Lrat−/− mice.
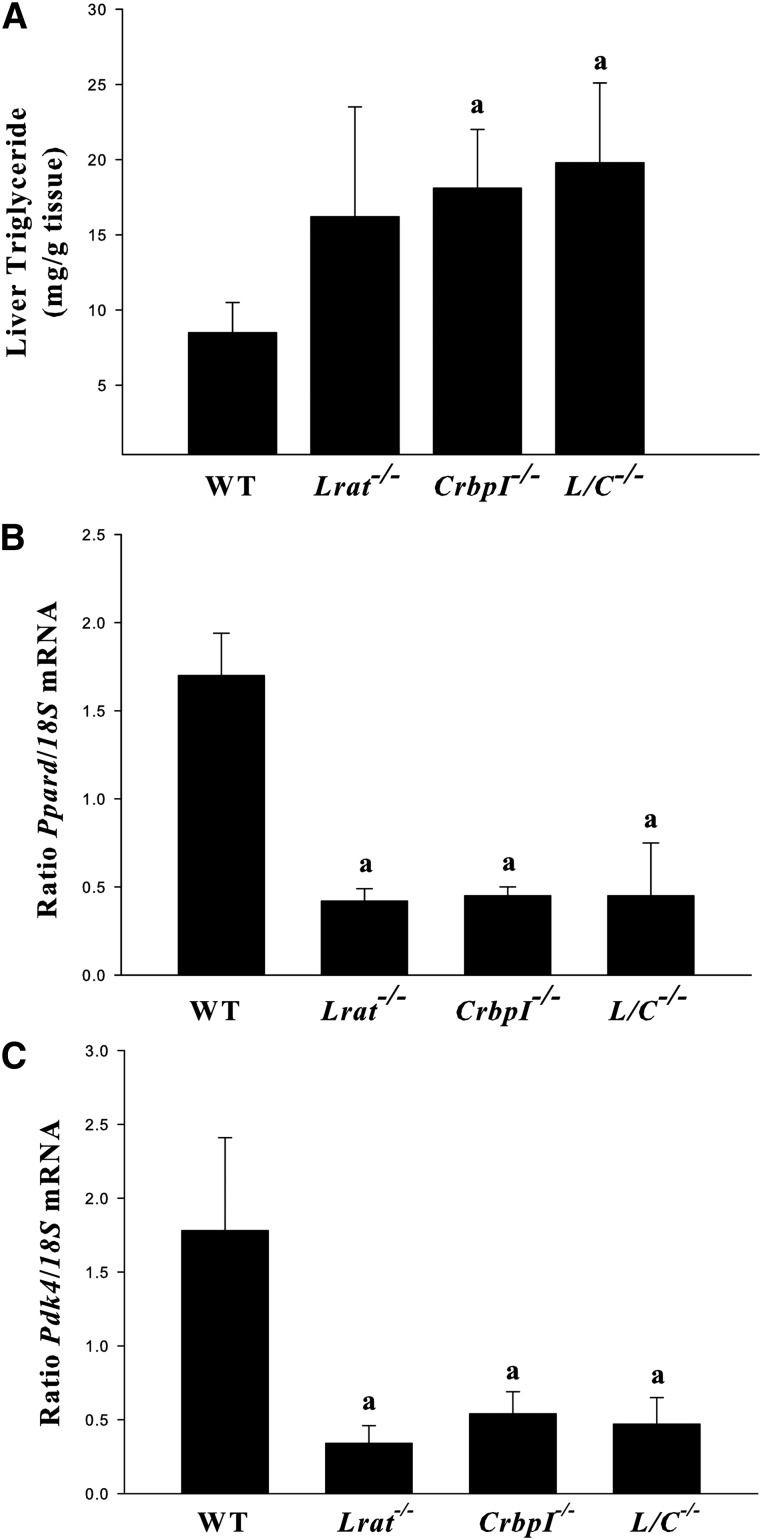
While studying the Lrat−/−/CrbpI−/− mice, we observed visually that these mice seemed to accumulate more hepatic fat than WT mice. We assessed this possibility in age- and diet-matched male WT, Lrat−/−, CrbpI−/−, and Lrat−/−/CrbpI−/− mice. Both CrbpI−/− and Lrat−/−/CrbpI−/− mice showed a statistically significant elevation in fasting triglyceride levels compared with WT mice (Fig. 6A). Although Lrat−/− mice tended to have higher hepatic fasting triglyceride concentrations than WT mice, statistical significance was not reached. To gain insight into the molecular basis for the elevated fasting triglyceride levels observed for CrbpI−/− and Lrat−/−/CrbpI−/− mice, we investigated expression of a number of key regulators of hepatic fat metabolism, Pparα, Pparγ, and Pparδ. As seen in Fig. 6B, Pparδ gene expression was significantly downregulated in the livers from Lrat−/−, Crbp1−/−, and Lrat−/−/CrbpI−/− mice. No significant differences in hepatic expression of either Pparα or Pparγ were observed for any of the mutants including the carbohydrate response element-binding protein (Chrebp), a regulator of glucose and lipid metabolism (data not shown). The body weights of age-, gender-, and diet-matched male WT, Lrat−/−, CrbpI−/−, and Lrat−/−/CrbpI−/− mice were not significantly different nor were the expression levels of Pparγ in adipose tissue obtained from these different genotypes (data not shown). We also examined possible changes in expression for genes involved in hepatic lipogenesis (Fas, Scd1, and Acc) and fatty acid oxidation (Cpt1) but observed no significant differences (data not shown). As shown in Fig. 6C, we observed a marked downregulation in expression of the key regulatory enzyme Pdk4, which is a known target gene for Pparδ transcriptional regulation (47).
Fig. 6.
A: Fasting triglyceride levels are significantly elevated in the livers of 3-month-old male chow-fed CrbpI−/− and Lrat−/−/CrbpI−/− (L/C−/−) mice compared with matched WT mice. Groups of WT, CrbpI−/−, Lrat−/−, and Lrat−/−/CrbpI−/− mice (n = 6 per strain) were fasted in the morning for 4 h after diet was removed from their housing prior to sacrifice. Statistical significance: a, P < 0.05 compared with WT mice. Livers (n = 6 per strain) were taken for RNA isolation and assessment of Pparδ (B) and Pdk4 (C) mRNA levels by qPCR. All values are given as means ± SD. Statistical significance: a, P < 0.01 compared with WT mice.
DISCUSSION
ARAT activities are not involved in RE synthesis in the liver
The literature indicates that ARATs are involved in the synthesis of hepatic REs (9–12, 28, 29). We have reported that DGAT1 can act as a physiologically significant ARAT in the mouse intestine (24) and Shih et al. (25) established that DGAT1 acts physiologically as an ARAT in mouse skin. It is well established that DGAT1 acts to facilitate triglyceride storage/metabolism and lipid droplet formation in the liver (19–21). Because DGAT1 is highly expressed in the liver, this raises a question as to whether DGAT1 might also act as an ARAT in the liver. Moreover, DGAT1 is expressed both in hepatocytes and in hepatic stellate cells (44), the cellular site in the liver where REs are stored and where LRAT is primarily expressed (48). Even though our earlier studies of Lrat−/− mice established that these mutant mice have very low levels of hepatic REs (<0.1% of matched WT levels) suggesting that LRAT is responsible for the preponderance of hepatic RE synthesis when mice are maintained on a standard chow diet (17), the literature suggests a role for an ARAT in hepatic RE formation. This extensive literature maintains that tissue ARAT activities may only become active when high levels of retinol are available and/or when the capacities of CRBPs like CRBPI and CRBPII to bind retinol and channel it to LRAT have been exceeded (27–29, 49). Indeed, our earlier work, which established DGAT1 as a physiologically relevant ARAT in the intestine, also established that one of the actions of CRBPII in the intestine was to channel retinol to LRAT for esterification (23). To directly address these possibilities, we employed a nutritional approach, feeding a 25-fold excess retinol diet for 4 weeks, coupled with a genetic approach, in an attempt to demonstrate LRAT-independent RE formation. Our data do not support the idea that an acyl-CoA-dependent ARAT enzyme(s) contributes to hepatic RE formation in vivo. Our data are consistent with the conclusion that LRAT is solely responsible for hepatic RE synthesis. This includes both RE storage in hepatic stellate cells and RE incorporation into nascent VLDLs in hepatocytes. Although DGAT1 is a physiologically relevant ARAT in the intestine and skin (24, 25), we were unable to obtain any evidence that it has this role in the liver. Moreover, contrary to what has been proposed by Yamaguchi et al. (44) from cell culture studies, we observed no relationship between Lrat and Dgat1 gene expression in the liver that suggests coordinated gene regulation as proposed by these authors.
CRBPI acts to prevent catabolism and loss of hepatic retinol
It has been proposed that CRBPI prevents retinol from being converted to REs by ARAT activities or exposure to nonspecific enzymes that may catalyze retinol oxidation (27–29, 34, 49, 50). Our data do not support the notion that CRBPI acts to prevent hepatic or adipose ARAT activities, like DGAT1, from catalyzing RE synthesis. Rather, our data are convincing that CRBPI prevents elimination or loss of retinol from the liver, and from adipose tissue as well (see Fig. 3). The absence of CRBPI from Lrat−/− livers (in Lrat−/−/CrbpI−/− mice), which possess no REs and hence hepatic retinol levels and metabolism can be very cleanly assessed, results in an 8- to 20-fold reduction in the level of hepatic retinol. Molotkov et al. (50) have proposed that hepatic CRBPI limits nonspecific oxidation of retinol by alcohol dehydrogenase 1 and proposed that this increases the ability of hepatic “esterifying enzymes” to produce REs for storage. Because retinol cannot be esterified in the livers of Lrat−/−/CrbpI−/− mice, our data establishes directly that hepatic CRBPI prevents loss of retinol from the liver. Interestingly, although the simple absence of CRBPI from adipose tissue does not affect the total retinol (retinol + REs) level found in adipose tissue (Fig. 5B), the absence of CRBPI from Lrat−/− mice results in a significant reduction of adipose total retinol. Total retinol levels present in Lrat−/− adipose tissue are approximately 2- or 3-fold elevated over those of age-, gender-, and diet-matched WT mice (17) (Fig. 5B). The absence of CRBPI from Lrat-deficient adipose tissue results in adipose tissue total retinol levels which are similar to those of matched WT mice. There are two possible bases for this observation. It is possible, that like in the liver, CRBPI prevents oxidation and loss of adipose retinol. However, because adipose total retinol levels are similar for WT and CrbpI−/− mice, we believe that this is unlikely. Alternatively, because the molecular identity of the enzyme(s) responsible for RE formation in Lrat−/−/Dgat1−/− adipose tissue is not known, possibly there is a previously unsuspected CRBPI-dependent retinol esterifying activity present in adipose tissue. This possibility needs to be explored in future research.
Elevated hepatic mRNA levels for known RA-responsive genes should not be taken to indicate that hepatic steady-state RA concentrations are elevated
Liu and Gudas (18) have demonstrated that Cyp26A1 mRNA expression is elevated in the livers of Lrat−/− mice. Earlier studies showed Cyp26A1 mRNA expression is induced either by acute loading with RA or long-term exposure to dietary retinoids, whereas expression was downregulated upon administration of a retinoid-deficient diet (51, 52). We have confirmed the published observation of Liu and Gudas (18) that Cyp26A1 expression is elevated in the livers of chow-fed Lrat−/− mice and have established further that expression of the retinoid-responsive transcription factor RARβ2 is also elevated in the livers of chow-fed Lrat−/− mice. Both the Cyp26A and Rarβ2 genes are known to contain RA response elements, which render the genes responsive to all-trans-RA availability (1–3, 53). This is usually taken to suggest that steady-state RA levels are elevated in tissues/cells expressing elevated levels of RA-responsive genes. However, as seen in Fig. 4C, D, serum and liver levels of all-trans-RA, assessed using very specific and sensitive LC/MS/MS protocols, were actually significantly lower in Lrat−/− compared with matched WT mice. This likely arises from the increased expression and catabolic actions of Cyp26A1, and possibly other RA metabolizing cytochromes. These data could be taken to suggest that the genes for Cyp26A1 and Rarβ2, and possibly other RA-responsive genes, may respond to increased fluxes of all-trans-RA (i.e., increased synthesis as well as increased degradation) rather than simply increased steady-state levels of this retinoid. However, there are other molecular processes that may explain increases in Cyp26A1 and Rarβ2 gene expression. These include possible differences in the half-lives of these mRNA species, effects of other transcription factors that modulate the expression of these genes, or possible differences in how all-trans-RA may be partitioned between the cytosol and nucleus within the cell. This later possibility though seems unlikely because we saw no differences in hepatic expression of CrabpI or CrabpII mRNA for the different mouse lines.
The LC/MS/MS protocols we used to measure all-trans-RA concentrations in liver extracts also allowed for separation and detection of purified 9-cis-RA. However, we did not detect much 9-cis-RA in any of our liver extracts, although we could readily detect 9-cis-RA when it was exogenously added to liver homogenates. Thus, we conclude that 9-cis RA is simply not present at substantial levels endogenously in the liver. Kane et al. (40) have reported a similar inability to detect 9-cis-RA in mouse tissues, also using state-of-the-art LC/MS/MS instrumentation. When 9-cis-RA originally was reported to be the natural ligand for the retinoid X receptors, its level in mouse liver was reported to be 4 ng/g wet weight (54). This reported level is clearly incorrect and likely too large by at least two orders of magnitude. Based on our data, we believe that relative to all-trans-RA there is very little 9-cis-RA in the mouse liver.
Increased hepatic RA-responsive gene expression is associated with elevated triglyceride levels
While undertaking our investigations aimed at understanding hepatic retinoid storage, we observed that the fasting livers of CrbpI−/− and Lrat−/−/CrbpI−/− mice accumulate significantly more triglyceride than matched chow-fed WT mice (Fig. 6). Livers from these strains also show elevated expression of RA-responsive gene expression. The literature suggests linkages between retinoid storage, metabolism, and actions and the development of fatty liver. Included in this literature are studies reporting ablation of hepatic retinoid receptor signaling resulting in hepatic steatosis (55), ablation of carotenoid-15,15′-oxygenase (which abolishes retinoid production from β-carotene) (56), studies of mice deficient in CRBPIII (57), nutritional studies carried out in mice or rats (58–61), studies of retinoid effects on hepatic endocannabinoid signaling (62), and human observational studies (63, 64). However, the specific mechanisms underlying these observations are not well-established. Given the focus of our research, we undertook only a limited survey to identify possible general molecular pathways that might be responsible for the observation. To this end, we examined by qPCR expression levels of a number of key regulators of hepatic lipid metabolism. We did not detect significant differences between matched mutant and WT mice in hepatic expression of regulatory genes commonly associated with hepatic steatosis, specifically Pparα and Pparγ. Strikingly though, Pparδ mRNA levels were downregulated by more than 75% and levels of the Pparδ target gene Pdk4 (47) were similarly downregulated in the livers of CrbpI−/−, Lrat−/−, and Lrat−/−/CrbpI−/− mice. Although it is generally believed that PPARδ exerts its effects on lipid metabolism primarily through actions in skeletal muscle (65), there is evidence that PPARδ also controls hepatic energy substrate homeostasis through coordinated regulation of glucose and fatty acid metabolism (66). Interestingly, all-trans-RA has been proposed to be a PPARδ agonist (4, 5). We believe that the observations of elevated hepatic triglyceride accumulation in CrbpI−/− and Lrat−/−/CrbpI−/− mice and elevated RA-responsive gene expression in these livers are directly related. However, further investigations will be needed before this possibility can be conclusively established.
Footnotes
Abbreviations:
- ARAT
- acyl-CoA:retinol acyltransferase
- CRABP(I or II)
- cellular-retinoic acid-binding protein (type I or II)
- CRBP(I, II, or III)
- , cellular retinol-binding protein (type I, II, or III)
- CrbpI−/−
- cellular retinol-binding protein, type I-deficient
- CYP26A1
- cytochrome 26A1
- CYP26B1
- cytochrome 26B1
- DGAT1
- diacylglycerol acyltransferase 1
- Dgat1−/−
- diacylglycerol acyltransferase 1-deficient
- LRAT
- lecithin:retinol acyltransferase
- Lrat−/−
- lecithin:retinol acyltransferase-deficient
- qPCR
- quantitative polymerase chain reaction
- RA
- retinoic acid
- RAR
- retinoic acid receptor
- Rarβ2
- retinoic acid receptor-β isoform 2
- RBP4
- retinol-binding protein
- Rbp4−/−
- retinol-binding protein-deficient
- RE
- retinyl ester
Retinol-binding protein is abbreviated in the literature either as RBP or using its gene nomenclature, RBP4. Throughout the article, we will employ solely RBP4 but the reader should be aware that RBP and RBP4 signify the identical protein.
We gratefully acknowledge the support of National Institutes of Health Grants R01 DK068437, R01 DK079221, and R21 AA021336 which allowed this work to be undertaken.
REFERENCES
- 1.Al Tanoury Z., Piskunov A., Rochette-Egly C. 2013. Vitamin A and retinoid signaling: genomic and nongenomic effects. J. Lipid Res. 54: 1761–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gudas L. J., Sporn M. B., Roberts A. B. 1994. Cellular biology and biochemistry of the retinoids. In The Retinoids: Biology, Chemistry, and Medicine. 2nd edition. M. B. Sporn, A. B. Roberts, and D. S. Goodman, editors. Raven Press, Ltd., New York. 443–520. [Google Scholar]
- 3.Balmer J. E., Blomhoff R. 2002. Gene expression by retinoic acid. J. Lipid Res. 43: 1773–1808 [DOI] [PubMed] [Google Scholar]
- 4.Shaw N., Elholm M., Noy N. 2003. Retinoic acid is a high affinity selective ligand for the peroxisome proliferator-activated receptor beta/delta. J. Biol. Chem. 278: 41589–41592 [DOI] [PubMed] [Google Scholar]
- 5.Berry D. C., Noy N. 2009. All-trans-retinoic acid represses obesity and insulin resistance by activating both peroxisome proliferation-activated receptor beta/delta and retinoic acid receptor. Mol. Cell. Biol. 29: 3286–3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross A. C. 2006. Vitamin A and carotenoids. In Modern Nutrition in Health and Disease. 10th edition. M. E. Shils, M. Shike, A. C. Ross, et al., editors. Williams & Wilkins, Baltimore. 351–375. [Google Scholar]
- 7.Blaner W. S., Olson J. A. 1994. Retinol and retinoic acid metabolism. In The Retinoids: Biology, Chemistry, and Medicine. 2nd edition. M. B. Sporn, A. B. Roberts, and D. S. Goodman, editors. Raven Press, Ltd., New York. 229–256. [Google Scholar]
- 8.O'Byrne S. M., Blaner W. S. 2013. Retinol and retinyl esters: biochemistry and physiology. J. Lipid Res. 54: 1731–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blomhoff R., Green M. H., Green J. B., Berg T., Norum K. R. 1991. Vitamin A metabolism: new perspectives on absorption, transport, and storage. Physiol. Rev. 71: 951–990 [DOI] [PubMed] [Google Scholar]
- 10.Senoo H., Kojima N., Sato N. M. 2007. Vitamin A-storing cells (stellate cells). Vitam. Horm. 75: 131–159 [DOI] [PubMed] [Google Scholar]
- 11.Ross A. C. 1982. Retinol esterification by rat liver microsomes. Evidence for a fatty acyl coenzyme A:retinol acyltransferase. J. Biol. Chem. 257: 2453–2459 [PubMed] [Google Scholar]
- 12.Drevon C. A., Blomhoff R., Rasmussen M., Kindberg G. M., Berg T., Norum K. R. 1985. Retinol esterification in cultured rat liver cells. Biochem. J. 230: 617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacDonald P. N., Ong D. E. 1988. A lecithin:retinol acyltransferase activity in human and rat liver. Biochem. Biophys. Res. Commun. 156: 157–163 [DOI] [PubMed] [Google Scholar]
- 14.Yost R. W., Harrison E. H., Ross A. C. 1988. Esterification by rat liver microsomes of retinol bound to cellular retinol-binding protein. J. Biol. Chem. 263: 18693–18701 [PubMed] [Google Scholar]
- 15.Saari J. C., Bredberg D. L. 1988. CoA- and non-CoA-dependent retinol esterification in retinal pigment epithelium. J. Biol. Chem. 263: 8084–8090 [PubMed] [Google Scholar]
- 16.Batten M. L., Imanishi Y., Maeda T., Tu D. C., Moise A. R., Bronson D., Possin D., van Gelder R. N., Baehr W., Palczewski K. 2004. Lecithin-retinol acyltransferase is essential for accumulation of all-trans-retinyl esters in the eye and in the liver. J. Biol. Chem. 279: 10422–10432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Byrne S. M., Wongsiriroj N., Libien J., Vogel S., Goldberg I. J., Baehr W., Palczewski K., Blaner W. S. 2005. Retinoid absorption and storage is impaired in mice lacking lecithin:retinol acyltransferase (LRAT). J. Biol. Chem. 280: 35647–35657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu L., Gudas L. J. 2005. Disruption of the lecithin:retinol acyltransferase gene makes mice more susceptible to vitamin A deficiency. J. Biol. Chem. 280: 40226–40234 [DOI] [PubMed] [Google Scholar]
- 19.Yu Y. H., Ginsberg H. N. 2004. The role of acyl-CoA:diacylglycerol acyltransferase (DGAT) in energy metabolism. Ann. Med. 36: 252–261 [DOI] [PubMed] [Google Scholar]
- 20.Yen C. L., Stone S. J., Koliwad S., Harris C., Farese R. V., Jr 2008. DGAT enzymes and triacylglycerol biosynthesis. J. Lipid Res. 49: 2283–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruggles K. V., Turkish A., Sturley S. L. 2013. Making, baking, and breaking: the synthesis, storage, and hydrolysis of neutral lipids. Annu. Rev. Nutr. 33: 413–451 [DOI] [PubMed] [Google Scholar]
- 22.Yen C. L., Monetti M., Burri B. J., Farese R. V., Jr 2005. The triacylglycerol synthesis enzyme DGAT1 also catalyzes the synthesis of diacylglycerols, waxes, and retinyl esters. J. Lipid Res. 46: 1502–1511 [DOI] [PubMed] [Google Scholar]
- 23.Orland M. D., Anwar K., Cromley D., Chu C. H., Chen L., Billheimer J. T., Hussain M. M., Cheng D. 2005. Acyl coenzyme A dependent retinol esterification by acyl coenzyme A: diacylglycerol acyltransferase 1. Biochim. Biophys. Acta. 1737: 76–82 [DOI] [PubMed] [Google Scholar]
- 24.Wongsiriroj N., Piantedosi R., Palczewski K., Goldberg I. J., Johnston T. P., Li E., Blaner W. S. 2008. The molecular basis of retinoid absorption: a genetic dissection. J. Biol. Chem. 283: 13510–13519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shih M. Y., Kane M. A., Zhou P., Yen C. L., Streeper R. S., Napoli J. L., Farese R. V., Jr 2009. Retinol esterification by DGAT1 is essential for retinoid homeostasis in murine skin. J. Biol. Chem. 284: 4292–4299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cases S., Smith S. J., Zheng Y-W., Myers H. M., Lear S. R., Sande E., Novak S., Collins C., Welch C. B., Lusis A. J., et al. 1998. Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA. 95: 13018–13023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ong D. E., MacDonald P. N., Gubitosi A. M. 1988. Esterification of retinol in rat liver. Possible participation by cellular retinol-binding protein and cellular retinol-binding protein II. J. Biol. Chem. 263: 5789–5796 [PubMed] [Google Scholar]
- 28.Ong D. E., Newcomer M. E., Chytil F. 1994. Cellular retinoid-binding proteins. In The Retinoids: Biology, Chemistry, and Medicine. 2nd edition. M. B. Sporn, A. B. Roberts, and D.S. Goodman, editors. Raven Press, Ltd., New York. 283–318. [Google Scholar]
- 29.Randolph R. K., Winkler K. E., Ross A. C. 1991. Fatty acyl CoA-dependent and -independent retinol esterification by rat liver and lactating mammary gland microsomes. Arch. Biochem. Biophys. 288: 500–508 [DOI] [PubMed] [Google Scholar]
- 30.Tsutsumi C., Okuno M., Tannous L., Piantedosi R., Allan M., Goodman D. S., Blaner W. S. 1992. Retinoids and retinoid-binding protein expression in rat adipocytes. J. Biol. Chem. 267: 1805–1810 [PubMed] [Google Scholar]
- 31.Vogel S., Mendelsohn C. L., Mertz J. R., Piantedosi R., Waldburger C., Gottesman M. E., Blaner W. S. 2001. Characterization of a new member of the fatty acid-binding protein family that binds all-trans-retinol. J. Biol. Chem. 276: 1353–1360 [DOI] [PubMed] [Google Scholar]
- 32.Ross A. C. 2003. Retinoid production and catabolism: role of diet in regulating retinol esterification and retinoic acid oxidation. J. Nutr. 133: 291S–296S [DOI] [PubMed] [Google Scholar]
- 33.Ross A. C., Cifelli C. J., Zolfaghari R., Li N. Q. 2011. Multiple cytochrome P-450 genes are concomitantly regulated by vitamin A under steady-state conditions and by retinoic acid during hepatic first pass metabolism. Physiol. Genomics. 43: 57–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghyselinck N. B., Båvik C., Sapin V., Mark M., Bonnier D., Hindelang C., Dierich A., Nilsson C. B., Håkansson H., Sauvant P., et al. 1999. Cellular retinol-binding protein I is essential for vitamin A homeostasis. EMBO J. 18: 4903–4914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith S. J., Cases S., Jensen D. R., Chen H. C., Sande E., Tow B., Sanan D. A., Raber J., Eckel R. H., Farese R. V., Jr 2000. Obesity resistance and multiple mechanisms of triglyceride synthesis in mice lacking Dgat. Nat. Genet. 25: 87–90 [DOI] [PubMed] [Google Scholar]
- 36.Quadro L., Blaner W. S., Salchow D. J., Vogel S., Piantedosi R., Gouras P., Freeman S., Cosma M. P., Colantuoni V., Gottesman M. E. 1999. Visual defect and impaired retinoid availability in mice lacking retinol-binding protein. EMBO J. 18: 4633–4644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.National Research Council. 2011. Guide for the Care and Use of Laboratory Animals. 8th edition. The National Academies Press, Washington, DC. [Google Scholar]
- 38.Reeves P. G., Nielsen F. H., Fahey G. C., Jr 1993. AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J. Nutr. 123: 1939–1951 [DOI] [PubMed] [Google Scholar]
- 39.Barua A. B., Furr H. C. 1998. Properties of retinoids. Structure, handling and preparation. Mol. Biotechnol. 10: 167–182 [DOI] [PubMed] [Google Scholar]
- 40.Kane M. A., Folias A. E., Wang C., Napoli J. L. 2008. Quantitative profiling of endogenous retinoic acid in vivo and in vitro by tandem mass spectrometry. Anal. Chem. 80: 1702–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wasan K. M., Subramanian R., Kwong M., Goldberg I. J., Wright T., Johnston T. P. 2003. Poloxamer 407-mediated alterations in the activities of enzymes regulating lipid metabolism in rats. J. Pharm. Pharm. Sci. 6: 189–197 [PubMed] [Google Scholar]
- 42.Johnston T. P. 2010. Poloxamer 407 as a general lipase inhibitor: its implications in lipid metabolism and atheroma formation in C57BL/6 mice. J. Pharm. Pharmacol. 62: 1807–1812 [DOI] [PubMed] [Google Scholar]
- 43.Millar J. S., Cromley D. A., McCoy M. G., Rader D. J., Billheimer J. T. 2005. Determining hepatic triglyceride production in mice: comparison of poloxamer 407 with Triton WR-1339. J. Lipid Res. 46: 2023–2028 [DOI] [PubMed] [Google Scholar]
- 44.Yamaguchi K., Yang L., McCall S., Huang J., Yu X. X., Pandey S. K., Bhanot S., Monia B. P., Li Y-X., Diehl A. M. 2008. Diacylglycerol acyltransferase 1 anti-sense oligonucleotides reduce hepatic fibrosis in mice with nonalcoholic steatohepatitis. Hepatology. 47: 625–635 [DOI] [PubMed] [Google Scholar]
- 45.Liang J. J., Oelkers P., Guo C., Chu P. C., Dixon J. L., Ginsberg H. N., Sturley S. L. 2004. Overexpression of human diacylglycerol acyltransferase 1, acyl-CoA:cholesterol acyltransferase 1, or acyl-CoA:cholesterol acyltransferase 2 stimulates secretion of apolipoprotein B-containing lipoproteins in McA-RH7777 cells. J. Biol. Chem. 279: 44938–44944 [DOI] [PubMed] [Google Scholar]
- 46.Yamazaki T., Sasaki E., Kakinuma C., Yano T., Miura S., Ezaki O. 2005. Increased very low density lipoprotein secretion and gonadal fat mass in mice overexpressing liver DGAT1. J. Biol. Chem. 280: 21506–21514 [DOI] [PubMed] [Google Scholar]
- 47.Degenhardt T., Saramäki A., Malinen M., Rieck M., Väisänen S., Huotari A., Herzig K. H., Müller R., Carlberg C. 2007. Three members of the human pyruvate dehydrogenase kinase gene family are direct targets of the peroxisome proliferator-activated receptor beta/delta. J. Mol. Biol. 372: 341–355 [DOI] [PubMed] [Google Scholar]
- 48.Nagatsuma K., Yoshihiro H., Hiroshi H., Hiroshi S., Kazuhiro M., Masaya S., Takahiro M., Tomoe L., Mitsugu T., Hideaki E., et al. 2009. Lecithin:retinol acyltransferase protein is distributed in both hepatic stellate cells and endothelial cells of normal rodent and human liver. Liver Int. 29: 47–54 [DOI] [PubMed] [Google Scholar]
- 49.Napoli J. L. 1999. Interactions of retinoid binding proteins and enzymes in retinoid metabolism. Biochim. Biophys. Acta. 1440: 139–162 [DOI] [PubMed] [Google Scholar]
- 50.Molotkov A., Ghyselinck N. B., Chambon P., Duester G. 2004. Opposing actions of cellular retinol-binding protein and alcohol dehydrogenase control the balance between retinol storage and degradation. Biochem. J. 383: 295–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ray W. J., Bain G., Yao M., Gottlieb D. I. 1997. CYP26, a novel mammalian cytochrome P450, is induced by retinoic acid and defines a new family. J. Biol. Chem. 272: 18702–18708 [DOI] [PubMed] [Google Scholar]
- 52.Ross A. C., Zolfaghari R. 2011. Cytochrome P450s in the regulation of cellular retinoic acid metabolism. Annu. Rev. Nutr. 31: 65–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shiota G., Kanki K. 2013. Retinoids and their target genes in liver functions and diseases. J. Gastroenterol. Hepatol. 28: 33–37 [DOI] [PubMed] [Google Scholar]
- 54.Heyman R. A., Mangelsdorf D. J., Dyck J. A., Stein R. B., Eichele G., Evans R. M., Thaller C. 1992. 9-cis retinoic acid is a high affinity ligand for the retinoid X receptor. Cell. 68: 397–406 [DOI] [PubMed] [Google Scholar]
- 55.Yanagitani A., Yamada S., Yasui S., Shimomura T., Murai R., Murawaki Y., Hashiguchi K., Kanbe T., Saeki T., Ichiba M., et al. 2004. Retinoic acid receptor alpha dominant negative form causes steatohepatitis and liver tumors in transgenic mice. Hepatology. 40: 366–375 [DOI] [PubMed] [Google Scholar]
- 56.Hessel S., Eichinger A., Isken A., Amengual J., Hunzelmann S., Hoeller U., Eiste V., Hunziker W., Goralczyk R., Oberhauser V., et al. 2007. CMO1 deficiency abolishes vitamin A production from beta-carotene and alters lipid metabolism in mice. J. Biol. Chem. 282: 33553–33561 [DOI] [PubMed] [Google Scholar]
- 57.Zizola C. F., Schwartz G. J., Vogel S. 2008. Cellular retinol-binding protein type III is a PPARgamma target gene and plays a role in lipid metabolism. Am. J. Physiol. Endocrinol. Metab. 295: E1358–E1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bonilla S., Redonnet A., Noël-Suberville C., Pallet V., Garcin H., Higueret P. 2000. High fat diets affect the expression of nuclear retinoic acid receptor in rat liver. Br. J. Nutr. 83: 665–671 [DOI] [PubMed] [Google Scholar]
- 59.Oliveros L. B., Domeniconi M. A., Vega V. A., Gatica L. V., Brigada A. M., Gimenez M. S. 2007. Vitamin A deficiency modifies lipid metabolism in rat liver. Br. J. Nutr. 97: 263–272 [DOI] [PubMed] [Google Scholar]
- 60.Kang H. W., Bhimidi G. R., Odom D. P., Brun P. J., Fernandez M. L., McGrane M. M. 2007. Altered lipid catabolism in the vitamin A deficient liver. Mol. Cell. Endocrinol. 271: 18–27 [DOI] [PubMed] [Google Scholar]
- 61.Amengual J., Ribot J., Bonet M. L., Palou A. 2010. Retinoic acid treatment enhances lipid oxidation and inhibits lipid biosynthesis capacities in the liver of mice. Cell. Physiol. Biochem. 25: 657–666 [DOI] [PubMed] [Google Scholar]
- 62.Mukhopadhyay B., Liu J., Osei-Hyiaman D., Godlewski G., Mukhopadhyay P., Wang L., Jeong W-I., Gao B., Duester G., Mackie K., et al. 2010. Transcriptional regulation of cannabinoid receptor-1 expression in the liver by retinoic acid acting via retinoic acid receptor-gamma. J. Biol. Chem. 285: 19002–19011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suano de Sousa F. I., Silverio Amancio O. M., Saccardo Sarni R. O., Sacchi Pitta T., Fernandes A. P., Affonso Fonseca F. L., Hix S., Ramalho R. A. 2008. Non-alcoholic fatty liver disease in overweight children and its relationship with retinol serum levels. Int. J. Vitam. Nutr. Res. 78: 27–32 [DOI] [PubMed] [Google Scholar]
- 64.Wu H., Jia W., Bao Y., Lu J., Zhu J., Wang R., Chen Y., Xiang K. 2008. Serum retinol binding protein 4 and nonalcoholic fatty liver disease in patients with type 2 diabetes mellitus. Diabetes Res. Clin. Pract. 79: 185–190 [DOI] [PubMed] [Google Scholar]
- 65.Narkar V. A., Downes M., Yu R. T., Embler E., Wang Y. X., Banayo E., Mihaylova M. M., Nelson M. C., Zou Y., Juguilon H., et al. 2008. AMPK and PPARdelta agonists are exercise mimetics. Cell. 134: 405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu S., Hatano B., Zhao M., Yen C-C., Kang K., Beilly S. M., Gangl M. R., Gorgun C., Balschi J. A., Ntambi J. M., et al. 2011. Role of peroxisome proliferator-activated receptor δ/β in hepatic metabolic regulation. J. Biol. Chem. 286: 1237–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]